Abstract
HIV-1 non-coding exon 3 can either be spliced to exons 4, 4a, 4b, 4c, and 5 to generate tat, rev, and nef mRNAs or remain unspliced to produce the 13a7 vpr mRNA. Here we show that serine- and arginine-rich proteins 55 and 75 (SRp55 and SRp75) inhibit splicing from the 5′-splice site of exon 3 thereby causing an accumulation of the partially unspliced 13a7 vpr mRNA. In contrast, serine- and arginine-rich protein 40 (SRp40) induces splicing from exon 3 to exon 4, thereby promoting the production of the 1347 tat mRNA. We demonstrate that SRp55 stimulates vpr mRNA production by interacting with the previously identified HIV-1 splicing enhancer named GAR and inhibiting its function. This inhibition requires both serine arginine-rich and RNA-binding domains of SRp55, indicating that production of HIV-1 vpr mRNA depends on the interaction of SRp55 with an unknown factor.
Keywords: RNA/Splicing, Viruses/HIV, Gene Regulation, Human Immunodeficiency Virus, RNA Processing, SR Protein, SRp55, Alternative Splicing, vpr
Introduction
To produce all the mRNAs that are needed for HIV-1 to be infectious it uses alternative splicing. The full-length 9-kb transcript contains several splice sites, including four 5′-splice sites, also called splice donors (SD1–4)2 and eight 3′-splice sites, also called splice acceptors (SA2–3, -4a–c, -5, and -7) (supplemental Fig. 1A), which can all be used in different combinations to create more than 35 differently spliced mRNAs (supplemental Fig. 1, B and C) (1–6). Cloning and expression of individual HIV-1 mRNAs have revealed that mRNAs spliced to SA3 produce Vpr, those spliced to SA4 produce Tat, those spliced to SA4a and -4b produce Rev and Nef, and those spliced to SA5 produce Nef (1, 2, 7, 8). mRNAs spliced to SA2 are believed to produce Vif (5, 7). Two additional mRNAs that are believed to produce Rev and Nef are spliced to SA4c or to SA7, respectively (4). To produce the correct amounts of all mRNAs, the splice acceptors in HIV-1 are sub-optimal because of short and interrupted polypyrimidine tracts, non-canonical branch points, and inhibitory sequences that down-regulate the usage of several splice sites (6, 9–13).
Enhancer or silencer sequences regulate alternative splicing by up-regulating or down-regulating the usage of a splice site (14). Generally, the family of serine- and arginine-rich proteins (SR proteins) target enhancer sequences, and silencer sequences are targeted by heterogeneous nuclear ribonucleoproteins (15). There are several enhancers and silencers on HIV-1 pre-mRNA (16). A silencer in exon 3 called ESSV inhibits the vpr splice acceptor SA3 (10, 17). Two silencers in exon 4, named ESS2 and ESS2p, inhibit splicing into exon 4 thereby inhibiting the production of tat mRNA (18, 19). SA7 is used by all mRNAs in the 2-kb class. In exon 7 a silencer called ESS3 blocks U2 small nuclear ribonucleoprotein, thereby suppressing SA7 (19–21). An intronic silencer that blocks SA7 is located in the intron, directly upstream of exon 7 (22). An enhancer element called ESE3 that activates SA7 is located close to ESS3 in exon 7 (9, 21, 23). An enhancer in exon 4 has been named ESE2, because it attenuates ESS2 and thereby enhances splicing into SA4 (24). In exon 5, an enhancer named GAR stimulates the usage of highly active SA5 and adjacent 3′-splice sites as well as SD5, which is used for production of the entire small mRNA size class (25, 26). HIV-1 vpr mRNA 13a7 contains an intron between exon 3 and exon 4 and is, therefore, partially spliced. Little is known about the regulation of vpr mRNA production.
The human SR proteins currently contain at least 10 known members, including SRp20, SRp30a–c, SRp40, SRp46, SRp54, SRp55, SRp75, and 9G8. SR proteins regulate alternative splicing by stimulating or repressing the usage of splice sites (27).
The complex splicing of the HIV-1 pre-mRNA indicates that HIV-1 is dependent on multiple splicing factors to generate all mRNAs. Recent studies have shown that the relative concentrations of SR proteins are of paramount importance for production of sufficient quantities of the various HIV-1 mRNAs (16). SR proteins have also been shown to be potential targets for novel therapy against HIV-1 infection (28). It is therefore of interest to identify the cellular splicing factors that regulate HIV-1 mRNA splicing.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
To construct the subgenomic plasmid pDP (Fig. 2A) HIV-1 sequences from position 246–1571 (all numbers refer to the HXB2R sequence) were PCR-amplified using oligonucleotides BSS and APAA (Table 1) and cloned into pCR2.1-TOPO (Invitrogen) and subsequently inserted into pNL13aE (7) using BssHII and SalI, resulting in pNL13aEBA. Nucleotide 3516–5348 was PCR-amplified using oligonucleotides MLUS and SALA (Table 1) and subcloned into pCR2.1-TOPO. This fragment was cloned into pNL13aEBA using MluI and SalI, creating pDP (Fig. 2A). pNL13a7 has been described previously (7). pNL13a7dG was constructed by PCR mutagenesis (246–5522 and 5556–8031) using oligonucleotides BSS, dGARas, dGARs, and BAMA (Table 1) and cloned into pNL13a7 (7) using SalI and BamHI. pNL13a7d was generated by PCR amplification of nucleotides 5393–8031 with oligonucleotides Downsa4s and BAMA (Table 1) and cloning into pCR2.1-TOPO. The fragment was excised with SalI and BamHI and cloned into pNL13a7. Plasmid pNL13a7d4 was constructed by PCR amplification of nucleotide 5480–8031 using oligonucleotides 5480s and BAMA (Table 1). The PCR fragment was subcloned into pCR2.1-TOPO and subsequently inserted into pNL13a7 using SalI and BamHI. pNL13a7sd1 was constructed by PCR amplification of nucleotides 245–569 using oligonucleotides BSS and sd1as (Table 1). The PCR fragment was subcloned into pCR2.1-TOPO and subsequently inserted into pNL13a7 using BssHII and EcoRI. Transfer of a SalI-BamHI fragment from pNL13a7dG to pNL13a7sd1 created pNL13a7sd1dG. To generate pNL13a7de3 PCR mutagenesis was performed with oligonucleotides exon1s, dexon3as, dexon3s, and sd5as (1–286 and 5007–5591) (Table 1). The mutated fragment was inserted into pNL13a7 using BssHII and SalI. Plasmid pNL13a7di3 was constructed by first PCR-amplifying nucleotides 245–289 and 4936–5055 using oligonucleotides BSS and di3sd3as (Table 1) followed by cloning into pNL13a7 (7) using BssHII and EcoRI. The plasmids pT1sd and pT1sde have been described previously (29). Plasmid pT1–5/GAR was generated by first PCR-amplifying HIV-1 sequences between nucleotides 5523 and 5591 using oligonucleotides AnnealGARBss/XbaS and AnnealGARBss/XbaAS (Table 1). The PCR fragment was inserted into pT1sd using BssHII and XbaI. Plasmid SRp55dRS was generated by PCR amplification of nucleotides 1–543 from SRp55 cDNA using oligonucleotides SRp55s and SRp55RRMAS (Table 1). The PCR fragment was cleaved with BssHII and XhoI and cloned into plasmid pCL086 (30). Recombinant plasmid gst-SRp55 was generated by first PCR-amplifying sequences from SRp55 cDNA using oligonucleotides SRp55gsts and SRp55gstas (Table 1). The PCR fragment was cleaved with BamHI and EcoRI and subcloned into pGEX-2T (Amersham Biosciences). SR protein expression plasmids have been described previously (31–33).
