Abstract
Ecm29 is a 200-kDa HEAT repeat protein that binds the 26 S proteasome. Genome-wide two-hybrid screens and mass spectrometry have identified molecular motors, endosomal components, and ubiquitin-proteasome factors as Ecm29-interacting proteins. The C-terminal half of human Ecm29 binds myosins and kinesins; its N-terminal region binds the endocytic proteins, Vps11, Rab11-FIP4, and rabaptin. Whereas full-length FLAG-Ecm29, its C-terminal half, and a small central fragment of Ecm29 remain bound to glycerol-gradient-separated 26 S proteasomes, the N-terminal half of Ecm29 does not. Confocal microscopy showed that Ecm-26 S proteasomes are present on flotillin-positive endosomes, but they are virtually absent from caveolin- and clathrin-decorated endosomes. Expression of the small central fragment of Ecm29 markedly reduces proteasome association with flotillin-positive endosomes. Identification of regions within Ecm29 capable of binding molecular motors, endosomal proteins, and the 26 S proteasome supports the hypothesis that Ecm29 serves as an adaptor for coupling 26 S proteasomes to specific cellular compartments.
Keywords: Endocytosis, Endosomes, Molecular Motors, Proteasome, Ubiquitin, Ecm29, Flotillin, HEAT Repeat
Introduction
The ubiquitin-proteasome system (UPS)4 encompasses a complex set of molecules, reactions, and pathways that have major impact on eukaryotic cell physiology. Although ubiquitin (Ub) modification can serve non-destructive purposes, degradation of intracellular proteins is the main function of the UPS (1, 2). Besides allowing cells to remove misfolded or otherwise abnormal proteins, the UPS degrades a vast array of normal cellular proteins. Selective proteolysis of key regulatory proteins by the UPS exquisitely controls a wide variety of physiological processes as divergent as cell cycle traverse (3, 4), transcription (5, 6), apoptosis (7), and circadian rhythm (7, 8). In higher eukaryotes selectivity is provided by hundreds of ubiquitin ligases that collaborate with fifty or more Ub carrier proteins to add chains of ubiquitin onto protein substrates (9–12). Chains can be formed through any of the seven lysine residues of ubiquitin (13, 14). Ub chains with appropriate linkages, e.g. Lys-48 or Lys-11, are recognized by a large ATP-dependent protease, and the substrate protein is degraded.
In contrast to the large numbers of UPS components involved in marking substrates, there is only a single enzyme, the 26 S proteasome, that degrades them (15, 16). But here too the situation is complicated, because the 26 S proteasome can associate with a variety of proteins, many of which are components of the Ub system. For example, some deubiquitylating enzymes associate with the 26 S proteasome (17–20). Likewise, several Ub ligases co-purify with 26 S proteasomes or interact with 26 S proteasome subunits (17, 21). An even larger number of proteins has been identified as interacting partners of 26 S proteasome subunits, especially its ATPases (22). Whereas some of these proteins may be substrates, others function to recruit substrates to the 26 S enzyme (23, 24) or to enhance proteolysis under conditions of stress (25). Thus, the 26 S proteasome consists of a central 20 S proteolytic core capped by one or two regulatory complexes (RCs) in dynamic equilibrium with a number of accessory proteins.
One of these accessory components is Ecm29, a protein first identified in a screen for yeast displaying cell wall defects (26). Ecm29 was later connected to the proteasome through large-scale proteomic screens in Saccharomyces cerevisiae (27, 28). Subsequent biochemical procedures confirmed the association of Ecm29 and proteasomes in both yeast and mammalian cells (29, 30). It has been proposed that yeast Ecm29 stabilizes the 26 S proteasome (29, 31). However, it is not clear that Ecm29 serves a similar function in mammalian cells, because levels of Ecm29 vary markedly among mouse organs (30). Moreover, multiple forms of Ecm29 are differentially distributed in mouse brain (32), and the axons of cultured cortical neurons contain different Ecm29 isoforms than those present in dendritic spines.5 Thus, it would seem that, in mammals, Ecm29 has biological functions beyond stabilizing the 26 S holoenzyme.
Ecm29 has been reasonably conserved during evolution, and all Ecm29 sequences are predicted to consist of numerous HEAT repeats, secondary structural motifs often present in proteins that function as adaptors (33, 34). Consistent with a possible adaptor function for Ecm29, Ecm proteasomes are localized on the endoplasmic reticulum (ER), on endosomes and at the centrosome in HeLa cells; based on its intracellular distribution we proposed that Ecm29 links 26 S proteasomes to these cellular compartments (30). Here we report that genome-wide two-hybrid screens and mass spectrometry (MS) analyses of affinity-purified Ecm29 complexes provide further support for the idea that Ecm29 is an adaptor in mammalian cells. Both approaches have identified molecular motors and endosome components as prominent members of a small set of Ecm29-interacting proteins. We also show that Ecm proteasome complexes are present on flotillin-positive endosomes, but they are virtually absent from clathrin- and caveolin-coated vesicles. We speculate that Ecm29 may recruit the 26 S proteasome to flotillin-positive endosomes for the degradation of vesicle-associated signaling proteins.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
See the supplemental Experimental Procedures for a list of materials, antibodies, and their sources. Conditions for the use of antibodies are listed in supplemental Table 1.
Genome-wide Yeast Two-hybrid Screens
High throughput genome-wide yeast two-hybrid screens using human brain libraries were performed as described (35–37). Briefly, cDNAs were generated from poly(A)+ human brain RNA by reverse transcription using random oligonucleotides with a common 5′ sequence, second strand synthesis, and ligation of an oligonucleotide to the 5′-end. The resulting cDNAs were amplified using the PCR and cloned into linearized prey and bait vectors by recombination in yeast. Transformed yeast were plated onto medium lacking uracil (prey constructs) or methionine (bait constructs) for ORF selection. Transformants expressing cDNA fragments fused to the markers URA4 or MET2 were then selected. Pooling the ORF-selected prey colonies into liquid medium-created prey libraries. Individual bait colonies were picked at random from ORF-selection plates and clonally expanded to ∼5 × 106 cells in 96-well plates. Aliquots of 5 × 106 cells from the prey libraries were added to each well and allowed to mate overnight. Matings were plated onto medium that selected simultaneously for the mating event, the expression of the ORF-selection markers, and the activity of the reporter genes ADE2 and HIS3. “Positive” colonies were grown on liquid medium and used as templates in PCR reactions that amplified both bait and prey cDNA inserts. The amplicons served in turn as templates in DNA-sequencing reactions from either the 5′ (DNA-binding domain inserts)- or 3′ (activation domain inserts)-end. The identities of inserts were determined by querying the vector-trimmed sequences against the annotated Homo sapiens genome. Statistical analysis of the resulting protein-protein associations to remove interactions involving fragments considered promiscuous was performed as described (35–37).
Identification of Functional Regions in Ecm29
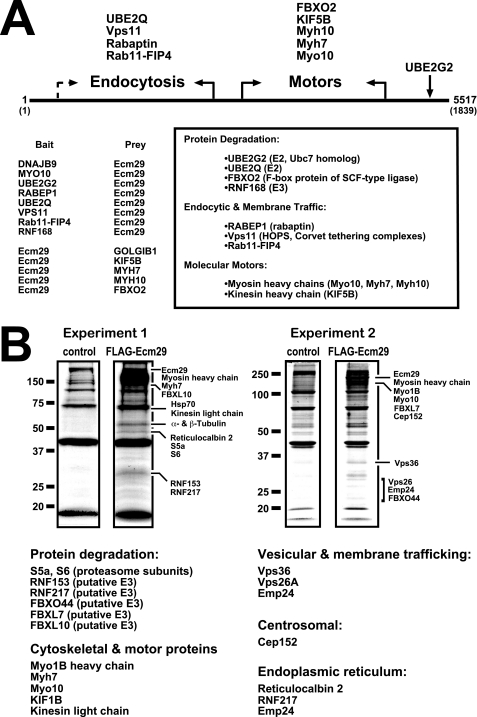
The cDNA libraries used in the two-hybrid assays were collections of fragmented cDNAs encoding on average, 200–300 amino acids (supplemental Table 3). By grouping the identified interactors into functional classes (i.e. endocytosis, transport, nuclear, etc.), we identified binding regions for each class along the Ecm29 sequence. The Ecm29 functional regions were defined on the basis of two criteria. First, both bait and prey Ecm29 cDNAs were isolated in two-hybrid assays in which the Ecm29 interactors belonged to the same functional class. Because the bait and prey cDNAs were, respectively, sequenced from the 5′- and 3′-end, a functional boundary along the Ecm29 sequence could easily be set. Second, we assumed that there was a higher probability that a given Ecm29 cDNA was fully sequenced when the sequenced portion significantly exceeded the average insert size within the respective cDNA library. For example, KIF5B, Myh7, and Myh10 were identified in screens that used Ecm29 cDNA fragments as bait. An Ecm29 fragment was also identified as prey in a screen that used a portion from myosin X (Myo10) as bait; a total of 1483 nucleotides of the prey Ecm29 construct were sequenced, twice the number of the average insert size within the corresponding cDNA library (supplemental Table 3). Given that in these four two-hybrid assays the Ecm29 cDNAs encoded sequences within the C-terminal half of the protein, we have concluded that the C-terminal half of Ecm29 binds kinesin and myosin. Conversely, the two-hybrid data showed that the endocytic components Vps11, Rab11-FIP4, and rabaptin bind the N-terminal portion of Ecm29 (Fig. 1A).
FIGURE 1.
Ecm29 interacting proteins. A, identification of Ecm29-binding proteins by yeast two-hybrid screens. The cDNA libraries used in the yeast two-hybrid screens were collections of fragmented cDNAs encoding on average, 200–300 amino acids (supplemental Table 3). Thus, it was possible to place binding sites for endocytic components in the N-terminal half of Ecm29 and for motor proteins in the C-terminal portion of the protein (see “Experimental Procedures” for details). When both bait and prey Ecm29 cDNAs were isolated for a given class of proteins, the functional boundary is indicated with solid arrows. The dashed arrow indicates an instance in which a functional boundary could not be firmly defined on the basis of available sequences (supplemental Table 3). B, identification of Ecm29-binding proteins by mass spectrometry. HEK293 cells expressing FLAG-Ecm29 were lysed in buffer containing Triton X-100, and the post-mitochondrial supernatants were incubated with M2TM anti-FLAG agarose beads. Bound proteins were eluted with 3× FLAG peptide. As controls, extracts from non-transfected cells were prepared and incubated with antibody beads. Samples (60 μl) of the FLAG-peptide-released proteins were separated by SDS-PAGE and stained with Colloidal Coomassie® Blue. Protein bands in the FLAG sample that were not present in the control lane were excised and subjected to in situ trypsin digestion followed by LC-MS/MS for identification. Corresponding regions in the control lanes were also excised and subjected to LC-MS/MS for comparison. Note that all of the listed Ecm29 interactors were absent in the control samples. A third experiment (not shown) confirmed the results presented above.
Expression of Tagged Versions of Mouse Ecm29 in Cultured Cells
A cDNA encoding mouse Ecm29 (96% identity to human Ecm29) cloned into the pXY vector was purchased from Open Biosystems and used as a template for PCR. The resulting DNA was cloned at the BamH1/SalI sites of the pFLAG-CMV2 (Sigma) vector, which places a single FLAG epitope at the N-terminal end of the encoded protein. HEK293 cells grown on T-75 flasks (Sarstedt) were transfected with 30 μg of pFLAG-CMV2-Ecm29 (or truncated versions thereof) using Lipofectamine 2000TM (Invitrogen) according to the manufacturer's instructions, and harvested 18 h after transfection. The cells were washed once with phosphate-buffered saline (PBS) and centrifuged, and the cell pellets were then frozen in liquid nitrogen and stored at −80 °C until used. For immunofluorescence, MEF cells grown on coverslips were transfected with 10 μg of plasmid DNA and 5 μl of GeneCarrier-2 reagent (Epoch Biolabs) and fixed and processed for confocal microscopy as described below.
