Abstract
The increasing prevalence of antibiotic-resistant pathogens reinforces the need for structures of antibiotic-ribosome complexes that are accurate enough to enable the rational design of novel ribosome-targeting therapeutics. Structures of many antibiotics in complex with both archaeal and eubacterial ribosomes have been determined, yet discrepancies between several of these models have raised the question of whether these differences arise from species-specific variations or from experimental problems. Our structure of chloramphenicol in complex with the 70S ribosome from Thermus thermophilus suggests a model for chloramphenicol bound to the large subunit of the bacterial ribosome that is radically different from the prevailing model. Further, our structures of the macrolide antibiotics erythromycin and azithromycin in complex with a bacterial ribosome are indistinguishable from those determined of complexes with the 50S subunit of Haloarcula marismortui, but differ significantly from the models that have been published for 50S subunit complexes of the eubacterium Deinococcus radiodurans. Our structure of the antibiotic telithromycin bound to the T. thermophilus ribosome reveals a lactone ring with a conformation similar to that observed in the H. marismortui and D. radiodurans complexes. However, the alkyl-aryl moiety is oriented differently in all three organisms, and the contacts observed with the T. thermophilus ribosome are consistent with biochemical studies performed on the Escherichia coli ribosome. Thus, our results support a mode of macrolide binding that is largely conserved across species, suggesting that the quality and interpretation of electron density, rather than species specificity, may be responsible for many of the discrepancies between the models.
Keywords: crystal structure, structure-based drug design
Antibiotics that target the translational machinery of bacterial cells are potent inhibitors of prokaryotic pathogens (1), but increasing rates of antibiotic resistance among bacteria often dampen the effectiveness of common, clinically used antibiotics (2–5). Recent work has shown that structure-based drug design exhibits the potential to overcome this problem by chemically linking together pairs of known classes of ribosomal inhibitors to generate novel antibiotics (6–8). The wealth of ribosomal antibiotics and corresponding resistance mutations suggests that many additional chemical derivatives of existing antibiotics might be created and could be useful in dealing with resistance, but conflicting crystallographically determined models for how some of these antibiotics are bound and oriented could be an impediment to the design of new hybrid molecules. For example, structures of several macrolide and ketolide antibiotics in complex with the large ribosomal subunits of Deinococcus radiodurans and Haloarcula marismortui (9–12) have shown significant differences in the orientations and conformations of the drugs, in particular with respect to the lactone rings and the contacts they make with the ribosome. Because it has been suggested that the discrepancies between these models result from species-specific differences in the ribosomal RNA (13), we chose to address these issues by determining the structures of the 70S ribosome from the eubacterium Thermus thermophilus in complex with the antibiotics erythromycin, azithromycin, and telithromycin. In addition, we decided to revisit the structure of chloramphenicol bound to the ribosome for several reasons: (i) only one such structure is available, and it originated from the same study that yielded the structure of a bacterial ribosome in complex with erythromycin that was inconsistent with later structural work at higher resolution; (ii) in contrast with the distinctive lactone ring of macrolides, chloramphenicol lacks obvious structural features that would facilitate its placement into a medium resolution electron density map; (iii) a number of minor inconsistencies between the sole available structural model and biochemical data warrant additional validation of the former (14–17). The structures of the erythromycin, azithromycin, telithromycin, and chloramphenicol complexes with the T. thermophilus 70S ribosome reported here are all inconsistent with the earlier published structural studies with the D. radiodurans large subunit, but the macrolide complexes are in complete agreement with the structural conclusions from the published H. marismortui large subunit complexes.
Results
Datasets for the chloramphenicol, erythromycin, and azithromycin ribosome complexes were collected from single crystals, whereas the data from the complex with telithromycin was obtained from three separate crystals. All crystals belonged to the primitive orthorhombic space group P212121 and diffracted to 3.1–3.3 Å. Statistics for the data can be seen in Table S1.
Chloramphenicol.
