Abstract
Recently, we found that divalent calcium has no detectable effect on the assembly of Mycobacterium tuberculosis FtsZ (MtbFtsZ), whereas it strongly promoted the assembly of Escherichia coli FtsZ (EcFtsZ). While looking for potential calcium binding residues in EcFtsZ, we found a mutation (E93R) that strongly promoted the assembly of EcFtsZ. The mutation increased the stability and bundling of the FtsZ protofilaments and produced a dominating effect on the assembly of the wild type FtsZ (WT-FtsZ). Although E93R-FtsZ was found to bind to GTP similarly to the WT-FtsZ, it displayed lower GTPase activity than the WT-FtsZ. E93R-FtsZ complemented for its wild type counterpart as observed by a complementation test using JKD7–1/pKD3 cells. However, the bacterial cells became elongated upon overexpression of the mutant allele. We modeled the structure of E93R-FtsZ using the structures of MtbFtsZ/Methanococcus jannaschi FtsZ (MjFtsZ) dimers as templates. The MtbFtsZ-based structure suggests that the Arg93-Glu138 salt bridge provides the additional stability, whereas the effect of mutation appears to be indirect (allosteric) if the EcFtsZ dimer is similar to that of MjFtsZ. The data presented in this study suggest that an increase in the stability of the FtsZ protofilaments is detrimental for the bacterial cytokinesis.
Keywords: Calcium, Cell Division, Protein Self-assembly, Protein-Protein Interactions, Site-directed Mutagenesis, Bacterial Cytokinesis, FtsZ Assembly Dynamics, GTPase, Mycobacterium tuberculosis FtsZ, Z-ring
Introduction
Cell division in almost all bacteria is primarily driven by FtsZ, a protein considered as the homologue of the eukaryotic cytoskeletal protein tubulin (1, 2). FtsZ is a highly conserved protein in prokaryotes and plays a key role in the spatiotemporal regulation of the divisome assembly (2). Polymerization of FtsZ results in the formation of a ring-like structure, known as the Z-ring. The Z-ring is a highly dynamic structure (3), and the dynamics of the Z-ring is attributed to the assembly and disassembly of FtsZ protofilaments (4). Perturbation of the FtsZ assembly has been reported to impair bacterial cell division (5–9).
Spatiotemporal regulation of the Z-ring at the midcell is tightly regulated by several effectors of FtsZ assembly (10–12). Although the arrangement of FtsZ polymers in the bacterial Z-ring has not yet been visualized, the reconstitution of the FtsZ polymers in vitro has contributed a lot to our understanding of the bacterial cell division. Recently, an attempt was made to reconstitute the Z-ring in a liposome in vitro, and it was found that neither FtsZ assembly nor the generation of the constriction force requires FtsA or other downstream division proteins (13). Further, FtsZ polymers have been found to have intrinsic ability to form cytoplasmic ringlike structures in yeast cells, indicating that FtsZ assembly into the ring may not require any other regulatory protein (14).
FtsZ protein of different bacteria exhibit different assembly properties in vitro. For example, the polymerization of Escherichia coli FtsZ (EcFtsZ)2 has been observed to be more rapid than that of the Mycobacterium tuberculosis FtsZ (MtbFtsZ) (15). Recently, we have shown that divalent calcium strongly influenced the assembly of EcFtsZ by promoting the stability and bundling of the protofilaments; however, it had no detectable effect on the assembly of MtbFtsZ (16). In vitro, divalent calcium bound to EcFtsZ with much higher affinity than to MtbFtsZ (16). To elucidate the mechanism of differential effects of calcium on the assembly of EcFtsZ and MtbFtsZ, we sought to identify the calcium binding site in EcFtsZ.
The crystal structure of MtbFtsZ (Protein Data Bank code 1RQ2) (17) and the three-dimensional structure of EcFtsZ predicted by comparative modeling using 3D-Jigsaw (18–20) were used to generate a secondary structure-based sequence alignment. Because the latter is the predicted three-dimensional structure, the crystal structure of Methanococcus jannaschi FtsZ (MjFtsZ; Protein Data Bank code 1W5B) (21) and Pseudomonas aeruginosa FtsZ (PaFtsZ; Protein Data Bank code 1OFU) (22) were also used for the sequence alignment. One criterion for finding the putative calcium-binding residues in EcFtsZ was to look for Asn, Asp, Gln, Glu, Ser, and Thr residues that are not conserved in MtbFtsZ because these amino acids are frequently found in the coordination sphere of calcium (23). Another criterion was whether such residues were at the lateral interface of EcFtsZ. We identified Asp84 and Glu93 in EcFtsZ as the putative residues fulfilling these criteria. In addition to these residues, Ser231 was also selected for mutation because this residue is near one of the loop regions predicted to be the calcium binding motif based on sequence similarity with calmodulin (24). We made three EcFtsZ constructs, namely D84A, E93R, and S231R, by site-directed mutagenesis; however, all three EcFtsZ mutants showed sensitivity toward divalent calcium, suggesting that these residues may not be critical for the binding of calcium to FtsZ. Interestingly, of the three mutants, E93R was found to assemble much more efficiently than the wild type (WT-FtsZ). The other two mutants showed polymerization ability similar to that of the WT-FtsZ.
In vitro, E93R-FtsZ formed bundles, and the polymers had higher stability than those formed by the WT-FtsZ. An analysis of the structure of EcFtsZ dimer modeled using MtbFtsZ/MjFtsZ dimers provided plausible explanations for the assembly-promoting effect of the mutation. The results suggest that an increase in the stability of FtsZ protofilaments can inhibit bacterial cell division and indicate that the assembly dynamics of the protofilaments is tightly regulated during bacterial cell division.
EXPERIMENTAL PROCEDURES
Materials
Pipes, GTP, GDP, EDTA, PMSF, lysozyme, and FITC were purchased from Sigma-Aldrich. TNPGTP was purchased from Molecular Probes, Inc. (Eugene, OR). Arabinose was obtained from Sisco Research Laboratories (Mumbai, India). Ammonium sulfate was from Merck (Mumbai, India). All other reagents used were of analytical grade.
Isolation and Purification of Mutant and Wild Type EcFtsZ
EcFtsZ protein was overexpressed and purified as described recently (25) and stored at −80 °C. FtsZ concentration was measured by the Bradford method using BSA as the standard (26). The concentration of FtsZ was finally adjusted using a correction factor of 1.2 for the BSA/FtsZ ratio (27). The purity of FtsZ was estimated to be ∼98% from Coomassie Blue-stained SDS-PAGE.
PCR-based site-directed mutagenesis of FtsZ was performed using the protocol supplied with the QuikChange site-directed mutagenesis kit (Stratagene). FtsZ expression plasmid pET11a was used as the template. The aspartate residue at position 84, glutamate residue at position 93, and serine residue at position 231 were replaced individually by alanine, arginine, and arginine residues, respectively, using primers 5′-AATGCGGCAGATGAGGCTCGCGATGCACTGCGT-3′ and 5′-ACGCAGTGCATCGCGAGCCTCATCTGCCGCATT-3′ for D84A, 5′-TTGCGTGCGGCGCTGAGAGGTGCAGACATGGTC-3′ and 5′-GACCATGTCTGCACCTCTCAGCGCCGCACGCAA-3′ for E93R, and 5′-GGTTCTGGCGTGGCGAGAGGTGAAGACCGTGCG-3′ and 5′-CGCACGGTCTTCACCTCTCGCCACGCCAGAACC-3′ for S231R substitutions. Further, three more mutations were made for substitution of Glu93 to alanine, valine, and lysine separately. For the E93A mutation, primers used were 5′-TTGCGTGCGGCGCTGGCAGGTGCAGACATGGTC-3′ and 5′-GACCATGTCTGCACCTGCCAGCGCCGCACGCAA-3′. For E93V, primers used were 5′-TTGCGTGCGGCGCTGGTAGGTGCAGACATGGTC-3′ and 5′-GACCATGTCTGCACCTACCAGCGCCGCACGCAA-3′). For E93K, primers used were 5′-TTGCGTGCGGCGCTGAAAGGTGCAGACATGGTC-3′ and 5′-GACCATGTCTGCACCTTTCAGCGCCGCACGCAA-3′. All mutations were confirmed by DNA sequencing (Macrogen). The underlined nucleotides represent mutation sites in the primer sequence. The mutated proteins were expressed in E. coli BL21 (DE3) and purified as described for WT-EcFtsZ protein.