M2TM Agarose Affinity Capture of Ecm29-binding Proteins
A volume of 0.5 ml of packed HEK293 cells expressing full-length FLAG-Ecm29 was lysed in 1 ml of 10 mm Tris-HCl, pH 7.5, 1 mm dithiothreitol (DTT), 0.2 mm EDTA, and 0.25% Triton X-100 containing 2× Complete® protease inhibitors (Roche Applied Science) plus 1 mm PMSF. The cell extract was centrifuged at 13,000 × g for 45 min, and the post-mitochondrial supernatant fraction (12.5 mg/ml total protein) was incubated with 0.3 ml of M2TM-agarose beads (Sigma) overnight at 4 °C. The beads were recovered by low speed centrifugation (3,000 × g) and washed three times with lysis buffer containing 1 mm PMSF. Bound proteins were eluted by incubation with 0.3 ml of 3× FLAG peptide (250 μg/ml) in lysis buffer for 1 h at 4 °C. As a control, a post-mitochondrial supernatant from non-transfected HEK293 cells was prepared and incubated with M2TM beads as described above. Sixty-microliter aliquots from both control and FLAG samples were separated by SDS-PAGE on a 10% high Tris (0.75 m Tris-HCl, pH 8.8) gel allowed to polymerize overnight. The gel was rinsed with deionized water and stained with Novex Colloidal Coomassie® Blue as recommended by the manufacturer (Invitrogen).
Mass Spectrometry Proteomic Analyses
For proteomic analyses following liquid chromatography-tandem mass spectrometry (LC-MS/MS), protein samples were digested overnight with tosylphenylchloromethyl ketone-modified trypsin (Promega, Madison, WI). In situ-gel tryptic digests of proteins were analyzed using positive-ion electrospray ionization LC-MS/MS using an Eksigent Nano LC-1D binary pump HPLC system (Eksigent Technologies, LLC) interfaced to a Finnigan LCQ Deca ion trap mass spectrometer (ThermoElectron Corp.) equipped with a Picoview Nanospray source (New Objective, Inc.). Tryptic digests were reconstituted in 5 μl of 5% acetonitrile with 0.1% formic acid, and then manually injected using a nano injector (Valco Instrument Co.) onto a self-packed homemade C18 nanobore column (75 mm inner diameter, 10 cm; Atlantis C18, Waters Corp., 3-μm particle). A 58-min gradient of 5–85% solvent B (A: 5% acetonitrile/0.1% formic acid; B: 80% acetonitrile/0.1% formic acid) was used at 400 nl/min; at 5% B for the first 3 min followed by a linear increase to 55% B in 50 min and finally maintained at 85% B for 5 min. Spectra were acquired in automated “tripleplay” mode for recording of full-scan MS, Zoom scan, and MS/MS data (Excalibur software, ThermoElectron). The scan range for full-scan spectra was set at m/z 400–2000 Da. Automated analysis of peptide fragmentation or product ion spectra (MS/MS) was performed with MASCOT (Matrix Science, London, UK) and/or SEQUEST (ThermoElectron) computer algorithms for protein database searching and protein identification. The MS data were also analyzed and confirmed using the PeptIdent/Aldente mass fingerprinting software tools.
Site-directed Mutagenesis and Truncated Versions of Mouse Ecm29
C-terminal truncated versions (N-terminal fragments) of mouse Ecm29 were constructed by introducing stop codons along the Ecm29 cDNA using the QuikChange® XL site-directed mutagenesis kit from Stratagene and pFLAG-CMV2-Ecm29 as template (see supplemental Experimental Procedures for oligonucleotide sequences). The resulting constructs were sequenced at the DNA Sequencing Core Facility of the University of Utah to verify the incorporation of the desired mutations. N-terminal truncated versions (C-terminal fragments) as well as two central fragments of Ecm29, Ecm29(882–1319), and Ecm29(1032–1319) were constructed by PCR using linearized pFLAG-CMV2-Ecm29 as template and Phusion® high fidelity DNA polymerase (Finnzyme-New England Bio Labs). The amplified cDNAs were cloned at the BamH1/SalI sites of the pEGFP-C2 (BD Biosciences Clontech) and pFLAG-CMV2 vectors and sequenced.
Glycerol Gradient Sedimentation, Gel Electrophoresis, and Western Blotting
HEK293 cells (∼0.4–0.5 ml of packed cells) expressing full-length FLAG-Ecm29 or truncated versions thereof were homogenized in 1 ml of 5 mm HEPES, pH 7.2, 0.25 m sucrose, 0.2 mm EDTA, 1 mm dithiothreitol plus 2× Complete® protease inhibitors and 1 mm PMSF with a Wheaton glass homogenizer equipped with a Teflon pestle. Three hundred-microliter aliquots of the post-mitochondrial supernatants were diluted to 0.4 ml with 50 mm sucrose in 5 mm HEPES, pH 7.2, 0.2 mm EDTA, 1 mm DTT and centrifuged on a 4.5-ml 10–30% glycerol gradient for 4 h at 48,000 rpm using a Beckman SW50.1 rotor. Fractions were assayed for peptidase activity in the presence or absence of ATP using suc-LLVY-MCA as substrate as previously described (38). Samples from each fraction were separated by either SDS-PAGE on 10% high Tris gels, or native gel electrophoresis for 800 V-h at 4 °C. Native gels were overlaid with 20 mm suc-LLVY-MCA for 1 h at 37 °C to visualize the 26 S and 20 S proteasomes as described previously (38). Proteins were transferred to nitrocellulose in 20 mm Tris base, 193 mm glycine, and 1% methanol for 20 h at 4 °C. The FLAG epitope was detected using a 1:250 dilution of monoclonal anti-FLAG antibody, which was visualized by enhanced chemiluminescence and horseradish-peroxidase (HRP)-labeled goat anti-mouse IgG secondary antibodies (diluted 1:500, ICN Biomedical). Regulatory complex subunit S7 was detected using rabbit polyclonal anti-S7 (1:500) and visualized with HRP-labeled goat anti-rabbit IgG (1:500, ICN Biomedical) and the SuperSignal® West Femto chemiluminescence reagent (Pierce Biotechnology).
To confirm the two-hybrid and mass spectrometry data, HEK cells transfected with full-length Ecm29 or FLAG-Ecm29(1–1039) or FLAG-Ecm29(1032–1840) were lysed in 10 mm Tris-HCl, pH 7.5, containing 0.25% Triton X-100, 1 mm DTT, 0.2 mm EDTA plus protease inhibitors. The post-mitochondrial supernatant fractions were then layered atop 4.5-ml 10–22.5% glycerol gradients and centrifuged for 5 h at 48,000 rpm. Fractions were assayed for peptidase activity as described above, and analyzed for anti-FLAG immunoreactivity by slot-blotting 20-μl samples onto nitrocellulose. The distribution of the FLAG epitope through the gradients was determined by densitometry, and pooled fractions were incubated with 0.3 ml of a 1:1 suspension of M2TM-agarose plus 1 mm PMSF overnight at 4 °C. The beads were collected by centrifugation and washed 5 × 10 min with Tris-buffered saline, pH 8.0, containing 0.05% Tween 20 (TBST). Bound proteins were released with 0.3 ml of 3× FLAG and analyzed by immunoblotting using a collection of antibodies to molecular motors, endocytic components, and 26 S proteasome subunits (see supplemental Table 1) using appropriate HRP-labeled secondary antibodies and chemiluminescence.
Purification of Microtubules and Microtubule-associated Endosomes
Microtubules were assembled in vitro, and enriched fractions were prepared as previously described with minor modifications (39). Briefly, 0.5 ml of packed HEK293 cells transfected with pFLAG-CMV2-Ecm29 were suspended in 2.5 volumes of ice-cold extraction buffer (EB, 100 mm PIPES, pH 6.9, containing 1 mm DTT, 1 mm MgSO4, 2 mm EGTA, 0.1 mm GTP, and 0.25 m sucrose plus 2× Complete® protease inhibitors and 1 mm PMSF) and homogenized using a Wheaton glass homogenizer equipped with a Teflon pestle. The cell homogenate was centrifuged at 35,000 × g for 30 min at 4 °C, and the supernatant fraction was incubated with 2 mm GTP and 20 μm Taxol for 45 min at 37 °C. The sample was diluted to 4 ml with 100 mm PIPES, pH 6.9, containing 1 mm DTT, 1 mm MgSO4, 2 mm EGTA, 2 mm GTP, 20 μm Taxol, and 50 mm sucrose plus 1× Complete® protease inhibitors and 1 mm PMSF, and layered atop a 1-ml cushion of 5% sucrose in 100 mm PIPES, pH 6.9, 1 mm DTT, 1 mm MgSO4, 2 mm EGTA, 2 mm GTP, and 20 μm Taxol. The sample was centrifuged at 150,000 × g for 1.5 h at 25 °C to recover the assembled microtubules and MT-associated proteins. The MT fraction was suspended in 0.5 ml of TBST containing 10 μg/ml nocodazole plus 1× Complete® protease inhibitors and 1 mm PMSF, and incubated on ice for 30 min to disassemble the microtubules. Endocytic vesicles enriched in FLAG-Ecm29 were further isolated by incubating the sample with 0.3 ml of a ∼1:1 suspension of M2TM-agarose beads overnight at 4 °C. The beads were recovered by centrifugation and washed three times for 10 min with 1 ml of TBST containing 1 mm PMSF. Bound proteins were then released from the beads with 3× FLAG peptide as described above.
Immunofluorescence
HeLa cell and MEF endocytic vesicles were labeled in the presence of 1 mg/ml Alexa® 568-dextran conjugate for 60 min at 37 °C in Dulbecco's-modified Eagle's (DME) medium containing 10% fetal calf serum. For most experiments, cells grown on coverslips were rinsed with phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde in PBS for 20 min at room temperature. Cells were washed briefly with PBS, permeabilized with −20 °C methanol for 10 min, incubated for 5 min in ice-cold 95% ethanol, and blocked with standard blocking buffer (PBS containing 10% normal goat serum, 2% bovine serum albumin (BSA), 0.2% gelatin, 0.1% Triton X-100, and 0.02% NaN3) overnight at 4 °C. Cells were labeled with primary antibodies in standard blocking buffer (diluted as indicated in supplemental Table 1) overnight at 4 °C, washed with TBST, and visualized with Alexa®-labeled secondary antibodies and confocal microscopy. Alternatively, HeLa cells on coverslips were incubated with PEM buffer (80 mm PIPES, pH 6.8, 5 mm EGTA, 1 mm MgCl2) containing 0.05% saponin on ice for 5 min. The samples were washed once with ice-cold PBS and fixed on ice with 3% paraformaldehyde in PBS for 15 min. Free aldehyde groups were quenched with 50 mm NH4Cl in PBS for 15 min at room temperature. The samples were briefly rinsed with PBS and incubated with primary antibodies in blocking buffer containing 0.05% saponin instead of Triton X-100 overnight at 4 °C. The samples were washed with PBS/saponin and visualized with Alexa®-labeled secondary antibodies diluted in PBS/saponin and confocal microscopy.
