Abstract
After DNA damage, cells must decide between different fates including growth arrest, DNA repair, and apoptosis. Both p53 and E2F1 are transcription factors involved in the decision process. However, the mechanism for cross-talk between the p53 and E2F1 pathways still remains unclear. Here, we proposed a four-module kinetic model of the decision process and explored the interplay between these two pathways in response to ionizing radiation via computer simulation. In our model the levels of p53 and E2F1 separately exhibit pulsatile and switching behaviors. Upon DNA damage, p53 is first activated, whereas E2F1 is inactivated, leading to cell cycle arrest in the G1 phase. We found that the ultimate decision between cell life and death is determined by the number of p53 pulses depending on the extent of DNA damage. For repairable DNA damage, the cell can survive and reenter the S phase because of the activation of E2F1 and inactivation of p53. For irreparable DNA damage, growth arrest is overcome by growth factors, and activated p53 and E2F1 cooperate to initiate apoptosis. We showed that E2F1 promotes apoptosis by up-regulating the proapoptotic cofactors of p53 and procaspases. It was also revealed that deregulated E2F1 by oncogene activation can make cells sensitive to DNA damage even in low serum medium. Our model consistently recapitulates the experimental observations of the intricate relationship between p53 and E2F1 in the DNA damage response. This work underscores the significance of E2F1 in p53-mediated cell fate decision and may provide clues to cancer therapy.
Keywords: Cell Cycle, Computer Modeling, DNA Damage, E2F Transcription Factor, p53, DNA Damage, E2F1, Cell Cycle, Computer Modeling, p53
Introduction
The tumor suppressor p53 has a crucial role in preventing tumorigenesis (1). Upon various stresses, p53 is stabilized and activated to function primarily as a transcription factor, regulating the expression of a large number of genes involved in cell cycle arrest, DNA repair, or apoptosis (2). Thus, p53 is at the hub of numerous signaling pathways triggered by various stresses. Previously, it was proposed that cell fate after DNA damage is governed by p53 levels, i.e. a low level of p53 leads to transient growth arrest and cell survival, whereas a high level promotes irreversible apoptosis (3). Recently, it has been reported that p53 levels can exhibit oscillations in response to DNA damage induced by ionizing radiation (IR) (4, 5). Whereas damped oscillations of p53 levels were observed at the population level (4), a series of undamped pulses was observed at the single-cell level (5). In such a digital mode, it is the number of p53 pulses rather than their amplitudes and duration that is related to the extent of DNA damage and determines cell fate (5). p53 pulses can be generated by negative feedback loops with time delay (6–8) or coupled positive and negative feedback loops (9, 10).
How stressed cells exploit p53 pulses to translate various stresses into different cellular outcomes is not completely understood. Several studies have explored the functional roles of p53 pulses in response to DNA damage. Tyson and co-workers (10) classified active p53 into three distinct forms according to its phosphorylation status and showed that p53 pulses subserve the decision between cell cycle arrest/repair and apoptosis. We developed an integrated model of the p53 signaling network to reveal the whole process from the generation of DNA damage to the choice of cell fate, stressing that two forms of phosphorylated p53 play distinct roles in cell fate decision (11). Batchelor et al. (12) proposed that p53 pulses can allow for a wide variety of temporal expression patterns of target genes. These studies suggest that the pulsatile response of p53 may represent a flexible and efficient mechanism by which cellular responses can be organized coherently. Notably, the different roles played by p53 are associated with its specific post-translational modifications.
Although p53 is pivotal to cell fate decision, cross-talk between p53 and other transcription factors also has important roles. Among them, the E2F family is best known for its ability to regulate entry into and progression through S phase of the cell cycle (13). Specifically, E2F1 can promote both cell cycle progression and apoptosis (14), and deregulated E2F1 can cooperate with p53 to trigger apoptosis (15–17). E2F1 can induce production of several p53 cofactors including p53DINP1,3 ASPP1, and ASPP2 (ASPP1/2 are collectively referred to as ASPP thereafter) (18). p53DINP1 promotes apoptosis by phosphorylating p53 at Ser-46 (19), while ASPP proteins enhance binding of p53 to the promoters of proapoptotic genes (20). On the other hand, the activation of p53 first induces G1 phase arrest by inhibiting E2F1 via p21 (21). Therefore, E2F1 competes with p53 for cell cycle control but cooperates with p53 in apoptosis induction. Moreover, E2F1 levels can behave like a bistable switch when driven by growth factors (22). An issue thus arises concerning how the combination of p53 pulsing and E2F1 switching contributes to the decision between cell survival and death after DNA damage. It is also interesting to explore the effect of p53 cofactors on cellular outcomes, which was seldom considered in the above theoretical models for p53 pulses.
Motivated by the above considerations, we developed an integrated model to explore how cell cycle progression and cell fate decision are well coordinated by p53 and E2F1 in the DNA damage response. The model is composed of four modules: a DNA repair module, an ATM sensor, a p53 pulse generator, and a cell fate decision module. The model can characterize the process from the generation of DNA damage to the choice of cell fate. We found that the cell fate is determined by the number of p53 pulses, which depends on the extent of DNA damage for each fixed concentration of growth factor. E2F1 potentiates p53-dependent apoptosis in two ways: 1) it induces p53 cofactors to bias p53 activity toward apoptosis, and 2) it up-regulates the levels of procaspases to make cells sensitive to death stimuli. We concluded that activation of E2F1 or p53 alone results in S-phase entry and G1 arrest, respectively, whereas concomitant activation of p53 and E2F1 initiates apoptosis.
EXPERIMENTAL PROCEDURES
The cellular response to DNA damage can be considered as a signal transduction process, and the signaling pathways involved are rather complicated. It is difficult to obtain all precise data to characterize in detail the whole process of DNA damage response. Here, we focus on exploring the essential mechanisms for cell fate decision. With limited experimental data available, our model was constructed based on established biological facts and reasonable simplifications. We also plotted schematic diagrams to depict cross-talk between the p53 and E2F1 pathways (23). The key points of the model are addressed as follows.
An Integrated Model for Cell Signaling Network in Response to DNA Damage
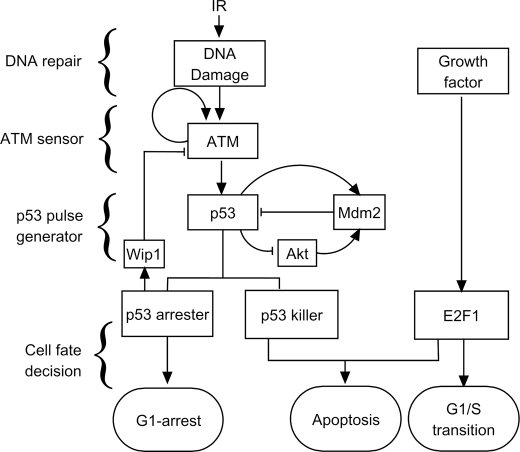
We developed an integrated model for the p53 network composed of four modules (Fig. 1). DNA damage is produced in cells exposed to IR. The DNA repair module characterizes the generation and repair of DNA damage, which is essentially stochastic. Upon IR, ATM is activated by autophosphorylation, acting as a sensor of DNA damage (6, 24). Subsequently, p53 is activated by phosphorylation (25), and interlinked positive and negative feedback loops involving p53 and Mdm2 underlie p53 pulses (10). Active p53 is distinguished between p53 arrester and p53 killer, which contribute to cell cycle arrest and apoptosis, respectively (10, 11). Once activated by growth factors, E2F1 can promote the transition from the G1 to S phase and cooperate with p53 killer to induce apoptosis. Moreover, p53-inducible Wip1 feeds back to inhibit ATM activity (8), enclosing a negative feedback loop from the fourth to the second module. In the following we present the details of each module sequentially.
