Abstract
Dendritic cells (DCs) are a group of professional antigen-presenting cells, and many genes are known to be associated with their maturation. We compared the transcriptional profiles of immature and mature mouse Langerhans cells using the suppressive, subtractive hybridization method and identified a novel gene of unknown function, termed herein transmembrane protein 123 (Tmem123), of which mRNA expression was enhanced in mature but not in immature Langerhans cells. Its expression was also enhanced in other mature DCs such as bone marrow-derived DCs (BMDCs) and splenic DCs. Interestingly, CD40 expression was up-regulated on mature BMDCs cultured with colchicine concurrently with the enhanced expression of Tmem123 compared with that of fresh BMDCs. Furthermore, the expression of CD40 was enhanced on Tmem123-transfected DC2.4 cells, a mouse BMDC-derived cell line, compared with that on mock-transfected DC2.4 cells. This enhancement of CD40 expression did not occur after deletion of lysosome/endosome targeting YXXφ motifs (where X is any amino acid and φ is a bulky hydrophobic amino acid) in the Tmem123 cytoplasmic tail. By stimulation with anti-CD40 monoclonal antibody, these transfectants secreted an increased amount of IL-12/23 p40 compared with mock-transfected DC2.4 cells. Thus, our study demonstrates that Tmem123 may be used as a new maturation marker in DCs and that this molecule may be closely associated with the cell surface expression of CD40.
Keywords: Antigen Presentation, Dendritic Cell, Immunology, Membrane Proteins, Transcription, CD40, Langerhans Cell, Maturation
Introduction
Dendritic cells (DCs)2 are a group of leukocytes specialized in antigen presentation to naïve T cells. DC maturation is associated with reduced phagocytic capacities, enhanced production of cytokines and chemokines, and the capacity to migrate from non-lymphoid to lymphoid tissues (1). Langerhans cells (LCs) are immature DCs localized in the epidermis. When inflammation occurs in the skin, LCs are exposed to various environmental and endogenous stimuli such as granulocyte macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor-α, and interleukin (IL)-1 and undergo a series of maturation processes including down-regulation of E-cadherin and Langerin expression, up-regulation of costimulatory molecules, and expression of CC chemokine receptor 7 (2). All of these processes are important for LCs to function as antigen-presenting cells; E-cadherin ligation keeps LCs in an immature state (3). Langerin acts as a nonconventional endocytic receptor (4). Costimulatory molecules provide essential signals to T cells for adaptive immune responses (1). CC chemokine receptor 7 is essential for LC migration into the regional draining lymph nodes (5). Thus, it is important to study the function of molecules related to LC maturation to comprehend cutaneous immune responses.
Transcription profiles indicate morphological, phenotypical, and functional changes in a cell induced by environmental factors (6). Transcriptome analyses of immature and mature DCs have been performed previously (7–10), but those of fresh and matured LCs have not yet been performed because of the difficulty in obtaining sufficient numbers of highly purified LCs from the skin. We succeeded in preparing highly purified LCs, which enabled us to carry out transcriptional profiling of immature and mature LCs. Using the polymerase chain reaction (PCR)-select complementary DNA (cDNA) subtraction method (called suppressive, subtractive hybridization (SSH)), we identified several genes regulated by maturation of LCs. Although the functions of some of the identified genes are not clear, we specifically focused on mouse transmembrane protein 123 (Tmem123), whose expression was up-regulated during maturation not only in LCs but also in other DCs. The possible role of Tmem123 in cell surface expression of CD40 was also investigated.
EXPERIMENTAL PROCEDURES
Animals
BALB/c female mice were purchased from Charles River Laboratories Japan, Inc. (Yokohama, Japan) and were maintained under specific pathogen-free conditions at the University of Tokyo Animal Facilities until use at the age of 8–12 weeks. All animal experiments were approved by the Animal Research Committee of the University of Tokyo.
Antibodies and Reagents
Mouse recombinant GM-CSF and recombinant IL-4 were purchased from Peprotech (Rocky Hill, NJ). Lipopolysaccharide (LPS; serotype 0111:B4) was purchased from Sigma-Aldrich. Colchicine was purchased from Wako Pure Chemical Industries (Osaka, Japan). For flow cytometry, immunoblotting, and immunostaining, the following antibodies were used: fluorescein isothiocyanate (FITC)-conjugated hamster anti-mouse CD40 monoclonal antibody (mAb), FITC-conjugated hamster anti-mouse CD80 mAb, FITC-conjugated rat anti-mouse CD86 mAb, FITC-conjugated mouse anti-mouse major histocompatibility complex (MHC) class II (I-Ad) mAb, FITC-conjugated mouse anti-mouse CD11c mAb, FITC-conjugated mouse IgG isotype control, FITC-conjugated hamster IgG isotype control, FITC-conjugated hamster IgM isotype control, FITC-conjugated rat IgG isotype control (BD Biosciences Pharmingen), phycoerythrin-conjugated goat anti-rabbit IgG antibody (Ab; Rockland Immunochemicals, Gilbertsville, PA), rabbit anti-hemagglutinin (HA) tag polyclonal Ab (pAb; Clontech), horseradish peroxidase-conjugated goat anti-rabbit IgG Ab (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-HA tag pAb, Living ColorsTM A.v. mAb (JL-8; Clontech), and rabbit γ-globulin (Jackson ImmunoResearch Laboratories, West Grove, PA). To detect Tmem123 protein expression, we used a custom-made rabbit anti-mouse Tmem123 pAb (anti-Tmem123 pAb) generated against a synthetic peptide corresponding to an 18-amino acid sequence (SRRGIRYRSIDEHDAIIC) of the C terminus of Tmem123 (Shima Laboratories, Tokyo, Japan). Rat anti-mouse CD40 mAb (no azide/low endotoxin, clone 3/23) used to stimulate DCs to produce IL-12 p40 and rat IgG isotype control were purchased from BD Biosciences Pharmingen.