Microtubule Staining
HeLa cells growing on coverslips were fed 5 mg/ml Alexa® 488-dextran conjugate for 1 h at 37 °C, washed briefly in PBS, and fixed in 0.5% glutaraldehyde in PBS for 10 min at 37 °C. Unreacted glutaraldehyde was reduced with 3 × 10 min incubations at room temperature with 0.1% NaBH4 in PBS. Washed cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min at room temperature and blocked overnight at 4 °C in standard blocking buffer. Cells were incubated with standard blocking buffer containing diluted anti-α-tubulin and ECM2 antibodies overnight at 4 °C, washed with TBST, and visualized with Alexa®-labeled secondary antibodies and confocal microscopy.
Confocal Microscopy
Fluorescence confocal laser scanning microscopy was performed on an Olympus FV300 confocal microscope equipped with argon 488 and helium/neon 543/633 lasers using a 60× PlanApo 1.4 numerical aperture oil objective and Fluoview 4.3 software.
RESULTS
Genome-wide Yeast Two-hybrid Screens and Mass Spectrometry Identify Endosomal Proteins, Molecular Motors, and Ubiquitin-Proteasome Components as Ecm29-interacting Proteins
Based on its intracellular distribution in HeLa cells we proposed that Ecm29 is an adaptor that recruits 26 S proteasomes to the endoplasmic reticulum and endosomal membranes as well as to the centrosome (30). If the adaptor hypothesis is correct, there should be proteins in these compartments that bind Ecm29. Genome-wide two-hybrid screens and mass spectrometry (MS) after FLAG-Ecm29 affinity capture were used to search for Ecm29-interacting proteins. Many of the Ecm29-interacting proteins identified by both techniques are components of the ubiquitin-proteasome system, molecular motors, or endosomal constituents (Fig. 1; see also supplemental Tables 2 and 4). Eleven of twenty-six Ecm29-binding proteins discovered by the two-hybrid screen fall into these categories; five of the additional fifteen proteins obtained in the genome-wide screen are actin-binding proteins (supplemental Table 2). Almost three-fourths of the Ecm29-interacting proteins identified by mass spectrometry are UPS components, molecular motors, or endosomal constituents (Fig. 1B and supplemental Table 4). The identified UPS components include Ub carrier proteins (UBE2Q and UBE2G2), a host of known or suspected Ub ligases (FBXO2, RNF168, FBXO44, RNF153, etc.), two proteasome subunits (S5a and S6), and the centrosomal protein Cep152. Both MS and the two-hybrid screens identified the molecular motors kinesin and myosin as Ecm29-interacting proteins. In addition, five endosomal components, Vps11, Vps26, Vps36, Rab11-FIP4, and rabaptin, were found by a combination of the two approaches.
N-terminal Portions of Ecm29 Bind Endosomal Components, whereas C-terminal Sequences Bind Molecular Motors
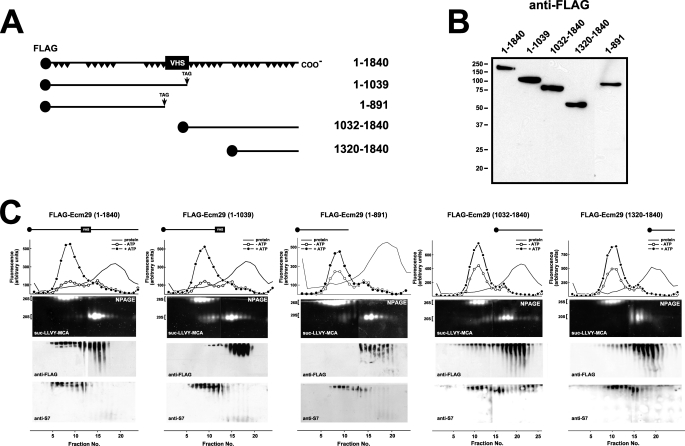
The genome-wide two-hybrid screens employed fragmented human cDNAs encoding, on average, 200–300 amino acids (supplemental Table 3). Their small size relative to the 1839-residue full-length Ecm29 allowed us to provisionally assign binding sites for endocytic components to the N-terminal half of Ecm29 and binding sites for motor proteins to the C-terminal portion of human Ecm29 (Fig. 1). To verify these assignments by co-precipitation methods we constructed FLAG-tagged versions of five N-terminal and three C-terminal mouse Ecm29 truncations. Two of the N-terminal fragments (FLAG-Ecm29(1–891) and FLAG-Ecm29(1–1039)), and two C-terminal constructs (FLAG-Ecm29(1032–1840) and FLAG-Ecm29(1320–1840)) expressed well in HEK293 cells (Fig. 2, A and B). We first asked whether any of the Ecm29 fragments are associated with the 26 S proteasome upon glycerol gradient sedimentation. Sucrose homogenates of HEK cells expressing FLAG-tagged, full-length, or truncated Ecm29 molecules were sedimented on 10–30% glycerol gradients, and fractions were assayed for peptidase activity and separated on native gels followed by immunoblotting for FLAG or for 26 S proteasome subunit S7. These analyses demonstrated that most full-length FLAG-Ecm29 molecules associated with 26 S proteasomes (far left panel in Fig. 2C). A fraction of Ecm29(1032–1840) also sedimented deep into the glycerol gradient suggesting that the C-terminal half of Ecm29 binds the 26 S enzyme (see the fourth panel in Fig. 2C). By contrast none of the N-terminal Ecm29 fragments or the FLAG-Ecm29(1320–1840) C-terminal truncation sedimented with the 26 S proteasome (see the second, third, and fifth panels in Fig. 2C).
FIGURE 2.
Truncated FLAG-Ecm29 constructs are not associated with the 26 S proteasome after glycerol gradient sedimentation. A, schematic representation of FLAG-tagged Ecm29 constructs. A black circle at the N terminus of each construct represents the FLAG epitope. Black triangles in full-length Ecm29 represent putative HEAT repeats, and the boxed VHS represents a putative VHS domain near the middle of the Ecm29 protein. B, expression of FLAG-tagged Ecm29 constructs in HEK293 cells. Transfected cells were lysed in buffer containing Triton X-100, and 40-μg samples were separated on 10% SDS-PAGE gels followed by immunoblotting with anti-FLAG monoclonal antibodies. The lane containing FLAG-Ecm29(1–891) was inserted from a longer exposure of the membrane as HEK293 cells consistently expressed 5- to 10-fold lower levels of this protein relative to the other constructs. C, glycerol gradient sedimentation of full-length and truncated versions of Ecm29. Post-mitochondrial supernatants (3–4 mg of total protein) from transfected HEK cells lysed in buffer containing sucrose were centrifuged on 4.5-ml 10–30% glycerol gradients and assayed for peptidase activity in the absence (○) or presence (●) of ATP using suc-LLVY-MCA as substrate (top panels). Samples were separated on duplicate native gels (NPAGE), overlaid with suc-LLVY-MCA (second panels), and immunoblotted with anti-FLAG (third panels) or an antibody to 19 S RC subunit S7 (bottom panels). Only one set of fluorescent substrate overlays for each gradient is shown. The immunoblots are composites, because two gels were required to analyze all the fractions from each gradient. Glycerol gradient/native gel analyses demonstrate that FLAG-Ecm29 truncations are not associated with the 26 S proteasome. However, some intact FLAG-Ecm29 molecules, present in fractions 12–18, were free of the 26 S proteasome presumably due to saturation of Ecm29 binding sites by excess recombinant molecules.
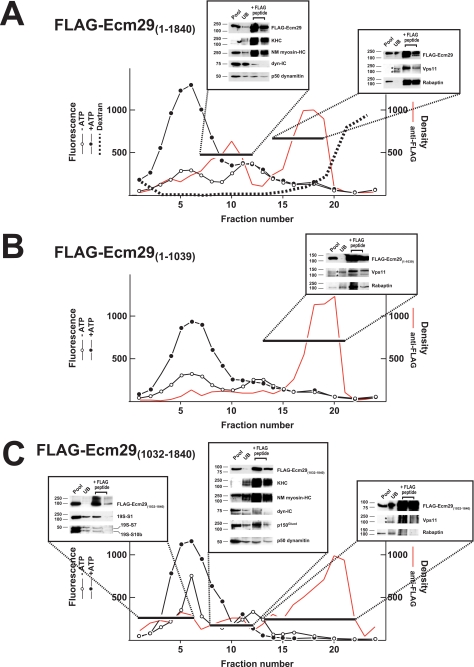
Previously we showed that, although Ecm29 is stably associated with the 26 S proteasome after homogenization in 0.25 m sucrose, it dissociates from the 26 S enzyme in extracts containing 0.25% Triton X-100 (30). Detergent-mediated dissociation of Ecm29 has proved useful, because the Ecm29-interacting proteins listed in Fig. 1 were identified by capture of FLAG-Ecm29 molecules that had been separated by glycerol gradient sedimentation from the 26 S proteasome following extraction of HEK293 cells with Triton X-100. As detergent treatment simplifies interpretation, HEK293 cells were transfected with plasmids encoding FLAG-tagged, full-length, or N- and C-terminal fragments and then extracted in buffer containing Triton X-100 prior to sedimentation of the extracts on 10–22.5% glycerol gradients. Full-length FLAG-Ecm29 was found in two regions of the glycerol gradient, fractions 7–12 and fractions 14–19, whereas the 26 S proteasome was centered at fraction 5. When fractions 7–12 were pooled and subjected to FLAG-affinity capture, SDS-PAGE/Western blotting demonstrated that substantial amounts of 160-kDa kinesin heavy chain (KHC) and non-muscle myosin heavy chain Myh10 (NM myosin-HC) were released upon FLAG-Ecm29 elution by FLAG peptide (boxed region at the left in Fig. 3A). In addition, the p50 dynamitin subunit of dynactin and trace amounts of the 70-kDa dynein intermediate chain (dyn-IC) were present in the peptide eluate from this region of the gradient. A similar analysis of pooled fractions 14–19 revealed that Vps11 and rabaptin were associated with full-length FLAG-Ecm29 (see right box in Fig. 3A), although the anti-FLAG beads failed to precipitate Rab11-FIP4 present in these fractions (not shown).
FIGURE 3.
Association of Ecm29 with molecular motors is mediated by the C-terminal half of the protein, whereas the N-terminal half of Ecm29 mediates its association with endocytic components. HEK cells transfected with FLAG-tagged full-length or truncated Ecm29 were lysed in buffer containing Triton X-100 and the post-mitochondrial supernatants (3.5–4.9 mg of total protein) were centrifuged on 4.5-ml of 10–22.5% glycerol gradients and assayed for peptidase activity in the absence (○) or presence (●) of ATP using suc-LLVY-MCA as substrate. The distribution of the FLAG proteins was determined by densitometry of 20-μl samples slot-blotted onto nitrocellulose and probed with anti-FLAG monoclonal antibodies (shown in red). Fractions were pooled as indicated by the black bars and incubated with 0.3 ml of a 1:1 suspension of M2TM-agarose plus 1 mm PMSF overnight at 4 °C. The beads were collected by centrifugation and washed 5 × 10 min with TBST. Bound proteins were released with 0.3 ml of 3× FLAG and analyzed by immunoblotting using a collection of antibodies to molecular motors, endocytic components, and 26 S proteasome subunits. A, HEK cells transfected with full-length FLAG-Ecm29 were exposed to 5 mg/ml Alexa®568-dextran conjugate for 1 h at 37 °C before lysis. The distribution of the fluorescent dextrans was determined at 560 nm excitation and 618 nm emission (dashed line). Two major peaks of anti-FLAG immunoreactivity were observed following expression of full-length FLAG-Ecm29. Fractions 7–12 contained Ecm29 bound to molecular motors, whereas the second peak (fractions 14–19) included Ecm29 complexed to Vps11 and rabaptin. B, only a single peak of anti-FLAG immunoreactivity was observed after expression of FLAG-Ecm29(1–1039), the N-terminal half of Ecm29. Whereas fractions 13–21 contained Ecm29 bound to Vps11 and rabaptin, they showed no reactivity for myosin heavy chain or the 160-kDa kinesin heavy chain (not shown). C, three major peaks of anti-FLAG immunoreactivity were observed upon gradient analysis following expression of Ecm29's C-terminal half. In fractions 1–6 truncated Ecm29 was associated with the 26 S proteasome. The second peak (fractions 8–12) included FLAG-Ecm29(1032–1840) bound to molecular motors, whereas the third peak (fractions 13–22) contained the uncomplexed C-terminal half of FLAG-Ecm29. The asterisks indicate two forms of Vps11 detected in the unbound (UB) fraction of the slowest sedimenting peak containing a FLAG-Ecm29 protein in each gradient. Note that, in the gradient presented in panel C, the nonspecific material detected in the FLAG peptide eluate of peak 3 does not migrate on the SDS gel with either form of Vps11. Presumably the apparent positive signal reflects streaking caused by contaminating anti-FLAG beads and a long film exposure required to detect Vps11 in the UB fraction. KHC, kinesin heavy chain; NM myosin-HC, non-muscle myosin heavy chain Myh10; and dyn-IC, dynein intermediate chain.
