Abstract
Background
The conformational conversion of the host-derived cellular prion protein (PrPC) into the disease-associated scrapie isoform (PrPSc) is responsible for the pathogenesis of transmissible spongiform encephalopathies (TSEs). Various single-point mutations in PrPCs could cause structural changes and thereby distinctly influence the conformational conversion. Elucidation of the differences between the wild-type rabbit PrPC (RaPrPC) and various mutants would be of great help to understand the ability of RaPrPC to be resistant to TSE agents.
Methodology/Principal Findings
We determined the solution structure of the I214V mutant of RaPrPC(91–228) and detected the backbone dynamics of its structured C-terminal domain (121–228). The I214V mutant displays a visible shift of surface charge distribution that may have a potential effect on the binding specificity and affinity with other chaperones. The number of hydrogen bonds declines dramatically. Urea-induced transition experiments reveal an obvious decrease in the conformational stability. Furthermore, the NMR dynamics analysis discloses a significant increase in the backbone flexibility on the pico- to nanosecond time scale, indicative of lower energy barrier for structural rearrangement.
Conclusions/Significance
Our results suggest that both the surface charge distribution and the intrinsic backbone flexibility greatly contribute to species barriers for the transmission of TSEs, and thereby provide valuable hints for understanding the inability of the conformational conversion for RaPrPC.
Introduction
The conformational conversion of the prion protein, from the normal cellular form (PrPC) to the abnormal scrapie isoform (PrPSc), is responsible for the pathogenesis of transmissible spongiform encephalopathies (TSEs) [1]. The “protein-only” hypothesis is supported by much compelling evidence presented recently [2]–[4]. To elucidate the detailed mechanism of the conformational change, understanding of the structural basis of the prion protein is required. The three-dimensional structures of PrPC among species have been extensively determined using NMR techniques so far [5]–[16]. Great effort has also been made to explore the architecture of PrPSc or PrPSc-like filaments [17]–[21].
Noticeably, all known forms of inherited human TSEs, including Creutzfeldt-Jakob disease (CJD), Gerstmann-Straussler-Scheinker syndrome (GSS) and fatal familial insomnia (FFI), are closely associated with dominant mutations in human PrPC (hPrPC) [22], [23]. In addition to apparently disease-causing mutations in humans, polymorphisms in sheep appear to exert substantial effects on variations of PrPC in disease susceptibility [24]–[27]. Particularly, rabbits are one of the few mammalian animals reported to be resistant to TSE agents [28]. Multiple amino acid residues throughout the rabbit PrPC (RaPrPC) sequence contribute to the inability of RaPrPC to form PrPSc. Several mouse PrPC (mPrPC) mutants, with one single residue substituted by the corresponding residue in RaPrPC, are completely inhibited to undergo the conversion to the disease-related isoform [29]. All these facts indicate a significant influence on the property of the PrPC structure as the result of one important residue mutation. Therefore, detailed comparisons of three-dimensional structures of wild-type PrPC and its mutants could provide valuable insights into the underlying molecular mechanism of prion conversions.
In our previous work we have demonstrated that the S173N substitution leads to distinct structural changes for RaPrPC [30]. However, whether similar changes could be observed in other single-point mutants remains to be addressed. Since mPrPC with the V214I substitution is prohibited to convert to the abnormal form [29], we thereby expect that the I214V substitution would cause structural changes more or less for RaPrPC. In this present work, we determined the solution structure of the I214V mutant of RaPrPC(91–228) using multi-dimensional heteronuclear NMR techniques. In addition, we performed 15N relaxation measurements to detect the backbone dynamics of its structured C-terminal domain (121–228). Furthermore, we investigated its structural stability using CD spectroscopy. Our results reveal significant structural changes caused by the single-residue mutation, which may be of benefit to understand the detailed molecular mechanism of the conformational conversion for prion proteins.
Results
Solution structure
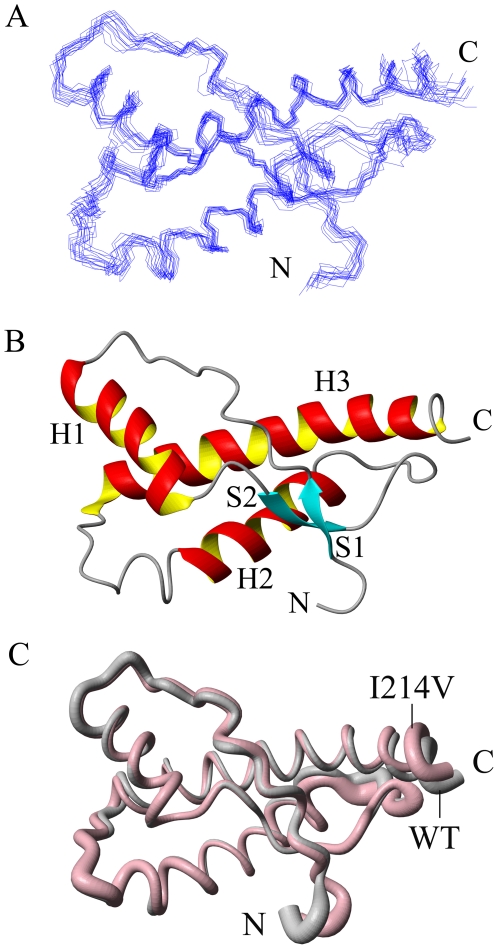
A family of 200 structures is calculated and the structural statistics are presented in Table 1. The diagram representing 15 lowest-energy structures for the I214V mutant of RaPrPC(91–228) in solution is shown in Fig. 1A, together with a ribbon cartoon of average secondary structure elements displayed in Fig. 1B. The I214 mutant contains two short antiparallel β-sheets (S1: 128–130, S2: 160–162) and three α-helices (H1: 144–155, H2: 175–186, H3:199–227), with a disulfide bond (C178–C213) stabilizing helices 2 and 3. The N-terminal loop 91–120 is highly disordered. Loop 165–172 is well defined owing to the long-range NOEs from residues at the end of helix 3. The overall structure of the I214V mutant appears to be identical to that of the wild-type (Fig. 1C).