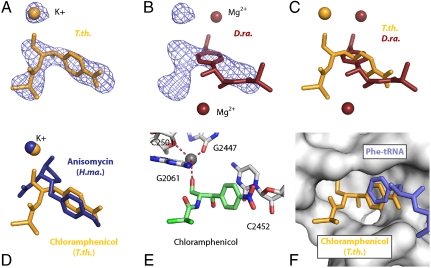
Chloramphenicol binds directly to the A-site crevice on the 50S ribosomal subunit, occupying the same location as the aminoacyl moiety of an A-site tRNA (Fig. 1) (18, 19). The placement of this drug in the immediate vicinity of the incoming A-site tRNA is consistent with biochemical experiments (20, 21). The location and orientation of chloramphenicol that we observe in our complex is completely different from the model that has been published for the D. radiodurans 50S subunit complex (11) (Fig. 1 A–C). Superimposition of the ribosomal RNA from the two structures shows that the two chloramphenicol models are related by a rotation of approximately 180° and that the planes of the benzyl rings are orthogonal to one another. Additionally, the ion(s) anchoring the molecule to the ribosome differ in their location, number, and identity; the D. radiodurans model posits two putative and as yet unproven magnesium ions holding the drug in place, whereas our data implicate a single potassium ion whose location in the peptidyl transferase center (PTC) is consistent with previous biochemical and crystallographic experiments (15, 22–24).
Fig. 1.
Interaction between chloramphenicol and the ribosome. (A) An unbiased 3.2-Å-resolution Fo-Fc difference electron density map contoured at +3σ calculated using amplitudes from T.th. 70S ribosome crystals soaked with 500 μM chloramphenicol. The chloramphenicol moiety is shown in orange, along with a potassium ion mediating the interaction between the drug and the ribosome. (B) The same electron density as shown in A, with the chloramphenicol moiety and two magnesium ions modeled in the D.ra. 50S structure (11) shown in red. The T.th. 70S ribosome and the D.ra. 50S subunit structures were superimposed using equivalent phosphate atoms in the 23S rRNA and the program Lsqman (54). The superimposed structures had an rmsd of 1.21 Å for 2,800 atoms. (C) Overlay of the chloramphenicol molecules modeled in the T.th. 70S (orange) and D.ra. 50S (red) complex structures. Both the model coordinates and the view are the same as in A and B. (D) Overlay of the chloramphenicol molecule modeled in the T.th. 70S complex (orange) and the anisomycin molecule modeled in H.ma. 50S complex (blue). The superimposed structures had an rmsd of 1.53 Å for 2,800 atoms. (E) Interactions between chloramphenicol and the T.th. 70S ribosome. The drug is shown as green sticks, and key interacting residues in the ribosome are shown in white. The potassium ion is displayed as a gray sphere. (F) A surface representation of the binding pocket for chloramphenicol in the T.th. 70S ribosome, showing surface complementarity between the drug (orange) and the A-site crevice in the ribosome as well as the aminoacyl group (blue) of bound phe-tRNA (37).
An overlay of the small molecule crystal structure of chloramphenicol and the model presented here shows good agreement (Fig. S1E). A similar alignment between the model of chloramphenicol bound to the D. radiodurans large subunit and the small molecule structure reveals major differences, particularly in the orientation of the nitro group that is nearly orthogonal relative to the plane of the benzene ring in the D. radiodurans model (Fig. S1F). In its lowest-energy orientation, the para-nitro group lies parallel to the conjugated pi system of the benzyl moiety (25, 26). This low-energy orientation, which is seen in both the T. thermophilus and small molecule crystal structures, is not reported in the D. radiodurans model.
Chlolramphenicol contacts the ribosome through a single pi-stacking interaction between its para-nitrobenzyl ring and the base of C2452 of the ribosomal 23S RNA, as well as through an interaction between a ribosome-bound potassium and the methylene hydroxyl group of chloramphenicol, which anchors the middle of the molecule. The potassium ion is coordinated by bases G2447, C2501, and G2061, with the lone pair of chloramphenicol’s hydroxyl group completing the tetrahedral coordination of the ion (Fig. 1E). The dependence of chloramphenicol binding on potassium has been demonstrated biochemically (15, 22, 27), and the position of the potassium ion has been verified crystallographically (24), suggesting that the interaction observed here is physiologically relevant.
Mutations of bases G2447, A2451, C2452, A2503, and U2504 have been shown to confer chloramphenicol resistance in mouse mitochondrial ribosomes (16, 28). These bases are in direct contact with the antibiotic or the coordinated potassium ion in our structure (Fig. S1C). Also, chemical protection studies have shown that A2451 and G2505 are occluded when chloramphenicol is bound, which is consistent with the structure we observe (29, 30) (Fig. S1D).