Whereas two peaks of full-length FLAG-Ecm29 were present in Fig. 3A, only a single peak of the FLAG-Ecm29 N-terminal region, fractions 16–20, was evident in the 10–22.5% glycerol gradient (Fig. 3B). FLAG-affinity capture of the pooled FLAG-Ecm29(1–1039) followed by SDS-PAGE and Western blotting revealed that Rab11-FIP4 (data not shown), Vps11, and rabaptin were associated with Ecm29's N-terminal half (see inset in Fig. 3B). Due to their large sizes, the molecular motors kinesin and myosin were not present in fractions containing FLAG-Ecm29(1–1039). More importantly, FLAG-Ecm29(1–1039) was not present in any appreciable amounts in fractions enriched in KHC and myosin heavy chain (Fig. 3B, fractions 7–12). We conclude from the absence of FLAG-Ecm29(1–1039) in fractions 7–12 that the N-terminal half of Ecm29 does not bind molecular motors.
The FLAG-tagged C-terminal half of Ecm29 produced the most complicated pattern after sedimentation, because it was present in three regions of the glycerol gradient (Fig. 3C). Like full-length FLAG-Ecm29, C-terminal FLAG-Ecm29 was present in fractions 7–12 and fractions 16–20. And like the full-length molecule, Ecm29(1032–1840), was also bound to the molecular motors kinesin and myosin as shown by FLAG-capture experiments, as well as the p150Glued and p50 dynamitin subunits of dynactin (Fig. 3C, central inset). However, unlike full-length Ecm29, the C-terminal half of Ecm29 did not bind the endosomal components, Vps11 or rabaptin (right inset, Fig. 3C). In addition to its presence in fractions 7–12 and 16–19, there was a small broad peak of FLAG-Ecm29(1032–1840) in fractions 1–6 of the gradient. This region of the gradient is enriched in 26 S proteasomes, and FLAG-capture analyses on these pooled fractions revealed that subunits S1, S7, and S10b of the 26 S proteasome were bound to the C-terminal half of Ecm29 (see left-most inset in Fig. 3C).
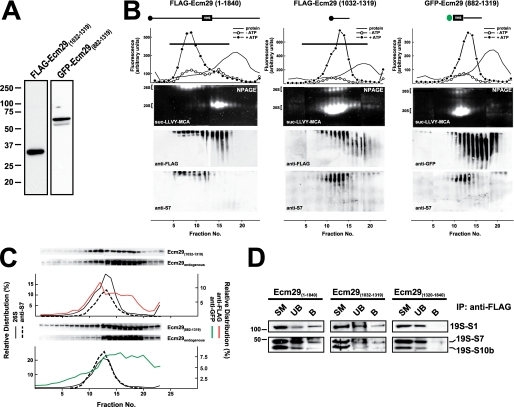
Expression of Small Central Regions of Ecm29 Displaces Endogenous Ecm29 from the 26 S Proteasome and Destabilizes the Enzyme
The weaker proteasome binding of Ecm29's C-terminal half relative to the intact molecule suggested that full proteasome binding might require sequences on both sides of the central truncation sites, residues 1032 and 1039, used to generate the two halves of Ecm29. To test this hypothesis, we constructed expression vectors encoding small central portions of Ecm29, GFP-Ecm29(882–1319), and FLAG-Ecm29(1032–1319), and expressed these small fragments in HEK cells. Expression of both central regions resulted in 26 S proteasomes that are dissociated on glycerol gradients (Fig. 4B). Sucrose homogenates of transfected HEK cells were sedimented on 10–30% glycerol gradients, assayed for peptidase activity, and analyzed on native gels. As shown in Fig. 4B, 26 S proteasomes in FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) homogenates sedimented much slower than 26 S proteasomes present in extracts expressing the full-length recombinant protein. Also, the activity of 26 S proteasomes separated on native gels was greatly diminished in samples from FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) gradients, whereas there was a concomitant increase in 20 S proteasomal activity (Fig. 4B). Moreover, larger amounts of 19 S RC were observed on Western blots of the FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) gradients using antibodies to proteasomal S7. Altogether these results indicate that expression of small central regions of Ecm29 in HEK cells results in 26 S proteasomes that are relatively unstable and readily dissociate into the 19 S RC and 20 S proteasome either within the transfected HEK cells or upon glycerol gradient sedimentation.
FIGURE 4.
Expression of FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) displaces endogenous Ecm29 and destabilizes the 26 S proteasome. A, expression of FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) in HEK293 cells. SDS-PAGE of Triton X-100 extracts from transfected HEK293 cells (30 μg of protein) were analyzed by immunoblotting with monoclonal antibodies to the FLAG epitope or green fluorescent protein (GFP). B, glycerol gradient sedimentation of FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319). The post-mitochondrial supernatants (2–3 mg of protein) from transfected 293 cells lysed in buffer containing sucrose were centrifuged on 4.5-ml 10–30% glycerol gradients and analyzed as shown in Fig. 2C except that one set of the duplicate native gels of the GFP-Ecm29(882–1319) gradient was immunoblotted with a monoclonal anti-GFP antibody (right). For comparison, the glycerol gradient for full-length Ecm29 (FLAG-Ecm29(1–1840)) shown in Fig. 2C has been placed next to the gradients of the two truncated versions of Ecm29. Only one set of fluorescent substrate overlays for each gradient is shown. The immunoblots are composites, because two gels were required to analyze all the fractions from each gradient. Note that the 26 S proteasome is labile and sediments much slower in FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) extracts. C, fractions from the FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) glycerol gradients were separated by SDS-PAGE and immunoblotted with either anti-FLAG (top), or anti-GFP (bottom), or anti-ECM3 (to detect full-length endogenous Ecm29). The relative distribution of the FLAG (red trace) or GFP (green trace) signals on the gradients was estimated by densitometry of the Western blots using the NIH image 1.63f software package, and is expressed as the percentage of the total amount of densitometric units. The relative distribution of the 26 S proteasome (% ATP-dependent activity, solid lines) and S7 (dashed lines) are shown for comparison. The black horizontal lines on the top panel indicate fractions that were pooled and further subjected to immunoprecipitation with M2TM agarose beads. D, full-length FLAG-Ecm29 and FLAG-Ecm29(1032–1319) immunoprecipitate subunits of the 19 S regulatory complex. The anti-FLAG immunoprecipitates of pooled fractions (panel B and data not shown) from the glycerol gradients of full-length FLAG-Ecm29 (fractions 1–14), FLAG-Ecm29(1320–1840) (fractions 1–14), and FLAG-Ecm29(1032–1319) (fractions 1–16) were analyzed by SDS-PAGE and immunoblotting with antibodies to the S1, S7, and S10b subunits of the 19 S RC. The figure shows that only full-length Ecm29 and Ecm29(1032–1319) associate with and precipitate the 19 S RC of the 26 S proteasome.
Fractions from the glycerol gradients in Fig. 4B were separated on SDS-PAGE gels followed by immunoblotting with antibodies to either tag (FLAG or GFP) or to Ecm29 itself. It is evident in Fig. 4C, that significant amounts of endogenous Ecm29 sedimented slower than the 26 S proteasome in homogenates from HEK cells expressing FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319). By contrast, significant amounts of each recombinant fragment co-sedimented with the 26 S enzyme. These results strongly suggest that FLAG-Ecm29(1032–1319) and GFP-Ecm29(882–1319) displace endogenous Ecm29 molecules from the 26 S proteasome. Presumably the small central fragments compete with full-length Ecm29 by directly binding the 26 S enzyme/19 S RC. We tested this presumption by FLAG-capture experiments. Fractions 1–14 from the FLAG-Ecm29 and FLAG-Ecm29(1320–1840) glycerol gradients, as well as fractions 1–16 from the FLAG-Ecm29(1032–1319) glycerol gradient, were pooled separately and incubated with M2TM-agarose beads. Proteins released from the beads with 3× FLAG peptide were resolved on SDS-PAGE gels, transferred to nitrocellulose, and probed with antibodies to 19 S RC subunits. Fig. 4D shows that the 19 S RC bound both full-length FLAG-Ecm29 and FLAG-Ecm29(1032–1319), but did not bind the FLAG-Ecm29(1320–1840) C-terminal fragment. Thus, these data indicate that, at a minimum, the short central region of Ecm29 binds the 26 S proteasome.
Ecm29 Is Present on Flotillin-decorated Endosomes That Are Also Positive for Its Interacting Components
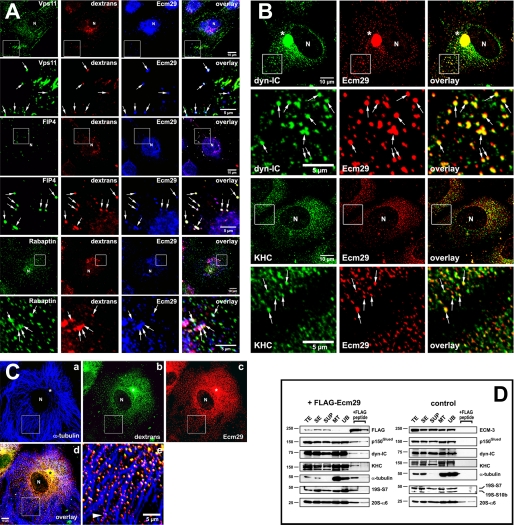
If the endocytic proteins listed in Fig. 1 are true Ecm29-binding partners, they should be found on endosomes along with Ecm proteasomes. To determine whether this is the case HeLa cells were exposed to Alexa®568-dextran conjugate for 60 min, fixed and examined by confocal microscopy after immunostaining with appropriate combinations of antibodies. We observed endosomes decorated by Ecm proteasomes and each of the endosomal proteins, Vps11 or Rab11-FIP4 or rabaptin (Fig. 5A; see also supplemental Fig. 1). We also observed the presence of Ecm29 on endosomes that stained positive for Vps26 and Vps36, two Ecm29-interacting proteins identified by FLAG-affinity capture and LC-MS/MS (supplemental Fig. 2). A sixth endosomal component, Arf6, was identified as an Ecm29-interacting protein in a proteomic screen conducted by Ewing et al. (40); it too was present on endosomes along with Ecm proteasomes (see supplemental Fig. 3). Similar experimental protocols revealed that Ecm proteasomes and the molecular motors, kinesin or dynein, were present together on endosomes (Fig. 5B and supplemental Fig. 5). Furthermore, Ecm29-positive endosomes were clearly associated with the microtubule network as expected for kinesin- or dynein-positive vesicles (Fig. 5, C and D and supplemental Fig. 4). Staining for the actin-based motor myosin mimicked the distribution of actin filaments within the cells, but revealed few clear Ecm29-decorated endosomes that were associated with myosin-HC or the actin cytoskeleton (data not shown). This may well reflect a transient interaction between Ecm29 and myosin-HC or, more likely, may be the result of the relatively long incubation of the HeLa cells with fluorescent dextran. Conceivably, endocytic vesicles internalized with the help of unconventional myosins and F-actin could quickly switch motors and “jump” onto microtubule tracks as they travel toward the centrosome (41–43).
