Abstract
Antiviral drugs targeting viral proteins often result in prompt selection for resistance. Moreover, the number of viral targets is limited. Novel antiviral targets are therefore needed. The unique characteristics of fusion between virion envelopes and cell membranes may provide such targets. Like all fusing bilayers, viral envelopes locally adopt hourglass-shaped stalks during the initial stages of fusion, a process that requires local negative membrane curvature. Unlike cellular vesicles, however, viral envelopes do not redistribute lipids between leaflets, can only use the energy released by virion proteins, and fuse to the extracellular leaflets of cell membranes. Enrichment in phospholipids with hydrophilic heads larger than their hydrophobic tails in the convex outer leaflet of vesicles favors positive curvature, therefore increasing the activation energy barrier for fusion. Such phospholipids can increase the activation barrier beyond the energy provided by virion proteins, thereby inhibiting viral fusion. However, phospholipids are not pharmacologically useful. We show here that a family of synthetic rigid amphiphiles of shape similar to such phospholipids, RAFIs (rigid amphipathic fusion inhibitors), inhibit the infectivity of several otherwise unrelated enveloped viruses, including hepatitis C and HSV-1 and -2 (lowest apparent IC50 48 nM), with no cytotoxic or cytostatic effects (selectivity index > 3,000) by inhibiting the increased negative curvature required for the initial stages of fusion.
Keywords: lipid bilayer fusion, DNA virus, RNA virus, Sindbis virus, nucleosides
Anti-infective drugs target differences between infectious agents and hosts. Targeting viral proteins, for example, confers specificity for infected cells. However, this method commonly leads to prompt selection for resistance (1). Even triple combinations of such drugs eventually select for resistance (2). The number of viral targets is also so limited that most clinical antivirals act on viral polymerases or proteases (3, 4), increasing the risk of cross-resistance. Moreover, the proteins of viruses, such as hepatitis C (HCV), have proven to be such difficult targets that the first drugs targeting them are in phase 3 clinical trials only now, 21 y after the HCV genome was sequenced. The only clinical HCV treatment (pegylated IFN-α with ribavirin) is effective for only ≈50% of patients infected with genotype-1 strains. Although novel compounds targeting viral proteins are continuously being developed (5, 6), novel antiviral strategies are still needed (7).
Drugs targeting viral glycoproteins or their receptors block the first events in the viral replication cycle and are less constrained by intracellular delivery or metabolism (6, 8, 9). Unfortunately, these drugs share the limitations of other drugs targeting viral proteins (2, 10, 11), such as prompt selection for resistance (12, 13). Drugs targeting cellular proteins may help to overcome these and related limitations (10, 14, 15), but have a greater risk of adverse side effects. For example, HCV IFN-α treatment is often discontinued because of such effects (6). Drugs targeting functions unique to viruses but provided by nonvirally encoded factors would minimize the potential for toxicity and for selection of resistant strains (7).
The envelopes of virions are unique metabolically inert extracellular lipid bilayers of relatively strong positive curvature. The radius of viral envelopes is typically 30 to 150 nm, in comparison with 3,500 to 7,000 nm for mammalian cells. Consequently, the area of the outer envelope leaflet of a model spherical virion (50-nm radius) is 11% larger than the inner leaflet, whereas the areas of the leaflets of plasma membranes of model spherical cells (2,500-nm radius) differ by only 0.1%.
Membrane curvature is partly determined by the membrane proteins and the molecular shape and charge distribution of membrane lipids (16, 17). Cylindrical lipids, with hydrophobic tails and hydrophilic heads of similar cross-sections, tend to form stable lamellar bilayers with no net curvature. Lipids with hydrophobic tails of larger cross-sections than their hydrophilic heads tend to adopt configurations with the headgroups bent toward each other (negative curvature). Lipids with hydrophobic tails of smaller cross-sections than their hydrophilic heads tend to adopt configurations with the headgroups bent away from each other (positive curvature). Outer membrane leaflets are relatively enriched in lipids of larger hydrophilic heads, favoring positive curvature.