The acetylation of chloramphenicol at the methylene hydroxyl position renders the drug ineffective against the bacterial ribosome, and the gene for chloramphenicol acetyl transferase is a widely used tool for the selection of cells in the presence of chloramphenicol (14, 31, 32). Our structure suggests that such acetylation would prevent chloramphenicol binding by preventing the methylene hydroxyl group from coordinating the potassium ion and by sterically inhibiting the drug from fitting into its binding pocket (Fig. S1B).
The binding of anisomycin to the H. marismortui ribosome mirrors that of chloramphenicol to the eubacterial ribosome (23) (Fig. 1D). In particular, the aromatic moieties of both antibiotics stack on the base of C2452 (Escherichia coli) and interact with a nearby potassium coordinated by G2447, C2501, and G2061. An overlay of the rRNA from the complex between anisomycin and the H. marismortui 50S with the eubacterial ribosome shows how a clash between anisomycin and U2504 prevents its binding, thereby explaining its specificity for eukaryotes. Moreover, the reorientation of U2504 and G2505 observed in archaea effectively widens the chloramphenicol binding pocket, suggesting that this rearrangement may be significant for the drug’s specificity toward the eubacterial ribosome (Fig. S1A).
Erythromycin.
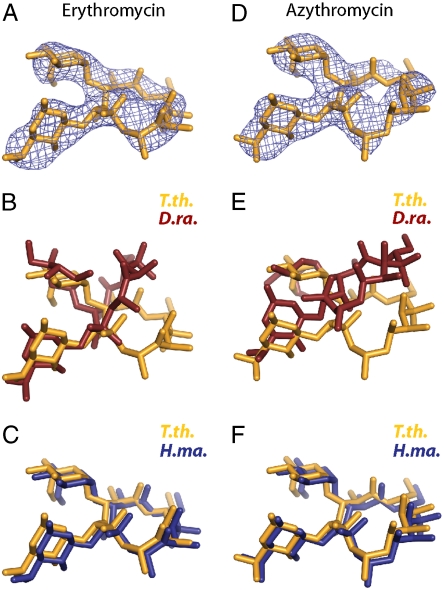
Macrolide antibiotics bound to the eubacterial ribosome occupy a position in the ribosomal exit tunnel near the PTC. Several structures of the macrolide erythromycin bound to the large subunit of the ribosome have been determined, including the H. marismortui and D. radiodurans large subunits (10, 11, 33). The structures of erythromycin and azithromycin bound to the G2099A mutant H. marismortui 50S are nearly identical to the structures of the complexes with the T. thermophilus 70S ribosome (Fig. 2 A and C). In contrast, the published D. radiodurans models of the 50S subunit complexes differ with regard to the positions of the lactone rings (Fig. 2 B). An undeposited model of the D. radiodurans complex also exists in which the antibiotic most likely occupies a position very similar to the one observed in this work (33). However, a detailed comparison cannot be undertaken in the absence of published coordinates.
Fig. 2.
Erythromycin and azithromycin, (A and D) Unbiased 3.2-Å Fo-Fc difference electron density maps contoured at +3σ calculated using amplitudes from T.th. 70S ribosome crystals soaked with 500 μM erythromycin (A) or azithromycin (D). (B, C, E, and F) Overlay of erythromycin or azithromycin molecules from the T.th. 70S ribosome (orange; B, C, E, and F), D.ra. 50S subunit (red; B and E) and H.ma. 50S subunit (blue; C and F) complex structures. The superimposed structures had an rmsd between 1.23 Å and 1.55 Å for 2,800 atoms.
The interactions between erythromycin and the surface of the exit tunnel of the T. thermophilus ribosome are identical to those seen in the G2099A (A2058 in E. coli) mutant large ribosomal subunit from H. marismortui. In both structures, three axial methyl groups belonging to the lactone ring of erythromycin rest on a surface formed by the bases of U2611, A2058, and A2059 (Fig. S2A). These interactions, along with a hydrogen bond between the 2′ OH group of the desosamine sugar of erythromycin and the N1 atom of A2058 appear to stabilize erythromycin in both the H. marismortui and T. thermophilus complex structures. As in the structure of the H. marismortui complex, the position of the lactone ring explains why an A to G mutation at position A2058 renders the ribosome insensitive to erythromycin. Even after desolvation, the N2 that is present in G (but absent in A) sterically prevents the tight packing of the lactone ring on the hydrophobic surface upon which the antibiotic rests (10).
