Abstract
Somatic mutations in the Jak2 protein, such as V617F, cause aberrant Jak/STAT signaling and can lead to the development of myeloproliferative neoplasms. This discovery has led to the search for small molecule inhibitors that target Jak2. Using structure-based virtual screening, our group recently identified a novel small molecule inhibitor of Jak2 named G6. Here, we identified a structure-function correlation of this compound. Specifically, five derivative compounds of G6 having structural similarity to the original lead compound were obtained and analyzed for their ability to (i) inhibit Jak2-V617F-mediated cell growth, (ii) inhibit the levels of phospho-Jak2, phospho-STAT3, and phospho-STAT5; (iii) induce apoptosis in human erythroleukemia cells; and (iv) suppress pathologic cell growth of Jak2-V617F-expressing human bone marrow cells ex vivo. Additionally, we computationally examined the interactions of these compounds with the ATP-binding pocket of the Jak2 kinase domain. We found that the stilbenoid core-containing derivatives of G6 significantly inhibited Jak2-V617F-mediated cell proliferation in a time- and dose-dependent manner. They also inhibited phosphorylation of Jak2, STAT3, and STAT5 proteins within cells, resulting in higher levels of apoptosis via the intrinsic apoptotic pathway. Finally, the stilbenoid derivatives inhibited the pathologic growth of Jak2-V617F-expressing human bone marrow cells ex vivo. Collectively, our data demonstrate that G6 has a stilbenoid core that is indispensable for maintaining its Jak2 inhibitory potential.
Keywords: Cell Division, Drug Action, Jak Kinase, STAT Transcription Factor, Tyrosine-protein Kinase (Tyrosine Kinase)
Introduction
Jak2 plays a critical role in animal development, because mice that are devoid of a functional Jak2 allele die during embryonic development because of a lack of hematopoiesis (1, 2). Deregulation of the Jak/STAT signaling pathway promotes cell growth and prevents apoptosis in a variety of solid tumors and hematological malignancies such as acute lymphoid leukemia and chronic myeloid leukemia (3–6). Additionally, a somatic Jak2 mutation (Jak2-V617F) is found in a high number of myeloproliferative neoplasm (MPN)2 patients including 90% of polycythemia vera patients and ∼50% of patients with essential thrombocythemia and primary myelofibrosis (7–11). MPNs are a group of heterogeneous diseases arising from a transformed hematopoietic stem cell and characterized by excessive numbers of one or more terminally differentiated blood cells of the myeloid lineage such as erythrocytes, thrombocytes, or white blood cells. A guanine to thymine mutation in hematopoietic stem cells results in a substitution of valine to phenylalanine at codon 617 in exon 14 of the JH2 pseudokinase domain of Jak2. It is believed that this mutation allows the kinase to evade negative feedback inhibition, thereby leading to a constitutively active Jak/STAT signaling pathway characterized by growth factor-independent cell growth (8, 10).
Given the critical role that Jak2 plays in the pathophysiology of MPNs, identification of specific Jak2 inhibitors has become an important step toward the development of an effective targeted therapy for these disorders. Using structure-based virtual screening, our group recently identified a novel small molecule inhibitor of Jak2 named G6 (12). We showed that G6 has a specific inhibitory effect on Jak2 kinase activity as measured by in vitro enzyme assays and an immunoassay ELISA (12). Examination of the chemical structure of G6 revealed the presence of a central stilbenoid core. Stilbenoids are a group of naturally occurring compounds having a wide range of biological activities. For example, resveratrol, piceatannol, 3,4,5,4′-tetramethoxystilbene, and 3,5,4′-trimethoxy-trans-stilbene are all stilbenoids and are known to have cytotoxic, anti-proliferative, pro-apoptotic, anti-angiogenic, or tumor-suppressive effects (13–17).
We hypothesized that the central stilbenoid core in the G6 structure has a critical role to play in mediating its Jak2 inhibitory potential. To test this, five derivative compounds of G6, namely D21, D23, D25, D28, and D30, having structural similarity to the original lead compound, were procured from the NCI, National Institutes of Health. Two of these compounds, D28 and D30, have a stilbenoid core present in their chemical structure similar to G6, whereas the three other compounds, D21, D23, and D25, lack the stilbenoid core. We report here that the core stilbenoid structure present in G6 is essential for maintaining its ability to inhibit Jak2 kinase activity.
EXPERIMENTAL PROCEDURES
Drugs
G6 and its five structurally related derivative compounds (D21, D23, D25, D28, and D30) were obtained from the National Institutes of Health NCI/Developmental Therapeutics Program, which maintains a repository of ∼140,000 compounds. Each compound was solubilized in dimethyl sulfoxide at a concentration of 10 mm and stored at −20°C.
Cell Culture
Human erythroleukemia (HEL) cells were purchased from the American Type Culture Collection. Ba/F3-EpoR-Jak2-V617F cells were created as described before (18). Both cell lines were cultured in RPMI 1640 (Mediatech) supplemented with 10% FBS, penicillin, streptomycin, and l-glutamine at 37°C and 5% CO2.
Cell Proliferation Assay
HEL cells or Ba/F3-EpoR-Jak2-V617F cells were plated in 96-well plates and treated with either 0.25% DMSO or varying concentrations of G6 and its derivatives for the indicated periods of time. Cell viability was assessed either by trypan blue exclusion staining or by MTS (Promega) as per the manufacturer's protocol.
ELISA
HEL cells were treated with either 0.25% DMSO or 25 μm of the inhibitor compounds for 48 h, and the cell lysates were analyzed by ELISA for detection of phospho-Jak2, phospho-STAT3, and phospho-STAT5. Jak2 (Tyr(P)1007/Tyr(P)1008), STAT3 (Tyr(P)705), and STAT5b (Tyr(P)699) ELISA kits were purchased from Invitrogen and used according to the manufacturer's protocol.