Following attachment and receptor binding, virion glycoproteins undergo conformational changes that expose their fusion peptides, bring virion and cell membranes into close apposition, and trigger fusion. In this latter process, the two membrane bilayers fuse to form a continuous bilayer. Experimental evidence and theoretical models indicate that the first fusion step involves the joining of the contacting outer leaflets of the membranes to form an intermediate stalk. At this stage, the external leaflets must form a curved structure, in which the packing of the external polar headgroups increases, whereas the internal ends of the hydrophobic acyl chains occupy a greater area. Such an arrangement is termed “negative curvature” and is the same kind of curvature that eventually results in the formation of HII phases (16, 17). The inner leaflets fuse next, creating a small pore that subsequently enlarges. Tilting and rearrangements of the hydrophobic tails of membrane lipids and diffusion of neutral lipids in the hydrophobic core of the fusing membranes minimize the activation energy required for fusion (18).
In cellular vesicles, lipid remodeling and translocations modify the normal relative enrichment in lipids with larger hydrophilic heads in outer leaflets. Such remodeling minimizes the activation energy for formation of negative curvature. Moreover, a variety of cellular proteins use ATP to bring the fusing leaflets together or induce different curvatures. In contrast, virions are metabolically inert; they cannot use ATP as an energy donor, or ATP-consuming proteins to modulate curvature, or remodel and rearrange envelope lipids. Only binding and rearrangements of virion glycoproteins releases energy to overcome the requirements for the formation of the negative curvature essential for fusion (19–22). Furthermore, outer leaflets of virion envelopes are commonly enriched in glycolipids with relatively large hydrophilic heads, such as gangliosides, which favor positive curvature (23, 24).
Virion infectivity or fusion can be modulated by changing the lipid composition of the virion envelope or target membranes (25, 26). Agents promoting positive curvature inhibit viral fusion (27, 28). For example, enrichment of lysophospholipids or lipophosphoglycans (with hydrophilic heads of much larger diameter than their hydrophobic tails) in external leaflets increases the energy required for formation of negative curvature. Such lipids consequently inhibit in vitro fusion between apposing membranes, including those of viruses and cells (29–32). Unfortunately lysophospholipids are not useful as drugs: they disrupt membranes, are cytotoxic, and, like all natural phospholipids, are metabolized too promptly. Small synthetic compounds of appropriate shape and polarity distribution may also be able to increase the activation energy for fusion beyond that provided by virion glycoproteins, but without disrupting membranes and still allowing for physiological fusions. Such compounds could be useful as antivirals or microbicides. Here, we show that synthetic rigid amphipathic compounds (rigid amphipathic fusion inhibitors, RAFIs) with hydrophilic heads of larger diameter than their hydrophobic tails target the virion envelope to inhibit infectivity of several otherwise unrelated enveloped viruses, including important human pathogens, by modulating the formation of the negative curvature required for fusion, and without lysing membranes or having cytotoxic effects.
Results
The RAFIs dUY11, aUY11, and ddUY11 Inhibit HSV-1 Infectivity.
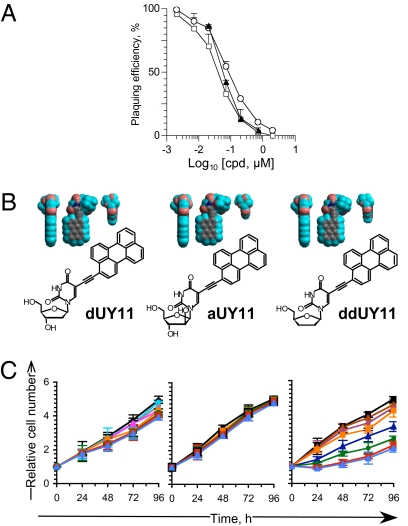
We tested the effects on viral infectivity of a family of nucleoside derivatives with appropriate amphipathicity and molecular shapes. Vero cells were infected with 200 infectious HSV-1 virions preexposed to the different compounds for 5 min at 37 to 40 °C. Infectivity was monitored by plaquing efficiency. Structure-activity relationship (SAR) studies indicated that amphipathicity of the compound, a hydrophilic head of larger diameter than the hydrophobic moiety, and planarity and rigidity of the hydrophobic moiety were all essential for antiviral activity (Fig. S1 and Table S1). We therefore named the compounds “rigid amphipathic fusion inhibitors.” The most potent RAFI, dUY11, was selected for further characterization; dUY11 inhibited HSV-1 infectivity with an apparent IC50 of 48 ± 12 nM (Fig. 1A and Table S1), independently of the cell-type infected (Fig. S2A and Table S2).
Fig. 1.