Cells and Cell Lines
LCs were isolated and purified from mouse skin using anti-mouse I-Ad mAb using a previously described panning method, and the purity of I-Ad+ LCs was consistently over 95% (2). The freshly isolated LCs (fresh LCs) demonstrated Birbeck granules and expressed a low level of costimulatory molecules, indicating that these cells were immature (2). BMDCs were obtained by culturing mouse bone marrow cells in Roswell Park Memorial Institute 1640 medium containing 5% fetal calf serum (FCS), 10 ng/ml GM-CSF, and 10 ng/ml IL-4 for 6 days. After culture, CD11c+ cells were separated by positive selection using microbead-conjugated hamster anti-mouse CD11c mAb and a magnetic cell separator according to the manufacturer's instructions (Miltenyi Biotec, Belgisch Gladbach, Germany). The purity of fresh CD11c+ BMDCs was consistently over 80% (data not shown). For splenic DCs (SpDCs), CD11c+ cells were separated by positive selection from a mouse spleen cell suspension as described above. The purity of fresh CD11c+ SpDCs was consistently over 80%. HeLa cells, WEHI164 cells, B16-F0 cells, EL4 cells, and COS-1 cells were purchased from the National Institute of Biomedical Innovation (Osaka, Japan). DC2.4 cells were a kind gift from Dr. Rock (Department of Pathology, University of Massachusetts Medical School). A20 cells were a kind gift from Dr. Fujimoto (Department of Dermatology, Kanazawa University Graduate School of Medical Science, Ishikawa, Japan). LCs, BMDCs, SpDCs, and DC2.4 cells were cultured in Roswell Park Memorial Institute 1640 medium containing 10% FCS. B16-F0, WEHI164, and COS-1 cells were cultured in Dulbecco's minimum essential Eagle's medium containing 10% FCS. HeLa cells were cultured in minimum essential Eagle's medium containing 10% FCS. The following reagents were added to the medium: 0.1% sodium pyruvate, 1% non-essential amino acid, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B.
Complementary DNA Subtraction and Library Screening
Fresh LCs and LCs cultured in GM-CSF (10 ng/ml) for 24 h were collected, and cDNA subtraction and library screening were entrusted to Clontech. Briefly, two cDNA pools were synthesized from poly(A)+ RNAs isolated from fresh and cultured LCs. These two cDNA pools were digested with RsaI before cDNA subtraction, and the cDNA of cultured LCs was subtracted using the cDNA of fresh LCs with a PCR-select cDNA subtraction kit (Clontech). The remaining cDNAs were randomly subcloned into a pAtlas vector (Clontech). One hundred and thirteen clones were sequenced from each cDNA pool, and their sequences were compared with those in the GenBankTM/EMBL/DDBJ database.
Preparation of Messenger RNA and cDNA
Cells and tissues were collected, and messenger RNA (mRNA) was extracted using the RNeasy minikit (Qiagen, Valencia, CA). Complementary DNA was synthesized using the SuperscriptTM III First-Strand Synthesis System for PCR (Invitrogen) and oligo(dT) primer (Invitrogen). For mouse Tmem123 tissue distribution studies, Rapid-Scan mouse cDNA panels (OriGene Technologies, Rockville, MD) and cDNA extracted from axial and popliteal lymph nodes was used.
Quantitative Real Time Reverse Transcription-PCR
The PCR was performed with a LightCycler® system (Roche Diagnostics) in 20-μl reaction volumes using 1× FastStart DNA Master SYBR Green I (Roche Diagnostics) in the presence of a 500 nm concentration of forward and reverse primers. For detection of Tmem123, the following primer pair was used: forward, 5′-CAA TGC AAG TGC TTC TCC AA-3′, and reverse, 5′-TGT TCG TCA ATG CTT CGG TA-3′ (Sigma-Genosys). The primer pair resulted in PCR products of 249 base pairs (bp) for Tmem123. pEGFP-C1-Tmem123, GAPDH Primer and Probe set for LightCycler (Nihon Gene Research Laboratories, Miyagi, Japan), and pcDNA3.1 containing mouse GAPDH cDNA were used for calibrating cDNA. The reactions were incubated at 95 °C for 10 min followed by 50 cycles of amplification (95 °C for 3 s, 65 °C for 3 s, and 72 °C for 12 s). The temperature ramp was set at 20 °C/s. The data were analyzed using LightCycler Software (Roche Diagnostics).
Sensitization and Elicitation of Contact Hypersensitivity (CHS)
Mice were sensitized with 50 μl of 0.5% 2,4-dinitrofluorobenzene (Sigma-Aldrich) in acetone and olive oil (4:1) on shaved abdominal skin for 2 consecutive days. Five days later, CHS was elicited by applying 20 μl of 0.25% 2,4-dinitrofluorobenzene on the right ear. As a negative control, the left ear was treated with acetone/olive oil alone. Complementary DNA was extracted from each ear 24 h after the elicitation, and PCR for Tmem123 was performed as described below.
Tmem123 Cloning and Cell Transfection
Complemantary DNA from bulk spleen cells was amplified with PCR (94 °C for 45 s, 60 °C for 1 min, and 72 °C for 1 min) with primers for Tmem123: forward, 5′-ATG AAT TCT ATG GCG CTG TGT GCG CGG GCC-3′, and reverse, 5′-GCG AAT TCT TAA ATG ATG GCG TCG TGT TCG-3′ (Sigma-Genosys). The PCR product was subcloned into the EcoRI site of pEGFP-C1 (Clontech) or the EcoRI-BglII site of pCMV-HA (Clontech), and the sequence was verified. ΔY176Y177Y184 was constructed as described previously (11) using pCMV-HA-Tmem123 as the template. COS-1 cells and HeLa cells were cultured for 24 h and transfected with plasmid DNA using FuGENE 6 (Roche Diagnostics) for 6 h according to the manufacturer's protocol. After transfection, cells were cultured for 18 h before the following assay. DC2.4 cells were transfected using the Dendritic Cell Nucleofector Solution and Nucleofector System (Amaxa, Koln, Germany) according to the manufacturer's protocol. Cells were cultured for 12 h before the following assay.
Flow Cytometry Analysis
Flow cytometry analysis was performed as described previously (2). For intracellular staining, cells were fixed and permeabilized with Cytofix/Cytoperm reagents (BD Biosciences Pharmingen) for 20 min at 4 °C, incubated with 0.1 μg/ml anti-Tmem123 pAb diluted in Perm/Wash Buffer for 30 min at 4 °C, incubated with 0.5 μg/ml phycoerythrin-conjugated goat anti-rabbit IgG Ab diluted in Perm/Wash Buffer for 30 min at 4 °C, and examined with flow cytometry. For double staining for both cell surface and intracellular molecules, after staining the cells with Abs against cell surface molecule as described previously (2), cells were incubated with LIVE/DEAD Reduced Biohazard Cell Viability kit 3 (red fluorescence) (Molecular Probes) for 20 min at room temperature to label dead cells, and intracellular flow cytometry analysis was performed. Dead cells were excluded from all analyses.
Immunoblotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed with NuPAGE Novex Bis-Tris gel (4–12%) and MES running buffer (Invitrogen). Resolved proteins were transferred to Immobilon-P membranes (Millipore, Bedford, MA), blotted with the indicated Ab, and visualized using SuperSignal Chemoluminescent Substrate (Pierce) in conjunction with horseradish peroxidase-detecting Ab.
Immunocytochemistry
Cells were cultured on chamber slides and fixed with acetone at 4 °C for 5 min followed by washing with phosphate-buffered saline (PBS) twice and blocking with 10% goat serum, PBS at room temperature for 20 min. In some experiments, cytospin slides were prepared as follows. The cell suspension was spun on a microscope slide using a cytocentrifuge (Shandon Elliot Cytospin, Shandon Elliot, Pittsburgh, PA), fixed with acetone at 4 °C for 5 min followed by washing with PBS twice, and blocking with 10% goat serum, PBS at room temperature for 20 min. These cell samples were incubated overnight with 0.1 μg/ml anti-Tmem123 pAb at 4 °C and stained with a VECTASTAIN ABC kit (rabbit IgG; Vector Laboratories Inc., Burlingame, CA) with diaminobenzidine (Wako Pure Chemical Industries).
IL-12/23 p40 Production from Stimulated DC2.4 Cells
DC2.4 cells were treated as indicated and cultured with anti-CD40 mAb (20 μg/ml) or rat IgG isotype control (20 mg/ml) for 48 h, and IL-12/23 p40 protein levels in culture supernatants were quantified with mouse IL-12/23 p40 immunoassay kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. Each sample was tested in duplicate.
Statistical Analysis
A paired two-tailed Student's t test was used to analyze the results, and a p value <0.05 was considered statistically significant.
RESULTS
Tmem123 as LC Maturation-related Gene
GM-CSF enhances maturation of mouse LCs and up-regulates CD80, CD86, and CD40 expression (2, 11). To detect novel genes associated with the maturation of LCs, we performed a PCR-select cDNA subtraction analysis using cDNA extracted from fresh LCs and LCs cultured with GM-CSF (10 ng/ml) for 24 h. The following criteria were used to select the target genes. 1) The gene was a mouse gene. 2) The function of the genes in the context of DCs had not yet been reported. 3) The length of the cloned cDNA fragment was more than 100 bp, and its sequence showed more than 95% identity to the sequence of the actual gene registered in the NCBI database. 4) More than three cDNA clones were detected in the subtraction analysis. The most frequently detected DNA in 226 cDNA clones was Langerin (14 clones), which was already known to be down-regulated during LC maturation. According to the selection criteria, seven genes of a total of 226 clones were picked up as target genes (Table 1). The frequency of the clone detected for each gene seemed to correlate well with the transcription credibility during LC maturation. Among the seven target genes, we picked Tmem123 for further analysis.
TABLE 1.
Target genes selected from result of PCR-select cDNA subtraction analysis using fresh LCs and LCs cultured with GM-CSF for 24 h
| GenBank accession no. | Gene name | Clone |
|---|---|---|
| Up-regulated genes | ||
| X82564 | 45 S pre-rRNA | 4 |
| BC010292 | Transmembrane protein 123 | 3 |
| NP032564 | Myristoylated alanine-rich protein kinase C | 3 |
| X66850 | Microsatellite DNA | 3 |
| Down-regulated genes | ||
| AC093445 | C57BL/6J chromosome 1 clone rp23-267j18 | 6 |
| AK017346 | Checkpoint suppressor 1 | 4 |
| BC004092 | Influenza virus NS1A-binding protein | 3 |
Characterization of Tmem123 Gene
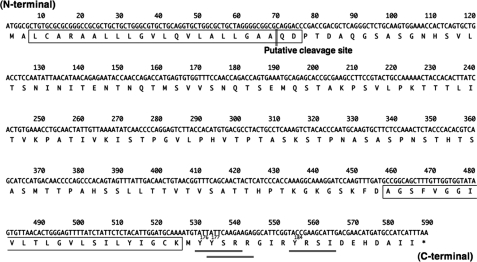
Tmem123 mRNA is 2864 bp of mRNA with a poly(A) tail containing 588 bp of open reading frame (ORF) encoding 195 amino acids. The predicted molecular mass of Tmem123 is ∼21 kDa, and the ORF encodes a type I membrane protein with one extracellular domain (127 amino acids), two hydrophobic transmembrane domains (23 amino acids each), and a cytoplasmic tail. In addition, there is a lysosome/endosome targeting motif, YXXφ (where X is any amino acid and φ is a bulky hydrophobic amino acid), in the cytoplasmic tail of Tmem123 (Fig. 1, doubly underlined).
FIGURE 1.
Tmem123 cDNA sequence and alignments. Nucleotide and predicted amino acid sequences of Tmem123 cDNA (GenBank accession number BC010292) are shown. The putative transmembrane domain is singly boxed. The lysosome/endosome targeting motif, YXXφ (where φ is a bulky hydrophobic amino acid) is doubly underlined. The target of anti-Tmem123 pAb is singly underlined.
