Abstract
The traditional tree of life shows eukaryotes as a distinct lineage of living things, but many studies have suggested that the first eukaryotic cells were chimeric, descended from both Eubacteria (through the mitochondrion) and Archaebacteria. Eukaryote nuclei thus contain genes of both eubacterial and archaebacterial origins, and these genes have different functions within eukaryotic cells. Here we report that archaebacterium-derived genes are significantly more likely to be essential to yeast viability, are more highly expressed, and are significantly more highly connected and more central in the yeast protein interaction network. These findings hold irrespective of whether the genes have an informational or operational function, so that many features of eukaryotic genes with prokaryotic homologs can be explained by their origin, rather than their function. Taken together, our results show that genes of archaebacterial origin are in some senses more important to yeast metabolism than genes of eubacterial origin. This importance reflects these genes’ origin as the ancestral nuclear component of the eukaryotic genome.
Keywords: endosymbiosis, gene essentiality, eukaryote origin, protein interaction network
As one of the three domains of cellular life, the eukaryotes are typically described as the sister group to the archaebacteria. This sister group relationship describes the evolutionary history of the “nuclear-cytoplasmic” component of eukaryotes, with mitochondria and plastids being of endosymbiotic bacterial origin (e.g., ref. 1). In this traditional scenario, the unique features of extant eukaryotes were gradually acquired in the eukaryote stem group before the endosymbiotic acquisition of the mitochondrion. Thus, the acquisition of the mitochondrion was an important, but not foundational, step in eukaryote origins, occurring subsequent to the evolution of many characteristic features of eukaryotic cell biology. Early molecular phylogenies of ribosomal RNA genes support this scenario (see refs. 2 and 3 for reviews), as do several other molecular markers. Many nuclear genes are more closely related to eubacterial homologs than to any known archaebacterial sequence (4, 5) and appear to have been transferred to the nucleus from the ancestral mitochondrial genome by a process known as endosymbiotic gene transfer (1, 6). A similar process occurred after other symbiotic events, for example, the introduction of many chloroplast-derived genes into the nuclei of green plants (6).
An alternative view of eukaryotic nuclear-cytoplasmic origins, first suggested by Lake (7–9) is that this lineage arose from within, rather than as a sister to, the archaebacteria. This view is supported by molecular phylogenies showing that many eukaryote genes actually derive from within the archaebacterial domain (7–11), including a recent reanalysis of informational genes with modern phylogenetic methods (10). It also has become clear that those eukaryotes that lack mitochondria either are derived from organisms that have mitochondria or themselves host hydrogenosomes or mitosomes, which are degenerate relicts of mitochondria (3, 12). Thus, all known eukaryotes possessed mitochondria at some point in their evolutionary history, suggesting either that the acquisition of the mitochondrion might have occurred early in eukaryote evolution (or at least that the characteristic features of extant eukaryotic cell biology arose after the initial mitochondrial endosymbiosis) or that many important lineages of primitively amitochondriate transitional “protoeukaryotes” have gone extinct. Various alternative scenarios have been proposed to explain the chimeric (archaebacterial and eubacterial) nature of eukaryotic genomes (3, 13, 14–16), some involving symbioses or “cell fusions” quite different in character from what we call the traditional scenario (5, 14, 17). These ideas remain somewhat controversial (18, 19), but appear to be supported by a growing body of empirical evidence (12, 20).
However they arose, eukaryotic nuclei clearly contain homologs to both eubacterial and archaebacterial genes, and a growing number of phylogenetic studies confirm that nuclear genes are derived from multiple sources (7, 12, 21, 22). Previous studies (23, 24) confirm that about half of the eukaryotic genes have homologs in prokaryotes, and that most of these homologs are eubacterial. Furthermore, archaebacterial and eubacterial homologs are known to fulfill broadly different functions in eukaryotic cells, with eubacterial homologs largely involved in “operational” metabolic processes and archaebacterial homologs largely involved in the “informational” processes of transcription, translation, and replication (23, 25). These different functions suggest that the different partners played different roles in the formation of the earliest eukaryotic cell. Here we reveal other fundamental differences between the contributions of the two partner genomes.