Rigid amphipathic nucleoside derivatives inhibit HSV-1 infectivity with no cytotoxic effects. (A) Plaquing efficiency of HSV-1 virions preexposed to increasing concentrations of dUY11 (□), aUY11 (○), or ddUY11(▲) (average ± SD; n ≥ 5). (B) Chemical structures and 3D models, displayed in three orthogonal perspectives, of dUY11, aUY11, and ddUY11. Gray, carbon; teal, hydrogen; pink, oxygen; blue, nitrogen. (C) Vero cell viability in the presence of dUY11, aUY11, or ddUY11. Relative numbers of viable cells plotted against time of treatment with 0 (black squares), 0.05 (sky blue circles), 0.15 (blue triangles), 0.5 (gray circles), 1.5 (plum circles), 5 (pink triangles), 7 (orange squares), 15 (blue triangles), 20 (green circles), 70 (red squares), or 150 (light blue triangles) μM dUY11, aUY11, or ddUY11, replenished every 24 h. Error bars, SD (n = 5, dUY11; n = 4, aUY11, ddUY11). RAFI dUY11 forms aggregates visible by optic microscopy after overnight incubation in DMEM at ≥70 μM.
We next modified the number of hydroxyl groups in dUY11 to produce aUY11 and ddUY11. Each of these compounds is chemically different, but they all have similar shapes (Fig. 1B). Both aUY11 and ddUY11 inhibited HSV-1 infectivity (IC50, 131.4 ± 34 and 87 ± 53 nM, respectively) (Fig. 1A and Table S1), confirming the importance of shape for antiviral activity. However, ddUY11 was cytostatic (Fig. 1C and Table S1); aUY11 was not cytotoxic or cytostatic and dUY11 was only marginally cytostatic (Fig. 1C and Table S1), even at 150 μM (selectivity index > 3,000).
RAFIs Inhibit Entry but Not Attachment or Binding.
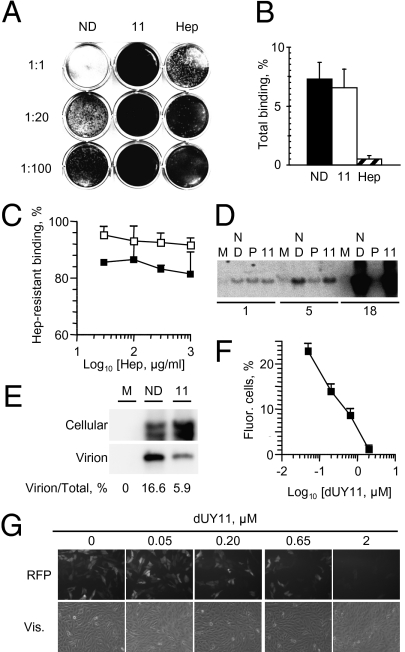
The effects of RAFIs on plaquing efficiency could result from inhibition of virion binding or viral replication. We first tested binding. The primary interactions between HSV-1 virions and cells are the attachment of the virion glycoprotein gC to cellular glycosaminoglycans. These interactions are blocked by heparin. HSV-1 virions were exposed to 7 μM dUY11, which inhibits plaquing by ≈100-fold (Fig. 2A). The virions exposed to dUY11 were adsorbed onto cells at 4 °C to allow binding but not fusion; dUY11 had no effect on binding. Nearly 7% HSV-1 virions exposed to 0 or 7 μM dUY11 bound to cells (Fig. 2B), as expected for untreated virions (33). In contrast, heparin equally inhibited HSV-1 infectivity and binding (by 10-fold) (Fig 2 A and B), as expected.
Fig. 2.
The RAFI dUY11 inhibits HSV-1 entry but not primary attachment or secondary binding or viral DNA replication. Photographic images and bar graphs showing infectivity (A), total gC-mediated attachment (B), or secondary gD-mediated binding (C), of HSV-1 virions exposed to dUY11, or intra- or extracellular HSV-1 DNA in infected cells treated with dUY11 (D and E), or RFP expression by cells infected with UV-inactivated and dUY11-exposed HSV-1 virions (F and G). (A) Infectivity of undiluted virions, or virions diluted 1:20 or 1:100. (B and C) Percent of virions bound to cells. (A–C) HSV-1 virions exposed to no drug (black in B or C), 7 μM dUY11 (white in B or C), or 100 μg/mL heparin (striped bars in B) were adsorbed onto Vero cells. Cells were washed and harvested (B), or further washed with heparin for 1 h (C). Southern blot analyses of cell-associated (D and E) or extracellular (E) HSV-1 DNA. Cells were infected with HSV-1 and treated with no drug, dUY11, or phosphonoacetic acid. Different exposures are shown for cellular and virion DNA (E). ND, no drug; 11, dUY11; Hep, heparin; P, phosphonoacetic acid. Error bars, SD (n ≥ 3). (F) Quantitation and (G) representative pictures of Vero cells containing an RFP reporter gene driven by VP16 and infected for 24 h with UV-inactivated HSV-1 virions preexposed to 0 to 2 μM dUY11.