Cell and Tissue Distribution of Tmem123 mRNA
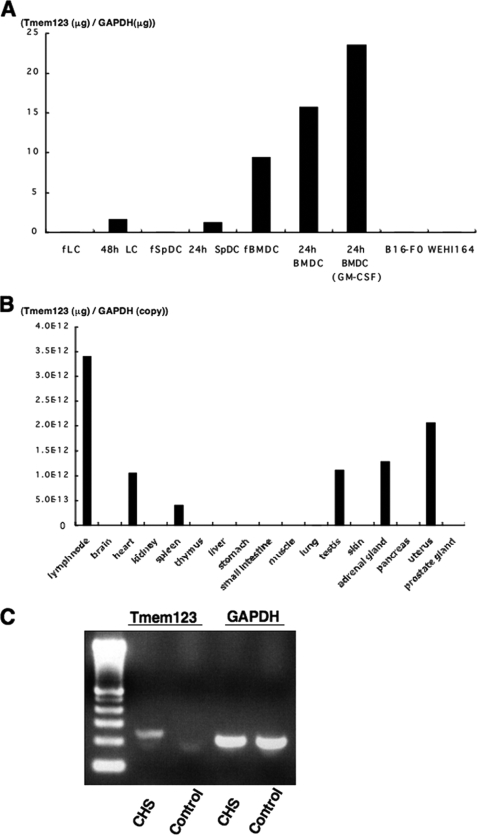
Quantitative reverse transcription-PCR was performed to assess expression levels of Tmem123 mRNA in different cell types and tissues. Fresh BMDCs expressed Tmem123 mRNA, whereas both fresh LCs and fresh SpDCs did not (Fig. 2A). Tmem123 mRNA expression was up-regulated after 24 h of culture in LCs, SpDCs, and BMDCs, and it was further up-regulated with GM-CSF-induced maturation. B16-F0 cells (a melanoma cell line) and WEHI164 cells (a fibrosarcoma cell line) did not express Tmem123 mRNA (Fig. 2A). Fig. 2B depicts the expression pattern of Tmem123 mRNA in various tissues. The strongest signal for Tmem123 mRNA was detected in lymph nodes, and a relatively lower level of Tmem123 expression was found in spleen but not in thymus or skin. Tmem123 mRNA expression was also detected in heart, testis, adrenal gland, and uterus. These findings indicated that the expression of mouse Tmem123 was not necessarily restricted to lymphoid organ but that it was most highly expressed in lymph nodes where mature DCs accumulated. Because Tmem123 mRNA was not detected in the skin, we examined whether Tmem123 mRNA is expressed in mouse ear skin after eliciting CHS where maturation of skin DCs is induced (12, 13). Complementary DNA was extracted from mouse ear skin 24 h after the elicitation at the time point when mature LCs are still found in the skin (14, 15). As expected, Tmem123 mRNA was detected in the ear skin after CHS elicitation but not in the control ear skin (Fig. 2C).
FIGURE 2.
Cell and tissue distribution of Tmem123 mRNA. A, comparative analysis of Tmem123 mRNA expression levels between fresh DCs (LCs, SpDCs, and BMDCs), mature DCs, and two other cell lines (B16-F0 and WEHI164 cells). Fresh DCs (fLC, fSpDC, and fBMDC) were cultured for 24 or 48 h with or without GM-CSF (10 ng/ml) to induce maturation. B, Tmem123 expression in various tissues. C, Tmem123 mRNA expression in the CHS-elicited ear skin and control ear skin. The result from a representative experiment is shown. A, n = 2; B, n = 3; C, n = 3.
Characterization of Tmem123 pAb
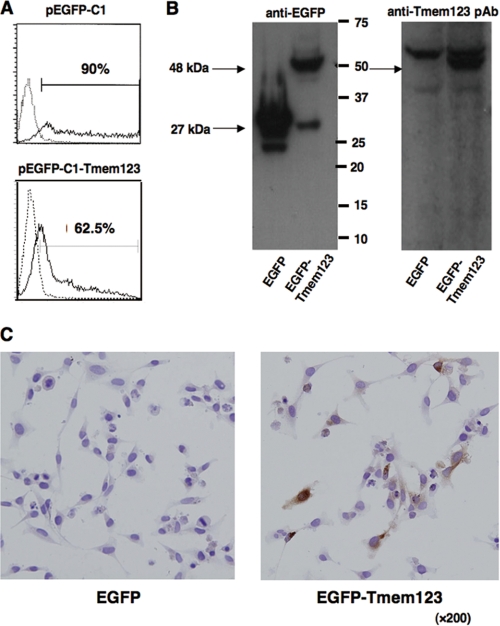
To study the pattern of Tmem123 protein expression in DCs, we used a custom-made anti-Tmem123 pAb generated against a synthetic peptide corresponding to the C terminus of Tmem123 (Fig. 1, singly underlined). An expression vector containing the ORF of Tmem123 (pEGFP-C1-Tmem123) or mock vector (pEGFP-C1) was introduced into COS-1 cells. Flow cytometry showed that more than 60% of COS-1 cells were transfected with pEGFP-C1-Tmem123 (Fig. 3A), and an immunoblot of cell lysate from these transfectants revealed that anti-Tmem123 pAb recognized an enhanced green fluorescent protein (EGFP)-Tmem123 of ∼48 kDa in accordance with the molecular mass predicted from the amino acid sequence (Fig. 3B). With immunohistochemistry, anti-Tmem123 pAb recognized Tmem123 protein expressed in pEGFP-C1-Tmem123-transfected HeLa cells, whereas anti-Tmem123 pAb did not stain mock-transfected HeLa cells (Fig. 3C). These data indicate that anti-Tmem123 pAb recognized Tmem123 protein with immunoblot and immunohistochemistry.
FIGURE 3.
Characterization of anti-Tmem123 pAb. A, flow cytometric analysis for EGFP expression in pEGFP-C1-transfected COS-1 cells (upper panel) and pEGFP-C1-Tmem123-transfected COS-1 cells (lower panel). Solid lines, transfected cells; dotted lines, non-transfected cells. B, cell lysates of pEGFP-C1-transfected COS-1 cells and pEGFP-C1-Tmem123-transfected COS-1 cells were blotted with Living Colors A.v. mAb (JL-8; left panel) or anti-Tmem123 pAb (right panel). The arrows indicate EGFP (∼27 kDa) and EGFP-Tmem123 (∼48 kDa). C, pEGFP-C1-transfected HeLa cells (left panel) and pEGFP-C1-Tmem123-transfected HeLa cells (right panel) were stained with anti-Tmem123 pAb.