Results
Our results are based on identifying prokaryote homologs of eukaryotic genes, examining every gene in the Saccharomyces cerevisiae genome. They support recent studies (23, 24) in showing that many eukaryotic genes are related to prokaryotic genes (2,460 of 6,704 genes), and that ∼75% of these have eubacterial affinities. For 1,980 yeast genes, the strongest BLAST hit is to a eubacterial gene, and for 480 yeast genes, the strongest hit is archaebacterial; 952 genes have only eubacterial homologs, showing no homology to any archaebacterial sequence, whereas 216 genes have only archaebacterial homologs. We carried out a number of phylogenetic analyses of 1,717 of these gene families, with only the very largest families not subjected to these analyses. The proportions of genes ascribed eubacterial ancestry and archaebacterial ancestry remained similar (see SI Results for details). These data confirm a significant bias toward archaebacterial homology for genes with informational functions [odds ratio (OR), 2.37; 95% confidence interval (CI), 1.59–3.52]. Although significant, this is not a clear-cut distinction, given that genes with archaebacterial homologs are involved in most of the biological processes of the yeast cell.
These absolute numbers of homologs suggest a larger role for genes with eubacterial homologs. Absolute numbers do not necessarily tell the whole story, however, given that genes may differ in function in many different ways, such as through different patterns of expression and involvement in different metabolic pathways. To explore this functional dimension, we mapped our homologs onto data from a comprehensive gene knockout study (26), identifying each gene as having either a lethal or a viable deletion phenotype. Lethal genes are more than twice as likely to have archaebacterial homologs than eubacterial homologs (OR, 2.23; 95% CI, 1.97–2.53). One possible explanation for this is that the informational functions of genes with archaebacterial homology are likely to be essential to cellular viability, and indeed informational genes are more often lethal than operational genes (OR, 2.98; 95% CI, 2.03–4.40). This does not explain our result, however, because both informational and operational genes with archaebacterial homologs are more likely to be lethal than those with eubacterial homologs. Furthermore, the greater propensity to lethality of archaebacterial genes is very similar across the two categories (for informational genes, OR, 2.01; 95% CI, 0.92–4.41; for operational genes, OR, 1.89; 95% CI, 1.43–2.47). Although the relatively small number of informational genes means that we cannot reject the null hypothesis of no association for this subset of the data, we note that the estimated strength of this effect is actually greater for informational genes than for operational genes, confirming that the lack of significance is due to a lack of power in the test for informational genes. Counts of genes in each category are given in SI Results.
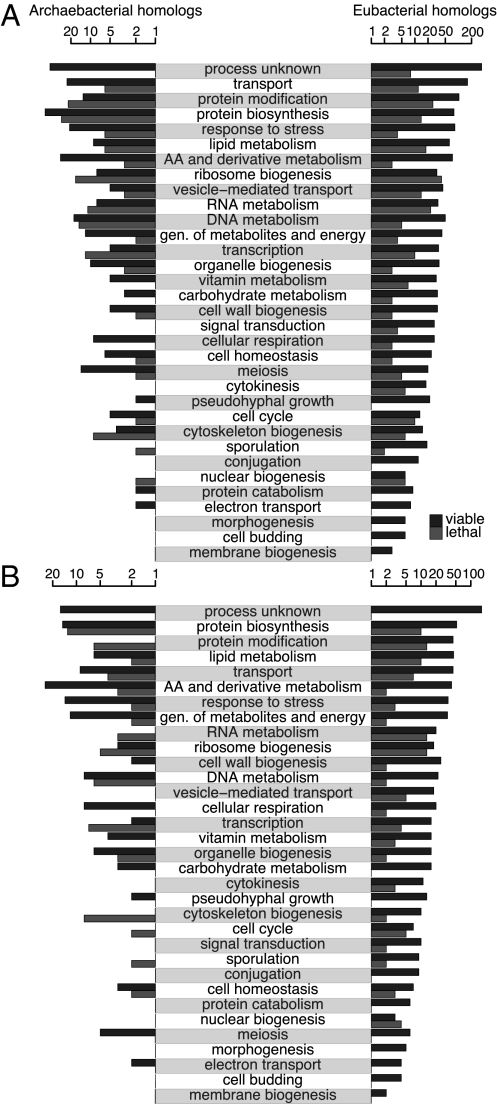