Cell Lysis and Immunoprecipitation
HEL cells were treated with the different drugs for the indicated periods of time. The cells (∼ 107) were then lysed in 0.8 ml of ice-cold radioimmunoprecipitation assay buffer, and protein concentration was determined using a Bradford assay (Bio-Rad). The cell lysates (∼2500 μg) were then immunoprecipitated by incubation with 2 μg of the appropriate antibody and 20 μl of protein A/G beads (Santa Cruz Biotechnology) for 4 h at 4 °C with constant shaking. The protein complexes were washed thrice with immunoprecipitation wash buffer (25 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Triton X-100) and then resuspended in SDS sample buffer. Immunoprecipitated proteins were separated by SDS-PAGE and then transferred onto nitrocellulose membranes. The anti-STAT3 and anti-STAT5 antibodies used for immunoprecipitation were from Santa Cruz Biotechnology. For whole cell protein lysates, ∼50 μg of soluble protein was separated via SDS-PAGE and then transferred to nitrocellulose membranes for analysis by Western blotting.
Western Blotting
Nitrocellulose membranes were first blocked with 5% milk/TBST solution and then probed with the different primary antibodies. The immunoreactive bands were then visualized using the enhanced chemiluminescence system (Western Lightning Ultra; PerkinElmer Life Sciences). The following antibodies were used at the indicated dilutions: phospho-STAT3 (Santa Cruz Biotechnology and Cell Signaling, 1:500), STAT3 (Santa Cruz Biotechnology, 1:1000), phospho-STAT5 (Cell Signaling, 1:500), STAT5 (Santa Cruz Biotechnology, 1:1000), and STAT1 (Santa Cruz Biotechnology, 1:1000). The following were all obtained from Cell Signaling and used at a 1:500 dilution; poly(ADP-ribose) polymerase, Bcl-2, Bax, Bim, and Bid.
Apoptosis Assay
Induction of apoptosis in HEL cells was determined with the FITC annexin V apoptosis detection kit (BD Pharmigen) per the manufacturer's protocol. The cells were incubated with 0.25% DMSO or 25 μm of inhibitors for 48 h and then analyzed using a FACSCalibur flow cytometer (BD Biosciences).
Real Time PCR Analysis
The mRNA levels of Bcl-xL and glyceraldehyde-3-phosphate dehydrogenase were measured by quantitative real time PCR analysis. Total RNA was extracted from HEL cells and treated with 25 μm of the different drugs for 8 and 24 h, using an RNeasy mini kit (Qiagen) as per the manufacturer's protocol. 2 μg of each RNA sample was reverse transcribed into cDNA in a final reaction volume of 20 μl using a high capacity cDNA reverse transcription kit (Applied Biosystems). TaqMan gene expression assays (Applied Biosystems) Hs02758991_g1 (glyceraldehyde-3-phosphate dehydrogenase) and Hs00236329_m1 (Bcl-xL) were used to detect the levels of expression of these genes. Glyceraldehyde-3-phosphate dehydrogenase gene expression was used as an internal loading control. Real time PCR was then performed with TaqMan universal PCR Master Mix (Applied Biosystems) in a final reaction volume of 20 μl in a StepOne real time PCR system as per the manufacturer's protocol.
Patient Sample
Bone marrow aspirates consisting of mononuclear cells were obtained from a de-identified Jak2-V617F-positive female diagnosed with polycythemia vera (World Health Organization criteria) at the University of Florida & Shands Teaching Hospital as per an institutional review board-approved protocol.
Colony Forming Unit-Erythroid Colony Formation Assay
Marrow-derived mononuclear cells were washed in Iscove's modified Dulbecco's medium and cultured in human methylcellulose complete medium without erythropoietin (R & D Systems) at a concentration of ∼4 × 105 cells/ml in the presence or absence of G6 or its structurally related derivatives. 0.9 ml of the cultures was placed in 35-mm Petri dishes and incubated at 37°C and 5% CO2 in a humidified atmosphere for 14 days, after which the number of erythroid colony-forming units was determined.
Computational Docking
The molecular docking program DOCK6 (19, 20) was used to study the interactions of Jak2 with ATP, the ATP analog ACP, G6, and the structurally related derivative compounds. The template used was the crystal structure of Jak2 kinase domain in complex with the Jak2 inhibitor 5B3 (Protein Data Bank code 3E64) (21). The coordinates for ATP, ACP, and each of the small molecules were generated and energy-minimized using PRODRG (22). Subsequently, Chimera (23) was used to add atomic charges to these small molecules. After removing the 5B3 molecule, the coordinates of the Jak2 kinase domain were saved in Protein Data Bank format. To prepare the protein for docking, hydrogen atoms and partial electrostatic charges were first added to the molecule. A molecular surface of the Jak2 kinase domain was then generated using the DMS tool in Chimera. The program SPHGEN was used to generate spheres around the active site for identification of the target pocket on the protein for small molecule docking. The GRID file, necessary for rapid GRID-based energy score evaluation, was then generated around the identified docking site using the GRID module in DOCK6. Each compound was docked in 1000 different orientations using flexible dock, and a net energy GRID score was generated for each, based on the van der Waal's and electrostatic interactions between the compound and the residues in the binding pocket. The favorable binding orientations were selected on the basis of the energy GRID scores generated for each orientation, wherein a favorable binding orientation would have a more negative score. Finally, the docking results were analyzed visually using COOT (24) and Chimera. Surface electrostatic potentials were calculated using APBS (25), and PyMol (26) was used to generate the figures.