Expression of Tmem123 Protein Was Up-regulated in Mature LCs and BMDCs
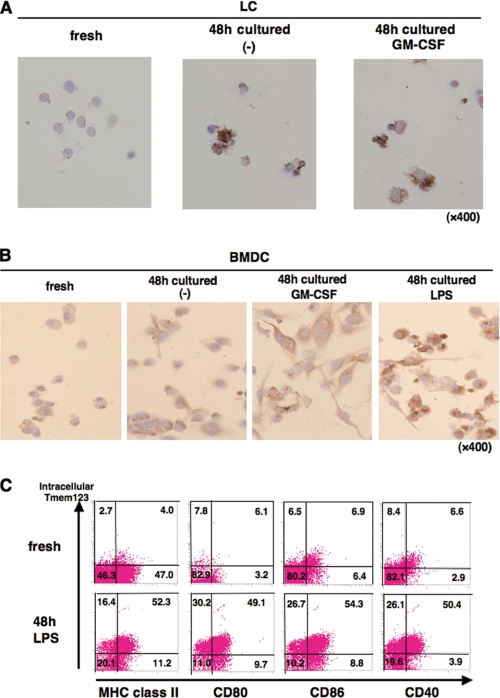
We next studied the regulation of Tmem123 protein in DCs. As shown in Fig. 4A, LCs cultured with or without GM-CSF for 48 h expressed a markedly increased level of Tmem123 protein compared with fresh LCs. Tmem123 protein expression was also up-regulated in BMDCs upon GM-CSF-induced maturation, and LPS further up-regulated its expression (Fig. 4B). Anti-Tmem123 pAb did not recognize cell surface expression of Tmem123, but it recognized the intracellular portion of Tmem123 after cell permeabilization as analyzed by flow cytometry (data not shown). Tmem123 protein expression was up-regulated in LPS-stimulated BMDCs compared with fresh BMDCs (Fig. 4C). Interestingly, Tmem123 expression correlated well with the cell surface expression of MHC class II and costimulatory molecules (CD80, CD86, and CD40; Fig. 4C).
FIGURE 4.
Tmem123 protein expression was up-regulated in mature LCs and mature BMDCs. A, fresh LCs were cultured in medium with or without GM-CSF (10 ng/ml) for 48 h. Fresh LCs and cultured LCs were stained with anti-Tmem123 pAb as described under “Experimental Procedures.” B, fresh BMDCs were cultured in medium only, with GM-CSF (10 ng/ml), or with LPS (1 μg/ml) for 48 h. Fresh BMDCs and cultured BMDCs were stained with anti-Tmem123 pAb as described under “Experimental Procedures.” C, fresh BMDCs (upper panels) and BMDCs cultured in LPS (1 μg/ml) for 48 h (lower panels) were first stained with FITC-conjugated anti-MHC class II mAb, anti-CD80 mAb, anti-CD86 mAb, and anti-CD40 mAb. After that, these cells were permeabilized and stained with anti-Tmem123 pAb followed by phycoerythrin-conjugated goat anti-rabbit IgG Ab and analyzed by flow cytometry. A representative of three independent experiments is shown.
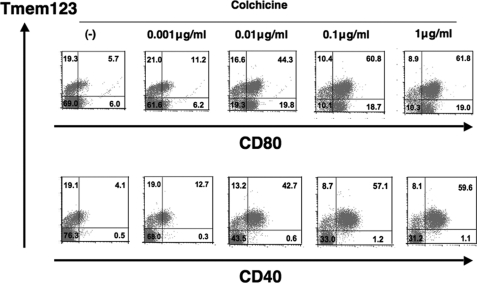
Co-expression of Tmem123 and CD40 in BMDCs Stimulated with Colchicines
Recently, colchicine has been reported to induce maturation in BMDCs (14). A double staining for Tmem123 and cell surface CD40 or CD80 was performed in BMDCs cultured with or without colchicine (0.001–1 μg/ml). As the colchicine concentration increased, the ratio of both Tmem123+/CD40+ and Tmem123+/CD80+ BMDC population increased. Interestingly, with a concentration of more than 0.1 μg/ml colchicine, virtually all CD40+ BMDCs co-expressed Tmem123, whereas two-thirds of CD80+ BMDCs co-expressed Tmem123 (Fig. 5). Thus, these results indicate that Tmem123 was closely associated with CD40 but not CD80.
FIGURE 5.
Co-expression of Tmem123 and CD40 in BMDCs stimulated with colchicine. BMDCs were cultured in medium only or with colchicine (0.001, 0.01, 0.1, or 1 μg/ml) for 48 h and stained with anti-CD80 mAb (upper panel) or with anti-CD40 mAb (lower panel). These cells were permeabilized and stained with anti-Tmem123 pAb followed by phycoerythrin-conjugated goat anti-rabbit IgG Ab and analyzed by flow cytometry. A representative of three independent experiments is shown.
Transfection with Tmem123 cDNA Up-regulated Cell Surface Expression of CD40 on DC2.4 Cells
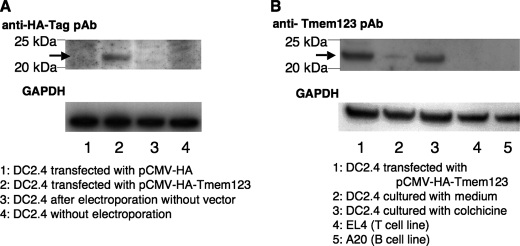
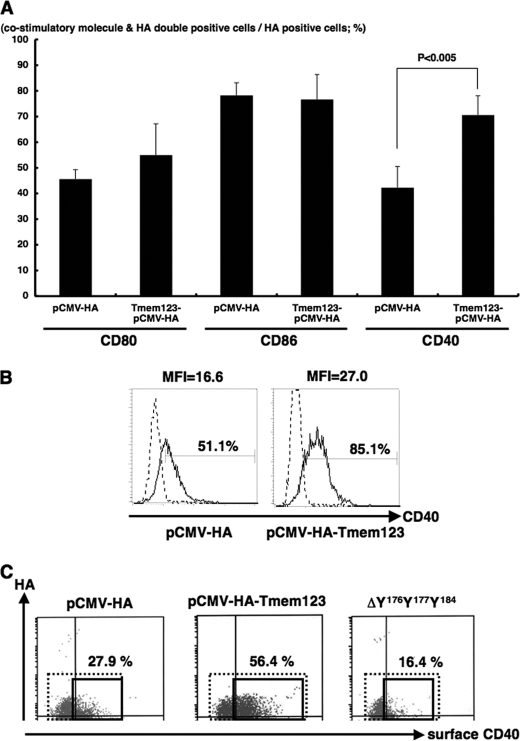
To further investigate the relation between Tmem123 and CD40 expression, we transfected mouse a BMDC cell line, DC2.4 cells (16), with either pCMV-HA or pCMV-HA-Tmem123 by electroporation. The HA-Tmem123 protein expression (∼22 kDa) in pCMV-HA-Tmem123-transfected DC2.4 cells was confirmed by immunoblot either with anti-HA pAb or with anti-Tmem123 pAb (Fig. 6, A and B). DC2.4 cells expressed a weak level of Tmem123 protein (∼21 kDa), and this expression was increased after stimulation with 0.05 μg/ml colchicine (Fig. 6B) as expected. A mouse T cell line, EL4 cells, and a mouse B cell line, A20 cells, did not express Tmem123 (Fig. 6B). The ratio of CD40+ DC2.4 cells was significantly higher when the cells were transfected with pCMV-HA-Tmem123 compared with those transfected with pCMV-HA (69.9 ± 7.7 versus 42.3 ± 6.7%, n = 3, p < 0.001), whereas the expression levels of CD80 and CD86 did not differ (Fig. 7A). The expression level of CD40 was also higher in pCMV-HA-Tmem123 transfectants compared with mock-transfected cells (Fig. 7B).