Table 1. Structural statistics of the I214V mutant of RaPrPC(91–228).
| Quantity | Value |
| Distance restraints | |
| Intraresidue (| i-j | = 0) | 703 |
| Sequential (| i-j | = 1) | 684 |
| Medium range (2<| i-j |<4) | 627 |
| Long range (| i-j |>5) | 717 |
| Total | 2731 |
| Dihedral angle restraints (Φ and Ψ)a | 134 |
| Hydrogen bond restraints | 54 |
| Restraints violations (15 structures) | |
| NOE distance violation (>0.3 Å) | 0 |
| Torsion angle violation (>5°) | 0 |
| RMSD from mean structure (Å) | |
| All residues (backbone atoms) | 0.87±0.22 |
| Secondary structures (backbone atoms) | 0.67±0.20 |
| All residues (heavy atoms) | 1.62±0.26 |
| Secondary structures (heavy atoms) | 1.43±0.24 |
| Ramachandran analysis (%) (124–228)b | |
| Residues in most favored regions | 77.9 |
| Residues in additionally allowed regions | 17.4 |
| Residues in generously allowed regions | 2.8 |
| Residues in disallowed regions | 1.9 |
The dihedral angle restraints are generated from secondary structure determined by the program CSI (David Wishart, Brian Sykes, Leigh Willard, Tim Jellard and reference [64]).
Residues included in the analysis are indicated in parentheses.
Figure 1. Solution structure of the I214V mutant of RaPrPC(91–228).
(A) Diagram of 15 lowest-energy conformations. (B) Ribbon cartoon showing the secondary structure elements of the mean structure. (C) Sausage diagram showing the superposition of structures of the wild-type (grey) and the mutant (pink). The structure was determined in acetate buffer at pH 4.5. The diagrams are generated using MolMol [61].
Electrostatic potential
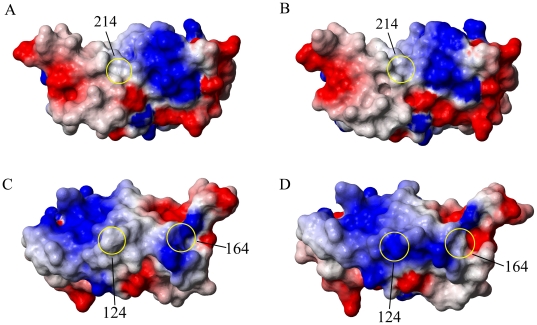
We evaluated the effect of the I214V substitution on the surface charge distribution of RaPrPC(91–228). Both the mutant and the wild-type carry neutral charge at the right substituted site 214 (Fig. 2A, B). The I214V substitution does not change the electrostatic potential at site 214 due to the similar non-charged feature of the two amino acids. Unexpectedly, significant changes are observed at many other unsubstituted sites. For example, the wild-type displays a neutral charge distribution at site 124, while the I214V mutant carries positive charge in the same position (Fig. 2C, D). Site 164 also alters electrostatic potential from positive to neutral after the mutation (Fig. 2C, D). The I214V substitution results in a prominent shift of the surface charge distribution.
Figure 2. Distributions of surface electrostatic potential.
Site 214 of (A) the wild-type and (B) the I214V mutant. Sites 124 and 164 of (C) the wild-type and (D) the I214V mutant. Positive, neutral and negative charges are colored blue, white and red, respectively. The surface diagrams are generated using MolMol.
Hydrogen bond
The hydrogen bond network usually makes a significant contribution to maintain secondary and tertiary structures of a protein. Stable hydrogen bonds are defined herein if they exist in at least 9 structures among the 15 lowest-energy structures. Totally, 41 hydrogen bonds are detected in the I214V mutant of RaPrPC(91–228), much less than the number 55 in the wild-type. In particular, only 4 hydrogen bonds participate in sustaining the tertiary structure of the mutant, compared with 8 in the wild-type (Table 2). As less hydrogen bonds are formed in the mutant, one could expect that the RaPrPC molecule is somehow readily to experience the conformational conversion after the mutation.