Light Scattering Assay
WT and mutant (D84A, E93R, and S231R) FtsZs were prepared in 25 mm Pipes buffer, pH 6.8, 50 mm KCl, and 10 mm MgCl2 (buffer A) on ice at the concentrations specified in individual experiments. After the addition of 1 mm GTP, the assembly milieu was immediately placed in a cuvette at 37 °C. The polymerization reaction was followed by monitoring 90° light scattering at 500 nm using a JASCO 6500 spectrofluorometer (28, 29).
Sedimentation Assay
WT and mutant FtsZs (D84A, E93R, and S231R) (7.2 μm) were polymerized in buffer A containing 1 mm GTP for specified times at 37 °C. The polymers were collected by centrifugation at 227,000 × g for 30 min at 30 °C. The protein concentration in the supernatant was measured by a Bradford assay using BSA as a standard (26). The amount of polymerized FtsZ was determined by subtracting the protein concentration in supernatant from the total protein concentration. The critical concentration for the assembly of the WT- or E93R-FtsZ was calculated by polymerizing different concentrations of the two proteins and measuring the amount of protein pelleted down after high speed centrifugation.
Size Exclusion Chromatography
A Superdex G-200 10/30 GL column (GE Healthcare) with a column bed volume of 23.5 ml in an AKTA FPLC system (GE Healthcare) was used to determine the oligomeric states of the WT- and E93R-FtsZs. The void volume of the column was estimated to be ∼9 ml by eluting blue dextran. The Superdex G-200 column was pre-equilibrated with 25 mm Pipes buffer (pH 6.8). 200 μl of human serum albumin, tubulin, WT-FtsZ, and E93R-FtsZ in 25 mm Pipes buffer, pH 6.8, were loaded individually onto a pre-equilibrated Superdex G-200 column. The proteins were eluted at a flow rate of 0.4 ml/min in 1-ml fractions each. Human serum albumin was taken as control, and the obtained elution profile was found to be similar to the standard. The protein concentration of each fraction was measured using the Bradford method (26).
Electron Microscopy
The polymer morphology of the WT or mutant FtsZ was analyzed by transmission electron microscopy (9, 28). FtsZ (7.2 μm) in buffer A in the presence of 1 mm GTP was polymerized at 37 °C. The FtsZ polymers in the samples were fixed with warm 0.5% glutaraldehyde. FtsZ polymeric suspension (50 μl) was placed on the carbon-coated copper grids (300 mesh size) and then blotted dry. The grids were subsequently subjected to negative staining by 2% uranyl acetate solution and air-dried. The samples were examined using a Philips FEI Tecnai-G2 12 electron microscope.
FITC Labeling of WT and Mutant Proteins
FtsZ was covalently modified with FITC using a reported protocol (30). Briefly, FtsZ (36 μm) was incubated with 300 μm FITC in 50 mm sodium phosphate buffer at pH 8 for 3 h at 25 °C. The labeling reaction was quenched by adding 5 mm tris(hydroxymethyl) aminomethane hydrochloride on ice for 30 min, and the complex was centrifuged for 10 min to remove any aggregates. Free FITC was removed from FtsZ-bound FITC in two steps: first by dialyzing the reaction mixture against 50 mm phosphate buffer (pH 6.8) at 4 °C and then by passing the solution through a size exclusion P4 column, previously equilibrated with 50 mm phosphate buffer (pH 6.8) at 4 °C. The concentration of FtsZ-bound FITC was determined from the absorbance at 495 nm using a molar extinction coefficient of 68,000 m−1 cm−1. The concentration of FtsZ was determined by the Bradford method (26). The incorporation ratio of FITC per mol of FtsZ was determined by dividing the bound FITC concentration by the FtsZ concentration.
Effect of E93R Mutation on the Binding of TNPGTP to FtsZ
TNPGTP, a fluorescent analogue of GTP, has been found to bind to FtsZ (9, 31). Therefore, TNPGTP was used to determine the effect of E93R mutation on the binding of GTP with FtsZ. WT- or E93R-FtsZ (7.2 μm) in 25 mm Pipes buffer (pH 6.8) was incubated with TNPGTP (50 μm) for 4 h on ice. Fluorescence spectra were recorded using 410 nm as the excitation wavelength.
Measurement of the GTPase Activity of FtsZ
The effect of the mutation (E93R) on the GTPase activity of FtsZ was determined using the standard malachite green ammonium molybdate assay (9, 32). Briefly, WT-FtsZ or E93R-FtsZ (7.2 μm) in buffer A and 1 mm GTP was kept for polymerization at 37 °C. The hydrolysis reaction was quenched at the desired time intervals by adding 10% (v/v) 7 m perchloric acid. The quenched reaction mixtures were kept on ice until samples of all of the time points were collected. Then the reaction mixtures were kept at room temperature for 10 min, and 40 μl of the reaction mixture were incubated with 900 μl of freshly prepared malachite green ammonium molybdate solution (0.045% malachite green, 4.2% ammonium molybdate, and 0.02% Triton X-100) at room temperature for 30 min, and the phosphate ions released were determined by measuring the absorbance of samples at 650 nm (9). An appropriate blank reading was subtracted from experimental data. A phosphate standard curve was prepared using sodium phosphate.
Dilution-induced Disassembly of FtsZ Polymers
WT-FtsZ or E93R-FtsZ (36 μm) was polymerized in buffer A with 1 m glutamate and 1 mm GTP at 37 °C for 30 min. The polymer suspension was then diluted 30 times to reach a final FtsZ concentration of 1.2 μm in warm Pipes buffer (pH 6.8) containing 50 mm KCl, 10 mm MgCl2, and 1 mm GTP. The diluted polymeric suspensions were kept at 37 °C for another 5 min. Polymers were sedimented at 227,000 × g for 30 min at 30 °C. The pellet obtained was dissolved in SDS loading buffer. The samples were analyzed by Coomassie Blue-stained 10% SDS-PAGE. The band intensity was analyzed using Image J software.
Circular Dichroism Spectroscopy Studies
FtsZ (4.8 μm) in 25 mm Pipes buffer (pH 6.8) was prepared. The far-UV CD spectra were monitored over the wavelength range of 190–260 nm using a 0.1-cm path length cuvette in a JASCO J810 spectropolarimeter. A spectral bandwidth of 1 nm and time constant of 1 s were used for all measurements. Each spectrum was recorded as an average of three scans. The spectral values were deconvoluted for the analysis of secondary structure content using CDNN CD spectra deconvolution software (JASCO, Tokyo, Japan).
Visualization of Bacterial Morphology
An overnight culture of E. coli BL21 (DE3) cells transformed with WT-FtsZ and E93R-FtsZ containing plasmids was used to inoculate tubes containing 10 ml of LB medium. Cells were grown until the A600 reached 0.6–0.8, and then the cells were grown for 3 h in the presence of 25 μm IPTG (9). The cells were examined using differential interference contrast microscopy.