FIGURE 6.
Tmem123 expression was up-regulated in colchicine-stimulated DC2.4 cells. A, DC2.4 cells were transfected by electroporation with pCMV-HA (lane 1), with pCMV-HA-Tmem123 (lane 2), or without vector (lane 3). These cells and untransfected DC2.4 cells (lane 4) were lysed and blotted with anti-HA pAb. The arrow indicates HA-Tmem123 protein (∼22 kDa). B, DC2.4 cells were transfected by electroporation with pCMV-HA-Tmem123 (lane 1) or cultured with 1 μg/ml colchicine for 48 h (lane 3). These cells were collected, lysed, and blotted with anti-Tmem123 pAb. DC2.4 cells cultured without stimulation (lane 2), EL4 cells (lane 4), and A20 cells (lane 5) were also collected, lysed, and blotted with anti-Tmem123 pAb. The arrow indicates HA-Tmem123 protein (∼22 kDa; lane 1) and Tmem123 protein (∼21 kDa; lanes 2 and 3).
FIGURE 7.
Transfection with Tmem123 cDNA up-regulated cell surface expression of CD40 on DC2.4 cells. DC2.4 cells were transfected with pCMV-HA or pCMV-HA-Tmem123 and stained with anti-CD80 mAb, anti-CD86 mAb, and anti-CD40 mAb. These cells were permeabilized and stained with anti-Tmem123 pAb followed by phycoerythrin-conjugated goat anti-rabbit IgG Ab and analyzed by flow cytometry. A, the ratio of CD80-, CD86-, and CD40-positive cells in the HA-Tmem123- or HA-positive cells is shown. The ratio of CD40-positive cells was significantly higher in the HA-Tmem123-positive transfectants compared with HA-positive transfectants (69.9 ± 7.7 versus 42.3 ± 6.7%, n = 3, p < 0.001), although the expression levels of CD80 and CD86 did not differ. Results are mean ± S.D. (n = 3). B, the level of cell surface expression of CD40 on DC2.4 cells transfected with pCMV-HA (left panel) and pCMV-HA-Tmem123 (right panel) is shown. Solid lines, CD40 mAb; dashed lines, isotype control (representative of three independent experiments). MFI, median fluorescence intensity. C, the cell surface expression of CD40 was higher in pCMV-HA-Tmem123-transfected cells than that in mock-transfected cells. There was no difference in cell surface CD40 expression between ΔY176Y177Y184-transfected cells and mock-transfected cells. A representative of three independent experiments is shown.
YXXφ Motifs Were Required for CD40 Up-regulation via Transfection with Tmem123 cDNA
We then investigated the role of YXXφ motifs of Tmem123 for CD40 up-regulation. To this end, we deleted tyrosines 176, 177, and 184 from Tmem123 (ΔY176Y177Y184; Fig. 1). The protein expression in ΔY176Y177Y184-transfected DC2.4 cells was confirmed by immunoblot (data not shown). As shown in Fig. 7D, the cell surface expression level of CD40 in ΔY176Y177Y184-transfected cells was the same as that in mock-transfected cells. Thus, the YXXφ motif of Tmem123 was required for cell surface expression of CD40.
IL-12/23 p40 Production Was Increased When DC2.4 Cells Were Transfected with Tmem123-encoding Vector
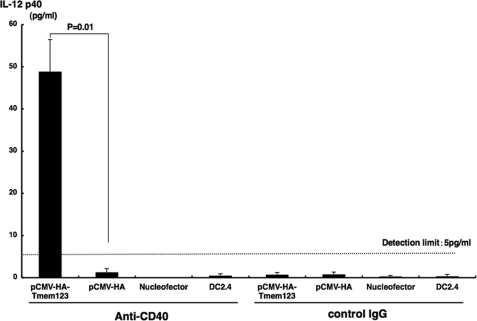
Finally, we investigated the effect of pCMV-HA-Tmem123 transfection on IL-12/23 p40 production. Because anti-CD40 mAb up-regulated IL-12/23 p40 secretion from DCs (11), we transfected DC2.4 cells with either pCMV-HA-Tmem123 or pCMV-HA and stimulated these cells for 48 h with anti-CD40 mAb (20 μg/ml). Then, we quantified IL-12/23 p40 secreted from these cells in the culture supernatant. As shown in Fig. 8, DC2.4 cells transfected with pCMV-HA did not produce a detectable amount of IL-12/23 p40 (<5 pg/ml), whereas DC2.4 cells transfected with Tmem123 expression vector secreted a significant amount of IL-12/23 p40 when stimulated with anti-CD40 mAb (48.8 ± 7.6 pg/ml, n = 3, p < 0.05).
FIGURE 8.
IL-12/23 p40 production was increased from pCMV-HA-Tmem123-transfected DC2.4 cells. DC2.4 cells were transfected with pCMV-HA, with pCMV-HA-Tmem123, or without vector. These cells and untransfected DC2.4 cells were cultured with anti-CD40 mAb (20 μg/ml) or isotype control (20 μg/ml) for 48 h. The supernatants were collected, and IL-12/23 p40 contents were measured by a mouse IL-12/23 p40 enzyme-linked immunosorbent assay kit. Results from three independent experiments are shown. Results are mean ± S.D. (n = 3).