Although dUY11 did not inhibit total attachment, it could still inhibit the secondary binding of glycoprotein gD in the HSV-1 virions to its receptors, such as HVEM or nectin-1. These secondary interactions are not affected by heparin. Virions were allowed to undergo the primary HSV-1 gC-mediated attachment and then to partially undergo secondary binding of HSV-1 gD to its receptors before adding heparin. Heparin then only outcompetes gC-glycosaminoglycans interactions, and not the interactions between gD and its receptors. Approximately 80% of untreated virions remained bound after washing with 0.03 mg/mL heparin, indicating they were already bound by gD receptor interactions. Almost 90% of virions preexposed to dUY11 also remained bound to the cells after washing with even 1 mg/mL heparin (Fig. 2C). Therefore, dUY11 inhibits plaquing after the transition from primary gC attachment to secondary gD binding.
We next tested the effects of dUY11 on virus replication. A subset of RAFIs had been proposed to inhibit HSV-1 replication, presumably by nonnucleosidic, herpes-specific mechanisms (34). We therefore tested whether dUY11 had any effects after infection. Cells previously infected with three untreated virions per cell were treated with 2 μM dUY11 (40-fold above IC50) between 1 and 24 h after infection (one replication round). DNA in cells or in extracellular (budded) virions was analyzed at 1 through 24 h. The RAFI dUY11 had no effect on HSV-1 DNA replication (Fig. 2D), and only inhibited virion release to the medium by 2.8-fold (Fig. 2E). As expected, HSV-1 DNA replication was inhibited by the DNA polymerase inhibitor phosphonoacetic acid, included as control (Fig. 2D).
RAFI dUY11 inhibits HSV-1 infectivity (Figs. 1A and 2A), but not attachment (Fig. 2 B and C) or DNA replication (Fig. 2 D and E). We therefore tested entry using reporter cells containing a red fluorescence protein (RFP) gene driven by a VP16-responsive promoter (35). VP16 is in the tegument of HSV-1 virions, a protein layer underneath the envelope but outside the capsid. When these cells are infected with UV-inactivated virions, no HSV-1 genes are expressed but the VP16 molecules in the virions activate RFP expression. RFP expression therefore indicates that VP16 has reached the nucleus and, indirectly, that the HSV-1 envelope has fused to the cell membranes.
Cells were infected with UV-inactivated HSV-1 virions exposed to 0 to 2 μM dUY11. Whereas 25.8 ± 1.7% of cells infected with virions exposed to no dUY11 expressed RFP, only 1.2 ± 0.9% cells infected with the virions exposed to 2.0 μM dUY11 expressed it (Fig. 2 F and G). As expected, inhibition of RFP expression was dose-dependent (Fig. 2 F and G). In summary, VP16 in virions preexposed to dUY11 does not enter the cells to activate its target promoters, indicating that dUY11 inhibits a step after attachment but before entry.
RAFIs Inhibit Infectivity of a Variety of Otherwise Unrelated Enveloped Viruses.