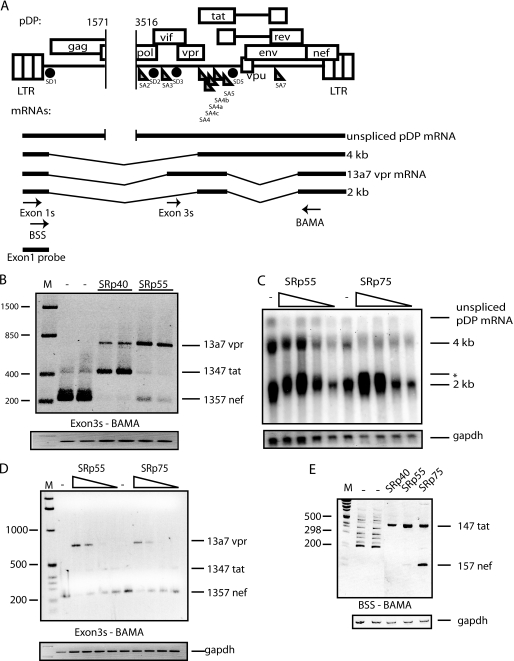
FIGURE 2.
A, schematic representation of the HIV-1 subgenomic plasmid pDP, which is derived from pNL43 (48) and contains a deletion between nucleotides 1571 and 3516 (numbers refer to HXB2R sequence). The three HIV-1 mRNA size classes, and vpr mRNA 13a7, are indicated below plasmid pDP. Arrows indicate oligonucleotides used for RT-PCR. The Northern blot probe is indicated. B, RT-PCR on cytoplasmic RNA extracted from HeLa cells transfected with pDP in the absence or presence of pCMVSRp40 or pCMVSRp55. Duplicate transfections are shown. The RT-PCR was performed with the oligonucleotides Exon3s and BAMA (A). The different bands represent the indicated mRNAs. C, Northern blot of cytoplasmic RNA extracted from HeLa cells transfected with pDP in the absence or presence of increasing concentrations of pCMVSRp55 or pCMVSRp75. The mRNA size classes are indicated. *, the shift in migration of 2-kb mRNAs. D, the RNAs in C were used for RT-PCR with oligonucleotides Exon3s and BAMA (A) that detect mRNAs containing exon 3. E, RT-PCR on cytoplasmic RNA extracted from HeLa cells transfected with pDP in the absence or presence of pCMVSRp40, pCMVSRp55, or pCMVSRp75. The RT-PCR was performed with the oligonucleotides BSS and BAMA (A) detecting all HIV-1 mRNAs. The bands representing mRNAs are indicated. M represents the molecular weight marker.
TABLE 1.
Oligonucleotides used for RT-PCR and plasmid construction
| Name | Sequence (5′→3′) |
|---|---|
| BSS | ggcttgctgaagcgcgcacggcaagagg |
| BAMA | gccaaggatccgttcactaatcgaatgg |
| MLUS | ggggcccggacgcgtatcagaagactgagttacaa |
| SALA | tattctgctatgtcgacaccc |
| APAA | ggtcgacggacgcgtgccctttttcctaggggccctgc |
| dGARs | ttaggcatctcctatggcaggaacagtcagactcatcaagcttctctatc |
| dGARas | gaagcttgatgagtctgactgttcctgccataggagatgcctaaggcttt |
| Downsa4s | agtcgacctagactagagccctggaag |
| 5480s | ggtcgacaagtttgtttcataacaaaagccttag |
| sd1as | ccgaattcacgcgtaatgatctaagttcttctgatcc |
| Exon1s | ggtctctctggttagaccagat |
| Exon1as | cagtcgccgcccctcgcctct |
| Exon3s | acatagttagccctaggtgtgaatatc |
| dexon3s | agaggcgaggggcgccgaaaggtaggatctctacaatacttgg |
| dexon3as | ttgtagagatcctacctttcgccgcccctcgcctcttgcc |
| sd5as | tgctttgatagagaagcttgat |
| di3sd3as | gaattcttggtgttattaatgctgctag |
| SRp55s | aagcgcgcaccatgccacgagtctacatagga |
| SRp55RRMas | ggctcgagttaatcttcaataagcctaatatt |
| T7gars | tctaatacgactcactataggggaagaagcggagacagcgacg |
| T7garas | tgatgagctcttcgtcgctgtctccgcttc |
| AnnealGARBss/XbaS | cgcgcgaagaagcggagacagcgacgaacagctcatcagaacagtcagactcatcaagcttctctatcaaagcat |
| AnnealGARBss/XbaAS | ctagatgctttgatagagaagcttgatgagtctgactgttctgatgagctcttcgtcgctgtctccgcttcttcg |
| SRp55gsts | ggggatccatgccgcgcgtctacataggac |
| SRp55gstas | gggaattcttaatctctggaactcgacctg |
| 757s | gtcgacggtatcgatcggttgtgcgtacaaagcacacacg |
| L1AM | cgctgggcagccacaggc |
| gapdhS | aggtcggagtcaacggatttgg |
| gapdhAS | acagtcttctgggtggcagtgatg |
Transfections
Transfections of plasmid DNA into HeLa cells were performed according to the FuGENE 6 method (Roche Molecular Biochemicals). When nothing else is stated, 1 μg of each plasmid was transfected into 60-mm diameter plates containing subconfluent HeLa cells. Transfected cells were harvested 24 h post-transfection.
RNA Extraction, Northern Blotting, and Radiolabeled DNA Probe Synthesis
Cytoplasmic RNA extraction was performed as previously described (34). Northern blot analysis was performed by size separation of 10 μg of cytoplasmic RNA on a 1.5% agarose gels containing 2.2 m formaldehyde followed by transfer of RNA to nitrocellulose filter and hybridization. The exon1 probe was generated by PCR amplification of nucleotides 1–289 of the HIV-1 genome (HXB2R) with oligonucleotides Exon1s and Exon1as (Table 1). The probe was [α-32P]dCTP-labeled by random priming with a Decaprime kit (Ambion Inc.) according to the manufacturer's instructions.
RT-PCR
Cytoplasmic RNA was reverse-transcribed at 37 °C for 50 min using random hexamers as described previously (35). 200 ng of RNA was used for each RT-PCR reaction in a total volume of 20 μl. A total of 1 μl of cDNA product was PCR-amplified using the oligonucleotides indicated for each experiment.
siRNA Analysis
Knockdown of protein was performed by using a pool of four siRNAs targeting SRp55 and four siRNAs targeting SRp75 (siGENOME SMARTpool, Dharmacon). A final concentration of 133 nm of each siRNA pool was reverse-transfected into HeLa cells at the time of passage. 48 h later, siRNA-transfected cells growing in 35-mm plates were transfected with 0.5 μg of pNL13a7 plasmid DNA as described in the previous section. Cells were harvested 24 h post plasmid transfection and subjected to RNA extraction and analysis.
Western Blotting
Western blotting was performed as previously described (36).
gst-SRp55 Purification
Purified gst-SRp55 was prepared by using glutathione-Sepharose (GS) beads as specified by the manufacturer (Amersham Biosciences). Briefly, a culture of Escherichia coli BL21 transformed with pGEX-SRp55 was induced with 0.1 mm isopropyl-β-d-thiogalactopyranoside for 2 h. The bacteria were pelleted and resuspended in ice-cold phosphate-buffered saline and lysed by sonication followed by incubation in 1% Triton X-100. Debris was pelleted, and gst-SRp55 was purified using the GS-beads.