Immunofluorescence Microscopy
The Z-ring and nucleoids of E. coli BL21 (DE3) cells containing WT-FtsZ or E93R-FtsZ plasmids were visualized as described recently (9). Briefly, E. coli cells were grown in LB medium for 12 h. The cells were diluted in LB medium and further grown until the A600 reached 0.6. FtsZ was induced by adding 25 μm IPTG and growing for an additional 3 h. Cells were fixed using 2.5% formaldehyde and 0.04% glutaraldehyde. FtsZ was stained with a polyclonal anti-FtsZ antibody followed by Cy3-conjugated goat anti-rabbit secondary antibody. Nucleoids were stained using 2.5 μg/ml DAPI and visualized under a fluorescence microscope (Eclipse TE2000-U, Nikon (Tokyo, Japan)).
Complementation Tests
Liquid and plate complementation tests were performed as described earlier (33). The complementation system, a gift from Dr. Harold Erickson, was used to study the effect of E93R mutation on the Z-ring formation in vivo. Briefly, WT and E93R ftsZ alleles were transferred to pJSB2 as described and were sequenced to confirm the proper orientation. Further, these plasmids were transformed into JKD7–1/pKD3 cells to form WT and E93R mutant strains. For plate complementation assays, plates containing 0.1, 0.2, and 0.3% arabinose were spread with an equal volume of diluted overnight culture grown in repression medium (containing 0.5% glucose). Repression plates containing 0.5% glucose were grown at 30 °C to compare the number of colonies in the plated volume. Induction plates containing different concentration of arabinose were grown at 42 °C. The number of colony-forming units was counted in each of these plates. The number of colonies formed in the induction plates (0.1, 0.2, and 0.3% arabinose) was normalized with the number of colonies formed in repression plates (0.5% glucose). The mutant was considered to complement the WT-FtsZ when the normalized value was ≥0.8 (33). Liquid complementation was performed as described earlier, and complementation was defined as the ability of the culture to grow to an A600 of 0.5 in 24 h at 42 °C (33). Immunostaining of the cells for FtsZ localization was done as described earlier (9).
Homology Modeling of E. coli FtsZ
Homology models were built using 3D-Jigsaw (18–20). Homology modeling was performed in the interactive mode by manually selecting a protein template and chain, followed by model building. The dimer was built by combining models generated using two chains of the same crystal structure. PyMOL was used for the visualization of protein structure and the creation of snapshots (34).
RESULTS
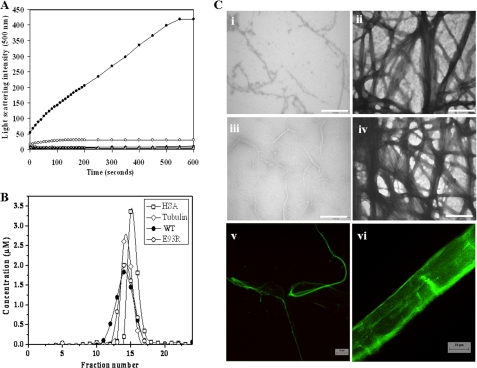
Under the assembly conditions used in this study, all four FtsZ constructs (WT and three mutated constructs, namely D84A, S231R, and E93R) displayed a time-dependent increase in light scattering intensities (Fig. 1A). D84A and S231R showed a light scattering pattern similar to that of the WT-FtsZ. In contrast, the assembly of E93R-FtsZ showed 15-fold higher light scattering intensity than that of the WT-FtsZ, indicating that E93R-FtsZ assembled much more efficiently than the WT and the other two FtsZ constructs (Fig. 1A).
FIGURE 1.
A, effect of E93R mutation on FtsZ assembly. WT and mutant FtsZ (14.4 μm) were polymerized at 37 °C. Shown are the light scattering traces of WT-FtsZ (○), E93R-FtsZ (●), D84A-FtsZ (♦), and S231R-FtsZ (△) in the presence of 1 mm GTP and E93R-FtsZ in the absence of 1 mm GTP (□). B, WT-FtsZ and E93R-FtsZ existed in the same oligomeric state in solution. E93R-FtsZ or WT-FtsZ was eluted through a size exclusion column (Superdex G-200). The elution profiles of WT- and E93R-FtsZs are similar. C, E93R mutant induced bundling of FtsZ protofilaments. E93R- and WT-FtsZ were polymerized at 37 °C in the assembly buffer containing 25 mm Pipes (pH 6.8), 50 mm KCl, 10 mm MgCl2, and 1 mm GTP. Shown are the electron micrographs of WT-FtsZ and E93R-FtsZ polymerized for 5 min (i and ii) and WT-FtsZ and E93R-FtsZ polymerized for 30 min (iii and iv). Scale bar, 1000 nm. Fluorescence images of FITC-labeled WT (v) and E93R (vi) FtsZ polymers (polymerized for 5 min) are also shown. Scale bar, 10 μm. HSA, human serum albumin.
There was no increase in the light scattering intensity of E93R-FtsZ in the absence of GTP (Fig. 1A). Like WT-FtsZ (35), E93R-FtsZ also did not polymerize in the presence of 1 mm GDP (data not shown). The results together suggested that the increase in the light scattering intensity of E93R-FtsZ in the presence of GTP is a result of GTP-dependent assembly and not due to nonspecific clumping or aggregate formation (Fig. 1A). Further, the elution profile of the E93R-FtsZ was found to be similar to that of the WT-FtsZ, suggesting that E93R-FtsZ does not show aggregate forming tendency under non-assembly conditions (Fig. 1B). Consistent with the previous finding (36), the results also indicated that FtsZ exists primarily as dimers in solution (Fig. 1B).
Further, the assembly kinetics of WT-FtsZ and E93R-FtsZ were monitored for 60 min. The light scattering data indicated that the disassembly of WT-FtsZ polymers started after ∼700 s, whereas the disassembly of E93R-FtsZ polymers started after ∼1500 s (supplemental Fig. S1). The results indicated that E93R-FtsZ forms polymers that are much more stable than WT-FtsZ.
The amount of FtsZ polymerized was determined by collecting polymers through sedimentation. Consistent with the light scattering experiment (Fig. 1A), E93R-FtsZ was found to polymerize more efficiently than WT-FtsZ. After 5 and 10 min of assembly, 27.8 ± 6.5 and 25.7 ± 8.1% of WT-FtsZ polymerized, whereas 71.7 ± 9.9 and 71.9 ± 7.4% of E93R were sedimented under similar conditions. However, the polymeric levels of the other two mutants (D84A and S231R) were found to be similar to the WT-FtsZ.
Morphologies of the polymers formed by the WT and mutant proteins were examined using two complementary techniques, namely transmission electron microscopy and fluorescence microscopy. Under similar assembly conditions, E93R-FtsZ showed a prevalence of thick bundles compared with WT-FtsZ, which showed dominance of small, thin protofilaments (Fig. 1C). The electron micrographs suggested that the increase in the light scattering intensity and the increase in the polymerized mass of E93R-FtsZ were due to the bundling of the FtsZ protofilaments and not due to the nonspecific aggregation of E93R-FtsZ monomers.
WT- and E93R-FtsZs were covalently labeled with the fluorescent probe, FITC, as described earlier (30) Fluorescence microscopic analysis of the assembly mixtures showed that the FITC-labeled E93R-FtsZ assembled to form large and thick bundles, whereas the FITC-labeled WT-FtsZ formed thin filamentous structures under similar assembly conditions (Fig. 1C).