Table 2. Hydrogen bonds maintaining the tertiary structures of RaPrPC(91–228) and the I214V mutant.
| Protein | Hydrogen bonds |
| RaPrPC(91–228) | M128HN-Y162O, G130HN-V160O, I138HN-Y149O, R155HH12-D201OD1, R155HH22-Y148O, H176HD1-E210OE2, H186HD1-R155O, S221HG-D166OD1 |
| I214V | G130HN-V160O, I138HN-Y149O, Y162HN- M128O, H186HD1-R155O |
Conformational stability
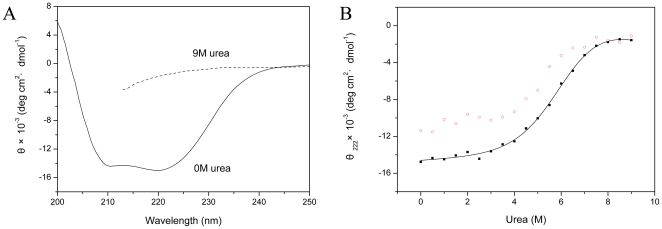
To access the conformational stability of the I214V mutant of RaPrPC(121–228), we performed urea-induced unfolding transition experiments using far-UV CD spectroscopy. The I214V mutant is rich in α-helix in the absence of urea (Fig. 3A), indicating a well-folded state. This mutant loses its secondary structure entirely under the condition of 9 M urea (Fig. 3A), implying a completely unfolded state. The mean residue ellipticity at 222 nm versus the urea concentration is shown in Fig. 3B, with a solid line showing the theoretical curve on the basis of a two-state mechanism. The denaturation was not reversible in our experimental condition (Fig. 3B), thus we determined the apparent thermodynamic parameters for the equilibrium unfolding of the I214V mutant (Table 3). 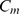 is the concentration of urea required to denature 50% of a protein, and
is the concentration of urea required to denature 50% of a protein, and 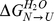 presents the apparent estimated free energy of unfolding extrapolated to zero concentration of denaturant. The measured values of
presents the apparent estimated free energy of unfolding extrapolated to zero concentration of denaturant. The measured values of 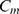 and
and 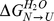 for the mutant are less than those for the wild-type (Table 3), indicating reduced conformational stability of RaPrPC after the I214V substitution. The coefficient m is also different from that for the wild-type (Table 3).
for the mutant are less than those for the wild-type (Table 3), indicating reduced conformational stability of RaPrPC after the I214V substitution. The coefficient m is also different from that for the wild-type (Table 3).
Figure 3. Urea-induced transitions of the I214V mutant of RaPrPC(121–228) characterized by CD spectroscopy.
(A) The folded protein in the absence of urea (solid line) compared with the unfolded protein in the presence of 9 M urea (dashed line). (B) Mean residue ellipticity measured at 222 nm (θ222) in the presence of urea of different concentration. θ222 in denaturation is indicated by solid squares (▪), while in renaturation by open circles (○). The solid line presents the fitting curve on the basis of a two-state mechanism.
Table 3. Apparent thermodynamic parameters for the equilibrium unfolding of RaPrPC(121–228) and the I214V mutant at 25°C.
| Protein |
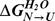 (kJ·mol−1) (kJ·mol−1) |
 (kJ·mol−1·M−1) (kJ·mol−1·M−1) |
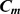 (M) (M) |
| RaPrPC(121–228) | 26.2±2.7 | −3.88±0.49 | 6.49±0.05 |
| I214V | 16.1±1.8 | −2.62±0.38 | 5.65±0.06 |
Note: 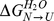 is an estimate of the free energy in the absence of denaturant, the parameter
is an estimate of the free energy in the absence of denaturant, the parameter  represents the cooperativity of the unfolding transition, and
represents the cooperativity of the unfolding transition, and 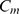 is the concentration of urea at the midpoint of unfolding. The determined parameters for the wild-type [30] are listed here to facilitate comparison.
is the concentration of urea at the midpoint of unfolding. The determined parameters for the wild-type [30] are listed here to facilitate comparison.