In Vitro Transcription and RNA Gel Shift Assay
In vitro transcription was performed on a PCR fragment containing bacteriophage T7 promoter upstream of GAR using T7 RNA polymerase (Ambion Inc.). The control RNA was transcribed from a T7 promoter-driven plasmid encoding a sequence from human papilloma virus type 1 named AUM/UM (34). The RNAs were labeled with [α-32P]CTP, and competitor GAR RNA was synthesized in the absence of radiolabeled nucleotide. RNA gel-shift assays were performed with gst-SRp55 or gst at room temperature in 20 μl of binding buffer (120 mm KCl, 10 mm HEPES (pH 7.6), 3 mm MgCl2, 1 mm dithiothreitol, and 5% (v/v) glycerol). After 20 min the RNA-protein complexes were resolved on non-denaturing 7.5% polyacrylamide gels (acrylamide-bisacrylamide, 29:1). The complexes were visualized by autoradiography.
RESULTS
SRp55 and SRp75 Inhibit Splicing from Exon 3 and Induce Production of 13a7 vpr mRNA
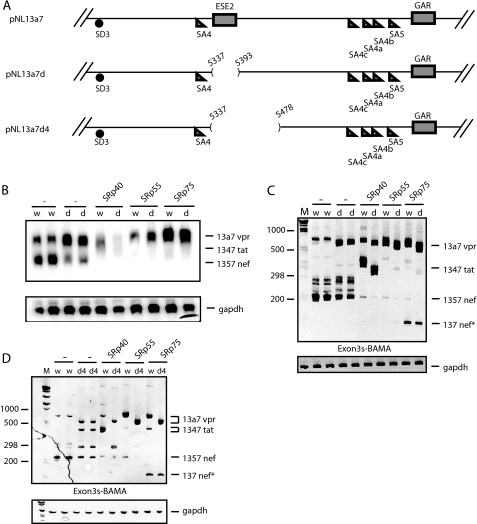
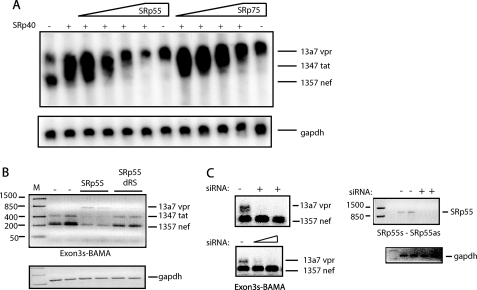
The HIV-1 vpr mRNA is encoded by mRNAs containing either exon 3a or 3aE (supplemental Fig. 1, A and B). vpr mRNA 13a7 is special in that it is incompletely spliced, but lacks the RRE and thus cannot be induced by Rev (supplemental Fig. 1C). To identify proteins that regulate vpr expression, we used a cDNA plasmid named pNL13a7 that expresses the 13a7 vpr mRNA (7) (Fig. 1A). However, the majority of this mRNA is further spliced from the unique 5′-splice site of exon 3 to exons 4, 4a, 4b, 4c, or 5 (7) (Fig. 1A). Northern blotting on cytoplasmic RNA extracted from cells transfected with pNL13a7 and SR protein expression plasmids revealed that SRp40 shifted the splicing of mRNAs generated by pNL13a7 to mRNAs migrating higher up in the gel (Fig. 1B). SRp55 and SRp75 totally inhibited splicing and promoted production of high levels of unspliced 13a7 vpr mRNA in the cytoplasm (Fig. 1B). In contrast, SRp20, SRp30c, and SC35 had an unspecific inhibitory effect on expression of the HIV-1 mRNAs (Fig. 1, B and C). RT-PCR with oligonucleotides Exon3s and BAMA (Fig. 1A) showed that SRp40 induced production of tat mRNAs spliced from exon 3 to exon 4 (Fig. 1C) and that SRp55 and SRp75 inhibited splicing. The latter proteins increased the levels of unspliced 13a7 vpr mRNA at the expense of all spliced HIV-1 mRNAs (Fig. 1C). Taken together, the results in Fig. 1, B and C, demonstrate that SRp40 enhances splicing from exon 3 to exon 4, whereas SRp55 and SRp75 inhibit splicing from exon 3 and promote production of 13a7 vpr mRNA. Western blot analysis showed expression of SR protein from the SR protein expression plasmids (Fig. 1D).
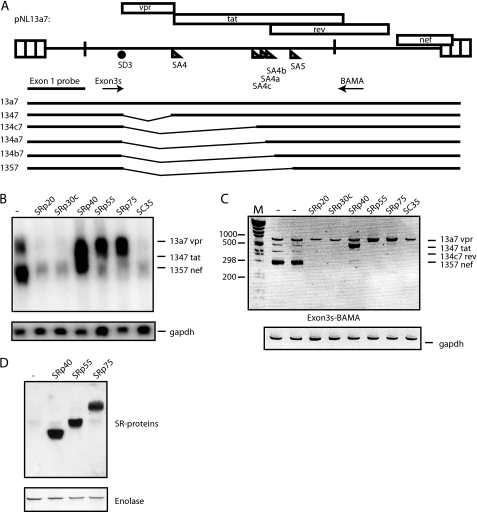
FIGURE 1.
A, schematic representation of the HIV-1 subgenomic plasmid pNL13a7 (7). The mRNAs produced from the plasmid are indicated below as well as the oligonucleotides used for RT-PCR and probe used for northern blotting. B, Northern blot of total cytoplasmic RNA extracted from HeLa cells transfected with pNL13a7 (7) in the absence or presence of CMV promoter-driven plasmids expressing SRp20, SRp30c, SRp40, SRp55, SRp75, or SC35. The blot was probed with exon 1 probe (A) detecting all HIV-1 mRNAs. Various mRNAs detected by the probe are indicated. C, RT-PCR on the RNA used in B performed with oligonucleotides Exon3s and BAMA (A). Bands representing mRNAs are indicated. M represents the molecular weight marker. D, SR proteins were detected by Western blot analysis using monoclonal antibody 1H4. Monoclonal antibody detection of enolase was used as loading control.
Overexpression of SRp55 and SRp75 Induces Production of 13a7 vpr mRNA in the Context of the Sub-genomic HIV-1 Plasmid pDP
We wished to study the effect of SR proteins on vpr mRNA in the context of all HIV-1 splice sites present. We constructed a plasmid named pDP (Fig. 2A) that contains the entire HIV-1 genome except for a 2-kb deletion in the pol gene. Oligonucleotide Exon3s was used in combination with oligonucleotide BAMA to specifically detect mRNAs that contain exon 3 and 3a (supplemental Fig. 1, C and A). The results revealed that SRp40 redirected splicing from production of primarily 1357 mRNA to 1347 tat mRNA, as expected (Fig. 2B). In contrast, SRp55 inhibited splicing from exon 3 and induced production of 13a7 vpr mRNA from pDP (Fig. 2B). Northern blotting of HIV-1 mRNAs produced from pDP in the absence or presence of serially diluted SRp55 revealed that high levels of SRp55 caused production of mRNAs migrating higher than the 2-kb mRNAs detected in the absence of exogenous SRp55 (Fig. 2C). SRp75 had a similar effect on the mRNAs produced from pDP (Fig. 2C). This was confirmed by RT-PCR analysis of the same RNA samples with the exon 3-specific oligonucleotides Exon3s and BAMA (Fig. 2D). These experiments demonstrated a robust increase in production of 13a7 vpr mRNA in an SRp55- or SRp75-dose-dependent manner (Fig. 2D).