FIGURE 1.
Schematic depiction of the integrated model. The model is composed of four modules: DNA repair, ATM sensor, p53 pulse generator, and cell fate decision modules. The model characterizes the whole process from the generation of DNA damage to the choice of cell fate. In the cell fate decision module, p53 and E2F1 coordinate to regulate expression of target genes, controlling cell cycle progression and cell fate.
DNA Repair Module
A double-strand break (DSB) is generally considered the typical form of DNA damage induced by IR (26). According to experimental observations, 1 Gy of IR may induce 25–40 DSBs per cell (27). DSB repair proceeds in a stochastic way. The stochasticity in the generation and repair of DSBs is transmitted downstream of this module (6); that is, there exists variability in cellular responses to the same stress signal. Non-homologous end joining is the predominant pathway for DSB repair, especially in the G1 phase (28). We simplified the repair process into a three-state process that is characterized by reversible binding of repair proteins to DSB, forming a complex (DSBC), and by an irreversible repair process from the complex to fixed DNA. Specifically, we adopted the two-lesion-kinetic model (29) and applied the Monte Carlo method proposed by Ma et al. (6) to mimic the repair process (see supplemental Method S1 and Fig. S1).
In simulations, we considered a population of 2000 cells that are exposed to the same IR. Because Poisson distribution is typically used to characterize the random induction of DSBs (30, 31), the initial numbers of DSBs are assumed to obey the Poisson distribution with a mean of 35 DSBs per Gy per cell. Because repair proteins are much fewer than DSBs in most cases (6), it is assumed that there are 20 repair proteins in each cell. p53 certainly plays a role in DNA repair, but its regulatory role in non-homologous end joining is complicated and controversial (32). Some studies revealed that p53 can promote the rejoining of DNA with lesions (33, 34), whereas inhibitory effects of p53 on non-homologous end joining were also reported (35, 36). For simplicity, we did not consider the effect of p53 on DNA repair.
ATM Sensor
The role of ATM as a sensor of DNA damage has been widely recognized (24, 37). In unstressed cells, ATM exists as a dimer, and its kinase activity is sequestered. Upon IR, ATM can be recruited by repair proteins, and intermolecular phosphorylation leads to rapid disassociation of dimers into monomers (37). There exists a positive feedback loop in which active ATM (i.e. phosphorylated monomer) further promotes the phosphorylation of inactive ATM (24). Thus, the activation of ATM is traditionally characterized by a switch (38, 39). However, pulses of ATM levels have recently been observed in human breast cancer MCF-7 cells, and a recurrent initiation mechanism was proposed based on the negative feedback loop between ATM and p53 via Wip1 (8). Here, we consider three forms of ATM: ATMd (inactive dimer), ATM (inactive monomer), and ATM* (active monomer). The total level of ATM is assumed to be constant (6) (see supplemental Fig. S2).
The dynamics of this module are characterized by Equations 1–3 in supplemental Method S2. The phosphorylation and dephosphorylation of ATM can be considered as enzyme-catalyzed reactions and assumed to follow the Michaelis-Menten kinetics (40). Due to the positive and negative feedback loops, the phosphorylation rate of ATM should be positively correlated with the number of DSBCs and ATM* levels, whereas the dephosphorylation of ATM* is promoted by Wip1. It is assumed that the dimerization rate of ATM is far smaller than its undimerization rate, so that ATM dimers are predominant in unstressed cells.
p53 Pulse Generator
Although a single negative feedback loop with time delay can produce periodic oscillations (6), it has been proposed that coupled negative and positive feedback loops can make oscillations more robust (9, 10). Indeed, several negative and positive feedback loops have been identified in the p53 pathway (41). The negative feedback between p53 and Mdm2 is the basis for p53 oscillation, while a double-negative feedback loop involving p53, Akt, and Mdm2 also has a role (9, 10). Akt promotes the nuclear translocation of Mdm2 to degrade p53 and inhibit its activity, whereas p53 can indirectly inactivate Akt through PTEN (42, 43). Moreover, as mentioned above, ATM* activates p53, but p53 inhibits ATM* via Wip1.
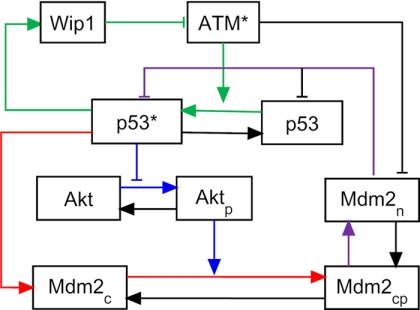
In our model the p53 pulse generator is composed of three coupled feedback loops (marked by lines with different colors in Fig. 2): 1) the negative feedback loop between p53 and Mdm2, 2) the positive feedback loop involving p53, Akt and Mdm2, and 3) the negative feedback loop between ATM and p53 via Wip1. Nuclear p53 is distinguished between inactive p53 (p53) and active p53 (p53*), whereas cytoplasmic p53 is ignored. Three forms of Mdm2 are introduced, namely Mdm2c (unphosphorylated cytoplasmic form), Mdm2cp (phosphorylated cytoplasmic form), and Mdm2n (nuclear form). Akt in the cytoplasm is differentiated between Akt (unphosphorylated form) and Aktp (phosphorylated form). For simplicity, the effect of nuclear Akt is not considered.
FIGURE 2.
p53 pulse generator model. There exist three feedback loops: the negative feedback loop between p53 and Mdm2 (red and purple), the positive feedback loop involving p53, Akt, and Mdm2 (blue and purple), and the negative feedback loop between ATM and p53 via Wip1 (green).
The dynamics of this module are characterized by Equations 4–13 in supplemental Method S2. The regulated transcription of Mdm2 by p53 is incorporated into the regulation of protein synthesis. The basal production rate of Mdm2 is set to be much smaller than the maximal p53-induced production rate (44). The Hill coefficient is set to 4 considering the cooperative binding of tetrameric p53 to DNA (45). Note that ATM can promote Mdm2 degradation and p53 activation by phosphorylation, and the two processes can be considered as enzyme-catalyzed reactions (25, 46). Consequently, both the degradation rate of Mdm2 and the activation rate of p53 are ATM-dependent and described in the form of the Michaelis-Menten function. Similarly, the degradation of p53 by Mdm2 is characterized by the Michaelis-Menten kinetics because Mdm2 acts as an E3 ubiquitin ligase in the ubiquitination of p53 (47).
Because there is no remarkable variation in the total level of Akt after irradiation (42), it is assumed to be constant. The phosphorylation of Akt should be phosphatidylinositol 3,4,5-trisphosphate (PIP3)-dependent (48), and the effect of PIP3 is reflected in the parameter kakt in the model (49). Similarly, the dephosphorylation of Aktp is phosphatidylinositol 4,5-biphosphate (PIP2)-dependent (48), and this effect is incorporated into the parameters kakts and k1akts, which are separately the rate constants of basal and p53-dependent dephosphorylation of Akt (49). Because p53-inducible PTEN can promote the conversion from PIP3 to PIP2 as well as Aktp dephosphorylation, this effect is simplified into the modulation of Aktp dephosphorylation by p53 with the rate constant of k1akts (49). We set kakts < kakt < k1akts to ensure that Aktp is predominant in unstressed cells and Akt becomes dominant in stressed cells. Moreover, we chose the values of other parameters in this module to ensure that the period of p53 pulses is between 4 and 7 h (5, 7).
Cell Fate Decision Module
Cell fate can be governed by signaling pathways involving p53 and E2F1. Both p53 and E2F1 function as transcription factors. It has been recognized that post-translational modifications and cofactors regulate the promoter selectivity of p53 (2). E2F1 can up-regulate the levels of p53 cofactors and proapoptotic proteins (18, 50). The schematics of this module are shown in Fig. 3, and its key points are addressed as follows.