Statistical Analysis
For statistical evaluation of time-dependent responses to the different inhibitor compounds, a two-way analysis of variance was used. For analysis of inhibition of phosphorylation, induction of apoptosis, modulation of gene expression, and suppression of pathologic cell growth ex vivo, a Student's t test was employed. The data were assumed to be statistically significant when p < 0.05.
RESULTS
A Stilbenoid Core Is Essential for Time- and Dose-dependent Inhibition of Jak2-V617F-dependent Cell Growth
The human erythroleukemia (HEL 92.1.7) cell line is homozygous for the Jak2-V617F mutation, and this gain-of-function mutation is responsible for its transformed phenotype (27, 28). Proliferation of HEL cells is mediated by the constitutively active Jak2-V617F signaling, which promotes a G1/S phase transition, thereby leading to increased cellular proliferation (29). G6 and its five structurally related derivatives were therefore first analyzed for their ability to inhibit the Jak2-V617F-dependent proliferation of HEL cells. Viable cell numbers were determined by trypan blue exclusion and hemocytometer after 72 h. Each sample was measured in triplicate. Inhibition by G6 was arbitrarily set at 100%, and the percentage of inhibition for all of the other compounds relative to G6 was defined as 1.00 − (Δ drug/Δ vehicle control). Supplemental Table S1 summarizes the percentage of growth inhibition for each of the six compounds. We found that the stilbene-containing derivatives (D28 and D30) had high growth inhibition potentials, whereas those compounds lacking the stilbenoid core (D21, D23, and D25) had low growth inhibition potentials.
To determine the ability of each of these compounds to inhibit Jak2-V617F-mediated HEL cell proliferation, the cells were treated either for varying periods of time or with increasing concentrations of G6 or its derivatives. Viable cell numbers for each treatment were determined. When compared with vehicle-treated cells, we found that G6 and its stilbenoid derivatives (D28 and D30) significantly reduced viable cell numbers in a time-dependent manner, whereas the non-stilbenoid derivatives (D21, D23, and D25) did not (Fig. 1A). We also found that G6 and the two stilbenoid derivatives (D28 and D30) markedly inhibited the growth of HEL cells in a dose-dependent manner (Fig. 1B), whereas the three non-stilbenoid derivatives (D21, D23, and D25) showed no growth inhibition of HEL cells (Fig. 1C).
FIGURE 1.
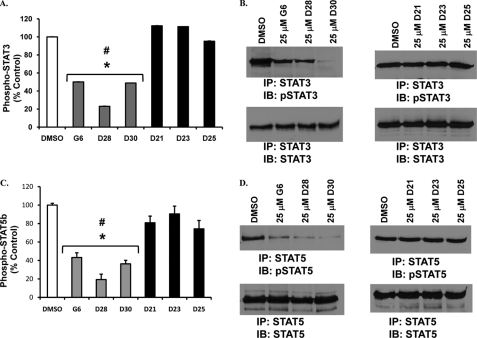
Time- and dose-dependent effect of G6 and its derivatives on the HEL cell proliferation and Jak2 phosphorylation. A, HEL cells were treated with either 0.25% DMSO or with 25 μm of G6 or its derivatives for 0, 24, 48, or 72 h. The number of viable cells in each sample was measured by trypan blue exclusion. Each sample was measured in triplicate. p = 1.22 × 10−10 (D23 versus G6). B and C, HEL cells were treated for 72 h either with DMSO or with 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 μm of the indicated compounds. The viable cell numbers for each treatment were determined in triplicate. Shown is one of two sets of representative results. D, HEL cells were treated with 25 μm of the different drugs for 48 h. The cells were then analyzed by ELISA for the detection of phospho-Jak2 (Tyr(P)1007/Tyr(P)1008). Each experiment was run in triplicate. Shown is one of two sets of representative results. *, p < 0.05 with respect to DMSO; #, p < 0.05 with respect to non-stilbenoids.
Phosphorylation of Jak2 at tyrosine residues 1007/1008 is concomitant with higher kinase activity and increased cellular proliferation (11). Therefore, we next wanted to determine whether the presence of the stilbenoid core is critical for reduction of phospho-Jak2 levels within treated cells. Phospho-Jak2 levels were measured 48 h after drug exposure rather than the 72 h used in Fig. 1 (A–C), because far fewer viable cells exist at the longer time point. Exposure of HEL cells to stilbenoid core-bearing compounds (G6, D28, and D30) significantly decreased the levels of phospho-Jak2 (Fig. 1D) when compared with those derivatives that lack the stilbenoid core (D21, D23, and D25).
We next wanted to determine whether the ability of G6 to inhibit Jak2-V617F-mediated cell proliferation was valid for other cell lines with constitutively active Jak2. For this, we studied the ability of these compounds to inhibit Ba/F3 cells stably expressing Jak2-V617F. The introduction of the Jak2-V617F cDNA into these cells via retroviral transduction confers cytokine-independent growth that is entirely Jak2-V617F-dependent (18). The cells were treated with various doses of the different drugs, and viable cell numbers were assessed after 72 h using an MTS assay. G6 and its stilbenoid derivatives (D28 and D30) showed significant inhibition of cell growth in a dose-dependent manner (Fig. 2A) when compared with the non-stilbenoid derivatives (D21, D23, and D25) (Fig. 2B). Collectively, these data demonstrate that the stilbenoid core is essential for maintaining the ability of G6 to inhibit Jak2-V617F-dependent cell growth, and the reduced cell growth correlates with significantly reduced levels of phospho-Jak2.
FIGURE 2.
Dose-dependent effect of G6 and its derivatives on the proliferation of Ba/F3-EpoR-Jak2-V617F cells. Ba/F3-EpoR-Jak2-V617F cells were treated for 72 hours either with DMSO or with 0.01, 0.03, 0.1, 0.3, 1, 3, 10, and 30 μm of the indicated compounds. A, treatment with the stilbenoids G6, D30, and D28. B, treatment with G6 and the non-stilbenoids D21, D23, D25. The viable cell numbers for each treatment were determined in triplicate using an MTS assay. Shown is one of two representative results.