FIGURE 5.
Presence of Ecm29 on endosomes decorated with its putative binding partners. A, Ecm29 is present on vesicles associated with the endocytic proteins Vps11, Rab11-FIP4, and rabaptin. HeLa cells growing on coverslips were exposed to 1 mg/ml Alexa® 568-dextran conjugate (dextrans) for 1 h at 37 °C and then incubated for 5 min in PBS/0.05% saponin prior to fixation with 3% paraformaldehyde. Cells were stained with rabbit anti-ECM2 (Ecm29) and either a mouse monoclonal antibody to Vps11, or a sheep polyclonal antibody to Rab11-FIP4 (FIP4), or a monoclonal antibody to rabaptin. White boxes enclose areas magnified in the panels directly below the full size images. The arrows indicate dextran-filled endosomes on which Ecm29 co-localizes with Vps11, Rab11-FIP4, or rabaptin. B, Ecm29-positive endosomes are bound to kinesin and cytoplasmic dynein. HeLa cells growing on coverslips were fixed with 0.5% glutaraldehyde at 37 °C and stained with ECM2 antibodies (Ecm29) and either a monoclonal antibody to the 70-kDa subunit of cytoplasmic dynein (dyn-IC) or an antibody to kinesin heavy chain (KHC). White boxes enclose areas magnified in the panels directly below the full size images. The arrows indicate examples of Ecm29-positive vesicles bound to the molecular motors. C, Ecm29 is present on endocytic vesicles bound to the microtubule cytoskeleton. HeLa cells growing on coverslips were labeled for 60 min at 37 °C with 5 mg/ml Alexa® 488-dextran conjugate (panel b) and fixed with 0.5% glutaraldehyde. Cells were stained with a mouse monoclonal antibody to α-tubulin (panel a) and anti-ECM2 (Ecm29, panel c) polyclonal antibodies. The merged image from panels a–c is shown in panel d. The arrowhead in panel e indicates a clear example of Ecm29-positive endosomes associated with a microtubule bundle. Samples in panels A–C were visualized with Alexa®-labeled secondary antibodies and confocal microscopy. Asterisk, centrosomal region. N, cell nucleus. Scale bars, 5 or 10 μm as indicated. D, immunoaffinity isolation of microtubule- and molecular motor-bound endosomes. HEK293 cells expressing FLAG-Ecm29 were lysed in buffer containing sucrose and a microtubule (MT)-enriched fraction was prepared in the presence of GTP and Taxol. The MT fraction was treated with nocodazole to disassemble the MTs prior to incubation with M2TM anti-FLAG-agarose beads overnight at 4 °C. The beads were washed, and bound proteins were released with 3× FLAG peptide. Samples of each fraction (20 μg of total protein, except for the samples released from the beads with FLAG peptide, which represent 60 μl or 20% of the total fraction) were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies (left). An MT-enriched fraction was prepared from non-transfected HEK cells as a control and treated as described above (right), except that Ecm29 in each sample was detected with polyclonal anti-ECM3 antibodies. The amount of FLAG-peptide-released protein was estimated by densitometry of the Western blots using the NIH image 1.63f software package and is expressed as the percentage of the amount of each protein found in the cell homogenate: FLAG-Ecm29, 8.7%; p150Glued, 0.5%; dynein intermediate chain (dyn-IC), 0.2%; kinesin heavy chain (KHC), 0.3%; α-tubulin, 1.2%; 19 S RC subunit 7, 1.8%. TE, cell homogenate; SE, soluble extract; SUP, supernatant fraction following centrifugation through a 5% sucrose cushion; MT, microtubule-enriched fraction; and UB, unbound fraction.
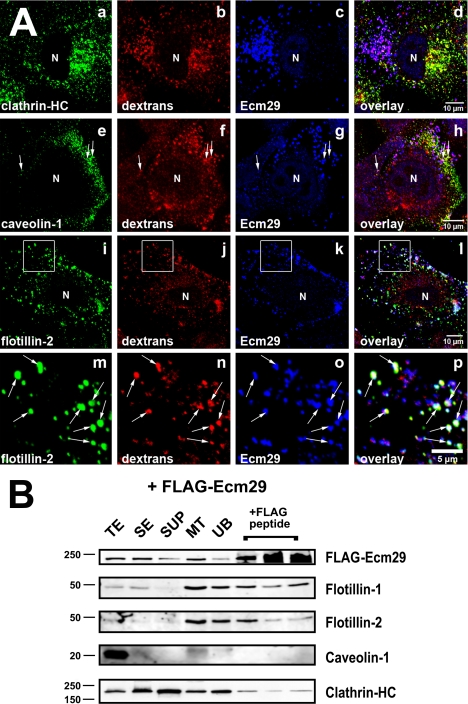
In an earlier publication employing fluorescent dextran labeling we showed that Ecm proteasomes are present on newly formed endosomes (30). During the past decade it has become apparent that many endocytic pathways exist in higher eukaryotic cells (see Refs. 44–46 for recent reviews). Therefore we asked whether Ecm proteasomes are restricted to a specific subclass of endosomes. Because early endosomes can be classified by their associated coat proteins, HeLa cells were exposed to Alexa®568-dextran conjugate and stained for Ecm29 and either clathrin, caveolin, or flotillin. It is clear from the confocal images in Fig. 6A that Ecm proteasomes are present almost exclusively on flotillin-positive endosomes (see also supplemental Figs. 6 and 7), and flotillins 1 and 2 are enriched in microtubule-bound vesicles isolated from cell extracts expressing FLAG-Ecm29 (Fig. 6B). Although an occasional dextran-filled endosome stained positive for Ecm29 and caveolin-1 (arrows in panels e–h of Fig. 6A), Ecm29 was not seen on newly formed clathrin-coated endosomes (Fig. 6A, panels a–d). Furthermore, no caveolin-1 and little clathrin heavy chain were recovered in the 3× FLAG eluates of MT-bound vesicles from FLAG-Ecm29-expressing cells (Fig. 6B). Possible reasons for their specific association with flotillin-positive endosomes are presented under “Discussion.”
FIGURE 6.
Ecm proteasomes localize to flotillin 2-positive endosomes. A, HeLa cells growing on coverslips were labeled with 1 mg/ml Alexa® 568-dextran conjugate (dextrans) for 1 h at 37 °C and fixed with 2% paraformaldehyde. Cells were stained with rabbit anti-ECM2 (Ecm29), and either monoclonal antibodies to clathrin heavy chain (clathrin-HC), or caveolin-1 (panel e), or a goat polyclonal antibody to flotillin 2 (panels i and m). Samples were visualized with Alexa®-labeled secondary antibodies and confocal microscopy. White boxes (panels i–l) enclose areas magnified in the images directly below each full-size image (panels m–p). Arrows identify examples of dextran-filled, flotillin-2-positive endosomes to which Ecm29 localizes (panels m–p). N, cell nucleus. Scale bars, 5 or 10 μm as indicated. B, Ecm29-positive endosomes bound to microtubules are enriched in flotillins. An MT fraction was prepared as described in the legend to Fig. 5 (see also “Experimental Procedures”); upon disassembly with nocodazole, the fraction was subjected to immunoprecipitation with M2TM anti-FLAG-agarose beads, and bound proteins were released with 3× FLAG peptide. Samples (20 μg of total protein, except for the samples released from the beads, which represent 60 μl or 20% of the total fraction) were analyzed by immunoblotting with the indicated antibodies. The amount of FLAG-peptide-released protein was estimated by densitometry of the Western blots using the NIH image 1.63f software package and is expressed as the percentage of the amount of each protein found in the cell homogenate: FLAG-Ecm29, 2.5%; flotillin-1, 4.3%; flotillin-2, 1.2%; clathrin heavy chain, 0.8%. TE, cell homogenate; SE, soluble extract; SUP, supernatant fraction following centrifugation through a 5% sucrose cushion; MT, microtubule-enriched fraction; and UB, unbound fraction.
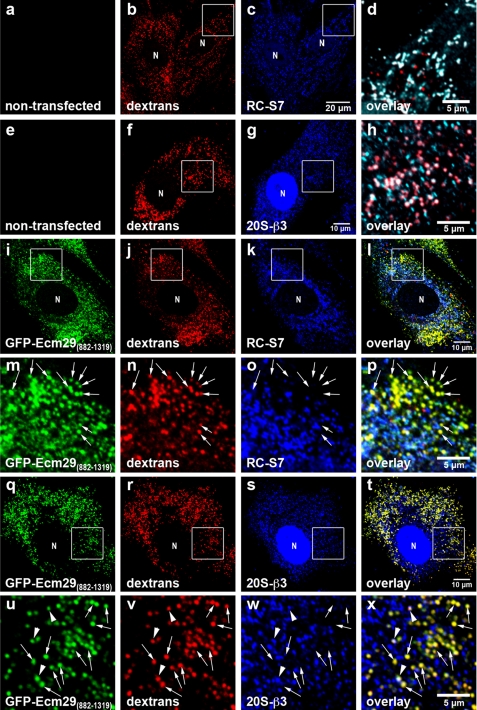
GFP-Ecm29(882–1319) Prevents Association of 26 S Proteasomes with Endosomes
The results shown in Fig. 4 suggest that GFP-Ecm29(882–1319) could function as a dominant negative derivative of Ecm29 and prevent the localization of 26 S proteasomes to endosomes. We tested this possibility by transiently transfecting the GFP-Ecm29(882–1319) construct in mouse embryonic fibroblasts (MEFs), because in our hands these cells have proved more suitable than HEK cells for imaging studies due to their larger size. Endosomes of transfected and non-transfected cells were labeled for 60 min with Alexa®568-dextran conjugate and fixed in 2% paraformaldehyde. The fixed cells were then stained with anti-S7 or with a monoclonal antibody to subunit β3 of the 20 S proteasome and visualized with Alexa®-labeled secondary antibodies and confocal microscopy. The images in Fig. 7 show that GFP-Ecm29(882–1319) produced a punctate pattern of staining that overlapped significantly with the fluorescent dextran consistent with the localization of the Ecm fragment to endosomes. The images also revealed that, although a substantial amount of 26 S proteasomes localized to endocytic vesicles (pseudocolored white in panels d and h) in non-transfected cells, 26 S proteasomes were virtually absent from endosomes of MEFs expressing GFP-Ecm29(882–1319). Moreover, 26 S proteasomes in MEFs expressing GFP alone (not shown), or GFP-Ecm29(1320–1840), were found on endosomes to the same extent as in non-transfected cells (compare Fig. 7, panels a–h and supplemental Fig. 8). These results indicate that high levels of the Ecm29(882–1319) fragment can prevent the 26 S proteasome from associating with flotillin-positive endosomes and should, therefore, be a useful reagent for future studies on Ecm29 function(s).
FIGURE 7.