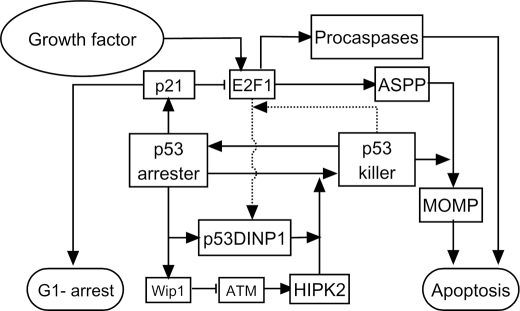
FIGURE 3.
Cell fate decision module model. Active p53 is divided into p53 arrester and p53 killer. p53 arrester is a primarily phosphorylated form of p53 on Ser-15 and Ser-20, whereas p53 killer is a further phosphorylated form of p53 on Ser-46. p53 arrester induces cell cycle arrest in the G1 phase by inducing p21, which inhibits E2F1 activity, whereas p53 killer induces expression of proapoptotic genes. The conversion between p53 arrester and p53 killer is controlled by Wip1 and p53DINP1. E2F1 can be activated by growth factors. p53 killer and E2F1 cooperate to transactivate p53DINP1. With the help of E2F1-induced ASPP, p53 killer induces expression of p53AIP1 and Bax, which results in mitochondrial outer membrane permeabilization (MOMP). Moreover, E2F1 promotes apoptosis by up-regulating several key proapoptotic factors including Apaf-1 and procaspase-9 and -3.
First, the phosphorylation of p53 modulates its selective expression of target genes (51). Based on phosphorylation on different residues, active p53 is divided into two forms in our model, p53 arrester and p53 killer, which promote cell cycle arrest and apoptosis, respectively (see Fig. 3 and supplemental Fig. S3). Here, p53 arrester refers to p53 primarily phosphorylated at Ser-15 and Ser-20 by ATM, whereas p53 killer is p53 further phosphorylated at Ser-46 by p53DINP1 and HIPK2 (homeodomain interacting protein kinase) (52). p53 arrester alone induces expression of p21 and Wip1 as they are induced by p53 before its phosphorylation at Ser-46 (53, 54). Although p53 arrester also regulates expression of p53DINP1, its synthesis is mainly determined by p53 killer and E2F1 (18). Wip1 and p53DINP1 have opposite effects on the accumulation of p53 killer by modulating p53 phosphorylation at Ser-46 (19, 55). Moreover, p21 can indirectly prevent the accumulation of p53 killer by inhibiting E2F1.
Second, cell cycle progression is mainly controlled by E2F1 and p53 arrester (see the schematic diagram in supplemental Fig. S4). In quiescent cells, E2F1 activity is sequestered by Rb (22). Sufficient growth stimulation, such as with serum, can up-regulate cyclin D, which forms a complex with Cdk4/6. This leads to hyperphosphorylation of Rb and the release of E2F1. Subsequently, E2F1 promotes production of cyclin E (CycE), which binds Cdk2 to further activate E2F1 and leads to S-phase entry (22). However, p53-induced p21 prevents E2F1 activation by inhibiting Cdk2/4 and arrests cell cycle in the G1 phase (21). For simplicity, our model focuses on the regulatory network involved in the G1/S transition, ignoring the oscillation events responsible for cell cycles. Due to the positive feedback loops in the Rb-E2F1 pathway, the G1/S transition can be characterized by a bistable switch (22). We incorporated the model proposed by Yao et al. (22) into our integrated model to characterize the switch.
Third, both p53 killer and E2F1 contribute to apoptosis induction (see the schematic diagram in supplemental Fig. S5). E2F1 induces expression of ASPP (18). Because both p53 phosphorylation at Ser-46 and ASPP can enhance the induction of proapoptotic genes by p53 (20, 53), we assume that p53 killer and ASPP cooperate to induce expression of several proapoptotic genes such as Bax, p53AIP1, and Apaf-1 (53, 56, 57). Moreover, E2F1 also up-regulates the levels of Apaf-1 and procaspase-3 and -9 through a transcriptional mechanism (50, 56). Considering that both Bax and p53AIP1 are significant for apoptosis induction (53, 57), it is assumed that Bax and p53AIP1 can cooperate to release mitochondrial cytochrome c (CytoC) into the cytoplasm. CytoC is then associated with Apaf-1 to form the apoptosome that acts as a platform for caspase activation (58). The caspase cascade is then activated by engaging caspase-9 (Casp9) and -3 (Casp3). Although other members of the Bcl-2 family and some inhibitors of apoptosis, such as PUMA, Bid, Bcl-2, Bcl-xL, and XIAP, also have a role in apoptosis, their effects are not explicitly modeled for simplicity (59, 60).
The dynamics of this module are represented by Equations 14–38 in supplemental Method S2. Equations 14–23 are mainly based on the model by Yao et al. (22). We assume that p21 is only induced by p53 arrester (Equation 14), linking the p53 pathway with the Rb-E2F1 pathway. The conversion between p53 arrester and p53 killer is described by Equations 24–28. Wip1 and p53DINP1 are important factors in this process (Equation 25). [Wip1] (the brackets denote the concentration of proteins) is only controlled by [p53 arrester], whereas[p53DINP1] is regulated by [p53 arrester], [p53 killer], and [E2F1] (18, 19). We assume that p53DINP1 production is divided into three parts: basal production with a rate of ksdinp11, p53 arrester-dependent production with a rate constant of ksdinp12, and p53 killer- and E2F1-dependent production with a rate constant of ksdinp13, satisfying ksdinp11 ≪ ksdinp12 < ksdinp13 (10). This ensures that p53DINP1 level slowly accumulates during pulses of p53 arrester and becomes marked after p53 killer is dominant. The synthesis of Bax and p53AIP1 is assumed to be controlled by p53 killer and ASPP (20, 53), while ASPP production is governed by E2F1 (18) (Equations 29–31). It is assumed that both p53 killer and E2F1 contribute to the production of Apaf-1 (56). Because the apoptosome is composed of seven Apaf-1 and CytoC molecules (58), the dynamics of the apoptosome are characterized by Equation 34. Due to limited experimental data, some parameters in Equations 24–38 are loosely chosen within the physiological ranges, provided that simulation results can be qualitatively consistent with the experimental observation that it takes only 10 min to activate the caspase cascade after mitochondrial outer membrane permeabilization (61).
Methods
Details of the equations, parameters, and methods for solving our model are presented in the supplemental Methods S1 and S2 and Tables S1 and S2. The generation and repair of DSBs was characterized by a Monte Carlo method. The differential equations were numerically solved using a second-order Runge-Kutta algorithm with a time step of 0.01 min. The bifurcation diagrams were plotted by Oscill8. The units of time and radiation dose are minutes and Gy, respectively, and the other variables are dimensionless.
RESULTS
An Overview of Signal Transduction in the Network
In our model the p53 network has been divided into four modules. To give an overview of signal transduction, the output of each module is illustrated in Fig. 4 for two IR doses. The number of DSB-repair protein complexes, nC, indicates the presence of DNA damage. Upon IR, all repair proteins quickly bind to DSBs, and nC remains the maximal number of repair proteins, i.e. nC = 20, until the number of DSBs decreases below 20.
FIGURE 4.
Overview of signal transduction in the model network. Shown is temporal evolution of the output of each module at the IR dose of 3 Gy (A) or 5 Gy (B). Upon IR, a number of DSBCs are produced, and ATM and p53 are activated. Consequently, the cell cycle is arrested in the G1 phase. With repairable DNA damage, only few pulses occur in ATM* and p53* levels before cells recover to normal proliferation, which is driven by activated E2F1. With irreparable DNA damage, ATM* is maintained at high levels after four pulses, whereas p53 level shows pulses in both phases. Activated E2F1 cooperates with p53 to activate caspase 3 and apoptosis ensues.