DISCUSSION
We analyzed transcriptomes of immature and mature LCs by SSH and cloned and characterized a novel gene, Tmem123. We found that Tmem123 was up-regulated in mature DCs overall and that Tmem123 was associated with CD40 expression.
We chose SSH for this analysis of transcriptomes because DNA microarray technology and serial analysis of gene expression are limited by insensitivity to transcripts of low abundance (17). SSH made it possible to detect some transcripts of low abundance due to a normalization step to equalize the abundance of cDNAs within the target population (18, 19). Therefore, transcriptional profiles analyzed with SSH may be different from those of DNA microarray or serial analysis of gene expression.
In GenBank, 19 genes containing Tmem123 sequence were previously cloned and registered from RCB-0527, CRL-1702, bone marrow macrophage, 16-day mouse neonate cerebellum, tongue, NOD-derived CD11c+ dendritic cells, C57BL/6-derived CD11c+ dendritic cells, and activated spleen. The human protein with the highest overall homology with Tmem123 was Porimin with 67% homology (20). Porimin cDNA was isolated from a Jurkat cell (human T lymphocyte cell line) cDNA library as type I transmembrane mucin, and it was shown to mediate cell adhesion and oncosis-like cell death (20, 21). We did not find any damage or loss of cell viability in cells transfected with Tmem123-encoding vector (data not shown); therefore, it is likely that murine Tmem123 has a different function from Porimin.
Within mouse tissues, the highest expression level of Tmem123 mRNA was found in peripheral lymph nodes, which are reported to have the highest ratio for mature DCs among lymphoid organs (22). Interstitial DCs reside in organs such as liver, heart, kidney, and adrenal gland (23). The significance of Tmem123 mRNA in organs other than lymph nodes and spleen remains to be explored. Although we could not detect Tmem123 mRNA in naïve skin, it was expressed after CHS elicitation, probably by inducing maturation of skin LCs/DCs. For T cell and B cell lines, we could not detect Tmem123 protein expression. These data altogether support the fact that Tmem123 expression is associated with mature DCs, although further study is necessary to determine whether or not its expression is indeed DC-specific.
The expression of Tmem123 was increased when DCs were stimulated for maturation using either GM-CSF, LPS, or colchicine. Colchicine is one of the inhibitors of microtubule polymerization, and it has recently been reported to induce full maturation of BMDCs (14). By culture of BMDCs with colchicine, intracellular Tmem123 expression increased and correlated quite well with that of CD40. When DC2.4 cells were transfected with Tmem123 expression vector, the expression level of CD40 on the cell surface was markedly enhanced. These in vitro data suggest that Tmem123 expression is associated with DC maturation. However, we still do not know whether cross-linking Tmem123 would generate a signal to the mature DC or not because the Tmem123 antibody we generated recognized only the intracellular portion of Tmem123, and we do not have any antibodies that can recognize the extracellular portion of Tmem123 or activate Tmem123 cross-linking. Furthermore, we still do not know the endogenous ligand for Tmem123.
Tmem123 has three YXXφ motifs, or tyrosine-containing lysosome/endosome targeting motifs, in its intracellular domain (24). YXXφ motif is reported to bind to adapter-protein complexes (known as AP1, AP2, AP3, and AP4), and these complexes are known to transport their binding protein into transport vesicles (25, 26). Tyrosine-containing signals are known to regulate post-Golgi transport events (25). Therefore, Tmem123 may help extracellular expression of CD40 by packaging CD40 into transport vesicles. In fact, when we stained both intracellular and extracellular CD40, there was no difference in total CD40 expression between pCMV-HA-Tmem123-transfected DC2.4 cells and pCMV-HA-transfected DC2.4 cells (data not shown). These data support our hypothesis that Tmem123 is not regulating CD40 protein synthesis but regulating its transportation to the cell surface. Furthermore, when we point deleted Tyr176, Tyr177, and Tyr184 from each YXXφ motif in Tmem123 and transfected DC2.4 cells with this ΔY176Y177Y184 Tmem123, CD40 up-regulation was impaired. These in vitro data also suggest that Tmem123 is associated with cell surface expression of CD40 and that YXXφ motif plays an important role probably by associating Tmem123 to intracellular organelles regulating CD40 expression. However, two other questions remain to be solved. First, it is not yet clear why Tmem123 contributes to cell surface expression of CD40 but not other costimulatory molecules. Second, an additional explanation is needed for why the relation between CD40 and Tmem123 expression is more prominent with colchicine than with other stimuli for maturation. Our result might indicate an involvement of microtubules in Tmem123 regulation of CD40 expression.
Mature DCs are required for efficient induction of primary T cell responses for adaptive immunity (1, 27), and it is essential to control DC maturation to generate DC-based vaccine or immunotherapy to prevent cancer and infection. CD40 is a member of the tumor necrosis factor receptor superfamily, and the interaction between CD40 and its cognate ligand, CD40 ligand (CD40L; CD154), is critical for a productive immune response (28). For DCs, CD40 influences DC survival (29), acts as a DC maturation stimulus in culture (30), enhances antigen presentation (31), and induces IL-12 production (32, 33). Indeed, Tmem123-transfected DC2.4 cells secreted IL-12/23 p40 when stimulated with anti-CD40 mAb, and this stimulation mimicked a mode of physiological DC stimulation by T cells (32, 33). Thus, CD40 is one of the attractive molecules to be engineered in DC-based immunotherapy. The principal transcription factors regulating CD40 expression were shown to be NFκB (34) and a splice variant of CD40. The latter reduces the level of surface full-length CD40 expression, and it is suggested to be involved in posttranslational regulation (35). However, there have been no reports of any molecules closely regulating cell surface expression of CD40. Our results indicate that Tmem123 may be an attractive molecule to engage in DC-based immunotherapy by up-regulating cell surface CD40 expression and promoting Th1 immune responses. We are now establishing Tmem123 knock-out mice, which will give further information on Tmem123 function in vivo.
Acknowledgment
We are grateful to Dr. Hideki Fujita for support.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to T. W., K. T., and T. Y.).