Induction of 13a7 vpr mRNA by SRp55 and SRp75 Is Dependent on SD3 in Exon 3
To investigate if SRp55 and SRp75 inhibited SD3 specifically, we replaced splice donor 3 (SD3) in pNL13a7 with splice donor 1 (SD1) from pDP, resulting in pNL13a7sd1 (Fig. 3A). SRp55 and SRp75 did not induce production of unspliced vpr mRNA from plasmid pNL13a7sd1 with SD1, whereas high levels of unspliced 13a7 vpr mRNA were induced from wild-type plasmid pNL13a7 with SD3, as expected (Fig. 3B). RT-PCR confirmed that SRp55 and SRp75 primarily induced production of unspliced 13a7 vpr mRNA in the presence of SD3 (Fig. 3C), whereas SRp55 and SRp75 induced production of 1347 tat mRNA in the presence of SD1 (Fig. 3C). Overexpression of SRp75 also strongly induced splicing to a cryptic splice acceptor in exon 5, in the presence of SD1 (Fig. 3C). SRp40 induced production of tat mRNA independently of SD1 or SD3 and did not inhibit splicing to produce vpr mRNA 13a7 as shown by RT-PCR (Fig. 3D). In conclusion, SRp55 and SRp75 stimulate splicing from SD1 to SA4 to generate 147 tat mRNA and inhibit splicing from SD3 to generate 13a7 vpr mRNAs. To confirm that SRp55 and SRp75 had different effects on SD1 and SD3 also in the context of the subgenomic HIV-1 expression plasmid pDP, in which all HIV-1 splice sites are present, we analyzed mRNAs produced from pDP in the absence or presence of SRp40, SRp55, and SRp75 with primers that were specific for mRNAs spliced at SD1 (BSS and BAMA) and with primers that are specific for mRNAs using SD3 (Exon3s and BAMA). The locations of the primers are shown in Fig. 2A. The results shown in Fig. 3 predicted that SRp40 should enhance splicing to exon 4 from both SD1 and SD3 to produce 147 and 1347 tat mRNAs, respectively. RT-PCR with primer pairs Exon3s and BAMA revealed that SRp40 induced splicing from SD3 to exon 4 to produce 1347 tat mRNA (Fig. 2B), and RT-PCR with primers BSS and BAMA revealed that SRp40 induced splicing from SD1 to exon 4 to produce 147 tat mRNA (Fig. 2E).
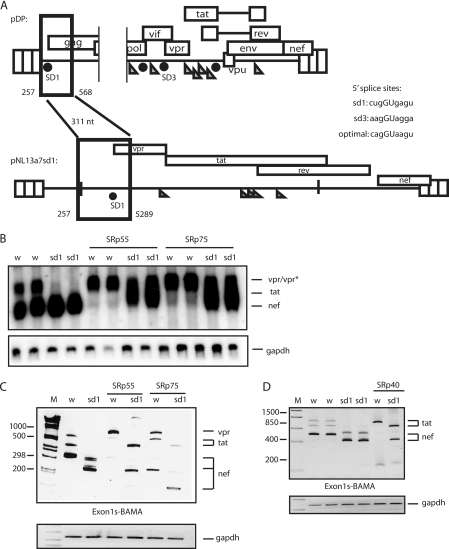
FIGURE 3.
A, schematic representation of the HIV-1 subgenomic plasmids pDP and pNL13a7sd1. Nucleotide positions of the deletions and insertions are indicated. The sequences of the different splice donors SD1 and SD3 are shown and compared with an optimal U1 snRNA binding site. B, Northern blot of cytoplasmic RNA extracted from HeLa cells transfected with pNL13a7 (w) (7) or pNL13a7sd1 (sd1) in the absence or presence of CMV promoter-driven plasmids expressing SRp55 or SRp75. The blot was probed with exon 1 probe (Fig. 2A), detecting all HIV-1 mRNAs. Identified mRNAs are indicated. C, the RNAs in B were subjected to RT-PCR with oligonucleotides Exon1s and BAMA (Fig. 1A) detecting all mRNAs. Bands representing mRNAs are indicated. M represents the molecular weight marker. Gapdh amplification was used as a control. D, RT-PCR performed on cytoplasmic RNA extracted from HeLa cells transfected with HIV-1 subgenomic plasmids pNL13a7 (w) or pNL13a7sd1 (sd1) in the absence or presence of CMV promoter-driven plasmid-expressing SRp40. Oligonucleotides Exon1s and BAMA (Fig. 1A) were used to detect all HIV-1 mRNAs. The bands representing mRNAs are indicated. M represents the molecular weight marker.
The results shown in Fig. 3 also predicted that SRp55 and SRp75 should enhance splicing to exon 4 from SD1 to produce 147 tat mRNAs, but should inhibit splicing from SD3, to generate the 13a7 vpr mRNA. This was indeed the case, as RT-PCR with primer pairs Exon3s and BAMA revealed that SRp55 and SRp75 inhibited splicing from SD3 to exon 4 to produce 13a7 vpr mRNA (Fig. 2, B and D), whereas RT-PCR with primers BSS and BAMA revealed that SRp55 and SRp75 induced splicing from SD1 to exon 4 to produce 147 tat mRNA (Fig. 2E). We concluded that high levels of SRp55 and SRp75 enhanced splicing from SD1 to exon 4 and inhibited splicing from SD3 to induce production of 13a7 vpr mRNA.
Inhibition of the 5′-Splice Site of Exon 3 by SRp55 and SRp75 Is Not Dependent on Sequences in Exon 3 or Intron 3
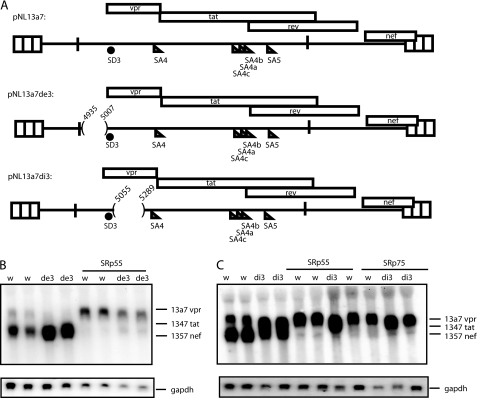
To identify HIV-1 sequences that were targeted by SRp55 and SRp75, deletions were introduced in exon 3 or intron 3 (sequences between SD3 and SA4) in pNL13a7 (Fig. 4A). Overexpression of SRp55 and SRp75 induced production of unspliced 13a7 vpr mRNA in the absence of exon 3 sequences (Fig. 4B) or intron 3 sequences (Fig. 4C). However, the inhibitory effect of SRp75 on vpr mRNA splicing was lower in the absence of exon 3 (data not shown). In addition, we observed that the deletion in exon 3 increased splicing efficiency (Fig. 4B), probably due to deletion of previously described splicing inhibitory sequences (10, 17). In contrast, the deletion in intron 3 reduced splicing efficiency (Fig. 4C), suggesting that splicing regulatory elements may be located downstream of exon 3. We concluded that SRp55 and SRp75 targeted other HIV-1 sequences than those of exon 3 or the intronic region between exon 3 and exon 4.
FIGURE 4.
A, schematic representation of HIV-1 subgenomic plasmids pNL13a7 (7), pNL13a7de3, and pNL13a7di3. Nucleotide positions of deletions are indicated. B and C, Northern blots with RNA extracted from HeLa cells transfected with pNL13a7 (w), pNL13a7de3 (de3), or pNL13a7di3 (di3) in the absence or presence of CMV promoter-driven plasmids expressing SRp55 or SRp75. The blots were probed with exon 1 probe (Fig. 1A) detecting all HIV-1 mRNAs. Detected mRNAs are indicated.