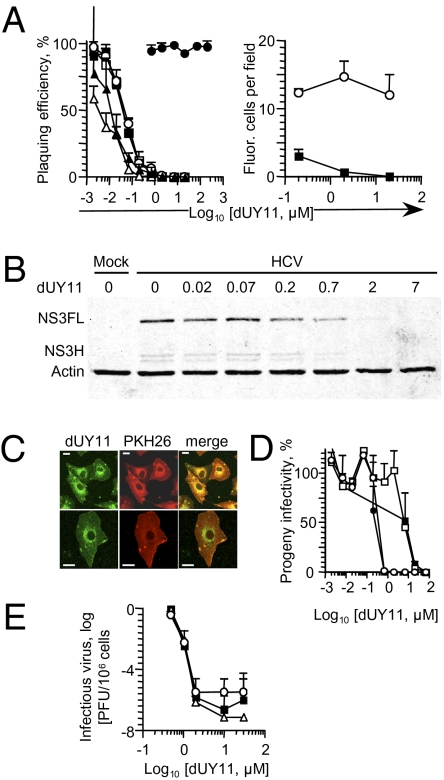
The RAFI dUY11 could target HSV-1 glycoproteins. We therefore tested dUY11 on two RNA viruses that have neither glycoprotein conservation nor common receptors with HSV-1, vesicular stomatitis (VSV) and Sindbis (SIN) virus. The glycoproteins of these viruses bind to yet unrecognized receptors or the high-affinity laminin receptor, respectively. In contrast to HSV-1, both are internalized by endocytosis and fuse with endosomal membranes following low pH-induced conformational changes. We also tested two nonenveloped viruses that are internalized by endocytosis (and perhaps also other routes), adenovirus (DNA genome) and poliovirus (RNA genome) (36). Although dUY11 inhibited the infectivity of all enveloped viruses tested, and with similar IC50, it did not that of the nonenveloped viruses (Fig. 3 A and B and Table S3). The targets of dUY11 are therefore conserved among viruses with no conserved glycoproteins, and which use different receptors and fuse to different cell membranes, but not in nonenveloped viruses that are internalized by endocytosis.
Fig. 3.
The RAFI dUY11 is active against HCV and other enveloped viruses, including drug-resistant mutant strains. (A) Infectivity of HSV-1 KOS (□), HSV-2 strains 186 (■), or 333 (○), vesicular stomatitis virus (△), Sindbis virus (▲); poliovirus (●) (Left), or GFP-expressing HSV-1 (■) or adenovirus (○) (Right) exposed to dUY11, tested by plaquing efficiency (Left) or fluorescence microscopy (Right). (B) HCV virions were exposed to dUY11 before infecting Huh7.5 cells with three genome copy equivalents per cell. Expression of HCV NS3 in the infected cells was evaluated 4 d later. NS3FL (full-length NS3 polypeptide) and NS3H (NS3 helicase domain), NS3 polypeptides; actin, loading control. (C) Confocal microscopy pictures of mock-infected cells treated with dUY11 and stained with a membrane dye. (Scale bars, 2.5 μM.) (D) Line graph showing infectivity associated with cells (black symbols) or supernatant (white symbols) of cells infected with HSV-1 and treated with dUY11 from 1 to 2h (squares) or from 1 to 23 h (circles), and harvested at 23 h after infection. (E) Infectivity of wild-type HSV-1 (■), or mutants resistant to phosphonoacetic acid (△) or acyclovir (○), produced by cells treated with dUY11 from 1 h after infection until harvest. Error bars, SD [n ≥ 3 independent experiments A (Left), D and E, or microscopic fields from one experiment representative of three, A (Right)].
RAFI dUY11 Inhibits the Infectivity of Important Human Pathogens.
We next tested dUY11 on two important human pathogens, HSV-2 and HCV. HSV-2 uses nectin-2 as a receptor more efficiently than HSV-1, but cannot use 3-O-sulfated heparan sulfates. HCV uses several different routes of internalization, requiring at least one of two receptors, SR-B1 or CD81, and fusion between virion envelopes and cell membranes. The RAFI dUY11 inhibited the infectivity of HSV-2 and HCV virions (Fig. 3 A and B) (IC50, 49 ± 9 or 55 ± 5 nM for HSV-2 strains 186 or 333, respectively, or 183 ± 1 nM for HCV) (Table S3).
The RAFI dUY11 Inhibits the Infectivity of Virions Produced by Cells Treated After Infection.
Envelope lipids are obtained from cellular membranes. We took advantage of the intrinsic fluorescence of dUY11 (37) to analyze its cellular distribution. The RAFI dUY11 is distributed to the plasma and intracellular membranes (Fig. 3C), from which budding virions acquire their envelopes. We therefore tested the ability of dUY11 to inhibit the infectivity of virions produced by cells treated after infection. Cells previously infected with untreated HSV-1 virions were treated with dUY11 from 1 to 2 h after infection (before expression of HSV-1 membrane glycoproteins). Cells were then washed and overlaid without drug for 22 h, when supernatants were harvested. The infectivity in the supernatants from cells treated with dUY11 was reduced to undetectable levels (Fig. 3D). As expected, the IC50 was higher than when the virions are directly exposed (Table S4). Also as expected, continuous dUY11 treatment of previously infected cells for 23 h resulted in lower IC50 than 1-h treatments (Table S4). Both 1- or 23-h treatments inhibited the infectivity of even the cell-associated virions (Fig. 3D).