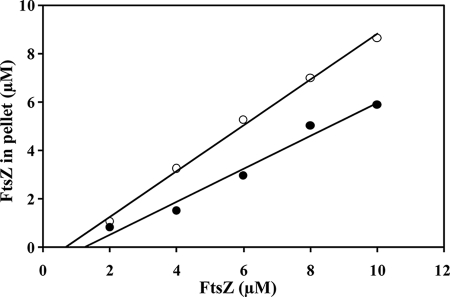
The critical concentrations of the WT-FtsZ and E93R-FtsZ assembly were determined to be 1.2 ± 0.1 and 0.7 ± 0.1 μm, respectively, suggesting that the E93R mutation enhanced the assembling ability of EcFtsZ (Fig. 2). The critical concentration of WT-FtsZ was found to be in agreement with the previously reported values (35, 37).
FIGURE 2.
E93R mutation decreased the critical concentration of EcFtsZ. Different concentrations of WT-FtsZ (●) and E93R-FtsZ (○) were polymerized at 37 °C for 10 min. Polymers were pelleted down by high speed centrifugation. The experiment was performed three times.
E93R Substitution Suppressed the GTPase Activity of FtsZ but It Did Not Affect the Binding of GTP to FtsZ
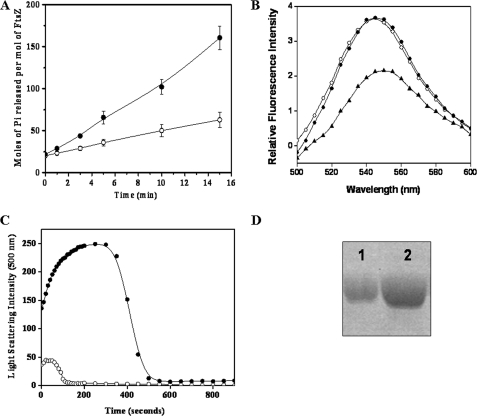
The GTPase activity of FtsZ is considered to be an important property through which FtsZ regulates its assembly dynamics (3, 12). The rate of Pi released/FtsZ molecule/min was calculated to be 9.1 and 2.9 for the WT and mutated FtsZ, respectively (Fig. 3A). The extent of the GTPase activity of E93R-FtsZ was considerably low as compared with that of WT-FtsZ. For example, after 10 min of hydrolysis reaction, 102 ± 9 and 50 ± 8 mol of inorganic phosphate were released per mol of the WT-FtsZ and E93R-FtsZ, respectively (Fig. 3A). The results suggested that the E93R mutation strongly reduced the GTPase activity of FtsZ.
FIGURE 3.
A, E93R mutation suppressed the GTPase activity of FtsZ. E93R- and WT-FtsZ were polymerized at 37 °C in the assembly buffer containing 25 mm Pipes (pH 6.8), 50 mm KCl, 10 mm MgCl2, and 1 mm GTP. Pi released was monitored by a standard malachite green ammonium molybdate assay. Shown are the mol of Pi released/mol of WT-FtsZ (●) and E93R-FtsZ (○). B, E93R mutation did not inhibit the TNPGTP binding to FtsZ. WT-FtsZ or E93R-FtsZ (6 μm) was incubated with 50 μm TNPGTP on ice for 4 h. Fluorescence spectra were taken using 410 nm as the excitation wavelength. The emission spectra of free TNPGTP (▴) and TNPGTP bound with WT (○) and E93R (●) FtsZ have been shown in the figure. C, E93R mutation increased the steady state duration of FtsZ assembly. WT and mutant FtsZ (18 μm) were polymerized in the limited amount of GTP (250 μm) at 37 °C. Shown are the light scattering traces of WT-FtsZ (○) and E93R-FtsZ (●). D, E93R mutation prevented the disassembly of preformed FtsZ polymers. WT- and E93R-FtsZ (36 μm) were polymerized in the presence of 1 m glutamate at 37 °C. Polymers were diluted 30-fold in warm 25 mm Pipes buffer and incubated for an additional 5 min at 37 °C as described under “Experimental Procedures.” Polymers were collected by centrifugation and loaded on SDS-PAGE. Shown in the figure are WT-FtsZ (lane 1) and E93R-FtsZ (lane 2) collected after centrifugation. The experiment was repeated three times. Error bars, S.D.
TNPGTP, a fluorescent analogue of GTP, was found to bind to FtsZ at the GTP site (9, 31). Therefore, we determined the effect of E93R mutation on the GTP binding of FtsZ using TNPGTP. The fluorescence intensities of TNPGTP upon binding to WT and to E93R-FtsZ were found to be similar, suggesting that the E93R mutation did not affect the binding of GTP to FtsZ (Fig. 3B).
E93R Mutation Increased the Steady State Duration of FtsZ Assembly
GDP-bound FtsZ polymers have been found to be comparatively unstable compared with the GTP-bound polymers (38, 39). Because the GTP hydrolysis rate of the mutant FtsZ was found to be slower than the WT-FtsZ, it is logical to think that under GTP-challenged conditions, E93R-FtsZ polymers would disassemble at a rate slower than that of the WT-FtsZ. To test this idea, E93R-FtsZ and WT-FtsZ were polymerized in the presence of a limited amount of GTP. The light scattering intensity of the WT-FtsZ assembly increased up to 40 s; it maintained the steady state level for 10 s and then decreased sharply to the basal level within ∼120 s, suggesting a rapid disassembly of the protofilaments (Fig. 3C). In contrast, the light scattering intensity of the E93R-FtsZ assembly increased until 200 s, and the light scattering intensity remained constant for the next 100 s (Fig. 3C). The light scattering intensity of the mutated FtsZ assembly started decreasing steadily after 300 s of the assembly and reached the minimum value at 500 s, indicating the disassembly of the polymers (Fig. 3C). This finding suggested that the E93R mutation of FtsZ increased the stability of the FtsZ protofilaments and decreased the subunit turnover rate.
Effect of E93R Mutation on the Dilution-induced Disassembly of Preformed FtsZ Polymers
WT or mutant FtsZ (36 μm) was assembled for 30 min in the presence of 1 m monosodium glutamate, 50 mm KCl, 10 mm MgCl2, and 1 mm GTP at 37 °C. The preformed polymers were diluted 30-fold in warm Pipes buffer (pH 6.8) containing 50 mm KCl, 10 mm MgCl2, and 1 mm GTP. After 5 min of incubation at 37 °C, the polymers were collected by sedimentation. The amount of FtsZ in the pellet was found to be substantially more with E93R-FtsZ compared with the WT-FtsZ. For example, the band intensities for WT and E93R-FtsZ were calculated to be 9.2 ± 3.7 and 23.9 ± 7.7, respectively, suggesting that E93R substitution prevented the disassembly of the FtsZ protofilaments (Fig. 3D).
E93R Substitution Minimally Altered the Secondary Structure of FtsZ
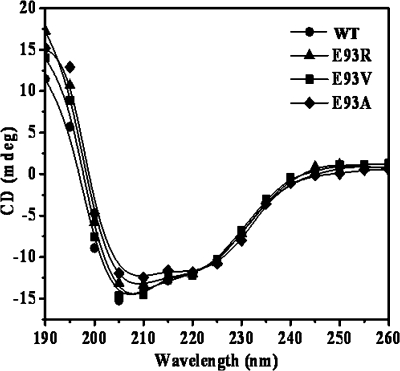
The effect of mutation on the secondary structure of FtsZ was determined by monitoring the far-UV CD spectra of the WT and mutated FtsZ proteins. The far-UV CD spectra of WT-FtsZ and E93R-FtsZ were not significantly different (Fig. 4). Further, a deconvolution analysis of the spectral data indicated that these proteins have similar secondary structure contents. For example, the α-helical content was found to be 24.9 ± 0.4 and 25.2 ± 0.2%, and the random coil content was 35.8 ± 0.4 and 36.2 ± 0.2% for the WT-FtsZ and E93R-FtsZ, respectively.