Expression of GFP-Ecm29(882–1319) inhibits association of 26 S proteasomes with endosomes. Transfected mouse embryonic fibroblasts were grown on coverslips for 48 h prior to 1-h labeling with 1 mg/ml Alexa® 568-dextran conjugate (dextrans) and processing for confocal microscopy. 26 S proteasomes were stained with antibodies to either RC subunit S7 (RC-S7) or 20 S proteasome subunit β3 (20S-β3). Enclosed areas are magnified in panels d and h or directly below the corresponding full-size images (panels m–p and u–x). The enclosed areas in panels c and g were pseudo-colored green (not shown) and then overlaid with the corresponding red (panels b and f) and blue images (panels c and g). Therefore, dextran-filled, proteasome-bound endosomes in panels d and h are shown white rather than pink and are clearly distinguished from dextran-filled endosomes that lack proteasomes (shown in red). Note that, unlike proteasomes in non-transfected cells (panels a–h), 26 S proteasomes in transfected MEF for the most part do not localize to dextran-filled early endocytic vesicles (shown in yellow; examples are indicated by the arrows in panels m–p and u–x). The arrowheads in panels u–x indicate a few dextran-filled vesicles to which both GFP-Ecm29(882–1319) and proteasomes localize. N, cell nucleus. Scale bars, 5 or 10 μm as indicated.
DISCUSSION
The 26 S proteasome is a stable assembly of at least 32 different subunits (47). In addition to these core components, there are a number of proteins that associate with the 26 S proteasome at substoichiometric levels and often transiently. A handful of proteasome activators bind the ends of the 20 S particle and provide access to its central proteolytic chamber (48). A much larger number of proteins bind the 26 S proteasome. Whereas some of these 26 S proteasome interacting proteins, dubbed PIPs by Verma et al. (49), are likely to be undegraded substrates, many PIPs are components of the UPS, e.g. Ub ligases, isopeptidases, and UbA/UbL proteins (50–54). Recent proteomic analyses have shown that cellular chaperones are present in preparations of affinity-purified 26 S proteasomes (55), and 26 S-specific assembly factors (56–59) have also been reported to persist in purified 26 S preparations (60).
Ecm29 does not readily fall into any of these classes of 26 S-associated components. Rather it binds an array of proteins with disparate cellular functions (see Fig. 1). This diversity supports our original suggestion that Ecm29 serves to localize 26 S proteasomes to specific compartments in higher eukaryotic cells (30). Ecm proteasomes, but not all 26 S proteasomes, are found on the ER, on endosomes, and at the centrosome. Consistent with this intracellular distribution, the Ecm29-interacting proteins identified in this study include members of the ER-associated degradation pathway (UBE2G2, FBXO2, FBXO44, Hsp70.1, and DNAJB9), other ER-resident proteins (reticulocalbin 2, emp24, and RNF217), a centrosomal protein (Cep152), and endocytic components (Vps11, rabaptin, Rab11-FIP4, Vps36, and Vps26A). Thus, it is reassuring that most of the Ecm29-interacting proteins listed in Fig. 1 are located in the same compartments as Ecm proteasomes.
The presence of myosins and kinesins (Myo10, Myh7, Myh10, Myo1B, KIF1B, and KIF5B) in the set of Ecm29-interacting proteins is somewhat unexpected. Fluorescence recovery after photobleaching analyses using GFP-labeled proteasomes indicated that proteasomes are freely diffusible in HT1080 cells (61). However, it is unlikely that diffusion can effectively disperse proteasomes in all cells, especially considering the extreme length of axons in many neurons. So Ecm29 may couple 26 S proteasomes directly to motors for transport as individual proteasome-motor complexes. Or another possibility, which is favored by us, is that Ecm29 may bind 26 S proteasomes only to endosomes or other vesicles to which motors are also attached. In this regard we have observed by immunofluorescence (IF) confocal microscopy Ecm proteasomes on vesicles linked to cytoplasmic dynein and kinesin (supplemental Fig. 5), as well as within the axons of primary mouse cortical neurons.6
We took the diversity among Ecm29-interacting proteins as support for the adaptor hypothesis. Others might propose that Ecm29 is simply a sticky protein and call into question the biological relevance of the observed interactions. There are, however, ample reasons to believe that most proteins listed in Fig. 1 are bona fide Ecm29-binding partners and, furthermore, to believe that their association with Ecm29 serves important physiological functions. First, Ecm29 is a large protein that contains numerous HEAT repeats (30, 33); these structural elements are present in other adaptor proteins. For example, Huntingtin, a founding member of the HEAT-repeat protein family, interacts with an equally diverse set of proteins (62, 63). Second, both two-hybrid screens and mass spectrometry identify proteins that, for the most part, fall into a limited set of functional categories, i.e. molecular motors, endosomal components, and UPS factors (see Fig. 1). Third, prior studies on the intracellular distribution of Ecm proteasomes localized them to the ER, to the endosomes, and at the centrosome (30). As mentioned, these are the very organelles in which the majority of the Ecm29-interacting proteins are also located. Thus we consider most of the identified Ecm29-interacting proteins to be true physiological partners.
The genome-wide two-hybrid screens revealed that the binding sites within Ecm29 for endosomal components, molecular motors, and the 26 S proteasome are distributed in a modular fashion (Fig. 1A). Endocytic components bind sequences from the N-terminal half of Ecm29, whereas molecular motors and cytoskeletal components bind C-terminal sequences. The co-immunoprecipitation experiments presented in Fig. 3 produced results consistent with such a modular distribution. N-terminal FLAG-Ecm29(1–1039) bound Vps11 and rabaptin while FLAG-Ecm29(1032–1840) bound kinesin, myosin heavy chains, and dynactin. A relatively short central region within Ecm29, residues 1032–1319, was found to bind the 26 S proteasome (Fig. 4). In contrast to the modularity exhibited for endosomal components and molecular motors, binding sites for UPS components are scattered along the sequence of Ecm29. For example, sequences that interact with the E2 ubiquitin carrier proteins, UBE2Q and UBE2G2, are located at opposite ends of Ecm29, and the binding site for the putative Ub ligase FBXO2 is located centrally. A summary of the observed interactions of endosomal components and molecular motors with Ecm29 is presented in Table 1.
TABLE 1.
Association of Ecm29 and Ecm29 fragments with the 26S proteasome, endocytic components and molecular motors
Association of Ecm29 or its derivativesa with putative binding partners was determined by immunofluorescence (IF), co-immunoprecipitation (IP), yeast two-hybrid screens (Y2H), co-purification (CP) and LC-MS/MS (MS) of affinity-captured FLAG-Ecm29 complexes.
| Binding partner | Full-length Ecm29 | Ecm29(1–1039) | Ecm29(1–891) | Ecm29(1032–1840) | Ecm29(1320–1840) |
|---|---|---|---|---|---|
| 26S Proteasome | IF, IP, CP, MS | IP(−)b, CP(−) | CP(−) | IP(−), CP(+) | IP(−), CP(−) |
| Vps11 | IF, IP | IP, Y2Hc | Y2H | IP(−) | NDd |
| Rab11-FIP4 | IF, IP(−)e | IPe, Y2H | Y2H | IP(−) | ND |
| Rabaptin | IF, IP | IP, Y2H | Y2H | IP(−) | ND |
| Arf6 | IF, IPe, MS (38) | ND | ND | ND | ND |
| Dynein | IF, IP | IP(−) | ND | IP | ND |
| Dynactin | IF, IP | IP(−) | ND | IP | ND |
| Kinesin HCf | IF, IP, MS | IP(−) | ND | IP, Y2H | Y2H |
| Myosin HC | IP, MS | IP(−) | ND | IP, Y2H | Y2H |
| Vps26A | IF, MS | ND | ND | ND | ND |
| Vps36 | IF, MS | ND | ND | ND | ND |
| UBE2G2 | ND | ND | ND | Y2H | Y2H |
| UBE2Q | ND | Y2H | Y2H | ND | ND |
| FBXO2 | ND | ND | ND | Y2H | ND |
| FBXO44 | MS | ND | ND | ND | ND |
| Flotillin-1 | IF, IP | ND | ND | ND | ND |
| Flotillin-2 | IF, IP | ND | ND | ND | ND |
| Caveolin-1 | IF(−), IP(−) | ND | ND | ND | ND |
| Clathrin | IF(−), IP | ND | ND | ND | ND |
a We have shown also that the small central fragment, FLAG-Ecm29(1032–1319), co-purifies with and immunoprecipitates subunits of the 26 S proteasome. The nucleotide sequence of this fragment is included in the sequenced regions from the Y2H assays encoding Ecm29 C-terminal fragments that bind kinesins and myosins (supplemental Table 3).
b The specified putative binding partner failed to co-immunoprecipitate with the corresponding FLAG-Ecm29 fusion protein.
c The specified FLAG-Ecm29 fragment includes the sequenced Ecm29 nucleotides from the yeast two-hybrid screens (see supplemental Table 3).
d ND, not done.
e Data not shown.
f Heavy chain.
Bioinformatic analyses on Ecm29 indicate that the protein consists of at least 27 HEAT repeats (30, 33). Tandemly arrayed HEAT repeats can form solenoids that wrap around binding partners as elegantly demonstrated by crystal structures of ternary complexes between Ran, nuclear export factors, and their cargoes (64, 65). Alternatively, HEAT repeats can collapse to form small domains as exemplified by four HEAT repeats in the C-terminal domain of eukaryotic translation initiation factor-5, which form a single domain that binds a number of other translation initiation factors (66).
Because the entire lengths of exportin polypeptides contact their binding partners, we hypothesized that during evolution the sequences of solenoid-forming HEAT-repeat proteins would be conserved throughout. By contrast, proteins containing tandem domains of collapsed HEAT repeats might exhibit stretches of high sequence conservation separated by less conserved regions. Based on this reasoning, we compared the sequence conservation in exportin-1, Ecm29, and the C-terminal HEAT domain of eIF5. As shown in supplemental Fig. 10, exportin-1 sequences are highly conserved over their entire length, whereas Ecm29 sequences exhibit a considerable “patchiness” in conservation. Moreover, the degree of sequence conservation is 2- to 3-fold greater in exportins than in Ecm29 or the C-terminal domain of eIF5. Based on this analysis we favor a collapsed domain model for Ecm29, although it is certainly not clear that the N- and C-terminal halves of Ecm29 form a series of collapsed domains.
What does seem clear is that the short central sequence in Ecm29, residues 1032–1319, comprises at most 6 HEAT repeats and is, therefore, too short to form a solenoid. This highly conserved stretch of 288 amino acids (see supplemental Fig. 9) is likely to form a discrete domain that binds one or more RC subunits, possibly S5a and/or S6 that were identified in the MS/MS experiments (Fig. 1B).
In a previous report we showed that, in both HeLa cells and in HEK cells expressing low levels of FLAG-Ecm29, Ecm proteasomes sediment as a single species following extraction in buffer containing sucrose (30). Those studies demonstrated that all endogenous Ecm29 molecules were associated with the 26 S enzyme. The data presented in Fig. 4B show that, following expression of high levels of FLAG-Ecm29, the recombinant Ecm29 molecules sediment in two regions of the glycerol gradient: a fast sedimenting species (fractions 5–10) that consists of Ecm proteasome complexes, and a slower sedimenting species centered at fraction 15 (Fig. 4B, left panel). The latter species migrated as a smear on a native gel, and immunoprecipitation analyses revealed it to be Ecm29 in association with other cellular components (data not shown). By contrast, the sedimentation profile of FLAG-Ecm29(1032–1319) (or GFP-Ecm29(882–1319); middle and right panels in Fig. 4B) reveals that neither the 26 S proteasome nor recombinant Ecm fragments are present in the fast sedimenting fractions (i.e. in fractions 5–10) of the glycerol gradient.
Whereas overexpression of full-length Ecm29 does not affect 26 S proteasome stability, overexpression of its central proteasome-binding region has a striking destabilizing effect. The assembled 26 S proteasome and its constituent subparticles, 19 S RC and 20 S proteasome, may be in dynamic equilibrium during sedimentation, because the active 20 S proteasome sediments two fractions deeper (fraction 12 versus 14 in Fig. 4B) when the central Ecm29 fragments are present.