Indispensability of the Stilbenoid Core in Decreasing Phosphorylation of STAT3 and STAT5
In the canonical Jak/STAT signaling pathway, an active Jak2 phosphorylates STAT proteins such as STAT3 and STAT5, which ultimately translocate to the nucleus and modulate gene transcription (10). Hence, we wanted to determine whether the stilbenoid core is critical for the inhibition of STAT phosphorylation. HEL cells were treated for 48 h with the different compounds, and phospho-STAT levels were determined. G6 and its stilbenoid derivatives (D28 and D30) significantly decreased phospho-STAT3 levels as measured by ELISA (Fig. 3A) and Western blot analysis (Fig. 3B). Similarly, only the stilbenoid-containing compounds (G6, D28 and D30) significantly decreased phospho-STAT5 levels as measured by ELISA (Fig. 3C) and Western blot analysis (Fig. 3D). As such, these data confirm the ability of G6 and its stilbenoid derivatives to down-regulate the phosphorylation of key signaling molecules involved in Jak2-dependent pathological cell growth, namely STAT3 and STAT5.
FIGURE 3.
Inhibition of phosphorylation of STAT3 and STAT5 by G6 and its derivatives. HEL cells were treated with 25 μm of the different drugs for 48 h. The cells were then analyzed for the detection of phospho-STAT3 and phospho-STAT5 by both ELISA (A and C) and Western blot analysis (B and D). Stilbenoid-containing compounds (G6, D28, and D30) effectively inhibited phosphorylation of STAT3 (A and B), and phosphorylation of STAT5b (C and D). Shown is one of two sets of representative results for each. *, p < 0.05 with respect to DMSO; #, p < 0.05 with respect to non-stilbenoids. IP, immunoprecipitation; IB, immunoblot.
Induction of Apoptosis in HEL Cells by G6 and Its Derivatives
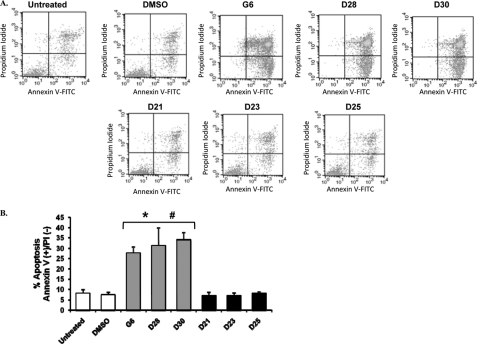
Deregulation of the Jak/STAT signaling pathway is known to promote cell proliferation and prevent apoptosis in several different cancers (30). Given that G6 and its stilbenoid core-containing derivatives inhibited Jak2-V617F-dependent cell proliferation and Jak/STAT activation, we next wanted to determine whether these drugs induce apoptotic death. HEL cells treated with G6 and the stilbenoid derivatives (D28 and D30) exhibited a significant increase in the percentage of cells in early apoptosis when compared with the DMSO or the non-stilbenoid-treated (D21, D23, and D25) cells (Fig. 4A). Fig. 4B is a quantitative graph of four independent experiments showing the amount of apoptosis plotted as a function of treatment condition. We observed that the percentage of cells in early apoptosis increased from 7.45% in the DMSO-treated control to 27.8% in G6-treated, 31.3% in D28-treated, and 34.2% in D30-treated HEL cells, whereas it remained almost unchanged for the non-stilbenoid-treated cells (Fig. 4B).
FIGURE 4.
Induction of apoptosis in HEL cells by G6 and its derivatives. HEL cells were treated with 25 μm of the different drugs for 48 h and then stained with annexin V-FITC and propidium iodide followed by flow cytometric analysis. A, shown are representative flow cytometry profiles from one of four independent results. B, quantification of the number of cells in early apoptosis (i.e. annexin V-positive and propidium iodide-negative). The data shown are the means ± S.D. from four independent experiments. *, p < 0.05 with respect to DMSO; #, p < 0.05 with respect to non-stilbenoids.
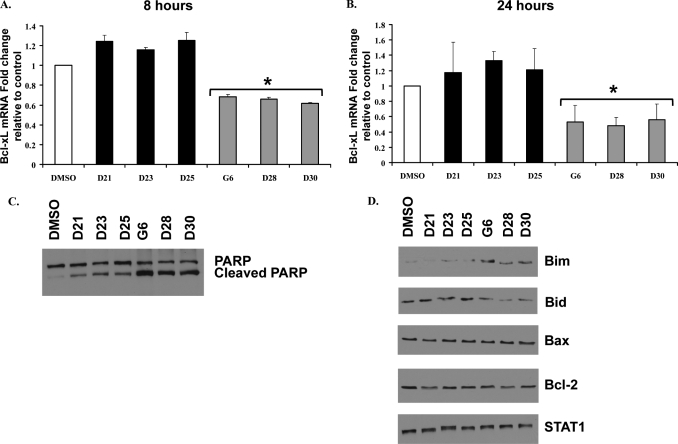
Jak2/STAT signaling is known to positively regulate cell growth by directly increasing expression of the anti-apoptotic marker, Bcl-xL, via STAT-binding elements present in its promoter region (31, 32). To determine whether the presence of the stilbenoid core correlates with reduced levels of Bcl-xL, we measured Bcl-xL mRNA levels in cells treated with the different compounds. The stilbenoids (G6, D28, and D30) significantly decreased Bcl-xL expression in HEL cells when compared with DMSO or the non-stilbenoids (D21, D23, and D25) at both 8 h (Fig. 5A) and 24 h (Fig. 5B) of treatment. As such, these data indicate that the presence of the stilbenoid core does in fact correlate with markedly reduced levels of the proliferative marker, Bcl-xL.