Relaxation rates R1, R2 and heteronuclear NOEs
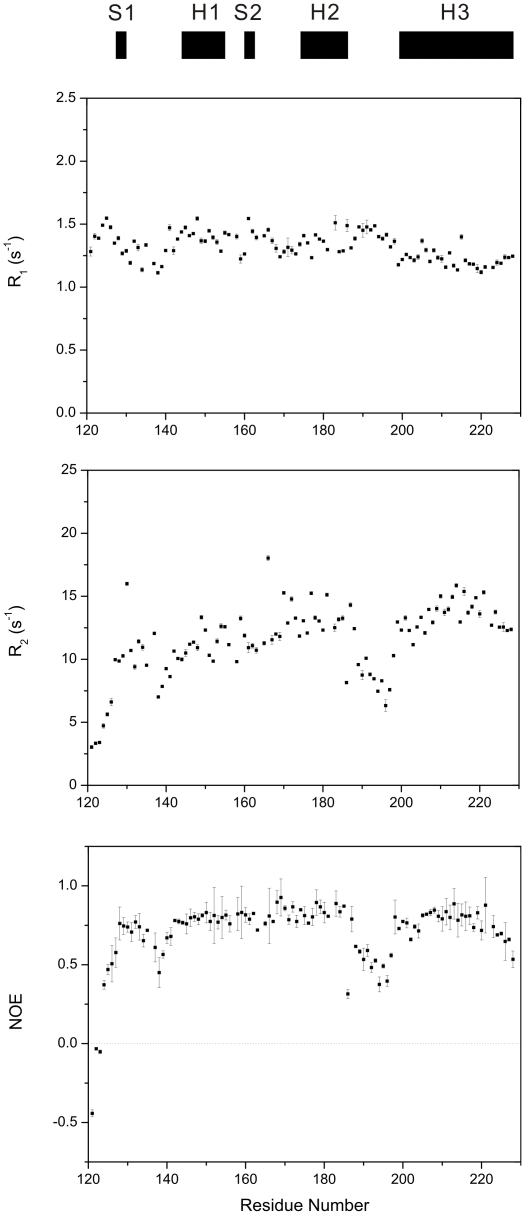
To analyze the backbone dynamics of the I214V mutant of RaPrPC(121–228), we performed 15N relaxation measurements using NMR spectroscopy. As a whole, 103 assigned residues are used except 5 residues with unobservable resonance signals. We utilized the peak height for curve fitting so as to avoid the effect of partially overlapped resonance peaks. The relaxation rates R1, R2 and heteronuclear NOEs versus residue number are shown in Fig. 4. The R1 values do not change much with the sequence, mostly between 1.1 and 1.5 s−1. Different from the R1 distribution, the R2 values are relatively variable with residue number, ranging from 3.0 to 18.1 s−1 approximately. The residue D166 shows the largest R2 value over 18.1 s−1, and G130 also displays distinctly large R2 value as high as 16.0 s−1. Almost all residues exhibit positive NOE values except the first three residues. Typically, NOE values for the two antiparallel β-sheets and three α-helices are obviously higher than those for the loop fragments encompassing residues 121–127, 137–141 and 188–197. The secondary structure elements display NOE values larger than 0.7, indicative of more restricted dynamics in these regions.
Figure 4. Relaxation rates R1, R2 and {1H}-15N heteronuclear NOEs of the I214V mutant of RaPrPC(121–228).
Regular secondary structure elements are indicated on the top. The relaxation constants and the experimental errors were extracted by a single exponential curve fitting of the peak heights using Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco).
Reduced spectral density mapping
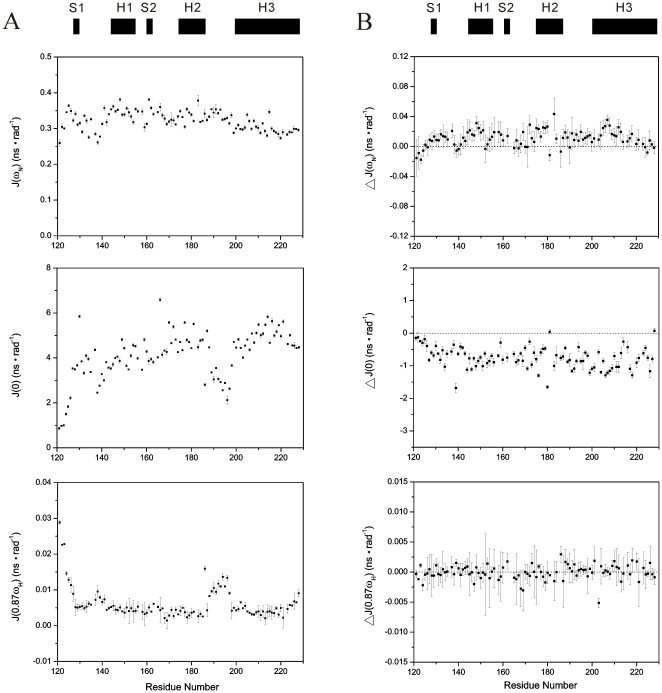
We adopted the spectral density function approach [31]–[33] to interpret the 15N relaxation data for the I214V mutant of RaPrPC(121–228). The calculated values of reduced spectral densities J(ωN), J(0) and J(0.87ωH) versus residue number are shown in Fig. 5A. The middle frequency spectral densities J(ωN) exhibit relatively invariable values ranging from 0.25 to 0.38 ns·rad−1, reflecting insensitivity to variations in backbone motion. Residues in helix 3 show a slight reduction in J(ωN) spectral densities compared to helices 1 and 2. It has been suggested that such a reduction reflects slightly anisotropic tumbling of PrPC [34].
Figure 5. Reduced spectral density functions analysis.
(A) Spectral densities of the I214V mutant of RaPrPC(121–228). (B) Differences of spectral densities between the wild-type and the I214V mutant. The difference is calculated as follows: ΔJ(ω) = J(ω)mutant−J(ω)wild-type. Regular secondary structure elements are indicated on the top. The notebook provided by Spyracopoulos [62] was used to execute the calculation.