At the low IR dose of 3 Gy, DNA damage is repairable in the cell, and only few pulses are evoked in ATM* and p53* levels. Accordingly, [E2F1] first remains at low levels during cell cycle arrest and then rises and gets saturated because of serum stimulation, whereas Casp3 is kept inactive. Thus, the cell only undergoes transient growth arrest and enters the S phase after DNA repair (Fig. 4A). By contrast, at the high IR dose of 5 Gy, [ATM*] is maintained at high levels after four pulses, whereas [p53*] exhibits sustained pulses. Consequently, [E2F1] rises after four p53 pulses, and E2F1 then cooperates with p53 to trigger apoptosis by activating Casp3 (Fig. 4B). Thus, our results suggest that p53 activation alone induces cell cycle arrest in repairable cells, whereas the synergic activation of p53 and E2F1 is necessary and sufficient for apoptosis induction in seriously damaged cells. Taken together, the signaling network can make a reliable cell fate decision relying on the extent of DNA damage.
Note that the entry into S phase occurs after growth arrest in the presence of serum. At low damage levels, serum stimulation drives cells to enter the S phase by activating E2F1 after DNA damage is fixed. By contrast, at high damage levels, serum stimulation overcomes p53-induced cell cycle arrest, and activated E2F1 cooperates with active p53 to induce apoptosis in the late stage of the cellular response. These results are consistent with experimental observations that E2F1 and p53 can cooperate to induce apoptosis (15). Therefore, cell cycle progression and cell fate decision are well coordinated by p53 and E2F1 in the DNA damage response.
ATM Dynamics with Two Phases
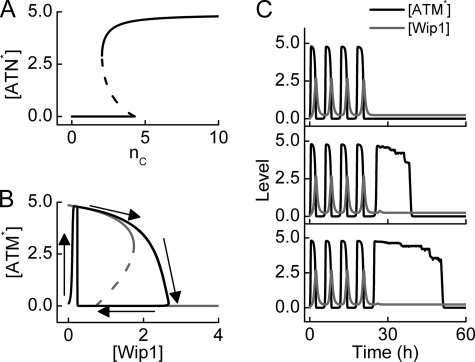
ATM is activated by DSBCs upon IR and functions as a sensor of DNA damage (37). The steady-state ATM* level versus nC is plotted in Fig. 5A. It is shown that ATM is so sensitive to DNA damage that five DSBs are sufficient to activate ATM. This is consistent with experimental observations that 0.1 Gy of IR can be detected by ATM (24). Moreover, ATM is inactivated only after the number of DSBs is reduced below 2. Thus, ATM is a sensitive and reliable detector for DNA damage.
FIGURE 5.
ATM dynamics with two phases. A, shown is a bifurcation diagram of ATM* level versus the number of DSBCs, nC. ATM is activated if nC > 4 and inactivated if nC < 2. B, bifurcation (gray) and phase (black) diagrams of ATM* level versus Wip1 level. The ATM* level either shows pulses or behaves as a switch, depending on Wip1 levels. C, shown are time courses of ATM* (black) and Wip1 (gray) levels for three individual cells at DIR = 5 Gy. Due to stochasticity in the generation and repair of DNA damage, there exists remarkable variability in cellular responses.
Notably, ATM activity can be inhibited by p53-inducible Wip1 (8). Fig. 5B plots the bifurcation and phase diagrams of [ATM*] versus [Wip1] with nC = 20. [ATM*] behaves as a bistable switch when [Wip1] is fixed between 0.74 and 1.75. Upon IR, [ATM*] quickly rises to a high level and then slowly decreases with increasing [Wip1]. If [Wip1] exceeds the upper threshold (1.75), [ATM*] quickly drops to nearly 0. Thus, [ATM*] repeatedly switches between the lower and higher levels when [Wip1] oscillates between 0.24 and 2.66 because of the negative feedback between ATM and Wip1.
Fig. 5C shows the temporal evolution of ATM* and Wip1 levels in three individual cells at DIR = 5 Gy. Due to stochasticity in the generation and repair of DNA damage, remarkable variability is manifested in protein levels. Upon IR, ATM is initially activated by DSBCs and is then dephosphorylated and inhibited by Wip1. This leads to the first pulse in ATM* and Wip1 levels. If the DNA damage is not fixed, ATM is reactivated by DSBCs and is then inactivated by Wip1 in a second round. This may extend to the fourth pulse (this number depends on the chosen set of parameter values). If DNA damage still exists after four pulses, [ATM*] switches to high levels and remains there because Wip1 is no longer induced by p53. This agrees with the experimental observation that removal of Wip1 leads to high levels of phosphorylated ATM without pulses (8). These results suggest that the presence of ATM pulses in the early stage may provide a flexible control mechanism, ensuring that ATM is quickly inactivated after DNA repair, whereas the presence of plateau levels in the late stage facilitates apoptosis induction. Therefore, the two-phase ATM dynamics represent a flexible and efficient mode for sensing DNA damage.
Dynamics of p53, Mdm2, and Akt Levels
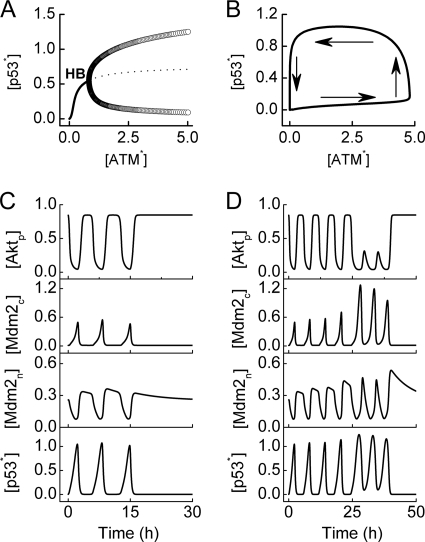
Activated ATM can stabilize and activate p53 through post-translational modification of both p53 and Mdm2. To reveal the initiation mechanism for p53 pulses, we plotted the bifurcation diagram of p53* level versus [ATM*] in Fig. 6A. There exists a Hopf bifurcation point (denoted by HB) at [ATM*] = 0.8. In unstressed cells, [ATM*] is close to zero, and thus [p53*] is kept at basal levels. Upon IR, [ATM*] quickly rises to high levels (2.9∼4.8), and [p53*] also quickly rises. If [ATM*] is kept at high levels, p53* level behaves as a limit-cycle oscillator.
FIGURE 6.
p53 pulses. A, shown is a bifurcation diagram of [p53*] versus [ATM*]. There is a Hopf bifurcation (HB). If [ATM*] is limited in a region between 2.9 and 4.8, the p53* level oscillates between the maximum and minimum of the limit cycle (open circle). B, shown is a phase diagram of [p53*] versus [ATM*]. When ATM is inhibited by Wip1, it switches between low and high levels; accordingly, the p53 level also undergoes oscillations. Displayed are time courses of [Aktp], [Mdm2c], [Mdm2n], and [p53*] at DIR = 3 (C) or 5 Gy (D).
The phase diagram of [p53*] versus [ATM*] is plotted in Fig. 6B to show the initiation of p53 pulses in the presence of ATM pulses. Due to inhibition by Wip1, [ATM*] decreases rapidly to nearly zero from its top, and [p53*] also drops to low levels after a delay. ATM is then reactivated by residual DNA damage, and p53 is activated. In this manner, [p53*] shows a series of pulses.