FIGURE 4.
Secondary structure analysis of WT and mutant FtsZ. Far-UV CD spectra of WT-FtsZ, E93R-FtsZ, E93V-FtsZ, and E93A-FtsZ (4.8 μm) were monitored over the wavelength range 190–260 nm using a 0.1-cm path length cuvette in a JASCO J810 spectropolarimeter. Shown are the far-UV CD spectra of WT-FtsZ (●), E93R-FtsZ (▴), E93V-FtsZ (■), and E93A-FtsZ (♦).
E93R-FtsZ Showed a Dominating Effect over the WT-FtsZ on Assembly
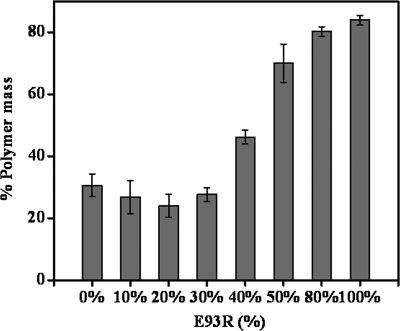
Increasing concentrations of E93R-FtsZ were mixed with the decreasing concentrations of the WT-FtsZ, keeping the total concentration of the FtsZ in the reaction mixtures constant (i.e. 7.2 μm) and were polymerized under similar assembly conditions. The amount of polymerized FtsZ was found to be 30.6 ± 3.7, 83.9 ± 1.6, and 70.0 ± 6.2% when WT-FtsZ (7.2 μm), E93R-FtsZ (7.2 μm), or a mixture of the WT and mutated FtsZ (3.6 μm each) was assembled under similar assembly conditions, respectively (Fig. 5). WT and mutant FtsZ (3.6 μm) were polymerized individually, and the amount of FtsZ polymerized was determined by sedimentation. Adding the polymeric fractions of each protein, the amount of FtsZ polymerized was calculated to be 43%, which was substantially lower than the amount polymerized when these proteins were polymerized together. This result indicated that E93R-FtsZ has a positive effect on the assembly of the WT-FtsZ.
FIGURE 5.
E93R mutant showed a dominating effect on assembly of WT-FtsZ. Increasing concentrations of E93R-FtsZ were mixed with decreasing concentrations of WT-FtsZ while keeping the total protein concentration in the reaction mixture constant (7.2 μm). Polymers were collected by centrifugation, and protein concentration in the supernatant was estimated by the Bradford method (26). Error bars, S.D.
E93R-FtsZ Complements FtsZ for Its Function
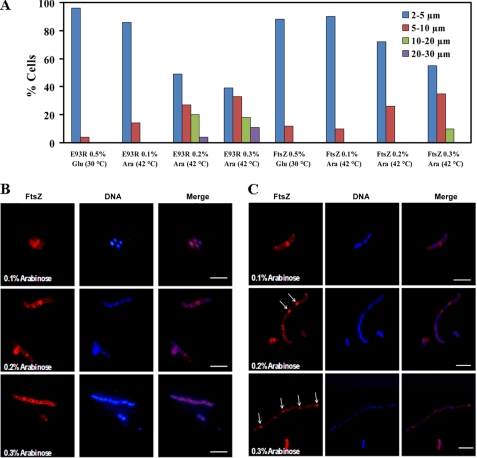
The E93R-ftsZ mutant allele and wild type ftsZ were inserted into pJSB2 and then transformed into the JKD7–1 strain containing pKD3-ftsZ. E. coli JKD7–1/pKD3 cells expressing WT-FtsZ and E93R-FtsZ plasmids did not show any elongation in repression medium (0.5% glucose), and the majority of cells were in the range of 2–5 μm cell length due to the suppression of the complementation system (Fig. 6A). At low concentrations of arabinose at 42 °C, E93R complemented WT-FtsZ in both liquid and plate complementation assays. For example, in liquid medium, cells grew normally in the presence of 0.1% arabinose, and the majority of the cells with E93R-FtsZ were in the 2–5-μm cell length range. Similarly, in a plate complementation assay, E93R-FtsZ was found to complement the WT-FtsZ at 0.1% arabinose, indicating that it functions properly at the normal expression levels (supplemental Fig. S2). However, E93R-FtsZ did not support cell division at high expression levels. For example, when the cells containing mutated FtsZ plasmids were grown at 0.2–0.3% arabinose, a huge increase in the cell length was observed, whereas under similar conditions, cells overexpressing WT-FtsZ did not show significant cell elongation. In cells expressing E93R-FtsZ, at 0.2% arabinose concentration, ∼20% of cells in the total cell population were in the range of 10–20 μm cell length, and at 0.3% arabinose concentration, ∼11% cells were in a 20–30-μm cell length range (Fig. 6A), whereas in the cells expressing WT-FtsZ, under similar conditions over 60–65% cells were in the normal cell length range (2–5 μm), and the rest of the cells did not show elongation beyond 10 μm (Fig. 6A). Thus, the overexpression of E93R-FtsZ resulted in a huge increase in the cell length of bacteria, which may be a direct consequence of the inhibition of formation of a functional Z-ring. To further probe this possibility, these cells were immunostained with antibody against FtsZ, and nucleoids were stained with DAPI. We found an intact Z-ring at the midcell of the majority of both WT- and E93R-FtsZ-expressing cells at 0.1% arabinose concentration (Fig. 6, B and C). Upon increasing the arabinose concentration (0.2%), a single prominent Z-ring was located at the center of the cells expressing WT-FtsZ, whereas the cells expressing E93R-FtsZ had no prominent Z-rings; instead, these cells had multiple smaller ringlike structures (Fig. 6, B and C). The results indicated that these mislocalized rings were unable to carry out cytokinesis.
FIGURE 6.
Effect of E93R mutation on the Z-ring assembly and nucleoid segregation in JKD7–1/pKD3 cells. WT-FtsZ and E93R-FtsZ genes were inserted in complementation vector pJSB2. JKD7–1/pKD3 cells were transformed with the recombinant plasmids. A, distribution of JKD7–1/pKD3 cell lengths in cells expressing WT-FtsZ and E93R-FtsZ in the presence of glucose (0.5%) at 30 °C and different concentrations (0.1, 0.2, and 0.3%) of arabinose at 42 °C. A length of 200 cells was measured in each case, and the percentage of cells was plotted as a function of cell length. WT-FtsZ (B) and E93R-FtsZ (C) were expressed in the presence of different concentrations of arabinose (0.1, 0.2, and 0.3%) at 42 °C in JKD7–1/pKD3 cells. FtsZ and DNA were stained with anti-FtsZ antibody and DAPI, respectively. Scale bar, 5 μm.