A2062 has been shown to play a pivotal role in erythromycin-dependent ribosome stalling (34). The position of this base when erythromycin is bound differs from its position on the T. thermophilus ribosome in the absence of the drug (35–37) and also differs from that seen in erythromycin complexes of both D. radiodurans and H. marismortui large subunits, suggesting that species specificity (or perhaps buffer composition) may modulate the orientation of this base in response to erythromycin binding. When complexed with the T. thermophilus ribosome, the desosamine sugar of erythromycin induces a rotation of A2062 by approximately 160° around the glycosidic bond and a slight translation (∼1 Å) of the ribose moiety. This movement pushes the base toward the PTC, placing the base within van der Waals contact of the amino group of P-site tRNA (Fig. S2B).
Azithromycin.
The binding of azithromycin closely mirrors that of erythromycin, both in this work and in the published structures of the H. marismortui complexes (23, 38) (Fig. 2 D and F). In contrast, the structure of the D. radiodurans complex with azithromycin differs from the T. thermophilus complex both in the placement of the lactone ring (11) (Fig. 2E) and in the number of azithromycin molecules bound (11, 33). In the T. thermophilus complex, only one molecule is observed, and its orientation is nearly identical to that observed in the H. marismortui mutant 50S subunit. Presumably the T. thermophilus complex with azithromycin is also similar to the undeposited structure of the D. radiodurans large subunit complexed with azithromycin (33). Additionally, biochemical experiments indicate that only one azithromycin molecule is bound to the E. coli ribosome (39).
Telithromycin.
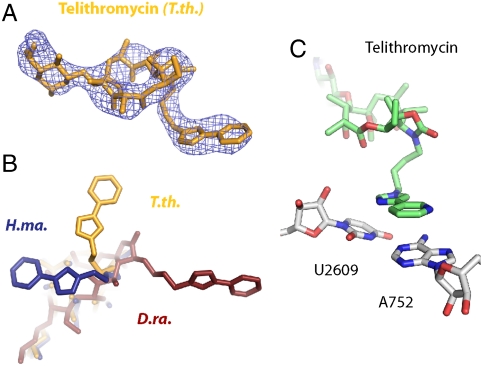
The lactone ring of telithromycin in its complex with the T. thermophilus ribosome occupies a position nearly identical to what has been observed in the H. marismortui mutant 50S complex, but somewhat different from that seen in the D. radiodurans complex (9, 10) (Fig. 3). The interactions anchoring the main ring are identical to those stabilizing erythromycin and azithromycin, with the bases of U2611, A2058, and A2059 providing a hydrophobic surface upon which the ring rests, whereas the base of A2058 hydrogen bonds with the deosamine sugar. As in the complexes with H. marismortui and D. radiodurans large subunits, the alkyl-aryl group tethered to the lactone ring of telithromycin stacks with nearby ribosomal bases. However, the position of the alkyl-aryl moiety in the T. thermophilus ribosome complex and the identities of the bases with which it interacts are different from both the H. marismortui and D. radiodurans large subunit complexes with telithromycin (Fig. 3B). The structure presented here shows the alkyl-aryl group stacking with the bases of U2609 and A752, consistent with biochemical experiments showing that these bases are chemically protected by telithromycin in the E. coli ribosome (39–41) (Fig. 3C).
Fig. 3.
Binding of telithromycin to the ribosome. (A) An unbiased 3.2-Å-resolution Fo-Fc difference electron density map contoured at +3σ calculated using amplitudes from T.th. 70S ribosome crystals soaked with 500 μM telithromycin. The drug is shown as orange sticks. (B) Overlay of the telithromycin moieties in the T.th. 70S (orange), D.ra. 50S (red), and H.ma. 50S (blue) complex structures. The superimposed H.ma. 50S and D.ra. 50S rRNA structures had an rmsd of 1.54 Å and 1.23 Å atoms for 2,800 atoms relative to the T.th. 70S structure, respectively. (C) Positioning of the alkyl-aryl moiety of telithromycin (green) relative to U2609 and A752 (E.coli numbering) from T.th. 23S rRNA (white).