Deletions in Exon 4, Upstream of Exon 5, Do Not Affect the Ability of SRp55 and SRp75 to Induce Production of Unspliced 13a7 vpr mRNA, But Affect the Ability of SRp40 to Induce Tat mRNA Splicing
A region in exon 4 described as a splicing enhancer (ESE2) (24) was deleted in pNL13a7, resulting in pNL13a7d (Fig. 5A). The deletion of ESE2 was analyzed by Northern blotting and RT-PCR, and the results revealed that deletion of ESE2 reduced splicing efficiency (Fig. 5, B and C) as expected. Northern blot experiments also showed that deletion of ESE2 in exon 4 did not affect the ability of SRp55 and SRp75 to induce production of 13a7 vpr mRNA (Fig. 5B). This finding was confirmed by RT-PCR (Fig. 5C). In addition, the ability of SRp40 to redirect splicing to exon 4 was largely independent of ESE2 (Fig. 5, B and C). However, introduction of a larger deletion in exon 4 (pNL13a7d4) (Fig. 5A) reduced the ability of SRp40 to induce 1347 tat mRNA (Fig. 5D). The same deletion did not affect the ability of SRp55 and SRp75 to induce the production of unspliced 13a7 vpr mRNA (Fig. 5D). We concluded that SRp40 was dependent on sequences in exon 4, whereas the target sequence of SRp55 and SRp75 most likely is located downstream of nucleotide position 5478 in exon 4.
FIGURE 5.
A, schematic representation of a segment of HIV-1 subgenomic plasmids pNL13a7 (7), pNL13a7d, and pNL13a7d4. Nucleotide positions of deletions are indicated. B, Northern blot with RNA extracted from HeLa cells transfected with pNL13a7 (w) or pNL13a7d (d) in the absence or presence of CMV promoter-driven plasmids expressing SRp40, SRp55, or SRp75. The Northern blot was probed with exon 1 probe (Fig. 1A) detecting all HIV-1 mRNAs. Detected mRNAs are indicated. C, RT-PCR with oligonucleotides Exon3s and BAMA (Fig. 1A) performed on the RNAs also analyzed in B. Bands representing mRNAs are indicated. M represents the molecular weight marker. D, RT-PCR on RNA extracted from HeLa cells transfected with pNL13a7 (w) or pNL13a7d4 (d4) in the absence or presence of plasmids expressing SRp40, SRp55, and SRp75. The oligonucleotides used were Exon3s and BAMA (Fig. 1A). Bands representing mRNAs are indicated. M represents the molecular weight marker.
Splicing to Exons 4a, 4b, 4c, and 5 in vpr cDNA 13a7 Is Reduced When a Previously Identified Splicing Regulatory Element in Exon 5, Termed GAR, Is Deleted
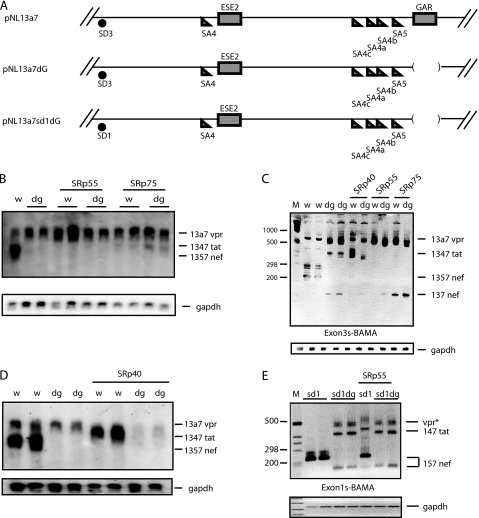
We had established that neither exon 3 nor intron 3, nor exon 4 sequences upstream of exon 5, were required for the splicing inhibitory effect of SRp55 and SRp75. Next we investigated if SRp55 or SRp75 acted on the remaining sequences of exon 4 that overlap with exon 5 and contain a previously identified exonic splicing enhancer named GAR (25, 26) (Fig. 6A). Deletion of GAR in pNL13a7, generating pNL13a7dG (Fig. 6A), resulted in mainly unspliced 13a7 vpr mRNA production, as can be seen by Northern blotting (Fig. 6B) and by RT-PCR (Fig. 6C). This deletion inhibited splicing from exon 3 to exons 4a, 4b, 4c, and 5, but enhanced splicing to exon 4 (Fig. 6, B and C). Overexpression of SRp40 with the GAR deletion mutant pNL13a7dG resulted in a very inefficient induction of 1347 tat mRNA, indicating that SRp40 was dependent on GAR (Fig. 6C), as expected from previously published results (25). Similar results were obtained by Northern blotting (Fig. 6D). However, because low levels of tat mRNA were detected in the presence of SRp40 (Fig. 6C), SRp40 may act also through other sequences than GAR. Overexpression of SRp55 and SRp75 did not alter the phenotype of the pNL13a7dG (Fig. 6B). However, RT-PCR analysis revealed that the induction of 1347 tat mRNA, caused by the deletion of GAR, was inhibited by SRp55 and SRp75 (Fig. 6C). Although SRp55 and SRp75 could still inhibit the 5′-splice site of exon 3 in the absence of GAR, the similar phenotype of the GAR deletion and overexpression of SRp55 and SRp75 indicated that SRp55 and SRp75 might act at least partly by inhibiting GAR.
FIGURE 6.
A, schematic representation of segments of the HIV-1 subgenomic plasmids pNL13a7 (7), pNL13a7dG, and pNL13a7sd1dG. B and D, Northern blot with RNA extracted from HeLa cells transfected with pNL13a7 (w) or pNL13a7dG (dg) in the absence or presence of CMV promoter-driven plasmids expressing SRp40, SRp55, or SRp75. The blots were probed with exon 1 probe (Fig. 1A). All detected mRNAs are indicated. C, RT-PCR with RNA extracted from HeLa cells transfected with pNL13a7 (w) or pNL13a7dG (dg) in the absence or presence of CMV promoter-driven plasmids expressing SRp40, SRp55, or SRp75. RT-PCR oligonucleotides Exon3s and BAMA (Fig. 1A) were used. The bands representing mRNAs are indicated. M represents the molecular weight marker. E, RT-PCR with RNA extracted from HeLa cells transfected with pNL13a7sd1 (sd1) (Fig. 3A) or pNL13a7sd1dG (sd1dg) in the absence or presence of CMV promoter-driven plasmid expressing SRp55. Oligonucleotides used for RT-PCR were Exon1s and BAMA (Fig. 1A). Bands representing mRNAs are indicated. M represents the molecular weight marker.
To investigate the relationship between GAR and SRp55 further, we replaced SD3 with SD1 in the GAR-deleted pNL13a7 plasmid, resulting in pNL13a7sd1dG (Fig. 6A). Previous results shown in Fig. 3, B and C, predicted that SRp55 should induce splicing from SD1 (the 5′-splice site of exon 1) to SA4 (the 3′-splice site of exon 4) and that it should not inhibit splicing from SD1. As predicted, SRp55 induced splicing to exon 4 in the presence of SD1 in pNL13a7sd1 but failed to affect splicing when GAR was deleted in the same plasmid (pNL13a7sd1dG) (Fig. 6E) supporting the idea that SRp55 acts through GAR.
The phenotype of pNL13a7sd1dG was similar to the phenotype of pNL13a7dG (Fig. 6, C and E). SRp55 did not alter the splicing of pNL13a7sd1dG, whereas induction of splicing from SD1 to SA4 was seen when GAR was present (pNL13a7sd1) (Fig. 6E). We concluded that GAR is required for splicing to exons 4a, 4b, 4c, and 5, and that at least part of the splicing inhibitory effect of SRp55 and SRp75 was through GAR.