FIGURE 5.
Treatment with G6 or its stilbenoid derivatives leads to HEL cell death via the intrinsic apoptotic pathway. HEL cells were treated with 25 μm of the different drugs for either 8 h (A) or 24 h (B). Bcl-xL mRNA levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase and plotted as the fold change over DMSO control. Each sample was run in duplicate. Shown is one of two sets of representative results. *, p < 0.05 with respect to DMSO. C, cells were treated with either DMSO or 25 μm of the different drugs for 24 h. The whole cell lysates were then analyzed by Western blotting with an anti-poly(ADP-ribose) polymerase (PARP) antibody. Shown is one of three sets of representative results. D, cells were treated with either DMSO or 25 μm of the different drugs for 24 h. Whole cell lysates were then serially analyzed by Western blot analysis for the indicated proteins. Shown is one of three sets of representative results.
The intrinsic apoptotic pathway is regulated by members of the Bcl-2 family (33). Therefore, we next monitored the expression of Bcl-2 family members in HEL cells treated with 25 μm of the different compounds for 24 h. As expected, the stilbenoid derivates (G6, D28, and D30) induced greater cleavage of poly(ADP-ribose) polymerase, a marker of apoptosis, when compared with the non-stilbenoids (D21, D23, and D25) (Fig. 5C). We also observed a stilbenoid-dependent increase in the protein levels of Bim, a pro-apoptotic member of the Bcl-2 family (Fig. 5D).
The pro-apoptotic protein, Bid, is present in the cytosol as an inactive precursor. Upon proteolytic cleavage, its active form is generated, which translocates to the mitochondria and induces mitochondrial damage and destabilization (34). We found that the stilbenoid compounds decreased the levels of the inactive precursor form of Bid, a hallmark of Bid cleavage and subsequent apoptosis. We found that the protein levels of Bax and Bcl-2 did not change with any treatment (Fig. 5D). Lastly, the samples were blotted with an anti-STAT1 antibody to demonstrate equal protein loading across all lanes (Fig. 5D). Overall, the data in Figs. 4 and 5 indicate that the stilbenoid core of G6 is critical for its ability to induce apoptotic cell death in HEL cells via the intrinsic apoptotic pathway by down-regulation of anti-apoptotic Bcl-xL, up-regulation of pro-apoptotic Bim, and cleavage of Bid.
Stilbenoid Core-bearing Derivatives of G6 Suppress Pathologic Cell Growth of Patient-derived Bone Marrow Cells ex Vivo
We next wanted to determine whether the stilbene core is also essential for its ability to inhibit pathologic cell growth of patient-derived bone marrow cells ex vivo. For this, we used a colony formation assay that measures the number of erythroid colony-forming units that are produced when marrow-derived stem cells are cultured ex vivo. Stem cells isolated from a normal individual will be unable to grow in the absence of exogenously added cytokine. However, the Jak2-V617F mutation confers cytokine-independent cell growth. Here, mononuclear cells isolated from the bone marrow of a Jak2-V617F-positive female polycythemia vera patient were cultured in medium lacking erythropoietin. DMSO or 5 μm of inhibitor was added as indicated. Our previous work has shown that 5 μm of G6 inhibits ∼50% of the cytokine-independent growth of Jak2-V617F-expressing, marrow-derived stem cells (12). The results here show that treatment of the primary cultures with either G6 or the stilbenoids (D28 and D30) significantly blocked cytokine-independent pathologic cell growth when compared with the DMSO and non-stilbenoid-treated (D21, D23, and D25) samples (Fig. 6). As such, the data in Fig. 6 demonstrate that the stilbenoid core of G6 is essential for reducing Jak2-dependent pathologic cell growth of human bone marrow mononuclear cells cultured ex vivo.
FIGURE 6.
Suppression of Jak2-V617F-mediated pathologic cell growth in patient-derived bone marrow cells by G6 and its derivatives ex vivo. Marrow-derived mononuclear cells were cultured in semisolid medium in the presence or absence of 5 μm G6 and its structurally related derivatives. At the end of 14 days of culture, the numbers of erythroid colony-forming units (CFU-E) were counted. Each condition was measured in duplicate. *, p < 0.05 with respect to DMSO; #, p < 0.05 with respect to non-stilbenoids.
Computational Docking of G6 and Its Derivatives into the ATP-binding Pocket of the Jak2 Kinase Domain
Using the known structure of the Jak2 kinase domain (21), ATP, the ATP analog ACP, G6, and each of its five structurally related derivatives were docked into the ATP-binding pocket. The goal was to analyze the potential interactions of these compounds with amino acids in this binding region.
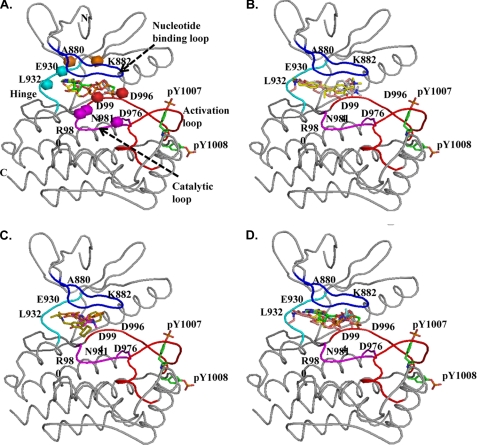
The ATP-binding pocket of Jak2 and the important residues clustered in this pocket have been described previously (35, 36). Generation of the electrostatic surface potential for the Jak2 kinase domain showed the predicted presence of acidic and basic patch residues clustered at the ATP-binding pocket (supplemental Fig. S1). We found that both ATP and ACP docked extremely well into this pocket (Fig. 7A) with GRID scores of −76.81 and −62.37 kcal/mol, respectively. Furthermore, the docked molecules showed excellent correlation with the known crystal structures of ACP and ATP complexed with the kinase domains of other proteins (37–40).