Limited Expression of the E93R-FtsZ Caused Elongation of the Bacterial Cells
Expression of WT- and E93R-FtsZ in E. coli BL21 (DE3) cells were induced with 25 μm IPTG. Western blot analysis of the cellular level of the two proteins in the absence and presence of 25 μm IPTG revealed a 3–4-fold increase in the expression of both the WT and mutant proteins. This showed that the expression level of EcFtsZ did not alter due to the E93R mutation (data not shown). Under limited expression conditions, the cells were observed by differential interference contrast microscopy. Cells containing the WT-FtsZ plasmid showed normal size in the absence and the presence of 25 μm IPTG. In contrast, E93R plasmid-containing cells showed elongated morphology in the presence of 25 μm IPTG (supplemental Fig. S3A). For example, the average length of E. coli cells in the absence and presence of E93R mutation were observed to be 2.1 ± 0.8 and 20 ± 15 μm, respectively. Although 70% of the cells overexpressing the WT-FtsZ were in the range of 2–4 μm, most of the E93R-expressing cells were larger than 4 μm; for example, 3% of cells corresponded to 4–6 μm, 25% were 6–8 μm, 18% cells were 8–10 μm, and the rest of the cells were more than 10 μm in length. The length of 12% of the bacterial cells expressing E93R-FtsZ reached even >40 μm (supplemental Fig. S3B). The data suggested that the overexpression of E93R-FtsZ inhibited the division of E. coli cells.
Expression of E93R-FtsZ in E. coli Inhibits Z-ring Formation and Perturbs the Segregation of Nucleoids in Bacteria
E. coli BL21 (DE3) cells harboring the WT and the E93R-FtsZ plasmids were grown in the absence and presence of 25 μm IPTG. Most of the cells containing the WT-FtsZ were found to contain an intact Z-ring at the middle of the cell along with the two properly segregated nucleoids both in the absence and presence of IPTG, showing that the limited expression of the WT-FtsZ did not produce an apparent phenotype (supplemental Fig. S4). In the case of E93R mutation, cells grown in the absence of IPTG showed normal morphology, as did the WT cells with a Z-ring at the midcell. When the E93R-FtsZ was overexpressed in the presence of 25 μm IPTG, the cells did not have the typical Z-ring (supplemental Fig. S4). For example, 74 and 13% of the cells overexpressing the WT and mutated FtsZs were found to contain a Z-ring, respectively.
In cells containing the WT-FtsZ, the Z-ring was observed to be intact and properly placed at the midcell site, whereas in cells harboring E93R-FtsZ, an intact Z-ring was not observed; instead spiral-like filaments of FtsZ were seen in all of the cells (supplemental Fig. S4J). It appeared that these spirals consisted of stable bundles of FtsZ protofilaments, which were unable to form functional Z-rings. Nearly 20% of the WT-FtsZ-overexpressing cells were found to contain a single nucleoid without any Z-ring, and the average length of these cells was determined to be 1.5 ± 0.3 μm. In contrast, cells expressing E93R-FtsZ contained multiple nucleoids, and none of these cells contained only one nucleoid, suggesting that E93R substitution hindered the cytokinesis of the bacterial cells.
In addition to its effect on the Z-ring assembly, the E93R mutation seems to exert a strong effect on the nucleoid segregation of the bacterial cells. For example, the frequencies of nucleoids per μm of the bacterial cell length were calculated to be 0.7 ± 0.1 and 0.14 ± 0.01 for cells that were overexpressing similar amounts of the WT-FtsZ and E93R-FtsZ, respectively. However, the number of nucleoids in each of the elongated cells was difficult to count accurately because the nucleoids in the elongated cells were diffused and were not properly separated from each other. The results indicated that the overexpression of the mutated (E93R) FtsZ affected the Z-ring assembly and the nucleoid segregation in bacterial cells, resulting in the filamentous morphology.
Substitution of Glu93 to Lysine, Alanine, or Valine
We examined whether the substitution of Glu93 with another positively charged residue, lysine, could influence the assembly of FtsZ like E93R substitution. The assembly of E93K-FtsZ produced light scattering intensity higher than that of the WT-FtsZ (supplemental Fig. S5A). However, E93K-FtsZ produced much lower light scattering intensity as compared with the E93R-FtsZ, indicating that the extent of assembly of E93K-FtsZ is lower than that of E93R-FtsZ (supplemental Fig. S5A). The results indicated that replacement of the negatively charged glutamate residue at the 93-position of EcFtsZ by a positively charged lysine residue increased the assembly of FtsZ; however, the effect was not as profound as that exerted by the arginine residue. Further, we made two more constructs substituting Glu93 by alanine, a neutral amino acid residue, and by valine, a hydrophobic residue. E93V-FtsZ displayed light scattering intensity similar to that of the WT-FtsZ, whereas E93A-FtsZ produced light scattering intensity significantly higher than that of the WT-FtsZ (supplemental Fig. S5B). However, the light scattering intensity of E93A-FtsZ was found to be nearly half that of E93R-FtsZ (supplemental Fig. S5B). Neither E93A nor E93V mutation induced the bundling of FtsZ protofilaments; both of the mutants (E93A and E93V) showed aggregate forming tendency under the assembly conditions used in this study (supplemental Fig. S5C). Thus, the increase in the light scattering intensity of the E93A mutant is due to the nonspecific aggregation of the protein and not due to the bundling of the protofilaments. However, E93K-FtsZ formed thin protofilaments as compared with the thick bundles of E93R-FtsZ (supplemental Fig. S5C and Fig. 1C). The far-UV CD spectra of these mutant proteins were found to be similar to that of the wild type FtsZ, suggesting that these mutations do not alter the secondary structure of FtsZ (Fig. 4). For example, the α-helical and random coil content of the WT-, E93A-, and E93V-FtsZ were estimated to be 24.9 ± 0.4, 26.1 ± 0.6, and 25.6 ± 1.0 and 35.8 ± 0.4, 34.9 ± 0.6, and 36.8 ± 1.0, respectively. The results indicated that the observed bundling effect of E93R-FtsZ is probably due to the presence of a positively charged arginine residue.
DISCUSSION
The substitution of glutamate (Glu93) by Arg in EcFtsZ caused a marked increase in the stability of FtsZ polymers. E93R-FtsZ could substitute WT-FtsZ in JKD7–1/pKD3 cells in the presence of a low concentration of the arabinose. However, it disrupted the division machinery when grown under high arabinose concentrations, whereas WT-FtsZ did not inhibit bacterial division under similar expression conditions. The results suggested that the disassembly of FtsZ protofilaments is important for bacterial cell division. In addition, BL21 cells were found to possess aberrant nucleoids when E93R-FtsZ was expressed, indicating that the FtsZ assembly dynamics might also play a role in nucleoid segregation, or the defective segregation could be an indirect consequence of the aberrant FtsZ assembly. Interestingly, several inhibitors of FtsZ assembly were found to perturb the formation of the Z-ring, but these agents did not alter nucleoid segregation (6, 9, 40).
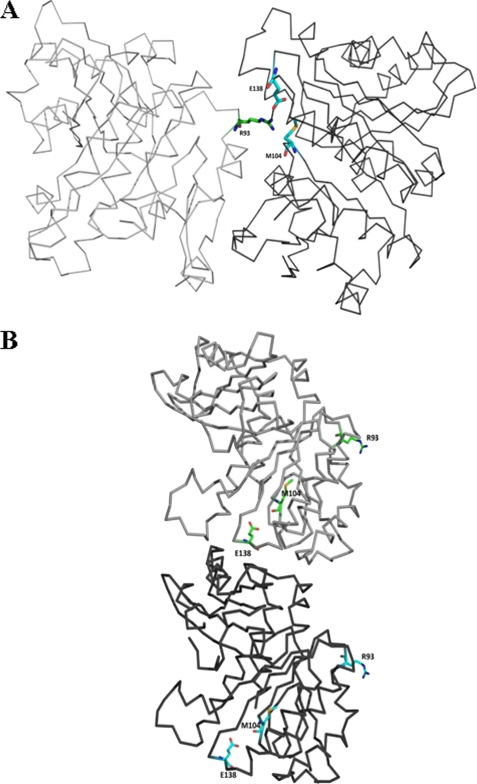
The Z-ring in E. coli is estimated to be 6–8 protofilaments thick, which suggests that the lateral interactions between protofilaments are important for Z-ring functions in vivo (41, 42). However, the mechanism of lateral association is poorly understood. Several possible mechanisms have been proposed, which include shielding of negatively charged FtsZ protofilaments by cations (29) and a conformational change in the monomer constituting the polymer exposing the binding site for lateral interaction. EcFtsZ protofilaments formed thick bundles upon substitution of a negatively charged residue glutamate (Glu93) by a positively charged arginine residue, probably indicating the possibility of enhanced protein-protein interactions. In order to explore this possibility, we modeled the three-dimensional structure of mutant EcFtsZ using the three-dimensional structures of MjFtsZ (Protein Data Bank code 1W59) (21) and of MtbFtsZ (Protein Data Bank code 1RQ7) (17) as templates. The protein-protein interfaces are different in these two crystal structures. We constructed the dimer by modeling EcFtsZ on both of the chains of template. It is evident that Arg93 is not part of MjFtsZ interface, whereas it is present at the interface in MtbFtsZ (Fig. 7, A and B).