As for HSV-1, dUY11 inhibited the infectivity of SIN virions released by cells treated for 1 or 23 h after infection (Fig. S2B and Table S4). Therefore, dUY11 most likely interacts with preexisting cellular factors that are incorporated into HSV-1 and SIN virions and are essential for infectivity, such as membrane lipids. Moreover, dUY11 in cellular membranes (Fig. 3C) inhibits the infectivity of the budded virions (Fig. 3D and Fig. S2B), but has no observable effects on the cells themselves (Figs. 1C and 3C).
RAFI dUY11 Inhibits the Infectivity of Drug-Resistant HSV-1 Mutants.
We next tested whether dUY11 was active against common drug-resistant viruses. We tested an HSV-1 point mutant in the DNA polymerase and a deletion mutant in the thymidine kinase genes, which are resistant to phosphonoacetic acid or acyclovir, respectively. Cells were treated with dUY11 from 1 to 24 h after infection, when they were harvested. Infectivity of the treated cells and their supernatants was a millionfold lower than that of untreated ones (Fig. 3E). Because dUY11 inhibits virion release by ≈2.8-fold (Fig. 2E), the infectivity of released virions was inhibited by ≈350,000-fold; dUY11 is therefore active against drug-resistant mutants.
RAFIs Target Viral Envelope Lipids.
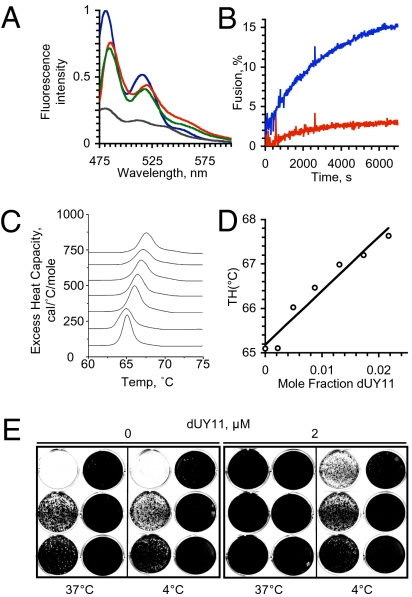
We next tested whether dUY11 targets virion lipids. VSV virions or liposomes were exposed to dUY11, which is intrinsically fluorescent (37). Fluorescence spectra are dependent on the polarity of the fluorochrome environment. We therefore tested whether the fluorescence spectra of dUY11 in virions was consistent with its placement in a hydrophobic environment. The RAFI dUY11 yielded most similar fluorescence spectra when mixed with VSV virions or liposomes (Fig. 4A). These spectra were distinct from the spectrum of dUY11 in aqueous environments, but very similar to that in octanol (Fig. 4A). Therefore, dUY11 was in similar organic environments, such as lipid membranes, in virions and liposomes.
Fig. 4.
The RAFI dUY11 targets lipid membranes. (A) Fluorescence spectra of dUY11 in octanol (blue), aqueous buffer (gray), or in aqueous buffer containing liposomes (green) or VSV virions (red); excitation at 460 nm. (B) Fluorescence dequenching of R18-labeled VSV virions preexposed to 15 nM dUY11 (red) or vehicle (blue) when fusing to Vero cells. (C) Differential scanning calorimetry of dUY11 in DEPE. Sections of heating scans showing the lamellar to hexagonal phase transition of membranes containing increasing mole fraction of dUY11 (0, 0.0021, 0.0048, 0.0086, 0.00129, 0.00172, or 0.0216, bottom to top traces). (D) Dose-response changes in transition temperature; the slope of the linear regression is 120 ± 13. (E) Vero cells were infected with 500,000 to 0.5 HSV-1 virions (in 10-fold dilutions from Top Left to Bottom Right in each panel) preexposed at 37 or 4 °C to 0 or 2 μM dUY11.