SRp55 Inhibits the Function of GAR in the Absence of Other HIV-1 Sequences
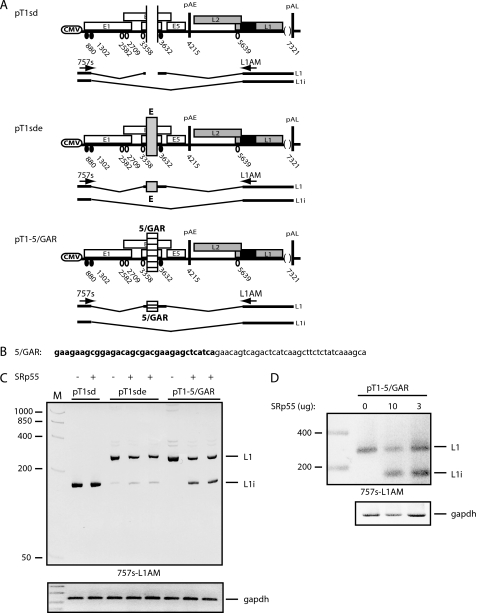
To further substantiate the finding that SRp55 inhibits the function of GAR specifically, we inserted a 69-nucleotide HIV-1 sequence encoding exon 5 and containing the GAR sequence (Fig. 7B) into an enhancerless human papillomavirus type 16 (HPV-16) plasmid named pT1sd (29) (Fig. 7A). This plasmid has the potential to produce HPV-16 L1 mRNAs, which contain an internal exon, and L1i mRNAs, which lack the internal exon (Fig. 7A). Transfection of pT1sd plasmid into HeLa cells gives rise to primarily L1i mRNA as the splicing enhancer in the internal exon has been deleted (Fig. 7C). If the HIV-1 GAR enhancer or the core HPV-16 enhancer (E) was added back into the internal exon of pT1sd to generate pT1–5/GAR and pT1sde (29), respectively (Fig. 7A), splicing shifted to L1 mRNA (Fig. 7C). These results confirmed previously published results that GAR acts as a splicing enhancer in a heterologous context (25, 26).
FIGURE 7.
A, schematic representation of the HPV-16 subgenomic plasmids pT1sd and pT1sde (29) as well as the HPV-16 subgenomic plasmid containing the HIV-1 exon 5 sequence, including the GAR sequence (25, 26), pT1–5/GAR. The L1 and L1i mRNAs produced from these plasmids are indicated. Arrows indicate PCR oligonucleotides 757s and L1AM used for RT-PCR. B, the HIV-1 sequence inserted in pT1sd is shown with the GAR sequence indicated in bold. C, RT-PCR, with oligonucleotides 757s and L1AM, on RNA extracted from HeLa cells transfected with pT1sd, pT1sde, or pT1–5/GAR in the absence or presence of CMV promoter-driven plasmid expressing SRp55. Bands representing L1 or L1i mRNAs are indicated. M represents the molecular weight marker. D, RT-PCR on RNA extracted from HeLa cells transfected with pT1–5/GAR in the absence or presence of various concentrations of SRp55 plasmid. M represents the molecular weight marker.
If SRp55 inhibits the function of GAR specifically, overexpression of SRp55 should restore production of L1i mRNA from pT1–5/GAR, but not from pT1sd or pT1sde. The three subgenomic HPV-16 plasmids were transfected in the absence or presence of SRp55 plasmid. The results revealed that SRp55 altered the splicing pattern of pT1–5/GAR by inducing L1i mRNAs (Fig. 7C). This is a result of exclusion of HPV-16 internal exon that is depending on a functional enhancer downstream of HPV-16 3′-splice site at nucleotide position 3358 (Fig. 7C). This effect was specific for the GAR-containing HPV-16 plasmid pT1–5/GAR, because the plasmids without GAR (pT1sd and pT1sde) were unaffected by SRp55 (Fig. 7C). The effect of SRp55 was concentration-dependent, but splicing did not shift completely from L1 to L1i (Fig. 7D), not even in the presence of 10 μg of co-transfected SRp55 plasmid. These results suggested that GAR acted very efficiently even in context of HPV-16. We concluded that SRp55 specifically inhibits the function of the HIV-1 GAR splicing enhancer.
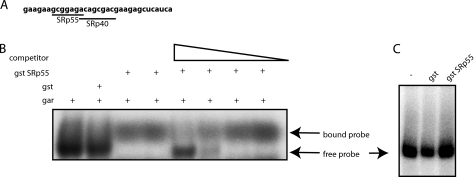
SRp55 Interacts Directly with the GAR Splicing Enhancer in Exon 5
We speculated that SRp55 binds directly to GAR. Indeed, the ESEfinder® (37, 38) predicted an SRp55 binding site in GAR (Fig. 8A) as previously reported (25). To determine if SRp55 was binding to GAR, an RNA gel shift assay was performed with gst-SRp55 and radiolabeled exon 5 GAR RNA (Fig. 8A). These experiments demonstrated that gst-SRp55 could bind directly to GAR (Fig. 8B). In competition experiments, unlabeled GAR RNA inhibited SRp55 binding to radiolabeled GAR RNA, indicating that the binding of SRp55 to the GAR enhancer was specific (Fig. 8B). Gst-SRp55 did not bind to a control RNA derived from human papillomavirus type 1 named AUM/UM (34) (Fig. 8C), further supporting that SRp55 binds specifically to HIV-1 GAR.
FIGURE 8.
A, representation of GAR sequence used as probe for RNA gel shift assay. Potential binding sites for SRp40 and SRp55 are indicated according to ESEfinder® (37, 38). B, RNA gel shift assay with radiolabeled GAR RNA and purified gst protein (gst) or purified gst SRp55 protein (gst SRp55). Increasing amounts of unlabeled GAR RNA (competitor). Free and bound probe are indicated. C, RNA gel shift assay with radiolabeled HPV-1 RNA named AUM/UM (34) and purified gst protein (gst) or purified gst SRp55 protein (gst SRp55). The AUM/UM RNA is 57 nucleotides and has an AU content of 76%, and the sequence of the AUM/UM mRNA is the following: AUACCUAUUAGUAGAUUACCUAUUAUAUAUUCCUAUAUUCCUAUACUUCCUAUACUU.
SRp55 and SRp75 Counteract the Effect of SRp40 on HIV-1 Splicing
Having established that SRp40 and SRp55 had different effects on HIV-1 vpr mRNA 13a7 and considering that SRp40 as well as SRp55 can act through GAR, we investigated if SRp40 and SRp55 were competing for GAR. Transfection of pNL13a7 with a constant, high amount of SRp40 and a serial dilution of either SRp55 or SRp75 showed that SRp40 induced production of tat mRNA 1347 in the presence of low levels of SRp55 and SRp75, as expected (Fig. 9A). However, increasing the levels of SRp55 or SRp75 resulted in a shift toward production of mainly unspliced vpr mRNA 13a7 (Fig. 9A). We concluded that production of tat mRNA 1347 or vpr mRNA 13a7 is determined by the relative concentration of SRp40 versus SRp55 or SRp75. If this competition was solely due to a competition for binding to GAR, an SRp55 mutant with retained RNA-binding domains, but without the RS domain, should function equally well or better than wild-type SRp55. Overexpression of SRp55-dRS, which lacks the RS domain, failed to inhibit splicing and production of 13a7 vpr mRNA (Fig. 9B). We concluded that the mechanism of inhibition of SD3 is more complex than simply competing with SRp40 for binding to GAR, because inhibition was dependent on the RS domain of SRp55. Taken together, our results suggested that SRp55 interacts with GAR and inhibits splicing from exon 3 by interacting with an unidentified factor.
FIGURE 9.