The low frequency spectral density J(0) is sensitive both to fast internal motions on the pico- to nanosecond (ps-ns) time scale and to slow motions (Rex) on the micro- to millisecond (µs-ms) time scale. Rapid internal motions tend to reduce the J(0) value, while slow internal motions usually lead to anomalously large J(0) values [35]. The plot of J(0) versus residue number shows the trend of R2 values change with the sequence since J(0) values are normally dominated by R2 values. In contrast to J(ωN), the spectral densities J(0) cover a wider range of values. Residues with lower J(0) values are mainly observed in loop regions 121–127, 138–141 and 189–197, implying sub-nanosecond flexibility of the N-H bond vector. Dissimilarly, loop 165–172 is not as flexible as the other three loop fragments. Residues in secondary structure elements have relatively higher J(0) values. However, the residue H186 in helix 2 displays a fairly small J(0) value, implying significant internal motion on the ps-ns time scale. D166 exhibits the largest J(0) value around 7.0 ns·rad−1, and G130 in β-sheet 1 also shows a remarkable J(0) value as large as 5.9 ns·rad−1, both indicative of slow µs-ms Rex motions. Furthermore, a few residues in helices 2 and 3 are likely to undergo slow Rex motions as well.
The high frequency spectral density J(0.87ωH) is sensitive only to fast internal motions on the sub-nanosecond time scale. Fast motions are reflected in relatively large values of J(0.87ωH). Aside from residues in loop regions 121–127, 138–141 and 189–197, the C-terminal end of helix 3 and the particular residue H186 show large J(0.87ωH) values, implying fast ps-ns motions. In addition, small J(0.87ωH) values and large J(0) values are observed in most residues in secondary structure elements, indicative of restricted motions on the fast ps-ns time scale.
The overall distributions of the reduced spectral densities J(ωN), J(0) and J(0.87ωH) for the I214V mutant are similar to those for the wild-type reported previously [30]. We compared the values of the spectral densities between the wild-type and the I214V mutant, and the differences versus residue number are shown in Fig. 5B. The most significant changes are observed in J(0). Almost all residues exhibit distinctly negative values in ΔJ(0). Generally, the smaller the value of J(0), the greater the sub-nanosecond flexibility of N-H bond vectors [36]. Thus the I214V mutation leads to increased internal dynamics.
Order parameter S2
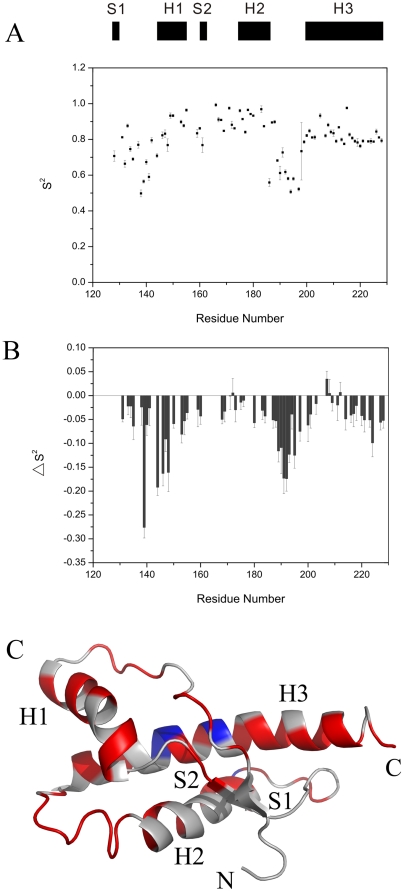
The modelfree formalism is usually used to analyze internal motions of a protein [37]–[39]. The D∥/D⊥ ratio of the rotational diffusion tensor for the I214V mutant of RaPrPC(121–228) was calculated to be 1.24±0.01 using the r2r1_diffusion program (kindly provided by Prof. A.G. Palmer 3rd), suggesting that the axially symmetric model is suitable for the data fitting. The calculated order parameter S2 versus residue number is plotted in Fig. 6A. This parameter describes the amplitude of the sub-nanosecond timescale motions (0≤S2≤1). Residues in loop 135–141 and 189–197 display the lowest S2 values, which imply that these regions are highly disordered. In contrast to these two loops, loop 165–172 show higher S2 values, indicating an ordered fragment. The S2 values for residues in secondary structure elements are relatively larger, which is consistent to the J(0) distribution. The mean values of S2 for helices 1, 2 and 3 are 0.88, 0.88 and 0.82, respectively. H186 in helix 2 shows a fairly small S2 value as low as 0.56, indicative of significant internal flexibility incompatible with the secondary structure property. We further compared the difference in S2 values between the wild-type and the mutant (Fig. 6B). Almost all residues show negative ΔS2 values, providing solid evidence that the I214V substitution significantly enhances the backbone flexibility of RaPrPC. The ΔS2 values are mapped onto the mean structure of the I214V mutant (Fig. 6C).
Figure 6. Modelfree analysis.
(A) Order parameters S2 of the I214V mutant of RaPrPC(121–228). Regular secondary structure elements are indicated on the top. The program Fastmodelfree [63] was used to perform the calculation. Unavailable S2 values for a few residues are due to the absence of data or failure in the data fitting. (B) Differences in S2 values between the wild-type and the mutant. The difference is calculated according to the equation: ΔS2 = S2 mutant−S2 wild-type. The absence of ΔS2 values for residues result from unavailable S2 values for either the wild-type or the mutant. (C) ΔS2 values are mapped onto the tertiary structures of the I214V mutant: blue for ΔS2≥0, red for ΔS2<0, and grey for ΔS2 unavailable. This ribbon diagram is generated by PyMol (kindly provided by Prof. DeLano WL).