FIGURE 7.
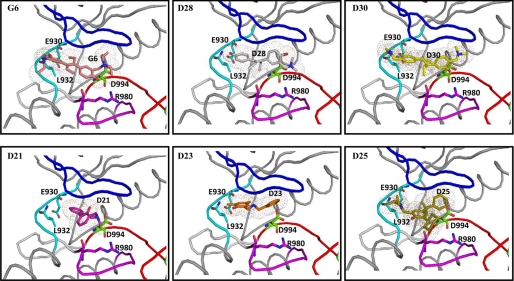
Molecular docking of G6 and its derivatives into the ATP-binding pocket of Jak2. ATP, the ATP analog ACP, G6, and each of its structurally related derivatives were docked into the ATP-binding pocket of the known crystal structure of Jak2 kinase domain (Protein Data Bank code 3E64). A, coiled representation of the structure of Jak2 with ATP and ACP docked into the ATP-binding pocket. The nucleotide binding loop is blue, the hinge region is cyan, the catalytic loop is magenta, and the activation loop is red. Residues within this pocket that are critical for interactions with ATP and other docked drugs have been represented as spheres. Phosphorylation of Tyr1007 within the activation loop is necessary for the activation of kinase activity of Jak2. ATP (yellow) and ACP (green) had strong interactions with the pocket as indicated by their highly negative GRID scores. B, G6 (pink), D28 (gray), and D30 (yellow) docked at the ATP-binding pocket of Jak2. C, D21 (orange), D23 (magenta), and D25 (green) docked at the ATP-binding pocket of Jak2. D, comparison of the docking of ATP (yellow), ATP analog ACP (green), and G6 (pink) with the crystal structure of a Jak2 inhibitor (5B3) (blue) in complex with Jak2 kinase domain showed good correlation (root mean square deviation of <2 Å) between the structures.
We found that the stilbenoids had higher binding affinities when compared with the non-stilbenoids as indicated by their more negative energy scores. G6, D28, D30, D21, D23, and D25 had energy scores of −75.46, −63.24, −72.26, −27.69, −38.90, and −48.91 kcal/mol, respectively. The docking orientations of G6, D28, and D30 into the pocket (Fig. 7B) were very similar to that of ATP/ACP (supplemental Fig. S1) and distinctly different from those of the non-stilbenoids (Fig. 7C). The docking conformation of each of the stilbenoid inhibitors into the ATP-binding pocket also correlated well (root mean square deviation of <2 Å) with the previously reported structure of another Jak2 inhibitor, 5B3, in complex with the Jak2 kinase domain (Fig. 7D).
Examination of the most favorable docking orientations for G6 and each of its derivatives showed the presence of hydrogen bonds (<3.5 Å) and van der Waal's interactions between the stilbenoid derivatives and the ATP-binding pocket of Jak2 (Fig. 8). Specifically, the stilbenoid core-containing derivatives exhibited van der Waal's interactions with many of the hydrophobic residues in the binding pocket such as Val863, Leu855, Leu983, Leu932, Tyr931, Ala880, and Val911. The stilbenoid-containing derivatives also formed hydrogen bond interactions with several important surrounding residues including Asp994, Arg980, Glu930, and Leu932. The non-stilbenoid derivatives, however, did not show any such hydrogen bond interactions with these critical residues of the Jak2 ATP-binding pocket. Overall, these data demonstrate the importance of the stilbenoid core structure for stronger docking interactions of the drug with the ATP-binding pocket of Jak2.
FIGURE 8.
Docking of G6 and its derivatives into the ATP-binding pocket of Jak2. G6 (pink) and its stilbenoid derivatives D28 (gray) and D30 (yellow) showed hydrogen bond interactions (<3.5 Å) with several critical residues (Glu930, Leu932, Arg980, and Asp994) that make up the ATP-binding pocket of Jak2. However, the nonstilbenoid derivatives D21 (magenta), D23 (orange), and D25 (yellow) show no such hydrogen bond interactions.
DISCUSSION
Somatic Jak2 mutations, such as the Jak2-V617F, which result in deregulated Jak/STAT signaling, have been identified in numerous patients with myeloproliferative neoplasms and are known to play a primary role in the pathogenesis of these diseases (7–11). Therefore, identification of novel compounds having inhibitory effects against hyperkinetic Jak2 has become an attractive therapeutic strategy in MPN. Using structure-based virtual screening, our group recently identified a novel Jak2 small molecule inhibitor called G6 (12).
In this report, we describe a structure-function correlation of this inhibitor compound. We demonstrate that it has a central stilbenoid core in its structure that is indispensable for maintaining its ability to inhibit Jak2 kinase activity. G6 and its stilbenoid core-containing derivatives (D28 and D30) effectively inhibit Jak2-V617F-mediated HEL cell proliferation in a time- and dose-dependent manner. Correspondingly, they are also capable of suppressing the phosphorylation of Jak2, STAT3, and STAT5 proteins. Furthermore, these stilbenoids significantly induce apoptosis in treated cells via the intrinsic pathway and possess the ability to block ex vivo pathologic cell growth of bone marrow cells isolated from a Jak2-V617F-positive MPN patient.