Discussion
In order to establish the source of the differences between the published crystallographic models of several antibiotics bound to eubacterial (9, 11, 12) versus archaeal ribosomes (10), we have reexamined the structures of several common, clinically relevant antibiotics bound to the 70S ribosome of the eubacterium T. thermophilus. The structures we have determined differ from several earlier models that were based on crystallographic studies of antibiotic complexes with the D. radiodurans large ribosomal subunit (9, 11, 12). In particular, we find that chloramphenicol occupies a similar but not identical location and a completely different orientation from the model that was derived from the studies of the D. radiodurans large subunit complex. The interactions between the ribosomal RNA and the drug that we report here are entirely different from the structural model that has been published (11); in contrast, they are in good agreement with a number of biochemical and genetic experiments (16, 22, 29). Chemical protection studies carried out on the E. coli ribosome in the presence of chloramphenicol identified protected bases that we observe to be in direct contact with the drug (29). Furthermore, mutations in mitochondrial rRNA that render the bacteria resistant to chloramphenicol are located either in the immediate vicinity of chloramphenicol or affect the coordination of the potassium ion that interacts with the bound drug (16, 28, 42). For example, mutation G2447A replaces the oxygen atom at position six of the guanosine base with a nitrogen atom, thereby disrupting the coordination of the potassium ion that binds the methylene hydroxyl group of chloramphenicol (Fig. 1). Several other mutations change the shape or interacting surface of the chloramphenicol binding site (SI Text).
In addition to mutational studies, biochemical experiments have shown that chloramphenicol binding depends on potassium, an observation that agrees with the potassium ion we observe interacting with the drug (15, 22, 27). Chloramphenicol acetyltransferase, an enzyme that confers chloramphenicol resistance in bacteria through the acetylation of hydroxyl groups on chloramphenicol, provides additional support for the model presented here, because acetylation of either hydroxyl would fatally disrupt chloramphenicol binding as we observe it (31, 32). The primary site of acetylation (at the methylene hydroxyl position) would be particularly deleterious because it would both disrupt the coordination of the potassium ion we observe bound to chloramphenicol and sterically hinder the drug from fitting into its binding pocket (Fig. S1).
Interestingly, chemically modified derivatives of chloramphenicol whose design was based on the D. radiodurans model have met with mixed success (43). The antibiotic was modified by replacement of the dichloroacetyl group with variable-length linkers attached to dicytosine moieties in order to make interactions that mimic the Watson–Crick base pairs between the CCdA-p-puromycin tRNA analog and residues G2252 and G2251 of the 23S RNA (44). If the structure of the D. radiodurans complex in which the dichloroacetyl tail of chloramphenicol points away from the exit tunnel and toward the CCA end of an incoming aminoacylated tRNA were correct, this modification could be expected to work. The failure of this chloramphenicol derivative to bind the bacterial ribosome suggests that the placement of the drug in the D. radiodurans model may be consistent with our conclusion that this model is incorrect. The structure we present shows that the dichloroacetyl moiety faces the opposite direction, implying that substitution at this position on the drug will not result in a productive CCA-like interaction with the ribosomal A site.
When considered together, these results underscore the critical role structure-based design can play in a drug discovery program. Before the adjacent locations of the ribosomal binding sites of macrolides (e.g., azithromycin) and phenyl propanoids (e.g., chloramphenicol) were revealed through crystallography, it appeared that the molecular target of chloramphenicol could not accommodate augmentation on the dichloroacetyl end of the molecule. Likewise, it seemed that little productive augmentation could be done on the exocyclic amino group of the macrolide’s desosamine ring (45). With the structures presented here, it is clear that—although the intervening space is tight—these two independent binding sites in fact reveal completely unique opportunities for an extended binding site. Indeed, as proof-of-concept, the discovery group at Rib-X Pharmaceuticals designed and prepared a chimera (RX-2102, Fig. S4) of a related phenyl propanoid (florfenicol) and a macrolide antibiotic (azithromycin). The binding of RX-2102 to the wild-type H. marismortui 50S was confirmed crystallographically (PDB ID code 3OW2), and its binding orientation aligns closely with our structures of azithromycin and chloramphenicol complexes (Fig. S3). Importantly, this chimera established that the clinical utility of the macrolide class could be restored through structure-based design: The antibacterial activity of RX-2102 against the most common macrolide-resistant streptococci was excellent, as shown in Table S2. This approach has been further validated and exemplified in Rib-X’s US Patent 7,091,196. Given the success of hybrid antibiotics in binding to ribosomes from drug-resistant species, the fusion of chloramphenicol to other ribosome-targeting drugs may yield effective therapeutics for dealing with antibiotic resistance (7, 46).