Discussion
Single-point mutation may result in functional alteration owing to the global or local structural change in the protein. Several mPrPC mutants exhibit local structural changes and show distinctly different behavior from the wild-type [29], [40]. Three single-residue mutants, including hPrPC(M166V), hPrPC(S170N) and hPrPC(R220K), show variations in the length and quality of definition of helix 3 [41]. In addition, loop 166–172, which is lack of backbone amide resonances in hPrPC and mPrPC, is well-defined in both hPrPC (S170N) and mPrPC (V166A) mutants [16], [41].
In this present work, we have determined the solution structure of the I214V mutant of RaPrPC(91–228). The three-dimensional structure of the mutant is almost identical to that of the wild-type (Fig. 1), however, both the altered hydrogen bond network and the changed surface charge distribution demonstrate that the I214V substitution could lead to marked structural changes (Fig. 2 and Table 2). Significant change in surface-restricted charges has also been observed in hPrPC (E200K) [42] and RaPrPC(S173N) mutants [30]. It is well known that electrostatic interaction could distinctly influence on both the binding specificity and affinity of a protein with substrates. Thus, the mutation-induced change of surface charge distribution is likely to affect the binding of PrPC with many potential chaperones, including nucleic acids [43], [44], protein X [45], [46], sugars, lipids, etc., which would modulate the conformational conversion. Interestingly, both hPrPC(E200K) [42] and RaPrPC(S173N) mutants [30] exhibit distinct charge alterations at the right substituted sites, whereas the I214V mutant described herein remains the neutral charge at site 214 as wild-type RaPrPC carries (Fig. 2). These observations suggest that the charge alteration at the substituted site is not a precondition for the global electrostatic potential shift. The change of surface charge distribution resulting from single-point mutation could be more significant than expected.
It has been demonstrated that both the tertiary structure and internal motions of a protein determine the protein function [47]. Therefore, evaluation of the differences in dynamics between wild-type PrPC and its mutants is important to address the molecular mechanism of prion conversions. Both molecular dynamics simulations and NMR relaxation measurement experiments have suggested that backbone flexibility of PrPC is associated with the conformational conversion [34], [48], [49], although some studies show that mutations do not lead to significant changes in backbone dynamics of proteins [40], [50]. Our results demonstrate that the I214V substitution leads to decreased J(0) and S2 values (Fig. 5 and 6), implicating an increase in backbone flexibility on the ps-ns time scale, similar to the S173N substitution [30]. Usually, fragments with high flexibility are indicative of low energy barrier to structural rearrangement. In addition, H186 in helix 2 displays a fairly small S2 value and a much lower J(0) value after the I214V mutation (Fig. 5 and 6), implying prominent internal motions which potentially make a significant contribution to destabilization of helix 2. Thus, it could be expected that the conformational change is somewhat easier to occur for the I214V mutant than for the wild-type. On the other hand, dynamics could modulate binding energy to maintain the moderate affinity required for biological function [51], [52]. The increased dynamics might enhance or inhibit the binding ability of PrPC with other chaperones, which would exert profound influence on prion conversions.
We have assessed the change of thermodynamic stability of RaPrPC after the I214V substitution. The coefficient m is implicated to be approximately proportional to the difference in solvent-accessible surface areas between the folded and unfolded states. Hydrophobic interactions stabilizing the protein native state can be interpreted, to a large extent, in terms of the change in accessible surface area upon protein folding [53]–[56]. The distinctly changed m value is indicative of differences in intramolecular hydrophobic interactions between the wild-type and the mutant (Table 3). Furthermore, similar to the S173 substitution [30], the I214V substitution leads to marked decreases in both 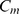 and
and 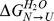 values during urea-induced transitions (Fig. 3 and Table 3), implying that the I214V mutant adopts a less stable conformation compared with the wild-type. Our experimental observations are supported by both molecular dynamics simulations reported recently [57] and the altered hydrogen bond network described above (Table 2). It has also been shown that internal dynamics especially the ps-ns motions has an intimate correlation with thermodynamics [58], [59]. The increased backbone flexibility, indicated by the decreased J(0) and S2 values (Fig. 5 and 6), implying the conformational instability of the I214V mutant. Therefore, one could expect that PrPC with low structural stability would be inclined to undergo conformational conversion.
values during urea-induced transitions (Fig. 3 and Table 3), implying that the I214V mutant adopts a less stable conformation compared with the wild-type. Our experimental observations are supported by both molecular dynamics simulations reported recently [57] and the altered hydrogen bond network described above (Table 2). It has also been shown that internal dynamics especially the ps-ns motions has an intimate correlation with thermodynamics [58], [59]. The increased backbone flexibility, indicated by the decreased J(0) and S2 values (Fig. 5 and 6), implying the conformational instability of the I214V mutant. Therefore, one could expect that PrPC with low structural stability would be inclined to undergo conformational conversion.
In conclusion, the I214V substitution results in distinct structural changes for RaPrPC. Both the unique surface charge distribution and the intrinsic backbone are likely to make significant contributions to species barriers for the transmission of TSEs. Our results provide valuable hints for understanding the inability of RaPrPC to form PrPSc, and shed light on the detailed molecular mechanism of the conformation conversion for prion proteins.