The foregoing results are robust to details of the data and analysis, but we emphasize that these are large-scale patterns rather than clear distinctions. Many archaebacterial homologs have operational functions with both viable and lethal deletion phenotypes, as do many informational eubacterial homologs. Homology, function, and phenotype are also not strongly associated with the metabolic pathway in which the genes are involved (Fig. 1 and Fig. S3). Most pathways contain both eubacterial and archaebacterial homologs, and the distribution of these within pathways shows no clear general pattern. Although we have not attempted a large-scale analysis of metabolic pathway structure or evolution, it is clear that some pathways (e.g., phospholipid and sphingolipid metabolism) are largely eubacterial, some have connected eubacterial and archaebacterial components (e.g., sterol synthesis), and others are a complex mixture of genes of different homologies (e.g., tyrosine, tryptophan, and phenylalanine metabolism). Three other example pathways are presented in SI Materials and Methods.
Fig. 1.
Distribution of homologs for yeast genes. Homologs are listed by homology domain, functional category, and deletion phenotype. (A) Best-hit domain. (B) Unambiguous hits, with homology only to one of the two prokaryotic domains. Light bars represent lethal genes and dark bars represent viable genes in each domain. Note that the number of genes is plotted on a log axis.
In an effort to explain the greater essentiality of archaebacteria-related genes, we examined other data that might shed light on the differing cellular functions of these genes and their protein products. Using data from RNAseq experiments (27), we found significantly greater expression of genes with archaebacterial homologs (Table 1). The average number of tags that could be attached to genes of archaebacterial origin was 164.64 (95% CI, 131.0–198.5), compared with 73.81 (95% CI, 61.03–86.46) for eubacteria. This is a >2-fold difference on average. No significant differences are seen between the expression levels of operational and informational gene categories (Table S4).
Table 1.
Functional correlates of prokaryote homology for yeast genes
| Data | Eubacterial | Archaebacterial | All | P value (arch ≤ bact) |
| Expression level: number of tags | 73.81 (61.03– 86.46) | 164.64 (131.0–198.5) | 85.89 (78.80–93.09) | < 0.0001 |
| Closeness centrality in interaction network | 0.314 (0.312–0.316) | 0.324 (0.321–0.327) | 0.316 (0.315–0.317) | < 0.0001 |
| Degree in interaction network | 15.91 (15.20–16.62) | 20.90 (19.33–22.48) | 18.02 (17.60–18.48) | < 0.0001 |
| Number of paralogs in yeast genome | 13.13 (12.09–14.16) | 8.02 (6.89–9.22) | 7.58 (7.14–8.04) | 1 |
Values are listed by domain of best BLAST hit, showing means and 95% bootstrap percentile CIs for the mean of each parameter (calculated using the nonparametric bootstrap). P values are bootstrap probabilities for the mean of the statistic in archaebacterial homologs being less than or equal to the mean in eubacterial homologs, based on 10,000 replicates.
Genes with archaebacterial homologs are more central and more highly connected in the yeast protein interaction network (28–30) (Fig. S4 and Table 1; see SI Materials and Methods for details on data and methods), which has been shown to reflect greater essentiality (29, 31). This difference is partly explained by the greater centrality and connectedness of informational genes, but a statistically significant difference is still observed for operational genes alone (Table S4). Furthermore, operational genes whose products interact directly with the products of genes with informational functions are more likely to have a lethal knockout phenotype compared with other operational genes; however, because this effect is similar for both archaebacterial and eubacterial homologs, the pattern of protein–protein interactions does not explain our main result (SI Results).