A, Northern blot with RNA extracted from HeLa cells transfected with plasmid pNL13a7 (7) (Fig. 1A) in the absence or presence of CMV promoter-driven plasmid expressing SRp40 and serially diluted CMV promoter-driven plasmids expressing SRp55 or SRp75. The blot was probed with exon 1 probe (Fig. 1A) detecting all mRNAs. Identified mRNAs are indicated. B, acrylamide gel showing RT-PCR with oligonucleotides Exon3s and BAMA (Fig. 1A) on cytoplasmic RNA from HeLa cells transfected with pDP (Fig. 2A) in the absence or presence of CMV promoter-driven plasmid expressing SRp55 or SRp55-dRS, which is lacking the RS domain. vpr, tat, and nef mRNAs are indicated. M represents the molecular weight marker. C, agarose gels showing RT-PCR reactions with oligonucleotides Exon3s and BAMA (Fig. 1A) on cytoplasmic RNA from HeLa cells transfected with pNL13A7 in the absence or presence of 133 nm siRNA against SRp55 and SRp75 (SMARTpool Dharmacon). The lower panel shows RT-PCR on cytoplasmic RNA from HeLa cells transfected with pNL13A7 in the absence or presence of either 133 nm or 27 nm of siRNAs against SRp55 and SRp75 (SMARTpool Dharmacon). The right panel shows RT-PCR on SRp55 mRNA in the absence or presence of siRNAs against SRp55 and -75.
To investigate if SRp55 and SRp75 were required for generation of the low levels of 13a7 vpr mRNA produced from pNL13a7 in the absence of exogenous SRp55 or SRp75, we performed an siRNA-mediated knockdown of endogenous SRp55 and SRp75 in cells transfected with pNL13a7. The results revealed that the levels of unspliced 13a7 vpr mRNA produced from the vpr cDNA pNL13a7 were reduced when SRp55 and SRp75 levels were knocked down (Fig. 9C). A scrambled siRNA and siRNAs against unrelated targets did not affect the levels of 13a7 vpr mRNA produced from pNL13a7 (data not shown). These results suggest that knockdown of SRp55 and SRp75 enhanced splicing of the 13a7 vpr mRNA and support a role for SRp55 and SRp75 in the production of partially spliced HIV-1 vpr mRNAs.
DISCUSSION
We show that overexpression of SRp55 specifically inhibits splicing from the 5′-splice site of HIV-1 exon 3. High levels of SRp55 override all splicing enhancers in exons 4 and 5, including ESE2 (24) and GAR (25, 26), causing splicing inhibition from exon 3 and production of the partially spliced 13a7 vpr mRNA. We demonstrate that SRp55 inhibits GAR, suggesting a model in which SRp55 interferes with the GAR-enhancer to inhibit splicing to exons 4a, 4b, 4c, and 5, at least from the 5′-splice site of exon 3. We also found that SRp55 simultaneously enhances splicing from exon 1 to exon 4 to produce tat mRNA, suggesting that production of tat mRNA is less GAR-dependent than rev and nef mRNAs and that SRp55 acts also on RNA elements in exon 4. We speculate that these interactions inhibit splicing when the 5′-splice sites is weak, as in exon 3 (Fig. 3A), whereas, in combination with the stronger 5′-splice site of exon 1, these interactions redirect splicing to exon 4. Although we identified GAR as one target for the splicing inhibitory effect of SRp55, it remains to be determined how SRp55 directs splicing from exon 1 to exon 4.
There are multiple splicing enhancer elements in exons 4 and 5 (16). Whereas SC35 binds to enhancer ESE2 (24), potential binding sites for ASF/SF2, SRp40, and SRp55 were identified in GAR (25). Previously published in vitro splicing experiments revealed that SRp40 and ASF/SF2 act through GAR (25, 26), whereas no enhancing effect was seen with SRp55 (25). Our experiments are in agreement with these published results and confirm that deletion of GAR in subgenomic HIV-1 plasmids abrogates production of rev and nef mRNAs, whereas tat mRNAs are unaffected (25). In addition, we confirm that GAR can act as a potent splicing enhancer on heterologous mRNAs. We showed that insertion of GAR downstream of the major HPV-16 3′-splice site SA3358 efficiently enhanced splicing in the context of HPV-16. Previously published results demonstrated that GAR was primarily dependent on ASF/SF2 and ASF/SF2-sites located in GAR and at SA5 (25), whereas the SRp40 site in GAR contributed less to GAR function. Other investigators have demonstrated that SRp40 enhances splicing to exon 4 (39). Our results confirm that overexpression of SRp40 enhances splicing to SA4, at the expense of SA4a, SA4b, SA4c, and SA5. These results suggest that SRp40 may act on multiple sites in exon 4, upstream of SA5. Deletions that we introduced in exon 4 lowered the effect of SRp40 (Fig. 5). So did the deletion of GAR (Fig. 6). One may speculate that SRp40 interacts with multiple sites in exon 4, including the site in GAR, to enhance production of exon 4 containing tat mRNAs.
SRp40 appears to induce production of exon 4-containing tat mRNA from various 5′-splice sites. Our experiments demonstrate that SRp55 overrides the induction of tat mRNA 1347 by SRp40. In the presence of high levels of SRp40 a strong induction of splicing to exon 4 (SA4) occurs. However, overexpression of SRp55 inhibits splicing from SD3 even at high concentrations of SRp40 and causes production of 13a7 vpr mRNA, demonstrating that SRp40 and SRp55 are competing for the HIV-1 pre-mRNA. Similarly, SRp55 overrides the potent GAR enhancer that likely functions so well in many cell types as a result of high levels of endogenous ASF/SF2. Although the predicted SRp55 binding site in GAR overlaps with the SRp40 binding site, and binding of SRp55 to this site may inhibit binding of SRp40, it is questionable if steric hindrance explains the inhibitory effect of SRp55. Although SRp55 binds to GAR and acts at least partly through GAR, its mechanism of splicing inhibition is likely to be more complex than mere physical hindrance of binding of other splicing factors to nearby sites. This is also reflected by the requirement for the SRp55 RS domain for inhibition of splicing. These results suggest that SRp55 interacts with an unknown factor to inhibit splicing from the 5′-splice site of exon 3. This factor could be another SR protein.
Overexpression of SR proteins with the infectious molecular clone pNL43 revealed that high levels of ASF/SF2 induced production of vpr mRNA, at the same time causing a dramatic decrease in gag, pol, and env proteins as well as virus production (40). These authors speculate that ASF/SF2 enhances splicing to the 3′-splice site of exon 3, either through a yet unidentified splicing enhancer, or by inhibiting a previously identified splicing silencer in exon 3 named ESSV (10, 17). This splicing silencer interacts with heterogeneous nuclear ribonucleoprotein A/B (10, 17, 41), and ASF/SF2 is known to counteract heterogeneous nuclear ribonucleoprotein A1, another member of the heterogeneous nuclear ribonucleoprotein family (42, 43). However, ASF/SF2 binds to the GAR enhancer and is necessary for splicing to exons 4a and 5 (25, 26). In contrast, overexpression of ASF/SF2 promotes production of the partially spliced vpr and overrides the function of GAR, much like SRp55 (39, 40). Because both ASF/SF2 and SRp55 can bind to exon 5, one may speculate that these interactions in combination with other binding sites only occupied in the presence of high levels of ASF/SF2 or SRp55, may inhibit splicing from exon 3 to exons 4, 4a, 4b, 4c, and 5 thereby creating the vpr mRNA. ASF/SF2 and SRp55 may induce production of vpr mRNA in a similar manner. In HIV-1-infected cells, levels of various SR proteins may change during the infection (44), or HIV-1 has adapted to the intracellular levels of SR proteins in T cells and monocyte/macrophages to optimize production of sufficient quantities of each alternatively spliced HIV-1 mRNA. Nevertheless, it is clear that HIV-1 alternative splicing and virus production are sensitive to minor changes in intracellular concentrations of various SR proteins (40, 45).