Comparison of anisomycin bound to the A site of the H. marismortui 50S ribosomal subunit and chloramphenicol in complex with the T. thermophilus ribosome reveals a similar mode of binding between the two drugs in spite of the fact that these molecules target ribosomes from different kingdoms of life (23). In both cases, the drugs stack with base C2452 in the A-site cleft and coordinate an adjacent potassium ion. Steric considerations explain why anisomycin is specific for the eukaryotic ribosome; U2504 blocks part of the region that is required for anisomycin to coordinate the A-site potassium ion with which it associates in H. marismortui. Conversely, the specificity of chloramphenicol for eubacteria may be explained by a relaxation of the A-site crevice in the archaeal ribosome as bases U2504 and G2505 widen the chloramphenicol binding pocket, disrupting the steric complementarity between the drug and the 23S RNA.
The structures of macrolide antibiotic complexes observed with the mutant H. marismortui large subunit are in complete agreement with the structures presented here. Critically, these structures all share a common set of hydrophobic interactions between the lactone ring of the macrolide and the hydrophobic surface of the ribosomal exit tunnel, comprised of bases U2611, A2058, and A2059 (10). The importance of this interaction is demonstrated by the fact that mutating A2058 to G or methylating A2058 (both of which alter the surface of the ribosomal exit tunnel, preventing the lactone ring from packing tightly against the wall of the exit tunnel) leads to erythromycin resistance (10, 47, 48). The deposited structures of erythromycin and azithromycin bound to the D. radiodurans large subunit lack most of these interactions (11, 12); the lactone rings in both of these complex structures are bent away from the surface of the exit tunnel. A refined redetermination of both these drugs bound to the D. radiodurans large subunit has been published in a review and appears to place them in a position similar to that observed in this work. However, a direct comparison with the present structure is not possible because the refined coordinates have not been deposited into the Protein Data Bank (33). Additionally, only one molecule of bound azithromycin is observed per ribosome in the T. thermophilus complex, unlike both the deposited and undeposited D. radiodurans structures (12, 33).
Although the lactone ring of telithromycin binds in relatively the same position and orientation in the D. radiodurans, H. marismortui, and T. thermophilus complexes, differences among the structures arise in the placement of the alkyl-aryl moiety that is tethered to the drug’s main ring. All three structures show varying positions for this group, though they share a common element in that each model features the alkyl-aryl substituent making pi-stacking interactions with a nearby base of the ribosomal RNA. The T. thermophilus complex structure presented here proposes that the alkyl-aryl motif interacts with the bases of U2609 and A752. This position is in perfect agreement with chemical protection experiments carried out in E. coli showing that both of these bases are shielded when telithromycin is bound (39–41).
Although species specificity no doubt modulates the interactions made by many antibiotics with the ribosome, this work suggests that macrolides bind identically to the G2099A mutant H. marismortui large ribosomal subunit and to the T. thermophilus 70S ribosome. The structure of telithromycin in complex with the T. thermophilus ribosome again corroborates this idea, showing that the lactone ring is oriented in a similar fashion among erythromycin, azithromycin, and telithromycin in both T. thermophilus and H. marismortui ribosome complexes (10). We conclude that the dramatic differences in the models of erythromycin and azithromycin bound to the D. radiodurans 50S subunit that were initially reported (11, 12) are due to an erroneous interpretation of the electron density maps, possibly as a result of low resolution. Finally, the structure of the T. thermophilus ribosome in complex with chloramphenicol disagrees dramatically with the existing model of chloramphenicol bound to the D. radiodurans large subunit (which we suggest is incorrect for the same reason), but shows a similar mode of binding to the drug anisomycin with the archaeal ribosome from H. marismortui (23).
Materials and Methods
70S ribosomes from T. thermophilus HB8 were prepared as described previously (35, 49). Crystallization experiments were also carried out under the same conditions, but sitting drop vapor diffusion was used in place of hanging drop. Crystals were cryoprotected by gradually increasing the concentration of 2-methyl-2,4-pentanediol while all other buffer components were held constant. Antibiotics were soaked into the cryo stabilized crystals overnight at a concentration of 500 μM, after which the crystals were flash frozen. Data were collected at Advanced Photon Source (APS) on beamlines 24-ID-C and 24-ID-E as well as at Brookhaven National Laboratory on beamlines X29 and X25. The collected data were processed using the program XDS (50), and the structures were solved by molecular replacement with PHASER (51) using the empty T. thermophilus 70S (36) as a search model. Structures were refined with the PHENIX (52) package and restraints for the antibiotic molecules were generated from high-resolution crystal structures of the drugs using the PRODRG2 server (53).