Finally, eubacterial homologs show more duplicate copies (paralogs) within the yeast genome, suggesting that a greater degree of genetic redundancy is protecting the cell against deletion of eubacterial homologs. Although there is a significant difference in the number of duplicate copies between operational genes and informational genes, the significant difference in the number of duplicates between archaeal and eubacterial homologs is consistent for both functional groups. However, unlike our other findings, this result is sensitive to the dataset used (SI Results), and other studies have found little evidence of a relationship between duplication and redundancy (32), which may vary with the function and mode of duplication of the genes and even between genomes (33).
Discussion
Genes of different origins play significantly different roles in eukaryotic cells that cannot be explained by the functional (operational vs. informational) distinction between sets of genes. Genes of archaebacterial origin and those of eubacterial origin differ significantly in many aspects, including essentiality, expression level, and centrality in protein interaction networks. This complex pattern suggests that this is a signal of the history of the yeast genome.
Our methods do not allow us to estimate the timing or exact source of the genes that we identify as having homology to genes from different prokaryotic domains. These genes could be found in the yeast genome as a result of more recent lateral gene transfer (LGT), rather than being a relict of mitochondrial endosymbiosis. Both pre-eukaryogenesis LGT events among and between groups of prokaryotes (34, 35) and LGT from either group to eukaryotes (21) could have affected some of our data. Although there are plenty of examples of prokaryote-to-eukaryote LGT, there is limited evidence of LGT being an important mode of genome evolution in most eukaryotes (36). Extensive investigation has found no conclusive evidence of prokaryotic genes in the human genome, and there appears to be little evidence of prokaryotic gene transfer into the yeast genome (37, 38), although there may be methodological problems with these studies (36). The statistically significant results of our analyses are even more surprising in light of these processes. Although it seems likely that recently acquired genes would occupy peripheral roles in cellular metabolism or regulation, we know of no proposed mechanism to explain the very different lethality of genes from archaebacterial and eubacterial sources if recent LGT is responsible for many of the prokaryotic homologs that we observe, unless there is some systematic difference in the timing of LGT from the two domains.
If most of the prokaryotic homologs that we observe are descended from the fusion of a eubacterium and archaebacterium to form the first eukaryotic cell, then our results can be interpreted in terms of the different roles of the two ancestors. In this scenario, genes from the archaebacterial host formed the original eukaryote nucleus and so have been cointeracting for a longer time and form a core part of metabolism. Incoming eubacterial genes, from genome fusion or from subsequent endosymbiotic gene transfer (our data cannot distinguish between the two scenarios), have more peripheral roles in the network of protein interactions that controls metabolism, because the archaebacterial genes that performed essential functions might have been more difficult to displace by the influx of eubacterial genes. Although genes of eubacterial affinity seem to have replaced large parts of this ancestral metabolism, our findings suggest that much of eukaryotic metabolism may have been built on an ancestral foundation that still plays a central role in the eukaryotic cell. Our results also support other ideas about genome evolution. For instance, the complexity hypothesis proposes that genes that encode proteins in large complexes are highly connected and thus less likely to experience LGT (39). Our findings add to the evidence indicating that the protein interaction network of yeast shows significant historical structure (40), confirming that subsequent evolution has not completely erased the effect of ancient evolutionary history on eukaryotic genomes.
Whatever the source of the prokaryote homologs that we have identified, our results demonstrate that whereas eubacteria have made a greater quantitative contribution to yeast metabolism, the archaebacteria made a different, arguably more important contribution. These results are compatible with previous findings (12, 20) and with some ideas about the origin of the eukaryotic cell (13, 41).
It is not clear that the historical process of eukaryogenesis should be able to help us understand the biology of modern eukaryotic cells, given that > 2.5 billion years (42) of evolution have shuffled genes between pathways, changed expression levels, and altered the interactions between gene products. For example, only half of eukaryotic genes have an identifiable prokaryotic homolog, and no large functional category consists solely of genes with homology to a sole prokaryotic domain. Rapid genomic changes are likely to have followed eukaryogenesis, as they did when genomes fused more recently (43), so it is remarkable that some of the original partners’ contributions might have persisted for > 1 billion years of evolution.