Fig. 6C displays the temporal evolution of [Aktp], [Mdm2c], [Mdm2n], and [p53*] at DIR = 3 Gy. Few pulses are produced during DNA repair. Because the activation rate of p53 and the degradation rate of Mdm2 vary with the progression of DNA repair, the intervals between successive pulses vary between 5 and 7 h, but the amplitudes are less variable. We also compared the series of p53 pulses with the experimental data from Geva-Zatorsky et al. (7) (see supplemental Fig. S6). Both have comparable periods, but the amplitudes of the pulses are less variable in our data as intrinsic noise in the signaling network is not considered in our model (8). After the cell recovers to normal proliferation, [p53*] and [Mdm2c] return to basal levels, whereas [Aktp] is maintained in the upper state, and [Mdm2n] is significantly larger than zero. Notably, these results agree with the experimental observation that Akt and p53 inhibit each other in a positive feedback loop (42).
At DIR = 5 Gy, the levels of those proteins exhibit complex dynamics with two phases (Fig. 6D). The first phase corresponds to the time interval comprising four p53 pulses. From the first phase to the second, the amplitudes of [p53*], [Mdm2c], and [Mdm2n] increase, whereas those of [Aktp] remarkably decrease due to the positive feedback between p53, Akt, and Mdm2. The difference between the two phases mainly results from the inhibitory effect of Wip1 on ATM, which nearly disappears in the second phase. Because the amplitudes of p53 pulses are positively correlated with [ATM*], they become larger in the second phase. The two-phase behavior of p53 level needs to be validated in future experiments.
Regulation of Cell Cycle Arrest and Reentry by the p53 and E2F1 Pathways
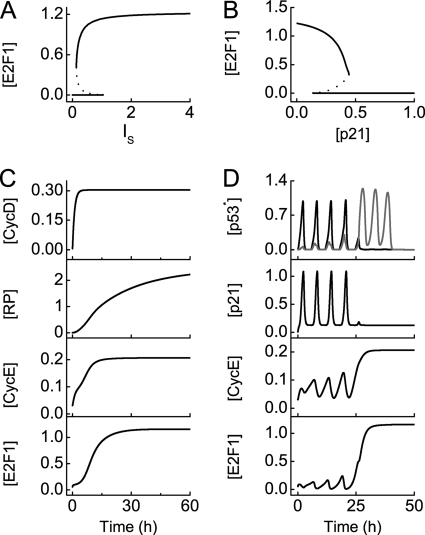
To clarify how E2F1 controls the G1/S transition, we first plotted the steady-state value of [E2F1] versus the concentration of serum, IS. [E2F1] can switch between two stable states over some range of IS (Fig. 7A). In quiescent cells (with IS = 0), [E2F1] is close to zero due to the inhibition by Rb. [E2F1] is kept in the upper stable state when IS is larger than the upper threshold (1.04), whereas [E2F1] returns to 0 when IS is smaller than the lower threshold (0.13). In simulations, IS is fixed at 10 unless specified elsewhere. Thus, E2F1 exhibits switch-like behaviors in controlling the G1/S transition.
FIGURE 7.
Cell cycle arrest and reentry into S phase. Shown is a bifurcation diagram of [E2F1] versus the concentration of serum, IS, (A) or [p21] (B). C, shown are time courses of [CycD], [RP], [CycE], and [E2F1] at the G1/S transition in unstressed cells with IS = 10. D, shown are time courses of [p53 arrester] (black) and [p53 killer] (gray), [p21], [CycE], and [E2F1] in stressed cells with IS = 10 and DIR = 5 Gy.
Moreover, p21 remarkably affects E2F1 activity. In the bifurcation diagram of [E2F1] versus [p21] (Fig. 7B), [E2F1] drops rapidly with increasing [p21]. When [p21] exceeds 0.45, the activity of E2F1 is fully inhibited, and cell growth is arrested in the G1 phase. This indicates that p21 can act as an efficient inhibitor of cell cycle progression.
Next, we explored how the cell cycle is controlled in unstressed and stressed cells, respectively. In unstressed cells, p53 is inactive, and [p21] is at basal levels. After growth factors are delivered at t = 0, the level of phosphorylated Rb (termed RP) gradually accumulates, whereas the levels of E2F1 and CycE increase relatively quickly and get saturated around t = 20 h, indicating the entry into S phase (Fig. 7C). At the IR dose of 5 Gy, the first four p53 pulses are predominated by p53 arrester, whereas the subsequent p53 pulses are dominated by p53 killer (Fig. 7D). Accordingly, four p21 pulses first develop, inhibiting the kinase activity of CycE-Cdk2, which suppresses the phosphorylation of Rb. Consequently, [E2F1] is kept at low levels, and cell cycle is arrested in the G1 phase. When p53 killer becomes dominant, [p21] drops to basal levels, and both CycE and E2F1 are activated. The cell then enters the S phase. On the other hand, with mild DNA damage, cells return to normal proliferation after transient growth arrest (see supplemental Fig. S7). Moreover, the duration of growth arrest depends on the extent of DNA damage. Therefore, p53 arrester induces cell cycle arrest via p21, whereas G1 arrest is relieved when p53 killer becomes dominant or DNA damage is fixed.
Role of E2F1 in Directing p53 Activity toward Apoptosis
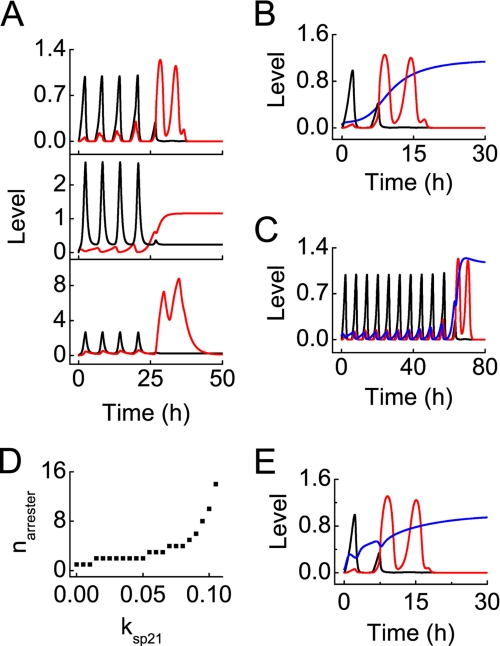
We explored how E2F1 biases p53 activity toward apoptosis, focusing on the role of E2F1 in promoting the conversion from p53 arrester to p53 killer. At DIR = 5 Gy, p53 arrester leads to induction of p21, which inhibits E2F1 (Fig. 8A). After four pulses of p53 arrester, the inhibitory effect of p21 on E2F1 is relieved, and E2F1 begins to accumulate. Meanwhile, E2F1 cooperates with p53 killer to induce p53DINP1, which further promotes accumulation of p53 killer because of the positive feedback. Our results reveal that E2F1 can direct p53 activity toward apoptosis by promoting the conversion from p53 arrester to p53 killer via p53DINP1.
FIGURE 8.
Conversion from p53 arrester to p53 killer after DNA damage. ksp21 is the p53-inducible synthesis rate of p21, and kre is the association constant between E2F1 and Rb. A, displayed is the conversion in the normal case (with ksp21 = 0.07 and kre = 2) at DIR = 5 Gy. Displayed are time courses of [p53 arrester] (black) and [p53 killer] (red) (top panel), [p21] (black) and [E2F1] (red) (middle panel), [Wip1] (black) and [p53DINP1] (red) (bottom panel). B, shown are time courses of [p53 arrester] (black), [p53 killer] (red), and [E2F1] (blue) with ksp21 = 0 and DIR = 2 Gy. C, the same conventions is used as in panel B with ksp21 = 0.1 and DIR = 12 Gy. D, shown is the number of p53 arrester pulses required for producing one pulse of p53 killer, narrester, as a function of ksp21. E, the same convention is used as in panel B with kre = 0.1, IS = 0.4 and DIR = 2 Gy.