Recently it was shown that splicing to exon 2A, another incompletely spliced HIV-1 exon that encodes the vif gene, appears to be positively regulated by SRp75 through the interaction of SRp75 with an enhancer in this region (46, 47). We show that SRp75 can induce production of vpr mRNA by inhibiting the 5′-splice site of exon 3. Our experiments revealed that deletions in exon 3 enhanced splicing from the 5′-splice site of exon 3 (Fig. 4). This is in line with previously published results that identified a splicing silencer named ESSV in exon 3 (10, 12). However, our results do not support an interaction between SRp75 and sequences in exon 3. It appears that splicing of the two partially spliced HIV-1 mRNAs 12a7 (vif) and 13a7 (vpr) is differently regulated.
Supplementary Material
Acknowledgments
We thank members of our group and the groups of Göran Akusjärvi, Göran Magnusson, and Catharina Svensson for providing plasmids and for comments and discussions at laboratory meetings. We also thank Monika Somberg, George N. Pavlakis, and Javier F. Cáceres for providing plasmids and Paul Jackman for participating in some experiments.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- SD1–4
- splice donors 1–4
- SR
- serine- and arginine-rich
- HPV-16
- human papillomavirus type 16.
REFERENCES
- 1.Schwartz S., Felber B. K., Benko D. M., Fenyö E. M., Pavlakis G. N. (1990) J. Virol. 64, 2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz S., Felber B. K., Fenyö E. M., Pavlakis G. N. (1990) J. Virol. 64, 5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo S. J., Weitsman S., Zack J. A., Chen I. S. (1990) J. Virol. 64, 4585–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell D. F., Martin M. A. (1993) J. Virol. 67, 6365–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arya S. K., Gallo R. C. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 2209–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guatelli J. C., Gingeras T. R., Richman D. D. (1990) J. Virol. 64, 4093–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S., Felber B. K., Pavlakis G. N. (1991) Virology 183, 677–686 [DOI] [PubMed] [Google Scholar]
- 8.Schwartz S., Felber B. K., Pavlakis G. N. (1992) Mol. Cell. Biol. 12, 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchand V., Mereau A., Jacquenet S., Thomas D., Mougin A., Gattoni R., Stevenin J., Branlant C. (2002) J. Mol. Biol. 323, 629–652 [DOI] [PubMed] [Google Scholar]
- 10.Bilodeau P. S., Domsic J. K., Mayeda A., Krainer A. R., Stoltzfus C. M. (2001) J. Virol. 75, 8487–8497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyhr-Mikkelsen H., Kjems J. (1995) J. Biol. Chem. 270, 24060–24066 [DOI] [PubMed] [Google Scholar]
- 12.Jacquenet S., Mereau A., Bilodeau P. S., Damier L., Stoltzfus C. M., Branlant C. (2001) J. Biol. Chem. 276, 40464–40475 [DOI] [PubMed] [Google Scholar]
- 13.O'Reilly M. M., McNally M. T., Beemon K. L. (1995) Virology 213, 373–385 [DOI] [PubMed] [Google Scholar]
- 14.Zheng Z. M. (2004) J Biomed Sci 11, 278–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.House A. E., Lynch K. W. (2008) J. Biol. Chem. 283, 1217–1221 [DOI] [PubMed] [Google Scholar]
- 16.Stoltzfus C. M., Madsen J. M. (2006) Cur HIV Res 4, 43–55 [DOI] [PubMed] [Google Scholar]
- 17.Madsen J. M., Stoltzfus C. M. (2005) J. Virol. 79, 10478–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Si Z., Amendt B. A., Stoltzfus C. M. (1997) Nucleic Acids Res. 25, 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amendt B. A., Hesslein D., Chang L. J., Stoltzfus C. M. (1994) Mol. Cell. Biol. 14, 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si Z. H., Rauch D., Stoltzfus C. M. (1998) Mol. Cell. Biol. 18, 5404–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staffa A., Cochrane A. (1995) Mol. Cell. Biol. 15, 4597–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tange T. O., Damgaard C. K., Guth S., Valcarcel J., Kjems J. (2001) EMBO J. 20, 5748–5758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amendt B. A., Si Z. H., Stoltzfus C. M. (1995) Mol. Cell. Biol. 15, 6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahler A. M., Damgaard C. K., Kjems J., Caputi M. (2004) J. Biol. Chem. 279, 10077–10084 [DOI] [PubMed] [Google Scholar]
- 25.Caputi M., Freund M., Kammler S., Asang C., Schaal H. (2004) J. Virol. 78, 6517–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kammler S., Leurs C., Freund M., Krummheuer J., Seidel K., Tange T. O., Lund M. K., Kjems J., Scheid A., Schaal H. (2001) RNA 7, 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long J. C., Caceres J. F. (2009) Biochem. J. 417, 15–27 [DOI] [PubMed] [Google Scholar]
- 28.Soret J., Bakkour N., Maire S., Durand S., Zekri L., Gabut M., Fic W., Divita G., Rivalle C., Dauzonne D., Nguyen C. H., Jeanteur P., Tazi J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rush M., Zhao X., Schwartz S. (2005) J. Virol. 79, 12002–12015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collier B., Oberg D., Zhao X., Schwartz S. (2002) J. Virol. 76, 2739–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cáceres J. F., Krainer A. R. (1993) EMBO J. 12, 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cáceres J. F., Stamm S., Helfman D. M., Krainer A. R. (1994) Science 265, 1706–1709 [DOI] [PubMed] [Google Scholar]
- 33.Screaton G. R., Cáceres J. F., Mayeda A., Bell M. V., Plebanski M., Jackson D. G., Bell J. I., Krainer A. R. (1995) EMBO J. 14, 4336–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiklund L., Sokolowski M., Carlsson A., Rush M., Schwartz S. (2002) J. Biol. Chem. 277, 40462–40471 [DOI] [PubMed] [Google Scholar]
- 35.Tan W., Felber B. K., Zolotukhin A. S., Pavlakis G. N., Schwartz S. (1995) J. Virol. 69, 5607–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan W., Schwartz S. (1995) J. Virol. 69, 2932–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith P. J., Zhang C., Wang J., Chew S. L., Zhang M. Q., Krainer A. R. (2006) Hum. Mol. Gen. 15, 2490–2508 [DOI] [PubMed] [Google Scholar]
- 38.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ropers D., Ayadi L., Gattoni R., Jacquenet S., Damier L., Branlant C., Stévenin J. (2004) J. Biol. Chem. 279, 29963–29973 [DOI] [PubMed] [Google Scholar]
- 40.Jacquenet S., Decimo D., Muriaux D., Darlix J. L. (2005) Retrovirology 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domsic J. K., Wang Y., Mayeda A., Krainer A. R., Stoltzfus C. M. (2003) Mol. Cell. Biol. 23, 8762–8772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eperon I. C., Makarova O. V., Mayeda A., Munroe S. H., Cáceres J. F., Hayward D. G., Krainer A. R. (2000) Mol. Cell. Biol. 20, 8303–8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayeda A., Helfman D. M., Krainer A. R. (1993) Mol. Cell. Biol. 13, 2993–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldarelli F., Xiang C., Chamoun G., Zeichner S. L. (1998) Virus Res. 53, 39–51 [DOI] [PubMed] [Google Scholar]
- 45.Jablonski J. A., Caputi M. (2009) J. Virol. 83, 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mandal D., Exline C. M., Feng Z., Stoltzfus C. M. (2009) J. Virol. 83, 6067–6078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Exline C. M., Feng Z., Stoltzfus C. M. (2008) J. Virol. 82, 3921–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.