Supplementary Material
Acknowledgments.
We thank David Keller for his help with crystal freezing and cryoprotection and the staff at the APS (beamline 24-ID) and National Synchrotron Light Source (beamlines X-29 and X-25) for the use of their facilities and for technical assistance with data collection. We also thank the team at Rib-X Pharmaceuticals for sharing their insights on structure-based design of next-generation antibiotics, the coordinates of the structure of RX-2102 in complex with the H. marismortui 50S subunit, and the antibacterial data for this proof-of-concept chimera. This work was supported by NIH Grant GM022778 awarded to T.A.S.
Footnotes
Conflict of interest statement: Thomas A. Steitz is a scientific advisor for Rib-X Pharmaceuticals, a product-driven small molecule drug discovery and development company focused on the structure-based design of unique classes of antibiotics.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates for the various ribosome-antibiotic complexes have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3OGE/3OGY/3OH5/3OH7 (chloramphenicol), 3OHC/3OHD/3OHJ/3OHK (erythromycin), 3OHY/3OHZ/3OI0/3OI1 (azithromycin), and 3OI2/3OI3/3OI4/3OI5 (telithromycin)].
See Commentary on page 17065.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008685107/-/DCSupplemental.
References
- 1.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 2.Ardanuy C, et al. Molecular characterization of macrolide- and multidrug-resistant Streptococcus pyogenes isolated from adult patients in Barcelona, Spain (1993–2008) J Antimicrob Chemother. 2010;65:634–643. doi: 10.1093/jac/dkq006. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins SG, Farrell DJ. Increase in pneumococcus macrolide resistance, United States. Emerg Infect Dis. 2009;15:1260–1264. doi: 10.3201/eid1508.081187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takaya A, et al. Mutational analysis of reduced telithromycin susceptibility of Streptococcus pneumoniae isolated clinically in Japan. FEMS Microbiol Lett. 2010;307:87–93. doi: 10.1111/j.1574-6968.2010.01962.x. [DOI] [PubMed] [Google Scholar]
- 5.Morozumi M, et al. Macrolide-resistant Mycoplasma pneumoniae: Characteristics of isolates and clinical aspects of community-acquired pneumonia. J Infect Chemother. 2010;16:78–86. doi: 10.1007/s10156-009-0021-4. [DOI] [PubMed] [Google Scholar]
- 6.Franceschi F, Duffy EM. Structure-based drug design meets the ribosome. Biochem Pharmacol. 2006;71:1016–1025. doi: 10.1016/j.bcp.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Zhou J, et al. Design at the atomic level: Generation of novel hybrid biaryloxazolidinones as promising new antibiotics. Bioorg Med Chem Lett. 2008;18:6179–6183. doi: 10.1016/j.bmcl.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Franceschi F, et al. Macrolide resistance from the ribosome perspective. Curr Drug Targets Infect Disord. 2004;4:177–191. doi: 10.2174/1568005043340740. [DOI] [PubMed] [Google Scholar]
- 9.Berisio R, et al. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185:4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tu D, et al. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 12.Schlunzen F, et al. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 13.Yonath A. Antibiotics targeting ribosomes: Resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz S, et al. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. 2004;28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Vogel Z, et al. Correlation between the peptidyl transferase activity of the 50 s ribosomal subunit and the ability of the subunit to interact with antibiotics. J Mol Biol. 1971;60:339–346. doi: 10.1016/0022-2836(71)90298-1. [DOI] [PubMed] [Google Scholar]
- 16.Kearsey SE, Craig IW. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature. 1981;290:607–608. doi: 10.1038/290607a0. [DOI] [PubMed] [Google Scholar]
- 17.Moazed D, Noller HF. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 18.Celma ML, et al. Substrate and antibiotic binding sites at the peptidyl transferase centre of E. coli ribosomes: Binding of UACCA-Leu to 50 S subunits. FEBS Lett. 1971;13:247–251. doi: 10.1016/0014-5793(71)80546-x. [DOI] [PubMed] [Google Scholar]
- 19.Ulbrich B, et al. Cooperative binding of 3′-fragments of transfer ribonucleic acid to the peptidyltransferase center of Escherichia coli ribosomes. Arch Biochem Biophys. 1978;190:149–154. doi: 10.1016/0003-9861(78)90262-x. [DOI] [PubMed] [Google Scholar]