To further assess the significance of E2F1 in induction of p53 killer, we investigated the conversion between the two forms of active p53 in p21-deficient cells at DIR = 2 Gy (Fig. 8B). In such cells, E2F1 is activated by growth factor without a marked delay, and p53DINP1 quickly accumulates. Consequently, p53 killer dominates after one pulse of p53 arrester. Thus, the proapoptotic activity of p53 can be evoked easily in p21-deficient cells. By contrast, when production of p21 is enhanced by increasing its synthesis rate, p53 killer is not induced until after 10 pulses of p53 arrester (Fig. 8C). This is consistent with the prosurvival role of p21, as shown experimentally (62, 63). Taken together, the prosurvival role of p21 partially results from its inhibitory effect on E2F1, whose activation is crucial for the induction of p53 killer.
Fig. 8D displays the number of pulses in [p53 arrester] required for producing one pulse of p53 killer, narrester, versus the p53-inducible synthesis rate of p21, ksp21. narrester rises from 1 to 10 with increasing ksp21. When ksp21 ≥ 0.12, it becomes impossible to induce p53 killer, i.e. apoptosis cannot be initiated in cells with overexpressed p21. These results suggest that p21 sets a threshold for activation of the proapoptotic activity of p53, whereas E2F1 competes with p21 to direct p53 activity toward apoptosis.
Moreover, E2F1 can be released from Rb by the oncoprotein E1A, which directly disrupts the E2F1/Rb complex by association with Rb (64). In simulations, we mimicked this effect by reducing the association constant between E2F1 and Rb (kre) to 0.1. Notably, [E2F1] rises remarkably, and p53 killer becomes dominant after one pulse of p53 arrester, even under low serum conditions (with IS = 0.4) (Fig. 8E). These results are consistent with experimental observations that activation of E1A deregulates E2F1 activity and induces expression of its proapoptotic target genes in low serum medium (18, 50). Therefore, oncoproteins (like E1A) may be used to kill tumor cells by inducing p53-dependent apoptosis via E2F1 activation in some cases.
Cooperation between p53 Killer and E2F1 in Apoptosis Induction
Fig. 9 describes the cooperation between p53 killer and E2F1 in the release of CytoC and caspase activation. In the late stage of the DNA damage response with DIR = 5 Gy, high levels of E2F1 induce synthesis of ASPP, which directs p53 killer to induce expression of Bax and p53AIP1 (Fig. 9A). Subsequently, Bax and p53AIP1 cooperate to trigger the release of CytoC. Meanwhile, E2F1 up-regulates the levels of Apaf-1, Procasp9, and Procasp3. CytoC and Apaf-1 then forms the apoptosome to activate Casp9. Finally, Casp3 is activated and apoptosis ensues. Our results are consistent with the experimental observation that apoptosis occurs within 10 min after the release of CytoC (61).
FIGURE 9.
Cooperation between p53 killer and E2F1 in apoptosis induction at DIR = 5 Gy. A, shown is the dynamic process of apoptosis induction by p53 killer and E2F1. Shown are time courses of [p53 killer] (black), [E2F1] (red), and [ASPP] (blue) (first panel), [Bax] (black) and [p53AIP1] (red) (second panel), [procasp9] (blue), [Apaf-1] (red), [Procasp3] (green), and [CytoC] (black) (third panel), and [Apops] (black), [Casp9] (red), and [Casp3] (blue) (fourth panel) (from top to bottom). B, shown are time courses of [Bax] (black), [p53AIP1] (red), [CytoC] (green), and [Casp3] (blue) in ASPP-deficient cells with ksaspp2 = 0. C, shown are time courses of [CytoC] (black), [Casp9] (red), and [Casp3] (blue) in Procasp9-deficient cells with kscasp92 = 0.
To verify the role of E2F1-induced ASPP in apoptosis induction, we simulated the cellular response without E2F1-inducible ASPP production. In this case, both Bax and p53AIP1 are insufficient to trigger the release of CytoC and activate Casp3 (Fig. 9B). To further validate the significance of E2F1 in caspase activation, we considered the case with Procasp9 deficiency resulting from that the promoter of Casp9 cannot be bound by E2F1. Notably, both the release of CytoC and activation of the caspase cascade are blocked (Fig. 9C). Similarly, the deficiency of either Apaf-1 or Procasp3 leads to inactivation of Casp3 (data not shown). These results indicate that E2F1-induced synthesis of ASPP and procaspases is required for apoptosis induction.
Cell Fate Decision in a Population of Cells
Because there is variability in the cellular response to DNA damage, the fraction of apoptotic cells within the population, FA, is used to characterize the relationship between cell fate and the IR dose at the population level. In the normal case, apoptosis first appears at DIR = 2 Gy, and FA rises with DIR until all cells are eliminated at DIR >7 Gy (Fig. 10A). In p21-deficient cells, marked apoptosis appears when DIR > 1.5 Gy, and almost all cells die when DIR > 3 Gy. Thus, p21-deficient cells become very sensitive to DNA damage. By contrast, in the presence of high p21 levels, apoptosis appears only when DIR > 5 Gy, and FA increases slowly with DIR, with more than 70% of cells still surviving at DIR = 10 Gy. Thus, overexpression of p21 makes cells resistant to DNA damage.
FIGURE 10.
Fraction of apoptotic cells within a population of 2000 cells, FA, versus DIR. A, shown are curves for cases with different synthesis rate of p21: ksp21 = 0.07 (rectangle), 0 (circle), or 0.1 (triangle). B, shown are curves for cases with different association constant between Rb and E2F1: ksp21 = 2 and IS = 10 (normal case, rectangle) or kre = 0.1 and IS = 0.4 (with E1A activation, triangle).
Moreover, in low serum media, the oncoprotein E1A can directly activate E2F1 by binding Rb (64), thereby promoting apoptosis induction. We simulated this effect by reducing kre to 0.1 with IS = 0.6. Compared with the normal case, FA becomes much larger at low damage levels, and all cells undergo apoptosis when DIR > 2.5 Gy (Fig. 10B). These results further verify the critical role of E2F1 in modulating the sensitivity of cellular response to DNA damage. Our results agree with the experimental observations that deregulated E2F1 activity promotes p53-mediated apoptosis (18).
DISCUSSION
In this work we explored how cell cycle progression and cell fate decision are coordinated in response to DNA damage in serum medium. An integrated model was constructed to characterize the process from the generation of DNA damage to the choice of cell fate. In our model, the coupled positive and negative feedback loops involving p53, Mdm2, and Akt are responsible for p53 pulses. Notably, ATM levels exhibit pulses or switch between two states, depending on Wip1 levels. At low damage levels, relatively few p53 pulses can induce transient G1 arrest, and serum-activated E2F1 can induce S-phase entry after DNA damage is fixed. At high damage levels, E2F1 is activated after four ATM pulses and then cooperates with p53 killer to induce apoptosis. We found that either p21 deficiency or disassociation of the Rb/E2F1 complex by E1A can counteract the protective effect of cell cycle arrest and potentiate p53-dependent apoptosis by activating E2F1. In addition, our results suggest that the induction of Apaf-1 and procaspase-3 and -9 is crucial for activation of the caspase cascade. Therefore, cross-talk between the p53 and E2F1 pathways ensures the coordination between cell cycle progression and cell fate decision, which is of great importance for tumor suppression.