Materials and Methods
The I214V mutants of RaPrPC(91–228) and RaPrPC(121–228) were constructed using site-directed mutagenesis PCR. Plasmid construction, protein expression and purification, NMR spectroscopy, structure calculation, 15N relaxation measurements, dynamics analysis and CD experiments were conducted as described previously [30], [60]. Nearly complete backbone and side-chain resonance assignments for the I214V mutant of RaPrPC(91–228) have been obtained. The chemical shift data are available at the Biological Magnetic Resonance Data Bank with accession number 16616. The atomic coordinates have been deposited into the Protein Data Bank with PDB ID 2JOM.
Acknowledgments
We would like to thank Prof. Daiwen Yang (National University of Singapore) for helpful discussions on dynamics analysis and Prof. Gengfu Xiao (Wuhan University) for providing RaPrPC gene.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Natural Science Foundation of China (Nos. 30730026, 30570352) and National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program,” China (No. 2009ZX09301-001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, et al. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 3.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, Wang X, Yuan CG, Ma J. Generating a Prion with Bacterially Expressed Recombinant Prion Protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, et al. NMR structure of the mouse prion protein domain PrP(121–231). Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 6.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, et al. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc Natl Acad Sci U S A. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci U S A. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riek R, Wider G, Billeter M, Hornemann S, Glockshuber R, et al. Prion protein NMR structure and familial human spongiform encephalopathies. Proc Natl Acad Sci U S A. 1998;95:11667–11672. doi: 10.1073/pnas.95.20.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Farr-Jones S, Ulyanov NB, Llinas M, Marqusee S, et al. Solution structure of Syrian hamster prion protein rPrP(90–231). Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 10.Lopez Garcia F, Zahn R, Riek R, Wuthrich K. NMR structure of the bovine prion protein. Proc Natl Acad Sci U S A. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zahn R, Liu A, Luhrs T, Riek R, von Schroetter C, et al. NMR solution structure of the human prion protein. Proc Natl Acad Sci U S A. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornemann S, Schorn C, Wuthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wuthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci U S A. 2005;102:646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, et al. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A. 2005;102:640–645. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christen B, Perez DR, Hornemann S, Wuthrich K. NMR structure of the bank vole prion protein at 20 degrees C contains a structured loop of residues 165–171. J Mol Biol. 2008;383:306–312. doi: 10.1016/j.jmb.2008.08.045. [DOI] [PubMed] [Google Scholar]
- 16.Christen B, Hornemann S, Damberger FF, Wuthrich K. Prion protein NMR structure from tammar wallaby (Macropus eugenii) shows that the beta2-alpha2 loop is modulated by long-range sequence effects. J Mol Biol. 2009;389:833–845. doi: 10.1016/j.jmb.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 17.Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, et al. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci U S A. 2002;99:3563–3568. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci U S A. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobb NJ, Sonnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci U S A. 2007;104:18946–18951. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, Wintrode PL, Surewicz WK. Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc Natl Acad Sci U S A. 2007;104:1510–1515. doi: 10.1073/pnas.0608447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh P, Simonetti K, Sharpe S. Core structure of amyloid fibrils formed by residues 106–126 of the human prion protein. Structure. 2009;17:417–426. doi: 10.1016/j.str.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 22.Prusiner SB. Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 23.Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 24.Hunter N. PrP genetics in sheep and the applications for scrapie and BSE. Trends Microbiol. 1997;5:331–334. doi: 10.1016/s0966-842x(97)01081-0. [DOI] [PubMed] [Google Scholar]
- 25.Westaway D, Zuliani V, Cooper CM, Da Costa M, Neuman S, et al. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 26.Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, et al. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 27.Clouscard C, Beaudry P, Elsen JM, Milan D, Dussaucy M, et al. Different allelic effects of the codons 136 and 171 of the prion protein gene in sheep with natural scrapie. J Gen Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs CJ, Jr, Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. 1973;182:67–68. doi: 10.1126/science.182.4107.67. [DOI] [PubMed] [Google Scholar]
- 29.Vorberg I, Groschup MH, Pfaff E, Priola SA. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J Virol. 2003;77:2003–2009. doi: 10.1128/JVI.77.3.2003-2009.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen Y, Li J, Yao W, Xiong M, Hong J, et al. Unique structural characteristics of the rabbit prion protein. J Biol Chem. 2010. (in press) [DOI] [PMC free article] [PubMed]
- 31.Peng JW, Wagner G. Mapping of the spectral densities of N-H bond motions in eglin c using heteronuclear relaxation experiments. Biochemistry. 1992;31:8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- 32.Peng JW, Wagner G. Mapping of Spectral Density Functions Using Heteronuclear NMR Relaxation Measurements. J Magn Reson. 1992;98:308–332. [Google Scholar]