Yeast metabolism, and presumably eukaryotic metabolism in general, is a complex tapestry of prokaryotic threads and eukaryotic innovations. Our analysis of the features of eukaryotic genes that have a prokaryotic history shows that a protein's group of origin plays an important role in defining its expression profile, likelihood of lethality, and position and connectivity in a protein interaction network independent of the actual function of the protein. This suggests that the roles of genes from the various partners in the eukaryotic cell differ in ways beyond the simple split between operational and informational functions.
Materials and Methods
Homology Search.
To produce a homology search that would be both sensitive and specific, we built a profile alignment of the amino acid sequence of a range of eukaryotic homologs for each yeast gene, then used PSI-BLAST (44) to search against a database of 197 eubacterial and 22 archaebacterial genome sequences. To build the profile alignments for PSI-BLAST, each protein-coding gene in the Saccharomyces cerevisiae genome sequence [downloaded from the Cogent database (45)] was compared with the protein-coding gene content of six other eukaryotic genomes (Caenorhabditis elegans, Arabidopsis thaliana, Schizosaccharomyces pombe, Neurospora crassa, Ashbya gossypii, and Trypanosoma cruzi) downloaded from the same source. For each yeast gene, a multiple sequence alignment of the yeast gene and the best (i.e., lowest e-value) hit with e < 0.001 from each of these genomes was constructed using the alignment program MUSCLE (46) with default settings. This alignment, of between one and seven sequences (depending on how many eukaryotic genomes had a hit with e < 0.001 for the yeast gene) was used as a seed profile for a PSI-BLAST search against the combined database of prokaryotic protein sequences, with an e-value cutoff of 1 × 10−6. Genes were classified as homologs to the prokaryotic domains in two different ways. In the least stringent case, genes were assigned to whichever domain their best BLAST hit sequence belonged, being considered ambiguous only if they had equally good best hits in both domains (Results and Fig. 1A). In the second case, genes were considered ambiguous unless all BLAST hits with an e-value below the cutoff were in the same domain.
Functional Comparisons.
Comparisons of domain homology and knockout phenotype, functional category, expression level, and interaction network position were carried out using Perl scripts (available from the authors on request). Genes annotated with Gene Ontology (GO) (47) terms “translation,” “transcription,” “DNA-dependent DNA replication” or any of their subterms were considered informational; all other genes were considered operational. Interaction network statistics were calculated using the Pajek (48) package. GO mappings were downloaded from the Saccharomyces Genome Database (49), RNAseq data were obtained from Nagalakshmi et al. (27), knockout phenotype data were downloaded from the comprehensive yeast genome database (50), and protein interaction data were obtained from BioGRID (30).
Statistical Analysis.
We describe the strength of associations between factors using ORs (51); for example, the odds of being archaebacterial for informational genes is calculated as the probability of an informational gene having an archaebacterial homolog, divided by the probability of the gene having a eubacterial homolog. We can similarly calculate the odds of being archaebacterial for operational genes. The OR is the ratio of these two odds. Thus, this statistic is not affected by the absolute sizes of the different categories. To test the significance of associations, we constructed a 95% CI for the OR under a normal approximation to the log OR (51). A significant association is one for which this CI does not overlap unity.
Supplementary Material
Acknowledgments
We thank the three anonymous referees for their comments, which greatly improved the manuscript. This research was funded by Science Foundation Ireland; the Irish Research Council for Science, Engineering and Technology; and a Research Councils UK Academic Fellowship. The computation was facilitated in part by the Irish Centre for High End Computing (ICHEC) and the NUI Maynooth computing centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000265107/-/DCSupplemental and http://bioinf.nuim.ie/supplementary/CottonMcInerneyPNAS_2010/tableS5.pdf.