Stilbenes are a group of compounds with a wide range of diverse biological activities. Stilbenoids, such as resveratrol, piceatannol, and diethylstilbestrol, are reported to have anti-proliferative, anti-oxidative, anti-neovascularization, and tumor-suppressive effects (13–15). Resveratrol has beneficial cardiovascular effects (41), whereas diethylstilbestrol is known to have estrogen activity (42). Piceatannol, a naturally occurring phenolic stilbenoid, is the only stilbenoid that is a known protein-tyrosine kinase inhibitor. It inhibits LMP2A, a viral tyrosine kinase implicated in diseases associated with the Epstein-Barr virus as well as the nonreceptor tyrosine kinases Syk and Lck (43, 44). Here, we report that G6, which is also a stilbene, has anti-Jak2 tyrosine kinase activity. More importantly, the trans-stilbenoid core identified in all of the active compounds is now being subjected to bioisosteric replacements (45) that would result in new Jak2 chemotypes with improved druglike properties.
Computational docking of G6 and its structurally related derivatives into the ATP-binding pocket of Jak2 revealed important interactions between the inhibitors and specific residues within Jak2. For example, Glu930 and Leu932 are part of the important hinge region of the Jak2 kinase domain and are known to be involved in adenine binding. We found that these residues were occupied by the stilbenoid-containing compounds but not the non-stilbenoids. Another amino acid that interacted with the docked stilbenoid inhibitors was Asp994, located in the highly conserved DFG motif. This motif is part of the critical activation loop of the kinase, and our group was the first to identify the importance of this residue as it relates to Jak2 kinase inhibition (12). We show here that the stilbenoid core-bearing inhibitors also had a hydrogen bond interaction with Arg980, which is a part of the catalytic loop and is known to be one of the residues involved in the coordination of magnesium ions (35, 36). Thus, the ability of the stilbenoid derivatives to simultaneously interact, in silico, with the hinge region, the DFG motif, and the catalytic loop suggests that they may interfere with both ATP-binding function and activation loop phosphorylation, thereby making them potent Jak2 inhibitors. In summary, our data collectively show that the central stilbenoid core structure is indispensable for maintaining anti-Jak2 kinase activity of the small molecule inhibitor G6.
Supplementary Material
Acknowledgments
We thank Nicholas Figueroa and Steve McClellan for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-HL67277. This work was also supported by a Biomedical Research Support Program for Medical Schools Award to the University of Florida College of Medicine by the Howard Hughes Medical Institute, a University of Florida Opportunity Fund Award, and a University of Florida/Moffitt Cancer Center Collaborative Initiative Award.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- MPN
- myeloproliferative neoplasm
- HEL
- human erythroleukemia
- DMSO
- dimethyl sulfoxide
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy-phenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- ACP
- adenosine-5′-[β,γ-methylene] triphosphate.
REFERENCES
- 1.Parganas E., Wang D., Stravopodis D., Topham D. J., Marine J. C., Teglund S., Vanin E. F., Bodner S., Colamonici O. R., van Deursen J. M., Grosveld G., Ihle J. N. (1998) Cell 93, 385–395 [DOI] [PubMed] [Google Scholar]
- 2.Neubauer H., Cumano A., Müller M., Wu H., Huffstadt U., Pfeffer K. (1998) Cell 93, 397–409 [DOI] [PubMed] [Google Scholar]
- 3.Peeters P., Raynaud SD., Cools J., Wlodarska I., Grosgeorge J., Philip P., Monpoux F., Van Rompaey L., Baens M., Van den Berghe H., Marynen P. (1997) Blood 90, 2535–2540 [PubMed] [Google Scholar]
- 4.Joos S., Granzow M., Holtgreve-Grez H., Siebert R., Harder L., Martín-Subero J. I., Wolf J., Adamowicz M., Barth T. F., Lichter P., Jauch A. (2003) Int. J. Cancer 103, 489–495 [DOI] [PubMed] [Google Scholar]
- 5.Reiter A., Walz C., Watmore A., Schoch C., Blau I., Schlegelberger B., Berger U., Telford N., Aruliah S., Yin J. A., Vanstraelen D., Barker H. F., Taylor P. C., O'Driscoll A., Benedetti F., Rudolph C., Kolb H. J., Hochhaus A., Hehlmann R., Chase A., Cross N. C. (2005) Cancer Res. 65, 2662–2667 [DOI] [PubMed] [Google Scholar]
- 6.Griesinger F., Hennig H., Hillmer F., Podleschny M., Steffens R., Pies A., Wörmann B., Haase D., Bohlander S. K. (2005) Genes Chromosomes Cancer 44, 329–333 [DOI] [PubMed] [Google Scholar]
- 7.Baxter E. J., Scott L. M., Campbell P. J., East C., Fourouclas N., Swanton S., Vassiliou G. S., Bench A. J., Boyd E. M., Curtin N., Scott M. A., Erber W. N., Green A. R. (2005) Lancet 365, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 8.James C., Ugo V., Le Couédic J. P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., Villeval J. L., Constantinescu S. N., Casadevall N., Vainchenker W. (2005) Nature 434, 1144–1148 [DOI] [PubMed] [Google Scholar]
- 9.Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C. (2005) N. Engl. J. Med. 352, 1779–1790 [DOI] [PubMed] [Google Scholar]