- 33.Farrow NA, Zhang O, Szabo A, Torchia DA, Kay LE. Spectral density function mapping using 15N relaxation data exclusively. J Biomol NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- 34.O'Sullivan DB, Jones CE, Abdelraheim SR, Brazier MW, Toms H, et al. Dynamics of a truncated prion protein, PrP(113–231), from (15)N NMR relaxation: order parameters calculated and slow conformational fluctuations localized to a distinct region. Protein Sci. 2009;18:410–423. doi: 10.1002/pro.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv Protein Chem. 2002;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 36.Lefevre JF, Dayie KT, Peng JW, Wagner G. Internal mobility in the partially folded DNA binding and dimerization domains of GAL4: NMR analysis of the N-H spectral density functions. Biochemistry. 1996;35:2674–2686. doi: 10.1021/bi9526802. [DOI] [PubMed] [Google Scholar]
- 37.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 38.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 39.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, et al. Deviations from the simple two-parameter model-free approach to the interpretation of nitrogen-15 nuclear magnetic relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 40.Kirby L, Agarwal S, Graham JF, Goldmann W, Gill AC. Inverse correlation of thermal lability and conversion efficiency for five prion protein polymorphic variants. Biochemistry. 2010;49:1448–1459. doi: 10.1021/bi901855z. [DOI] [PubMed] [Google Scholar]
- 41.Calzolai L, Lysek DA, Guntert P, von Schroetter C, Riek R, et al. NMR structures of three single-residue variants of the human prion protein. Proc Natl Acad Sci U S A. 2000;97:8340–8345. doi: 10.1073/pnas.97.15.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Swietnicki W, Zagorski MG, Surewicz WK, Sonnichsen FD. Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases. J Biol Chem. 2000;275:33650–33654. doi: 10.1074/jbc.C000483200. [DOI] [PubMed] [Google Scholar]
- 43.Cordeiro Y, Machado F, Juliano L, Juliano MA, Brentani RR, et al. DNA converts cellular prion protein into the beta-sheet conformation and inhibits prion peptide aggregation. J Biol Chem. 2001;276:49400–49409. doi: 10.1074/jbc.M106707200. [DOI] [PubMed] [Google Scholar]
- 44.Lima LM, Cordeiro Y, Tinoco LW, Marques AF, Oliveira CL, et al. Structural insights into the interaction between prion protein and nucleic acid. Biochemistry. 2006;45:9180–9187. doi: 10.1021/bi060532d. [DOI] [PubMed] [Google Scholar]
- 45.Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 46.Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, et al. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci U S A. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kay LE. Protein dynamics from NMR. Nat Struct Biol. 1998;5(Suppl):513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]
- 48.Viles JH, Donne D, Kroon G, Prusiner SB, Cohen FE, et al. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry. 2001;40:2743–2753. doi: 10.1021/bi002898a. [DOI] [PubMed] [Google Scholar]
- 49.Blinov N, Berjanskii M, Wishart DS, Stepanova M. Structural domains and main-chain flexibility in prion proteins. Biochemistry. 2009;48:1488–1497. doi: 10.1021/bi802043h. [DOI] [PubMed] [Google Scholar]
- 50.Bae SH, Legname G, Serban A, Prusiner SB, Wright PE, et al. Prion proteins with pathogenic and protective mutations show similar structure and dynamics. Biochemistry. 2009;48:8120–8128. doi: 10.1021/bi900923b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kay LE, Muhandiram DR, Wolf G, Shoelson SE, Forman-Kay JD. Correlation between binding and dynamics at SH2 domain interfaces. Nat Struct Biol. 1998;5:156–163. doi: 10.1038/nsb0298-156. [DOI] [PubMed] [Google Scholar]
- 52.Tzeng SR, Kalodimos CG. Dynamic activation of an allosteric regulatory protein. Nature. 2009;462:368–372. doi: 10.1038/nature08560. [DOI] [PubMed] [Google Scholar]
- 53.Privalov PL, Makhatadze GI. Heat capacity of proteins. II. Partial molar heat capacity of the unfolded polypeptide chain of proteins: protein unfolding effects. J Mol Biol. 1990;213:385–391. doi: 10.1016/S0022-2836(05)80198-6. [DOI] [PubMed] [Google Scholar]
- 54.Alonso DO, Dill KA. Solvent denaturation and stabilization of globular proteins. Biochemistry. 1991;30:5974–5985. doi: 10.1021/bi00238a023. [DOI] [PubMed] [Google Scholar]
- 55.Dill KA, Shortle D. Denatured states of proteins. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 56.Privalov PL, Makhatadze GI. Contribution of hydration and non-covalent interactions to the heat capacity effect on protein unfolding. J Mol Biol. 1992;224:715–723. doi: 10.1016/0022-2836(92)90555-x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J. Studies on the structural stability of rabbit prion probed by molecular dynamics simulations of its wild-type and mutants. J Theor Biol. 2010;264:119–122. doi: 10.1016/j.jtbi.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 58.Akke M, Bruschweiler R, Palmer AG., 3rd NMR Order Parameters and Free Energy: An Analytical Approach and Its Application to Cooperative Ca2+ Binding by Calbindin D9k. J Am Chem Soc. 1993;115:9832–9833. [Google Scholar]
- 59.Yang D, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 60.Yin SM, Zheng Y, Tien P. On-column purification and refolding of recombinant bovine prion protein: using its octarepeat sequences as a natural affinity tag. Protein Expr Purif. 2003;32:104–109. doi: 10.1016/S1046-5928(03)00195-5. [DOI] [PubMed] [Google Scholar]
- 61.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55, 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 62.Spyracopoulos L. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J Biomol NMR. 2006;36:215–224. doi: 10.1007/s10858-006-9083-0. [DOI] [PubMed] [Google Scholar]
- 63.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 64.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]