We have previously proposed that cell fate is determined by the number of p53 pulses (11), but we did not explore the cross-talk between p53 and other transcription factors. It is important to clarify functional roles of cell cycle control in cell fate decision. For example, Pfeuty et al. (65) recently investigated the decision between G0 arrest, G1 arrest, S-phase entry, and apoptosis in a coarse-grained model of the G1 regulatory network. They mainly characterized the interaction between the p53-p21 and Rb-E2F1 pathways in cell cycle progression and simply considered the cooperation between p53 and E2F1 in Apaf-1 induction. By comparison, the present study probed in detail how cell cycle progression and cell fate decision are coordinated by the p53 and E2F1 pathways.
It is already established that E2F1 is a key mediator of S-phase entry (13) and can cooperate with p53 to induce apoptosis (15). In this work we focused on the role of E2F1 in the p53-mediated cellular response to DNA damage in serum medium. Note that E2F1-inducible ARF can also stabilize and activate p53 by inhibiting its degradation upon oncogene activation (66). Upon DNA damage, however, p53 is activated by ATM. Thus, here we did not consider the direct activation of p53 by E2F1.
In our model, cross-talk between the p53 and E2F1 pathways is mainly reflected in two aspects. On the one hand, E2F1 modulates the conversion between the two forms of active p53, i.e. p53 arrester and p53 killer. p53 arrester induces cell cycle arrest, whereas growth signals can drive cells to overcome cell cycle arrest and activate E2F1. E2F1 then induces expression of p53DINP1, which promotes the accumulation of p53 killer. On the other hand, the up-regulation of ASPP1/2 and procaspases by E2F1 is crucial for p53-dependent apoptosis. Together, our results reveal that E2F1 can promote p53-mediated apoptosis by both biasing p53 activity toward apoptosis and ensuring the commitment of apoptosis. These results are in good agreement with experimental observations (18, 50).
Our results suggest that the phase of the cell cycle is associated with distinct cell fates; both processes are mediated by p53 and E2F1. We showed that p53-targeted p21 induces transient G1 arrest by inhibiting E2F1 activity, allowing time for DNA repair and facilitating cell survival. Thus, p21-induced G1 arrest provides a protective mechanism for cells with mild damage. This is different from the notion that the prosurvival role of p21 is linked with its ability to suppress apoptosis by inactivating caspase-3 or -9 (62, 67). In the present study, we emphasized that it is important for p21 to promote cell survival by inhibiting the transcriptional activity of E2F1. It was demonstrated that prolonged cell cycle arrest by incremental p21 pulses can make cells resistant to death signals. On the other hand, our results suggest that S-phase entry may be required for p53-dependent apoptosis in response to IR-induced DNA damage. Accelerated entry into S phase due to p21 deficiency or oncogene activation makes cells sensitive to DNA damage. Indeed, it has been reported that oncogene activation is linked to S-phase entry as well as deregulated E2F1 activity (15–17). In addition, ultraviolet light-induced apoptosis is also associated with S-phase entry (68). Nevertheless, the mechanism for the correlation of S-phase entry with apoptosis induction still needs to be explored further.
Our work may provide clues to the diagnosis and treatment of cancers. It is noted that E2F1 is frequently deregulated in cancer cells due to Rb deficiency (69). At the same time, p53 is inactivated by mutation in more than 50% of human tumors (3). Thus, cancer cells with deregulated E2F1 and inactive p53 may be killed by p53 reactivation (70). It is expected that reactivation of p53 should induce either transient growth arrest in normal cells or apoptosis in cancer cells. Thus, the treatment not only eliminates tumor cells but also reduces the side effects in normal cells. Moreover, our results suggest that some oncogene proteins, such as E1A, may be exploited to reactivate E2F1 so as to kill the tumor cells with overexpressed p21 in concert with p53. Nevertheless, it is still a challenge to develop more effective, less toxic treatments that fully take advantage of cross-talk between the p53 and E2F1 pathways.
In the present work we have reported the coordination of cell cycle progression and cell fate decision by p53 and E2F1 in response to DNA damage. In unstressed cells, E2F1 is activated to promote cell proliferation, while Akt is activated to inhibit p53 activity. In stressed cells, Akt is repeatedly deactivated by p53. Transient p53 pulses induce cell cycle arrest before E2F1 activation, whereas overcoming cell cycle arrest by growth factors leads to the activation of E2F1, which cooperates with p53 to initiate apoptosis. Therefore, this work clarifies the link between cell cycle progression and cell fate decision and may provide clues to cancer treatment.
Supplementary Material
This work was supported by the National Basic Research Program of China (2007CB814806), the National Natural Science Foundation of China (10604028), the Natural Science Foundation of Jiangsu Province (SBK200910089), the Jiangsu Planned Projects for Postdoctoral Research Funds (0204003443), the Program for New Century Excellent Talents in Universities (NCET-08-0269), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods S1 and S2, Tables S1 and S2, and Figs. S1–S7.
- p53DINP1
- p53-dependent damage inducible nuclear protein 1
- ATM
- ataxia telangiectasia mutated
- Rb
- retinoblastoma protein
- Wip1
- wild-type p53-induced phosphatase 1
- ASPP
- apoptosis-stimulating protein of p53
- p53AIP1
- p53-regulated apoptosis-inducing protein 1
- Apaf-1
- apoptotic protease activating factor-1
- Gy
- gray
- DSB
- double-strand break
- DSBC
- DSB complex
- CycE
- cyclin E
- CytoC
- cytochrome c
- Casp
- caspase.
REFERENCES
- 1.Meek D. W. (2009) Nat. Rev. Cancer 9, 714–723 [DOI] [PubMed] [Google Scholar]
- 2.Murray-Zmijewski F., Slee E. A., Lu X. (2008) Nat. Rev. Mol. Cell Biol. 9, 702–712 [DOI] [PubMed] [Google Scholar]
- 3.Vousden K. H., Lane D. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 4.Lev Bar-Or R., Maya R., Segel L. A., Alon U., Levine A. J., Oren M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11250–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A. J., Elowitz M. B., Alon U. (2004) Nat. Genet. 36, 147–150 [DOI] [PubMed] [Google Scholar]
- 6.Ma L., Wagner J., Rice J. J., Hu W., Levine A. J., Stolovitzky G. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14266–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geva-Zatorsky N., Rosenfeld N., Itzkovitz S., Milo R., Sigal A., Dekel E., Yarnitzky T., Liron Y., Polak P., Lahav G., Alon U. (2006) Mol. Syst. Biol. 2, 0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchelor E., Mock C. S., Bhan I., Loewer A., Lahav G. (2008) Mol. Cell 30, 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciliberto A., Novak B., Tyson J. J. (2005) Cell Cycle 4, 488–493 [DOI] [PubMed] [Google Scholar]
- 10.Zhang T., Brazhnik P., Tyson J. J. (2007) Cell Cycle 6, 85–94 [DOI] [PubMed] [Google Scholar]