References
- 1.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 4.Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pühler G, et al. Archaebacterial DNA-dependent RNA polymerases testify to the evolution of the eukaryotic nuclear genome. Proc Natl Acad Sci USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lake JA. Evolving ribosome structure: Domains in archaebacteria, eubacteria, eocytes and eukaryotes. Annu Rev Biochem. 1985;54:507–530. doi: 10.1146/annurev.bi.54.070185.002451. [DOI] [PubMed] [Google Scholar]
- 8.Lake JA. Prokaryotes and archaebacteria are not monophyletic: Rate- invariant analysis of rRNA genes indicates that eukaryotes and eocytes form a monophyletic taxon. Cold Spring Harb Symp Quant Biol. 1987;52:839–846. doi: 10.1101/sqb.1987.052.01.091. [DOI] [PubMed] [Google Scholar]
- 9.Lake JA. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988;331:184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- 10.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 12.van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- 13.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 14.Hartman H. The origin of the eukaryotic cell. Speculations Sci Technol. 1984;7:77–81. [PubMed] [Google Scholar]
- 15.Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 16.Searcy DG. Origins of mitochondria and chloroplasts from sulfur based symbiosis. In: Hartman H, Matsuno K, editors. The Origin and Evolution of the Cell. Singapore: World Scientific; 1992. pp. 47–78. [Google Scholar]
- 17.Doolittle WF. Some aspects of the biology of cells and their evolutionary significance. In: Roberts DM, Sharp P, Alderson G, Collins M, editors. Evolution of Microbial Life. Vol. 54. Cambridge, UK: Cambridge Univ Press; 1995. pp. 1–21. [Google Scholar]
- 18.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 19.Cavalier-Smith T. Predation and eukaryote cell origins: A coevolutionary perspective. Int J Biochem Cell Biol. 2009;41:307–322. doi: 10.1016/j.biocel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 21.Doolittle WF, et al. How big is the iceberg of which organellar genes in nuclear genomes are but the tip? Philos Trans R Soc Lond B Biol Sci. 2003;358:39–57. doi: 10.1098/rstb.2002.1185. discussion 57–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–1630. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esser C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 24.Horiike T, Hamada K, Kanaya S, Shinozawa T. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria is revealed by homology-hit analysis. Nat Cell Biol. 2001;3:210–214. doi: 10.1038/35055129. [DOI] [PubMed] [Google Scholar]
- 25.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95:6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 27.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 29.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 30.Stark C, et al. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- 32.Wagner A. Robustness against mutations in genetic networks of yeast. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 33.Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3:180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 37.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 38.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin H, Lu HHS, Wu WB, Li WH. Evolution of the yeast protein interaction network. Proc Natl Acad Sci USA. 2003;100:12820–12824. doi: 10.1073/pnas.2235584100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Searcy DG. Rapid hydrogen sulfide consumption by Tetrahymena pyriformis and its implications for the origin of mitochondria. Eur J Protistol. 2006;42:221–231. doi: 10.1016/j.ejop.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Brocks JJ, Logan GA, Buick R, Summons RE. Archean molecular fossils and the early rise of eukaryotes. Science. 1999;285:1033–1036. doi: 10.1126/science.285.5430.1033. [DOI] [PubMed] [Google Scholar]
- 43.Soltis DE, Soltis PS. Polyploidy: Recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- 44.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janssen PJ, et al. COmplete GENome Tracking (COGENT): A flexible data environment for computational genomics. Bioinformatics. 2003;19:1451–1452. doi: 10.1093/bioinformatics/btg161. [DOI] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner M, et al. The Gene Ontology Consortium Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Batagelj V, Mrvar A. Pajek—Program for Large Network Analysis. Connections. 1998;21:47–57. [Google Scholar]
- 49.Nash R, et al. Expanded protein information at SGD: New pages and proteome browser. Nucleic Acids Res. 2007;35(Database issue):D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Güldener U, et al. CYGD: The Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33(Database issue):D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agresti A. Categorial Data Analysis. 2nd Ed. Hoboken, NJ: Wiley; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.