Leggett et al. proposed that Ecm29 stabilizes yeast 26 S proteasomes based on dissociation of the 26 S proteasome during purification from an Ecm29-deletion strain (29). Previously we argued that mammalian Ecm29 was unlikely to serve such a function for two reasons: HeLa 26 S proteasomes remain assembled even after Ecm29 is dissociated by Triton X-100, and several mouse organs are devoid of Ecm29 indicating that Ecm29 is not an obligate clamp for 19 S and 20 S particles (30). However, the results in Fig. 4 show that expression of the central fragment of Ecm29 displaces full-length Ecm29 from the 26 S proteasome and destabilizes the enzyme. Thus these newer results provide some evidence that Ecm29 may stabilize association of the 19 S RC with the 20 S proteasome.
It is clear from the IF and immunoprecipitation analyses presented in Fig. 6 (see also Table 1) that Ecm proteasomes are present on flotillin-positive endosomes and are almost completely absent from newly formed clathrin- or caveolin-coated vesicles. The IF images in Fig. 7 show that, whereas expression of GFP-Ecm29(882–1319) prevents the association of the 26 S proteasome with endosomes, this effect is not a property of all Ecm29 fragments, because expression of GFP-Ecm29(1320–1840), which does not bind the 26 S proteasome (see Figs. 2C and 4D), did not prevent association of the 26 S proteasome with endosomes (supplemental Fig. 8).
Fig. 6B shows that a small amount of clathrin-HC was recovered in the 3× FLAG-eluates of MT-bound vesicles from FLAG-Ecm29-expressing cells. Clathrin in this eluate possibly reflects the presence of Ecm proteasomes on sorting, recycling, and late endosome/multivesicular body-derived vesicles bound to the microtubule network (Fig. 6B). Sorting endosomes are decorated by flat, bilayered clathrin microdomains where ubiquitylated cargoes are concentrated on their way to the lysosome (67, 68). These clathrin domains are also the points in which cargoes engage the “endosomal sorting complex required for transport” (ESCRT) machinery prior to inclusion into luminal vesicles of the multivesicular body (see Ref. 69 for a recent review). Interestingly, proteasomes have been reported to function in late endocytic events involving transfer of growth factor receptors to multivesicular bodies or lysosomes (70–78). The MS/MS experiments shown in Fig. 1 identified Vps36, a component of the ESCRT-II complex (79) as an Ecm29-interacting protein. The IF images in supplemental Fig. 2 provide confirmation that both Ecm29 and Vps36 are present in late endosomes lending support to the notion that proteasomal degradation plays a role in lysosomal targeting of membrane receptors. Another membrane-associated component identified in similar analyses was Vps26A, which IF images show is present on a subset of Ecm29-positive vesicles (Fig. 1, supplemental Fig. 2, and supplemental Table 4). Vps26A is a component of the retromer complex that regulates retrograde protein transport from endosomes to the trans-Golgi network on specific clathrin-coated carriers linked to the adaptors AP1 and epsinR (80–85). Thus, a role for Ecm proteasomes at late stages of endocytosis could explain the release of small amounts of clathrin-HC with 3× FLAG peptide observed in Fig. 6B.
The confocal images in Fig. 6 raise an obvious question: why are Ecm proteasomes restricted to flotillin-positive early endosomes? Conceivably proteasomes remodel the surfaces of flotillin-positive vesicles to promote downstream fusion/conversion events. We favor an alternate hypothesis, namely that Ecm proteasomes mediate the down-regulation of signaling molecules that end up exclusively on flotillin-positive endosomes. Flotillin is a member of the prohibitin family of membrane proteins that include prohibitin, stomatin, and erlin (86–89). Their intracellular location varies from mitochondria (prohibitin) to the ER (erlin) to plasma membrane/endosome/lipid droplets (flotillin and stomatin), and they are considered lipid raft markers, because all are resistant to detergent extraction. Several members of the prohibitin family have been implicated in receptor tyrosine kinase (RTK) signaling. For example, prohibitin is needed for effective Raf activation by Ras (90). Likewise, flotillin binds CAP (91–93), an adaptor that couples the Ub ligase c-Cbl to RTK downstream targets like IRS-1 and the Src-family kinases Fyn and Lyn (91, 92). IRS-1 and Fyn are rapidly degraded by proteasomes upon RTK activation (94–98), and in results not shown, we have confirmed that internalization of flotillin-positive vesicles is markedly stimulated by RTK activation (99, 100). So while most RTKs are ubiquitylated, internalized in clathrin-positive endosomes and degraded within lysosomes (69, 101, 102), we suggest that RTK-activated “downstream signaling” proteins, e.g. IRS-1, Fyn, and possibly Ras (103), are internalized on the cytosolic face of flotillin-positive endosomes where they are degraded by proteasomes. We speculate that Ecm29 couples proteasomes to these endosomes to increase proteolytic efficiency, providing at the protein level an example of metabolic channeling.
We have considered why Ecm proteasomes are restricted to flotillin-positive endosomes. A related question is how are they restricted to these endosomes? The confocal fluorescence micrographs presented in Figs. 5–7 and in supplemental Figs. 1–3, 6, and 7 show co-localization of Ecm29 and a variety of interacting proteins on dextran-positive vesicles. It is also clear in most of these micrographs that there is not a strict one-to-one correspondence between Ecm29 and any of its binding partners. For example in Fig. 5A Ecm29 is located on vesicles that contain Vps11 or rabaptin, but not on all vesicles containing either of these proteins. This raises the possibility that the localization of Ecm proteasomes to a specific compartment requires the simultaneous association of Ecm29 with several of its binding partners. Perhaps Ecm proteasomes associate with vesicles only when Ecm29 binds both one of its endosomal partners and a molecular motor. If the HEAT repeats in Ecm29 form tandem, small domains, association of Ecm proteasomes with a specific vesicle might necessitate binding to several endosomal factors at the same time. An underlying combinatorial mechanism for the recruitment of Ecm proteasomes to specific compartments would explain the lack of a strict correspondence between the localization of Ecm29 and its binding partners. Determining whether the N- and C-terminal halves of Ecm29 form solenoids or multiple domains and determining how many interacting partners can bind Ecm29 simultaneously should test such combinatorial hypotheses.
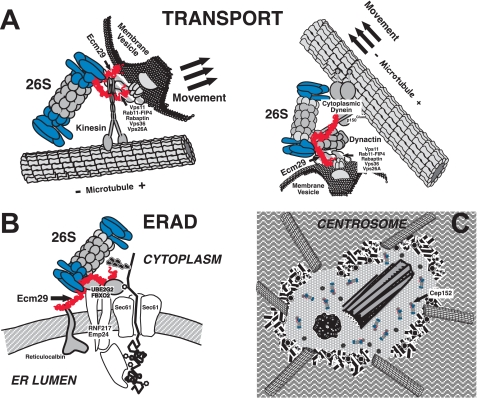
Although the one or more mechanisms by which Ecm29 recruits 26 S proteasomes to specific cellular compartments remain to be discovered, we consider the evidence to be quite substantial that Ecm29 in higher eukaryotes functions as an adaptor for such recruitment. As discussed above, three methods of analysis, yeast two-hybrid screens, co-immunoprecipitation followed by MS/MS analyses or Western blotting, and immunofluorescence co-localization, reinforce the connection between Ecm29 and components on or at specific subcellular compartments. The adaptor hypothesis is presented diagrammatically in Fig. 8 where we depict Ecm29's function in the localization of the 26 S proteasome on endosomes, on the ER membrane, and at the centrosome.
FIGURE 8.
Proposed model of the function of Ecm29 as an adaptor that couples the 26 S proteasome to molecular motors, endocytic vesicles, the endoplasmic reticulum, and the centrosome. A, we propose that Ecm29 binds simultaneously to endocytic components, molecular motors, and the 26 S proteasome. The C-terminal half of Ecm29 binds kinesin (left) or dynactin (right) or myosin (not shown), whereas the N-terminal half of the protein binds endocytic components (Vps11, Rab11-FIP4, rabaptin, Vps36, or Vps26A). A conserved central region of Ecm29 binds the 26 S proteasome. The complexes thus assembled may function to attenuate downstream secondary signaling from receptor tyrosine kinases (see “Discussion” for details) or may be transported on endocytic vesicles along the microtubule network (or the actin cytoskeleton (not shown)) to promote membrane fusion/conversion events. B, Ecm29 couples the 26 S proteasome to the ER-associated degradation pathway by binding the Ub-conjugating E2 enzyme UBE2G2 and ER-associated Ub-ligases or E3s such as FBXO2 or RNF217. C, Ecm29 localizes the 26 S proteasome to the centrosome by binding centrosomal components such as Cep152.
Supplementary Material
Acknowledgments
We thank Chad Nelson and Krishna Parsawar for performing LC-MS/MS proteomic analyses. We are indebted to Chris Rodesch for help and advice in performing confocal laser scanning microscopy. We are thankful to Harald Stenmark (University of Oslo, Norway) for his immunofluorescence protocol for staining endocytic vesicles. We also thank Matt Mulvey (Dept. of Pathology, University of Utah) for helpful advice and reagents and Jody Rosenblatt (Huntsman Cancer Institute, University of Utah) for advice on staining microtubules. We appreciate the gifts of anti-Rab11-FIP4 from Dr. Gwyn Gould (University of Glasgow, UK), anti-S12 from Dr. Wolfgang Dubiel (Charité-Universitätsmedizin Berlin, Germany), anti-arf6 from Dr. Julie Donaldson (NHLBI, National Institutes of Health), and anti-20 S proteasome subunit α6 from Dr. Klavs Hendil (University of Copenhagen, Denmark).
This work was supported, in whole or in part, by National Institutes of Health Grant NS042892 (to M. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Figs. 1–10, and Tables 1–5.