- 11.Zhang X. P., Liu F., Cheng Z., Wang W. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12245–12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batchelor E., Loewer A., Lahav G. (2009) Nat. Rev. Cancer 9, 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L., Timmers C., Maiti B., Saavedra H. I., Sang L., Chong G. T., Nuckolls F., Giangrande P., Wright F. A., Field S. J., Greenberg M. E., Orkin S., Nevins J. R., Robinson M. L., Leone G. (2001) Nature 414, 457–462 [DOI] [PubMed] [Google Scholar]
- 14.Hallstrom T. C., Nevins J. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Levine A. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3602–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin X. Q., Livingston D. M., Kaelin W. G., Jr., Adams P. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 10918–10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeGregori J., Leone G., Miron A., Jakoi L., Nevins J. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko T., Chaussepied M., Oren M., Ginsberg D. (2005) Cell Death Differ. 12, 377–383 [DOI] [PubMed] [Google Scholar]
- 19.Okamura S., Arakawa H., Tanaka T., Nakanishi H., Ng C. C., Taya Y., Monden M., Nakamura Y. (2001) Mol. Cell 8, 85–94 [DOI] [PubMed] [Google Scholar]
- 20.Samuels-Lev Y., O'Connor D. J., Bergamaschi D., Trigiante G., Hsieh J. K., Zhong S., Campargue I., Naumovski L., Crook T., Lu X. (2001) Mol. Cell 8, 781–794 [DOI] [PubMed] [Google Scholar]
- 21.He G., Siddik Z. H., Huang Z., Wang R., Koomen J., Kobayashi R., Khokhar A. R., Kuang J. (2005) Oncogene 24, 2929–2943 [DOI] [PubMed] [Google Scholar]
- 22.Yao G., Lee T. J., Mori S., Nevins J. R., You L. (2008) Nat. Cell Biol. 10, 476–482 [DOI] [PubMed] [Google Scholar]
- 23.Elkon R., Vesterman R., Amit N., Ulitsky I., Zohar I., Weisz M., Mass G., Orlev N., Sternberg G., Blekhman R., Assa J., Shiloh Y., Shamir R. (2008) BMC Bioinformatics 9, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakkenist C. J., Kastan M. B. (2003) Nature 421, 499–506 [DOI] [PubMed] [Google Scholar]
- 25.Stommel J. M., Wahl G. M. (2004) EMBO J. 23, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fei P., El-Deiry W. S. (2003) Oncogene 22, 5774–5783 [DOI] [PubMed] [Google Scholar]
- 27.Al Rashid S. T., Dellaire G., Cuddihy A., Jalali F., Vaid M., Coackley C., Folkard M., Xu Y., Chen B. P., Chen D. J., Lilge L., Prise K. M., Bazett Jones D. P., Bristow R. G. (2005) Cancer Res. 65, 10810–10821 [DOI] [PubMed] [Google Scholar]
- 28.Burma S., Chen B. P., Chen D. J. (2006) DNA Repair 5, 1042–1048 [DOI] [PubMed] [Google Scholar]
- 29.Stewart R. D. (2001) Radiat. Res. 156, 365–378 [DOI] [PubMed] [Google Scholar]
- 30.Bonner W. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4973–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rief N., Löbrich M. (2002) J. Biol. Chem. 277, 20572–20582 [DOI] [PubMed] [Google Scholar]
- 32.Sengupta S., Harris C. C. (2005) Nat. Rev. Mol. Cell Biol. 6, 44–55 [DOI] [PubMed] [Google Scholar]
- 33.Yang T., Namba H., Hara T., Takmura N., Nagayama Y., Fukata S., Ishikawa N., Kuma K., Ito K., Yamashita S. (1997) Oncogene 14, 1511–1519 [DOI] [PubMed] [Google Scholar]
- 34.Tang W., Willers H., Powell S. N. (1999) Cancer Res. 59, 2562–2565 [PubMed] [Google Scholar]
- 35.Akyüz N., Boehden G. S., Süsse S., Rimek A., Preuss U., Scheidtmann K. H., Wiesmüller L. (2002) Mol. Cell. Biol. 22, 6306–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bill C. A., Yu Y., Miselis N. R., Little J. B., Nickoloff J. A. (1997) Mutat. Res. 385, 21–29 [DOI] [PubMed] [Google Scholar]
- 37.Lee J. H., Paull T. T. (2005) Science 308, 551–554 [DOI] [PubMed] [Google Scholar]
- 38.Mouri K., Nacher J. C., Akutsu T. (2009) PLoS. ONE 4, e5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chickarmane V., Ray A., Sauro H. M., Nadim A. (2007) SIAM J. Appl. Dyn. Syst. 6, 61–78 [Google Scholar]
- 40.Kholodenko B. N. (2006) Nat. Rev. Mol. Cell Biol. 7, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris S. L., Levine A. J. (2005) Oncogene 24, 2899–2908 [DOI] [PubMed] [Google Scholar]
- 42.Gottlieb T. M., Leal J. F., Seger R., Taya Y., Oren M. (2002) Oncogene 21, 1299–1303 [DOI] [PubMed] [Google Scholar]
- 43.Mayo L. D., Donner D. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landers J. E., Cassel S. L., George D. L. (1997) Cancer Res. 57, 3562–3568 [PubMed] [Google Scholar]
- 45.Jeffrey P. D., Gorina S., Pavletich N. P. (1995) Science 267, 1498–1502 [DOI] [PubMed] [Google Scholar]
- 46.Prives C. (1998) Cell 95, 5–8 [DOI] [PubMed] [Google Scholar]
- 47.Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vivanco I., Sawyers C. L. (2002) Nat. Rev. Cancer 2, 489–501 [DOI] [PubMed] [Google Scholar]
- 49.Wee K. B., Aguda B. D. (2006) Biophys. J. 91, 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 51.Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 52.Tomasini R., Samir A. A., Carrier A., Isnardon D., Cecchinelli B., Soddu S., Malissen B., Dagorn J. C., Iovanna J. L., Dusetti N. J. (2003) J. Biol. Chem. 278, 37722–37729 [DOI] [PubMed] [Google Scholar]
- 53.Oda K., Arakawa H., Tanaka T., Matsuda K., Tanikawa C., Mori T., Nishimori H., Tamai K., Tokino T., Nakamura Y., Taya Y. (2000) Cell 102, 849–862 [DOI] [PubMed] [Google Scholar]
- 54.Fiscella M., Zhang H., Fan S., Sakaguchi K., Shen S., Mercer W. E., Vande Woude G. F., O'Connor P. M., Appella E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6048–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takekawa M., Adachi M., Nakahata A., Nakayama I., Itoh F., Tsukuda H., Taya Y., Imai K. (2000) EMBO J. 19, 6517–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moroni M. C., Hickman E. S., Denchi E. L., Caprara G., Colli E., Cecconi F., Müller H., Helin K. (2001) Nat. Cell Biol. 3, 552–558 [DOI] [PubMed] [Google Scholar]
- 57.Cory S., Adams J. M. (2002) Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 58.Bao Q., Shi Y. (2007) Cell Death Differ. 14, 56–65 [DOI] [PubMed] [Google Scholar]
- 59.Chipuk J. E., Fisher J. C., Dillon C. P., Kriwacki R. W., Kuwana T., Green D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20327–20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schimmer A. D., Dalili S., Batey R. A., Riedl S. J. (2006) Cell Death Differ. 13, 179–188 [DOI] [PubMed] [Google Scholar]
- 61.Green D. R. (2005) Cell 121, 671–674 [DOI] [PubMed] [Google Scholar]
- 62.Sohn D., Essmann F., Schulze-Osthoff K., Jänicke R. U. (2006) Cancer Res. 66, 11254–11262 [DOI] [PubMed] [Google Scholar]
- 63.Garner E., Raj K. (2008) Cell Cycle 7, 277–282 [DOI] [PubMed] [Google Scholar]
- 64.Nevins J. R. (1992) Science 258, 424–429 [DOI] [PubMed] [Google Scholar]
- 65.Pfeuty B., David-Pfeuty T., Kaneko K. (2008) Cell Cycle 7, 3246–3257 [DOI] [PubMed] [Google Scholar]
- 66.Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., Vousden K. H. (1998) Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]
- 67.Gartel A. L., Tyner A. L. (2002) Mol. Cancer Ther. 1, 639–649 [PubMed] [Google Scholar]
- 68.McKay B. C., Becerril C., Spronck J. C., Ljungman M. (2002) DNA Repair 1, 811–820 [DOI] [PubMed] [Google Scholar]
- 69.Chau B. N., Wang J. Y. J. (2003) Nat. Rev. Cancer 3, 130–138 [DOI] [PubMed] [Google Scholar]
- 70.Stanelle J., Pützer B. M. (2006) Trends Mol. Med. 12, 177–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.