FIGURE 7.
Homology model of E93R-FtsZ built using MtbFtsZ (Protein Data Bank code 1RQ7; top) and MjFtsZ (Protein Data Bank code 1W59; bottom) as templates. Homology modeling was carried out using the automated 3D-Jigsaw server. The alignment reported by the server did not require any tweaking because MtbFtsZ, EcFtsZ, and MjFtsZ share high sequence similarity. A, the two chains of a dimer are shown as gray and black wire frames. Residues Arg93, Met104, and Glu138 are shown as green or cyan sticks. The dimer was constructed by combining models built on both templates. The dimers were built to mimic the dimerization in their respective templates. B, the salt bridge between Arg93:Nη and Glu138:Oϵ in E93R-FtsZ is shown by a black dashed line (lower panel); the distance between these two atoms is 2.8 Å.
An analysis of the protein-protein interface of MtbFtsZ indicated two salt bridges formed by Arg91 of one chain with Glu102 and Glu136 of another chain at the interface of the MtbFtsZ crystal structure (supplemental Fig. S6A). When these residues were traced on the sequence alignment of EcFtsZ and MtbFtsZ (supplemental Fig. S6B), it was observed that Glu136 is conserved in EcFtsZ (as Glu138), but Glu102 (Met104 in EcFtsZ) and Arg91 (Glu93 in EcFtsZ) are not conserved. Incidentally, when we replaced Glu93 of EcFtsZ by Arg, the formation of a salt bridge between the Arg93 of one FtsZ subunit and Glu138 of the neighboring FtsZ subunit could be restored (Fig. 7B), which ultimately resulted in the formation of lateral interactions between the two protofilaments.
After leucine, arginine is found to be the most abundant residue at protein-protein interfaces, implying a crucial role for this residue in such interfaces (43–45). In fact, mutation of a single, interfacial arginine residue (Arg181) was found to block the US2 (unique short protein-2)-mediated degradation of HLA-A2 (human leukocyte antigen-A2) by disrupting the interaction between these two proteins (46). In the present study, we have replaced Glu93 by an arginine residue (MtbFtsZ has arginine at this position). The mutation produced a strong phenotype, suggesting that the introduced arginine residue plays a crucial role in the lateral interaction of FtsZ protofilaments, thereby enhancing bundle formation. E93K-FtsZ also displayed enhanced assembly and bundling of FtsZ polymers, albeit weakly as compared with E93R-FtsZ. If the positive charge is the main contributing factor for the enhanced bundling of FtsZ protofilaments, then replacement of Glu93 by a hydrophobic residue or a neutral residue should not produce the bundling effect. To explore this argument, Glu93 of EcFtsZ was replaced with either an alanine or a valine residue. We found that neither E93A nor E93V showed a bundling effect; instead, both of the mutants formed aggregates upon polymerization, suggesting that the positive charge contributed to the enhanced lateral interactions. Therefore, the bundling of E93R-FtsZ may be due to the cumulative effects of the presence of a positively charged residue as well as the presence of arginine at the site.
The crystal structure of MtbFtsZ showed a protein-protein interface (17) different from that of the MjFtsZ (21). Although the physiological relevance of this interface is not yet known, it has been suggested that the phenotypic effects of the Asp86 and Asp96 mutants of EcFtsZ can be explained using this interface. This interface has been suggested to be important for lateral interaction (17), whereas the head-to-tail interactions are necessary for polymerization (47).
If the E93R-FtsZ dimer is like that of MjFtsZ, then the effect of the mutation is indirect. The effects are “transmitted” to the “distant” interface. There are also other examples of such “allosteric” mutations reported in the literature. 3CLPro is the main protease of severe acute respiratory syndrome causing coronavirus, SARS-CoV. Mutation of Ser147 totally inhibited dimerization, although this residue is located 9 Å away from the dimer interface (48). Our mutation study suggests that this interface might be physiologically relevant with the assumption that the observed effect of E93R mutation is due to enhanced protein-protein interactions.
Erickson and co-workers (49) have explored the longitudinal or polymerization interface of FtsZ by site-directed mutagenesis of several interfacial residues. The residues were classified as “top” or “bottom” by reference to the structure of the tubulin protofilament. Few “bottom” mutants, such as D212G, displayed reduced GTPase activity and also showed dominant negative effects when overproduced (49). Incidentally, such residues are part of the synergy or T7 loop, which contacts the GTP of the subunit below it. In this study, we have identified an interfacial residue that is important for the lateral interactions. E93R-FtsZ bound to GTP similarly to the WT-FtsZ (Fig. 3B) and E93R-FtsZ polymers also disassembled under challenged GTP conditions, suggesting that the reduction in the GTP hydrolysis rate of E93R-FtsZ is unlikely to be due to its inability to bind or hydrolyze GTP (Fig. 3C). The bundling may suppress polymer ↔ monomer turnover, thereby causing a decrease in the rate of GTP hydrolysis (38, 50, 51). Alternatively, the trapping of inorganic phosphate between the FtsZ bundles might reduce the rate of phosphate release, leading to the suppression of the GTPase activity.
The turnover rate of the Z-ring has been reported to be reduced 10-fold in the cells with a FtsZ84 mutation, which possesses reduced GTPase activity in vitro (3, 52). Microtubule dynamics are thought to be properly regulated for the division and viability of mammalian cells (53, 54). Similarly, the Z-ring dynamics has been considered to be properly regulated for the faithful division and survival of bacterial cells (12). Here, we propose that the mutation, due to the increased stability of the FtsZ polymers, suppresses the Z-ring dynamics and ultimately results in a reduction in cytokinesis and cell viability.
The GTPase activity of MtbFtsZ is slower than that of EcFtsZ, which probably makes MtbFtsZ protofilaments less dynamic than EcFtsZ (15, 16). In the present study, a strong increase in the assembly and bundling of FtsZ protofilaments was observed when the negatively charged residue Glu93 of EcFtsZ was replaced by the positively charged residue arginine present in MtbFtsZ at the same location. The sequence identity between EcFtsZ and MtbFtsZ peptide sequences is ∼45% (supplemental Fig. S6B). Residues that are different in MtbFtsZ from EcFtsZ might be playing a role in the differential rate of cell division of the two bacteria. We believe that we have found one such mutation, which formed stabilized polymers and suppressed the GTP hydrolysis rate of EcFtsZ. Indeed, there would be many more factors playing their parts in bacterial cell division, but it is tempting to speculate that the Glu93 residue of FtsZ is important for E. coli cell division.