C. Gorbea, M. Rechsteiner, and S. Rogers, unpublished data.
C. Gorbea and S. Rogers, unpublished observation.
- UPS
- ubiquitin-proteasome system
- dyn-IC
- cytoplasmic dynein intermediate chain
- Ecm proteasome
- Ecm29–26 S proteasome complex
- eIF5
- eukaryotic translation initiation factor 5
- ESCRT
- endosomal sorting complex required for transport
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein from Aequorea victoria
- IF
- immunofluorescence
- KHC
- kinesin heavy chain
- MEF
- mouse embryonic fibroblast
- MT
- microtubule
- NM-myosin-HC
- non-muscle myosin heavy chain
- PIP
- 26 S proteasome-interacting protein
- RC
- 19 S regulatory complex
- RTK
- receptor tyrosine kinase
- Ub
- ubiquitin
- UbA
- ubiquitin associated domain
- UbL
- ubiquitin-like domain
- Vps
- vacuolar protein sorting homolog.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 3.Ang X. L., Wade Harper J. (2005) Oncogene. 24, 2860–2870 [DOI] [PubMed] [Google Scholar]
- 4.Sullivan M., Morgan D. O. (2007) Nat. Rev. Mol. Cell. Biol. 8, 894–903 [DOI] [PubMed] [Google Scholar]
- 5.Muratani M., Tansey W. P. (2003) Nat. Rev. Mol. Cell. Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 6.Bour G., Lalevée S., Rochette-Egly C. (2007) Trends Cell Biol. 17, 302–309 [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula S. M., Ashwell J. D. (2008) Mol. Cell 30, 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Virshup D. M., Forger D. B. (2007) Cell 129, 857–859 [DOI] [PubMed] [Google Scholar]
- 9.Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell. Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 10.Peters J. M. (2002) Mol. Cell 9, 931–943 [DOI] [PubMed] [Google Scholar]
- 11.Rodrigo-Brenni M. C., Morgan D. O. (2007) Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 12.Deshaies R. J., Joazeiro C. A. (2009) Ann. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 13.Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell. Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 16.Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borodovsky A., Kessler B. M., Casagrande R., Overkleeft H. S., Wilkinson K. D., Ploegh H. L. (2001) EMBO J. 20, 5187–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., Finley D. (2006) Cell 127, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 19.Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., Cohen R. E. (2006) Nat. Cell Biol. 8, 994–1002 [DOI] [PubMed] [Google Scholar]
- 20.Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y., Varshavsky A. (2002) Nat. Cell Biol. 4, 1003–1007 [DOI] [PubMed] [Google Scholar]
- 22.Ferrell K., Wilkinson C. R., Dubiel W., Gordon C. (2000) Trends Biochem. Sci. 25, 83–88 [DOI] [PubMed] [Google Scholar]
- 23.Hartmann-Petersen R., Gordon C. (2004) Curr. Biol. 14, R754–756 [DOI] [PubMed] [Google Scholar]
- 24.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell. Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 25.Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 26.Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., Southard S. B., Horenstein C. I., Chen-Weiner J., Ram A. F., Kapteyn J. C., Roemer T. W., Vo D. H., Bondoc D. C., Hall J., Zhong W. W., Sdicu A. M., Davies J., Klis F. M., Robbins P. W., Bussey H. (1997) Genetics 147, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 28.Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 29.Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 30.Gorbea C., Goellner G. M., Teter K., Holmes R. K., Rechsteiner M. (2004) J. Biol. Chem. 279, 54849–54861 [DOI] [PubMed] [Google Scholar]
- 31.Kleijnen M. F., Roelofs J., Park S., Hathaway N. A., Glickman M., King R. W., Finley D. (2007) Nat. Struct. Mol. Biol. 14, 1180–1188 [DOI] [PubMed] [Google Scholar]
- 32.Gorbea C., Kaufmann A. G., Pratt G., Rechsteiner M., Rogers S. W. (2006) Is. J. Chem. 46, 207–217 [Google Scholar]
- 33.Kajava A. V., Gorbea C., Ortega J., Rechsteiner M., Steven A. C. (2004) J. Struct. Biol. 146, 425–430 [DOI] [PubMed] [Google Scholar]
- 34.Andrade M. A., Petosa C., O'Donoghue S. I., Müller C. W., Bork P. (2001) J. Mol. Biol. 309, 1–18 [DOI] [PubMed] [Google Scholar]
- 35.LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005) Nature 438, 103–107 [DOI] [PubMed] [Google Scholar]
- 36.Kaltenbach L. S., Romero E., Becklin R. R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., Cha G. H., Ukani L., Chepanoske C. L., Zhen Y., Sahasrabudhe S., Olson J., Kurschner C., Ellerby L. M., Peltier J. M., Botas J., Hughes R. E. (2007) PLoS. Genet. 3, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell R., Hubbard A., Chettier R., Chen D., Miller J. P., Kapahi P., Tarnopolsky M., Sahasrabuhde S., Melov S., Hughes R. E. (2009) PLoS. Genet. 5, e1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman L., Pratt G., Rechsteiner M. (1992) J. Biol. Chem. 267, 22362–22368 [PubMed] [Google Scholar]
- 39.Vallee R. B. (1982) J. Cell Biol. 92, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart II, Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woolner S., Bement W. M. (2009) Trends Cell Biol. 19, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9616–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroeder H. W., 3rd, Mitchell C., Shuman H., Holzbaur E. L. F., Goldman Y. E. (2010) Curr. Biol. 20, 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miaczynska M., Stenmark H. (2008) J. Cell Biol. 180, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayor S., Pagano R. E. (2007) Nat. Rev. Mol. Cell. Biol. 8, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandvig K., Torgersen M. L., Raa H. A., van Deurs B. (2008) Histochem. Cell Biol. 129, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K. (2009) Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rechsteiner M., Hill C. P. (2005) Trends Cell Biol. 15, 27–33 [DOI] [PubMed] [Google Scholar]
- 49.Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., Murata S. (2006) EMBO J. 25, 4524–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besche H. C., Haas W., Gygi S. P., Goldberg A. L. (2009) Biochemistry 48, 2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bousquet-Dubouch M. P., Baudelet E., Guérin F., Matondo M., Uttenweiler-Joseph S., Burlet-Schiltz O., Monsarrat B. (2009) Mol. Cell Proteomics 8, 1150–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su V., Lau A. F. (2009) Cell Mol. Life Sci. 66, 2819–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaake R. M., Milenković T., Przulj N., Kaiser P., Huang L. (2010) J. Proteome Res. 9, 2016–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 60.Thompson D., Hakala K., DeMartino G. N. (2009) J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reits E. A., Benham A. M., Plougastel B., Neefjes J., Trowsdale J. (1997) EMBO J. 16, 6087–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharyya N. P., Banerjee M., Majumder P. (2008) FEBS J. 275, 4271–4279 [DOI] [PubMed] [Google Scholar]
- 63.Caviston J. P., Holzbaur E. L. (2009) Trends Cell Biol. 19, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsuura Y., Stewart M. (2004) Nature 432, 872–877 [DOI] [PubMed] [Google Scholar]
- 65.Monecke T., Güttler T., Neumann P., Dickmanns A., Görlich D., Ficner R. (2009) Science 324, 1087–1091 [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto Y., Singh C. R., Marintchev A., Hall N. S., Hannig E. M., Wagner G., Asano K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clague M. J. (2002) Curr. Biol. 12, R529–531 [DOI] [PubMed] [Google Scholar]
- 68.Sachse M., Urbé S., Oorschot V., Strous G. J., Klumperman J. (2002) Mol. Biol. Cell 13, 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams R. L., Urbé S. (2007) Nat. Rev. Mol. Cell Biol. 8, 355–368 [DOI] [PubMed] [Google Scholar]
- 70.Yu A., Malek T. R. (2001) J. Biol. Chem. 276, 381–385 [DOI] [PubMed] [Google Scholar]
- 71.Malik B., Schlanger L., Al-Khalili O., Bao H. F., Yue G., Price S. R., Mitch W. E., Eaton D. C. (2001) J. Biol. Chem. 276, 12903–12910 [DOI] [PubMed] [Google Scholar]
- 72.Deng L., He K., Wang X., Yang N., Thangavel C., Jiang J., Fuchs S. Y., Frank S. J. (2007) Mol. Endocrinol. 21, 1537–1551 [DOI] [PubMed] [Google Scholar]
- 73.Melman L., Geuze H. J., Li Y., McCormick L. M., Van Kerkhof P., Strous G. J., Schwartz A. L., Bu G. (2002) Mol. Biol. Cell 13, 3325–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alwan H. A., van Zoelen E. J., van Leeuwen J. E. (2003) J. Biol. Chem. 278, 35781–35790 [DOI] [PubMed] [Google Scholar]
- 75.Hammond D. E., Urbé S., Vande Woude G. F., Clague M. J. (2001) Oncogene. 20, 2761–2770 [DOI] [PubMed] [Google Scholar]
- 76.Longva K. E., Blystad F. D., Stang E., Larsen A. M., Johannessen L. E., Madshus I. H. (2002) J. Cell Biol. 156, 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patrick G. N., Bingol B., Weld H. A., Schuman E. M. (2003) Curr. Biol. 13, 2073–2081 [DOI] [PubMed] [Google Scholar]
- 78.van Kerkhof P., Alves dos Santos C. M., Sachse M., Klumperman J., Bu G., Strous G. J. (2001) Mol. Biol. Cell 12, 2556–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. (2002) Dev. Cell 3, 283–289 [DOI] [PubMed] [Google Scholar]
- 80.Bonifacino J. S., Rojas R. (2006) Nat. Rev. Mol. Cell Biol. 7, 568–579 [DOI] [PubMed] [Google Scholar]
- 81.Lauvrak S. U., Torgersen M. L., Sandvig K. (2004) J. Cell Sci. 117, 2321–2331 [DOI] [PubMed] [Google Scholar]
- 82.Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. (2000) EMBO J. 19, 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Medigeshi G. R., Schu P. (2003) Traffic 4, 802–811 [DOI] [PubMed] [Google Scholar]
- 84.Saint-Pol A., Yélamos B., Amessou M., Mills I. G., Dugast M., Tenza D., Schu P., Antony C., McMahon H. T., Lamaze C., Johannes L. (2004) Dev. Cell 6, 525–538 [DOI] [PubMed] [Google Scholar]
- 85.Popoff V., Mardones G. A., Tenza D., Rojas R., Lamaze C., Bonifacino J. S., Raposo G., Johannes L. (2007) J. Cell Sci. 120, 2022–2031 [DOI] [PubMed] [Google Scholar]
- 86.Babuke T., Tikkanen R. (2007) Eur. J. Cell Biol. 86, 525–532 [DOI] [PubMed] [Google Scholar]
- 87.Langhorst M. F., Reuter A., Stuermer C. A. (2005) Cell Mol. Life Sci. 62, 2228–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Browman D. T., Hoegg M. B., Robbins S. M. (2007) Trends Cell Biol. 17, 394–402 [DOI] [PubMed] [Google Scholar]
- 89.Morrow I. C., Parton R. G. (2005) Traffic 6, 725–740 [DOI] [PubMed] [Google Scholar]
- 90.Rajalingam K., Wunder C., Brinkmann V., Churin Y., Hekman M., Sievers C., Rapp U. R., Rudel T. (2005) Nat. Cell Biol. 7, 837–843 [DOI] [PubMed] [Google Scholar]
- 91.Limpert A. S., Karlo J. C., Landreth G. E. (2007) Mol. Cell. Biol. 27, 5686–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu J., Deyoung S. M., Zhang M., Dold L. H., Saltiel A. R. (2005) J. Biol. Chem. 280, 16125–16134 [DOI] [PubMed] [Google Scholar]
- 93.Stuermer C. A., Langhorst M. F., Wiechers M. F., Legler D. F., Von Hanwehr S. H., Guse A. H., Plattner H. (2004) FASEB J. 18, 1731–1733 [DOI] [PubMed] [Google Scholar]
- 94.Kaabeche K., Lemonnier J., Le Mée S., Caverzasio J., Marie P. J. (2004) J. Biol. Chem. 279, 36259–36267 [DOI] [PubMed] [Google Scholar]
- 95.Ghosh A. K., Reddi A. L., Rao N. L., Duan L., Band V., Band H. (2004) J. Biol. Chem. 279, 36132–36141 [DOI] [PubMed] [Google Scholar]
- 96.Pederson T. M., Kramer D. L., Rondinone C. M. (2001) Diabetes 50, 24–31 [DOI] [PubMed] [Google Scholar]
- 97.He F., Stephens J. M. (2006) Biochem. Biophys. Res. Commun. 344, 95–98 [DOI] [PubMed] [Google Scholar]
- 98.Lee A. V., Gooch J. L., Oesterreich S., Guler R. L., Yee D. (2000) Mol. Cell. Biol. 20, 1489–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langhorst M. F., Reuter A., Jaeger F. A., Wippich F. M., Luxenhofer G., Plattner H., Stuermer C. A. (2008) Eur. J. Cell Biol. 87, 211–226 [DOI] [PubMed] [Google Scholar]
- 100.Neumann-Giesen C., Fernow I., Amaddii M., Tikkanen R. (2007) J. Cell Sci. 120, 395–406 [DOI] [PubMed] [Google Scholar]
- 101.Clague M. J., Urbé S. (2006) Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 102.Mills I. G. (2007) Semin. Cell Dev. Biol. 18, 459–470 [DOI] [PubMed] [Google Scholar]
- 103.Porat-Shliom N., Kloog Y., Donaldson J. G. (2008) Mol. Biol. Cell 19, 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.