- 10.Levine R. L., Wadleigh M., Cools J., Ebert B. L., Wernig G., Huntly B. J., Boggon T. J., Wlodarska I., Clark J. J., Moore S., Adelsperger J., Koo S., Lee J. C., Gabriel S., Mercher T., D'Andrea A., Fröhling S., Döhner K., Marynen P., Vandenberghe P., Mesa R. A., Tefferi A., Griffin J. D., Eck M. J., Sellers W. R., Meyerson M., Golub T. R., Lee S. J., Gilliland D. G. (2005) Cancer Cell 7, 387–397 [DOI] [PubMed] [Google Scholar]
- 11.Zhao R., Xing S., Li Z., Fu X., Li Q., Krantz S. B., Zhao Z. J. (2005) J. Biol. Chem. 280, 22788–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss R., Polgár T., Kirabo A., Sayyah J., Figueroa N. C., List A. F., Sokol L., Zuckerman K. S., Gali M., Bisht K. S., Sayeski P. P., Keseru G. M. (2009) Bioorg. Med. Chem. Lett. 19, 3598–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Corre L., Chalabi N., Delort L., Bignon Y. J., Bernard-Gallon D. J. (2005) Mol. Nutr. Food Res. 49, 462–471 [DOI] [PubMed] [Google Scholar]
- 14.Larrosa M., Tomás-Barberán F. A., Espín J. C. (2004) Eur. J. Nutr. 43, 275–284 [DOI] [PubMed] [Google Scholar]
- 15.Kimura Y. (2005) In Vivo 19, 37–60 [PubMed] [Google Scholar]
- 16.Gosslau A., Chen M., Ho C. T., Chen K. Y. (2005) Br. J. Cancer 92, 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belleri M., Ribatti D., Nicoli S., Cotelli F., Forti L., Vannini V., Stivala L. A., Presta M. (2005) Mol. Pharmacol. 67, 1451–1459 [DOI] [PubMed] [Google Scholar]
- 18.Pradhan A., Lambert Q. T., Griner L. N., Reuther G. W. (2010) J. Biol. Chem. 285, 16651–16663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. (1982) J. Mol. Biol. 161, 269–288 [DOI] [PubMed] [Google Scholar]
- 20.Lang P. T., Brozell S. R., Mukherjee S., Pettersen E. F., Meng E. C., Thomas V., Rizzo R. C., Case D. A., James T. L., Kuntz I. D. (2009) RNA 15, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antonysamy S., Hirst G., Park F., Sprengeler P., Stappenbeck F., Steensma R., Wilson M., Wong M. (2009) Bioorg. Med. Chem. Lett. 19, 279–282 [DOI] [PubMed] [Google Scholar]
- 22.Schüttelkopf A. W., van Aalten D. M. (2004) Acta. Crystallogr D. Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 23.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. (2004) Acta. Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLano W. (2002) PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 27.Martin P., Papayannopoulou T. (1982) Science 216, 1233–1235 [DOI] [PubMed] [Google Scholar]
- 28.Quentmeier H., MacLeod R. A., Zaborski M., Drexler H. G. (2006) Leukemia 20, 471–476 [DOI] [PubMed] [Google Scholar]
- 29.Walz C., Crowley B. J., Hudon H. E., Gramlich J. L., Neuberg D. S., Podar K., Griffin J. D., Sattler M. (2006) J. Biol. Chem. 281, 18177–18183 [DOI] [PubMed] [Google Scholar]
- 30.Bromberg J. (2002) J. Clin. Invest. 109, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker S. J., Rane S. G., Reddy E. P. (2007) Oncogene. 26, 6724–6737 [DOI] [PubMed] [Google Scholar]
- 32.Socolovsky M., Fallon A. E., Wang S., Brugnara C., Lodish H. F. (1999) Cell 98, 181–191 [DOI] [PubMed] [Google Scholar]
- 33.Antonsson B., Martinou J. C. (2000) Exp. Cell Res. 256, 50–57 [DOI] [PubMed] [Google Scholar]
- 34.Gross A., Yin X. M., Wang K., Wei M. C., Jockel J., Milliman C., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. (1999) J. Biol. Chem. 274, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 35.Lindauer K., Loerting T., Liedl K. R., Kroemer R. T. (2001) Protein Eng. 14, 27–37 [DOI] [PubMed] [Google Scholar]
- 36.Lucet I. S., Fantino E., Styles M., Bamert R., Patel O., Broughton S. E., Walter M., Burns C. J., Treutlein H., Wilks A. F., Rossjohn J. (2006) Blood 107, 176–183 [DOI] [PubMed] [Google Scholar]
- 37.Biondi R. M., Komander D., Thomas C. C., Lizcano J. M., Deak M., Alessi D. R., van Aalten D. M. (2002) EMBO J. 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Favelyukis S., Till J. H., Hubbard S. R., Miller W. T. (2001) Nat. Struct. Biol. 8, 1058–1063 [DOI] [PubMed] [Google Scholar]
- 39.Lougheed J. C., Chen R. H., Mak P., Stout T. J. (2004) J. Biol. Chem. 279, 44039–44045 [DOI] [PubMed] [Google Scholar]
- 40.Till J. H., Ablooglu A. J., Frankel M., Bishop S. M., Kohanski R. A., Hubbard S. R. (2001) J. Biol. Chem. 276, 10049–10055 [DOI] [PubMed] [Google Scholar]
- 41.Pace-Asciak C. R., Hahn S., Diamandis E. P., Soleas G., Goldberg D. M. (1995) Clin. Chim. Acta. 235, 207–219 [DOI] [PubMed] [Google Scholar]
- 42.Dodds E. C., Goldberg L., Lawson W., Robinson R. (1938) Nature 141, 247–248 [Google Scholar]
- 43.Geahlen R. L., McLaughlin J. L. (1989) Biochem. Biophys. Res. Commun. 165, 241–245 [DOI] [PubMed] [Google Scholar]
- 44.Swanson-Mungerson M., Ikeda M., Lev L., Longnecker R., Portis T. (2003) J. Antimicrob. Chemother. 52, 152–154 [DOI] [PubMed] [Google Scholar]
- 45.Akwabi-Ameyaw A., Bass J. Y., Caldwell R. D., Caravella J. A., Chen L., Creech K. L., Deaton D. N., Jones S. A., Kaldor I., Liu Y., Madauss K. P., Marr H. B., McFadyen R. B., Miller A. B., Iii F. N., Parks D. J., Spearing P. K., Todd D., Williams S. P., Wisely G. B. (2008) Bioorg. Med. Chem. Lett. 18, 4339–4343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.