Supplementary Material
Acknowledgments
We thank Dr. Harold Erickson for providing the complementation system used in this study. We thank the Centre for Research in Nanotechnology and Science, Indian Institute of Technology Bombay, for the electron microscopy facility.
This work was supported by a grant from the Department of Science and Technology, Government of India (to D. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- EcFtsZ
- E. coli FtsZ
- MtbFtsZ
- M. tuberculosis FtsZ
- MjFtsZ
- M. jannaschi FtsZ
- TNPGTP
- 2′-(or 3′-)-O-(trinitrophenyl) guanosine 5′-triphosphate
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1.Goehring N. W., Beckwith J. (2005) Curr. Biol. 15, R514–526 [DOI] [PubMed] [Google Scholar]
- 2.Margolin W. (2005) Nat. Rev. Mol. Cell Biol. 6, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stricker J., Maddox P., Salmon E. D., Erickson H. P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michie K. A., Löwe J. (2006) Annu. Rev. Biochem. 75, 467–492 [DOI] [PubMed] [Google Scholar]
- 5.Addinall S. G., Lutkenhaus J. (1996) J. Bacteriol. 178, 7167–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuria T. K., Santra M. K., Panda D. (2005) Biochemistry 44, 16584–16593 [DOI] [PubMed] [Google Scholar]
- 7.Dai K., Mukherjee A., Xu Y., Lutkenhaus J. (1994) J. Bacteriol. 176, 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haydon D. J., Stokes N. R., Ure R., Galbraith G., Bennett J. M., Brown D. R., Baker P. J., Barynin V. V., Rice D. W., Sedelnikova S. E., Heal J. R., Sheridan J. M., Aiwale S. T., Chauhan P. K., Srivastava A., Taneja A., Collins I., Errington J., Czaplewski L. G. (2008) Science 321, 1673–1675 [DOI] [PubMed] [Google Scholar]
- 9.Jaiswal R., Beuria T. K., Mohan R., Mahajan S. K., Panda D. (2007) Biochemistry 46, 4211–4220 [DOI] [PubMed] [Google Scholar]
- 10.Adams D. W., Errington J. (2009) Nat. Rev. Microbiol. 7, 642–653 [DOI] [PubMed] [Google Scholar]
- 11.Kapoor S., Panda D. (2009) Expert Opin. Ther. Targets 13, 1037–1051 [DOI] [PubMed] [Google Scholar]
- 12.Romberg L., Levin P. A. (2003) Annu. Rev. Microbiol. 57, 125–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osawa M., Anderson D. E., Erickson H. P. (2008) Science 320, 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan R., Mishra M., Wu L., Yin Z., Balasubramanian M. K. (2008) Genes Dev. 22, 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White E. L., Ross L. J., Reynolds R. C., Seitz L. E., Moore G. D., Borhani D. W. (2000) J Bacteriol. 182, 4028–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal R., Panda D. (2009) J. Biochem. 146, 733–742 [DOI] [PubMed] [Google Scholar]
- 17.Leung A. K., Lucile White E., Ross L. J., Reynolds R. C., DeVito J. A., Borhani D. W. (2004) J. Mol. Biol. 342, 953–970 [DOI] [PubMed] [Google Scholar]
- 18.Bates P. A., Kelley L. A., MacCallum R. M., Sternberg M. J. (2001) Proteins 5, 39–46 [DOI] [PubMed] [Google Scholar]
- 19.Bates P. A., Sternberg M. J. (1999) Proteins 3, 47–54 [DOI] [PubMed] [Google Scholar]
- 20.Contreras-Moreira B., Bates P. A. (2002) Bioinformatics 18, 1141–1142 [DOI] [PubMed] [Google Scholar]
- 21.Oliva M. A., Cordell S. C., Löwe J. (2004) Nat. Struct. Mol. Biol. 11, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 22.Cordell S. C., Robinson E. J., Lowe J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7889–7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding M. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 849–859 [DOI] [PubMed] [Google Scholar]
- 24.Löwe J., Amos L. A. (1998) Nature 391, 203–206 [DOI] [PubMed] [Google Scholar]
- 25.Santra M. K., Panda D. (2007) Proteins 67, 177–188 [DOI] [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Lu C., Stricker J., Erickson H. P. (1998) Cell Motil. Cytoskeleton 40, 71–86 [DOI] [PubMed] [Google Scholar]
- 28.Jaiswal R., Panda D. (2008) Protein Sci. 17, 846–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee A., Lutkenhaus J. (1999) J. Bacteriol. 181, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santra M. K., Panda D. (2003) J. Biol. Chem. 278, 21336–21343 [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee A., Santra M. K., Beuria T. K., Panda D. (2005) FEBS J. 272, 2760–2772 [DOI] [PubMed] [Google Scholar]
- 32.Geladopoulos T. P., Sotiroudis T. G., Evangelopoulos A. E. (1991) Anal. Biochem. 192, 112–116 [DOI] [PubMed] [Google Scholar]
- 33.Stricker J., Erickson H. P. (2003) J. Bacteriol. 185, 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano W. L. (2002) The PyMOL Molecular Genetics Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 35.Mukherjee A., Lutkenhaus J. (1998) EMBO J. 17, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sossong T. M., Jr., Brigham-Burke M. R., Hensley P., Pearce K. H., Jr. (1999) Biochemistry 38, 14843–14850 [DOI] [PubMed] [Google Scholar]
- 37.Dajkovic A., Mukherjee A., Lutkenhaus J. (2008) J. Bacteriol. 190, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffers D. J., den Blaauwen T., Driessen A. J. (2000) Mol. Microbiol. 35, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 39.Small E., Addinall S. G. (2003) Microbiology 149, 2235–2242 [DOI] [PubMed] [Google Scholar]
- 40.Rai D., Singh J. K., Roy N., Panda D. (2008) Biochem. J. 410, 147–155 [DOI] [PubMed] [Google Scholar]
- 41.González J. M., Jiménez M., Vélez M., Mingorance J., Andreu J. M., Vicente M., Rivas G. (2003) J. Biol. Chem. 278, 37664–37671 [DOI] [PubMed] [Google Scholar]
- 42.Löwe J., Amos L. A. (1999) EMBO J. 18, 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2003) Proteins 53, 708–719 [DOI] [PubMed] [Google Scholar]
- 44.Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 45.Pednekar D., Tendulkar A., Durani S. (2009) Proteins 74, 155–163 [DOI] [PubMed] [Google Scholar]
- 46.Thilo C., Berglund P., Applequist S. E., Yewdell J. W., Ljunggren H. G., Achour A. (2006) J. Biol. Chem. 281, 8950–8957 [DOI] [PubMed] [Google Scholar]
- 47.Scheffers D. J., de Wit J. G., den Blaauwen T., Driessen A. J. (2002) Biochemistry 41, 521–529 [DOI] [PubMed] [Google Scholar]
- 48.Barrila J., Bacha U., Freire E. (2006) Biochemistry 45, 14908–14916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redick S. D., Stricker J., Briscoe G., Erickson H. P. (2005) J. Bacteriol. 187, 2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santra M. K., Dasgupta D., Panda D. (2005) Proteins 61, 1101–1110 [DOI] [PubMed] [Google Scholar]
- 51.Yu X. C., Margolin W. (1997) EMBO J. 16, 5455–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson D. E., Gueiros-Filho F. J., Erickson H. P. (2004) J. Bacteriol. 186, 5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohan R., Panda D. (2008) Cancer Res. 68, 6181–6189 [DOI] [PubMed] [Google Scholar]
- 54.Panda D., Samuel J. C., Massie M., Feinstein S. C., Wilson L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9548–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.