- 20.Barta A, et al. Identification of a site on 23S ribosomal RNA located at the peptidyl transferase center. Proc Natl Acad Sci USA. 1984;81:3607–3611. doi: 10.1073/pnas.81.12.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noller HF. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- 22.Bayfield MA, et al. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc Natl Acad Sci USA. 2001;98:10096–10101. doi: 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaha G, et al. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008;379:505–519. doi: 10.1016/j.jmb.2008.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein DJ, et al. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boese R, et al. Low-temperature crystal and molecular-structure of nitrobenzene. Struct Chem. 1992;3:363–368. [Google Scholar]
- 26.Borisenko KB, Hargittai I. Barrier to internal rotation of the nitro group in ortho-nitrophenols from gas-phase electron diffraction. J Mol Struct. 1996;382:171–176. [Google Scholar]
- 27.Xaplanteri MA, et al. Effect of polyamines on the inhibition of peptidyltransferase by antibiotics: Revisiting the mechanism of chloramphenicol action. Nucleic Acids Res. 2003;31:5074–5083. doi: 10.1093/nar/gkg686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanc H, et al. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3′ end of the large ribosomal RNA. Proc Natl Acad Sci USA. 1981;78:3789–3793. doi: 10.1073/pnas.78.6.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Fonseca C, et al. Fine structure of the peptidyl transferase centre on 23 S-like rRNAs deduced from chemical probing of antibiotic-ribosome complexes. J Mol Biol. 1995;247:224–235. doi: 10.1006/jmbi.1994.0135. [DOI] [PubMed] [Google Scholar]
- 31.Leslie AG, et al. Structure of chloramphenicol acetyltransferase at 1.75-A resolution. Proc Natl Acad Sci USA. 1988;85:4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw WV, Brodsky RF. Characterization of chloramphenicol acetyltransferase from chloramphenicol-resistant Staphylococcus aureus. J Bacteriol. 1968;95:28–36. doi: 10.1128/jb.95.1.28-36.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson DN, et al. Species-specific antibiotic-ribosome interactions: Implications for drug development. Biol Chem. 2005;386:1239–1252. doi: 10.1515/BC.2005.141. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Laslop N, et al. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 35.Blaha G, et al. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 37.Voorhees RM, et al. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen JL, et al. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 39.Xiong L, et al. Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob Agents Chemother. 2005;49:281–288. doi: 10.1128/AAC.49.1.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garza-Ramos G, et al. Binding site of macrolide antibiotics on the ribosome: New resistance mutation identifies a specific interaction of ketolides with rRNA. J Bacteriol. 2001;183:6898–6907. doi: 10.1128/JB.183.23.6898-6907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen LH, et al. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–631. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 42.Blanc H, et al. Different nucleotide changes in the large rRNA gene of the mitochondrial DNA confer chloramphenicol resistance on two human cell lines. Nucleic Acids Res. 1981;9:5785–5795. doi: 10.1093/nar/9.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson D, et al. Design, synthesis and ribosome binding of chloramphenicol nucleotide and intercalator conjugates. Bioorg Med Chem Lett. 2005;15:2079–2083. doi: 10.1016/j.bmcl.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 44.Nissen P, et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 45.Or YS, et al. Design, synthesis, and antimicrobial activity of 6-O-substituted ketolides active against resistant respiratory tract pathogens. J Med Chem. 2000;43:1045–1049. doi: 10.1021/jm990618n. [DOI] [PubMed] [Google Scholar]
- 46.Skripkin E, et al. R chi-01, a new family of oxazolidinones that overcome ribosome-based linezolid resistance. Antimicrob Agents Chemother. 2008;52:3550–3557. doi: 10.1128/AAC.01193-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovalic D, et al. Methylation of minimalist 23S rRNA sequences in vitro by ErmSF (TlrA) N-methyltransferase. Biochemistry. 1995;34:15838–15844. doi: 10.1021/bi00048a029. [DOI] [PubMed] [Google Scholar]
- 49.Stanley RE, et al. The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome. Nat Struct Mol Biol. 2010;17:289–293. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W. Xds. Acta Crystallogr D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 54.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.