- DC
- dendritic cell
- LC
- Langerhans cell
- BMDC
- bone marrow-derived DC
- SpDC
- splenic DC
- SSH
- suppressive, subtractive hybridization
- EGFP
- enhanced green fluorescent protein
- Ab
- antibody
- pAb
- polyclonal Ab
- CHS
- contact hypersensitivity
- Tmem123
- transmembrane protein 123
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Banchereau J., Steinman R. M. (1998) Nature 392, 245–252 [DOI] [PubMed] [Google Scholar]
- 2.Salgado C. G., Nakamura K., Sugaya M., Tada Y., Asahina A., Fukuda S., Koyama Y., Irie S., Tamaki K. (1999) J. Invest. Dermatol. 113, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 3.Riedl E., Stöckl J., Majdic O., Scheinecker C., Knapp W., Strobl H. (2000) Blood 96, 4276–4284 [PubMed] [Google Scholar]
- 4.Valladeau J., Ravel O., Dezutter-Dambuyant C., Moore K., Kleijmeer M., Liu Y., Duvert-Frances V., Vincent C., Schmitt D., Davoust J., Caux C., Lebecque S., Saeland S. (2000) Immunity 12, 71–81 [DOI] [PubMed] [Google Scholar]
- 5.Saeki H., Moore A. M., Brown M. J., Hwang S. T. (1999) J. Immunol. 162, 2472–2475 [PubMed] [Google Scholar]
- 6.Staudt L. M., Brown P. O. (2000) Annu. Rev. Immunol. 18, 829–859 [DOI] [PubMed] [Google Scholar]
- 7.Granucci F., Vizzardelli C., Virzi E., Rescigno M., Ricciardi-Castagnoli P. (2001) Eur. J. Immunol. 31, 2539–2546 [DOI] [PubMed] [Google Scholar]
- 8.Türeci O., Bian H., Nestle F. O., Raddrizzani L., Rosinski J. A., Tassis A., Hilton H., Walstead M., Sahin U., Hammer J. (2003) FASEB J. 17, 836–847 [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto S. I., Suzuki T., Nagai S., Yamashita T., Toyoda N., Matsushima K. (2000) Blood 96, 2206–2214 [PubMed] [Google Scholar]
- 10.Cho M., Ishida K., Chen J., Ohkawa J., Chen W., Namiki S., Kotaki A., Arai N., Arai K., Kamogawa-Schifter Y. (2008) Int. Immunol. 20, 155–164 [DOI] [PubMed] [Google Scholar]
- 11.Tada Y., Asahina A., Nakamura K., Tomura M., Fujiwara H., Tamaki K. (2000) J. Immunol. 164, 5113–5119 [DOI] [PubMed] [Google Scholar]
- 12.Nuriya S., Yagita H., Okumura K., Azuma M. (1996) Int. Immunol. 8, 917–926 [DOI] [PubMed] [Google Scholar]
- 13.Toebak M. J., Gibbs S., Bruynzeel D. P., Scheper R. J., Rustemeyer T. (2009) Contact Dermatitis 60, 2–20 [DOI] [PubMed] [Google Scholar]
- 14.Mizumoto N., Gao J., Matsushima H., Ogawa Y., Tanaka H., Takashima A. (2005) Blood 106, 3082–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoitzner P., Holzmann S., McLellan A. D., Ivarsson L., Stössel H., Kapp M., Kämmerer U., Douillard P., Kämpgen E., Koch F., Saeland S., Romani N. (2003) J. Invest. Dermatol. 120, 266–274 [DOI] [PubMed] [Google Scholar]
- 16.Shen Z., Reznikoff G., Dranoff G., Rock K. L. (1997) J. Immunol. 158, 2723–2730 [PubMed] [Google Scholar]
- 17.Evans S. J., Datson N. A., Kabbaj M., Thompson R. C., Vreugdenhil E., De Kloet E. R., Watson S. J., Akil H. (2002) Eur. J. Neurosci. 16, 409–413 [DOI] [PubMed] [Google Scholar]
- 18.Diatchenko L., Lau Y. F., Campbell A. P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E. D., Siebert P. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao W., Epstein C., Liu H., DeLoughery C., Ge N., Lin J., Diao R., Cao H., Long F., Zhang X., Chen Y., Wright P. S., Busch S., Wenck M., Wong K., Saltzman A. G., Tang Z., Liu L., Zilberstein A. (2004) BMC Genomics 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma F., Zhang C., Prasad K. V., Freeman G. J., Schlossman S. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9778–9783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., Xu Y., Gu J., Schlossman S. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 6290–6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson N. S., El-Sukkari D., Belz G. T., Smith C. M., Steptoe R. J., Heath W. R., Shortman K., Villadangos J. A. (2003) Blood 102, 2187–2194 [DOI] [PubMed] [Google Scholar]
- 23.Hart D. N. (1997) Blood 90, 3245–3287 [PubMed] [Google Scholar]
- 24.Mallabiabarrena A., Jiménez M. A., Rico M., Alarcón B. (1995) EMBO J. 14, 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trowbridge I. S., Collawn J. F., Hopkins C. R. (1993) Annu. Rev. Cell Biol. 9, 129–161 [DOI] [PubMed] [Google Scholar]
- 26.Moody D. B., Porcelli S. A. (2003) Nat. Rev. Immunol. 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 27.Dhodapkar M. V., Steinman R. M., Krasovsky J., Munz C., Bhardwaj N. (2001) J. Exp. Med. 193, 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quezada S. A., Jarvinen L. Z., Lind E. F., Noelle R. J. (2004) Annu. Rev. Immunol. 22, 307–328 [DOI] [PubMed] [Google Scholar]
- 29.Josien R., Li H. L., Ingulli E., Sarma S., Wong B. R., Vologodskaia M., Steinman R. M., Choi Y. (2000) J. Exp. Med. 191, 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores-Romo L., Björck P., Duvert V., van Kooten C., Saeland S., Banchereau J. (1997) J. Exp. Med. 185, 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delamarre L., Holcombe H., Mellman I. (2003) J. Exp. Med. 198, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch F., Stanzl U., Jennewein P., Janke K., Heufler C., Kämpgen E., Romani N., Schuler G. (1996) J. Exp. Med. 184, 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cella M., Scheidegger D., Palmer-Lehmann K., Lane P., Lanzavecchia A., Alber G. (1996) J. Exp. Med. 184, 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukamoto N., Kobayashi N., Azuma S., Yamamoto T., Inoue J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tone M., Tone Y., Fairchild P. J., Wykes M., Waldmann H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1751–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]