We next tested whether dUY11 inhibits fusion by analyzing lipid mixing between virions and target cells. VSV virions labeled with self-quenching concentrations of R18 were exposed to 15 nM dUY11 before mixing with target cells. To allow binding without triggering fusion, virus and cells were incubated on ice in a minimal volume at pH 7.4 for 30 min. The mix of cells and virus was then diluted 17-fold, the temperature was raised, and lipid bilayer fusion was triggered by decreasing the pH to 5.5 (Fig. 4B). Fusion was analyzed by the dequenching of R18 fluorescence. Fluorescence was dequenched by 15% when virions exposed to vehicle were induced to fuse to target cells. In contrast, it was dequenched by only 2.5% when virions exposed to dUY11 were induced to fuse under the same conditions. The RAFI dUY11 therefore inhibits lipid mixing by otherwise fusing lipid bilayers.
Therefore, dUY11 interacts with a hydrophobic component of the virions to inhibit lipid mixing with the target cells. The activation energy for fusion is partly required to overcome the negative curvature required during stalk formation. We therefore tested whether dUY11 inhibits the formation of lipid structures with negative curvature. Dielaidoylphosphatidylethanolamine (DEPE) lamellar phases were reconstituted with increasing concentrations of dUY11. The transition from the flat lamellar to the negatively curved hexagonal phase was evaluated by differential scanning calorimetry (Fig. 4C). Less than 2% dUY11 in DEPE increased the transition temperature from the lamellar to the hexagonal phase by 2 °C (Fig. 4D). These effects on the temperature required for the formation of an inverted phase with negative monolayer curvature, characterized by a regression of 120 ± 13, are indicative of dUY11 disfavoring negative membrane monolayer curvature.
Therefore, dUY11 interacts with hydrophobic structures in virions and inhibits the formation of the negative curvature required for fusion. We next tested whether interactions with the lipid envelope were required for inhibition of infectivity. Most eukaryotic lipid bilayers, including the envelopes of influenza virus (38), are rigid at 4 °C. We reasoned that such rigidity would prevent the insertion of hydrophobic compounds into envelopes (but not potential interactions with exposed glycoproteins). Virions exposed to dUY11 at 4 °C retained their infectivity (Fig. 4E), indicating that the inhibitory interactions between dUY11 and virions require envelope fluidity.
Discussion
We describe the discovery of unique inhibitors of infectivity of HCV, HSV-1 and 2, drug-resistant HSV-1 mutants, and other enveloped viruses. The inhibitors target the lipids in viral envelopes. Individually, each of the results could be explained by different mechanisms. However, the (i) molecular shape, amphipathicity, and rigidity requirements; (ii) inhibition of infectivity when only virions are exposed; (iii) lack of inhibition of primary attachment or secondary binding; (iv) inhibition of entry of tegument proteins into the target cells; (v) inhibition of infectivity of several otherwise unrelated enveloped viruses that bind to unrelated receptors and have no glycoprotein homology; (vi) lack of inhibition of infectivity of nonenveloped viruses that are internalized by endocytosis; (vii) interaction with hydrophobic component of virions; (viii) inhibition of lipid mixing in fusion assays; (ix) inhibition of transition from lamellar to hexagonal phase (i.e., from flat to negative curvature); (x) no inhibition when virions with rigid membranes are exposed; and (xi) no obvious envelope lytic effects (Fig. S3), are altogether the most consistent with RAFIs inhibiting viral infectivity by interacting with the lipids in the virion envelope to inhibit the increased negative curvature required for stalk formation, the first step in membrane fusion. Also consistently with this model, we have not been able yet to select for any RAFI-resistant HSV-1 mutants.
Although the targets of RAFIs are of cellular origin, their antiviral and cytotoxic effects are not related. For example, dUY9 has no antiviral effects but is cytotoxic, whereas aUY11 and Pv-ddUY11 are among the most potent and least cytotoxic RAFIs (Table S1). Some RAFIs are cytotoxic or cytostatic at IC50, others at concentrations 10-fold higher than IC50, and others are not cytostatic at 200-fold higher (Fig. S2 C and D). The RAFI dUY11 itself has no cytotoxic effects (Fig. 3C); it does not inhibit cell duplication (Fig. 1C), for example, a process that requires extensive fission and fusion of intracellular vesicular compartments, as well as fission of the plasma membrane. Several mechanistic details unique to viral fusion may result in such differential effects on virus infectivity. Virion envelopes are unique, metabolically inert, small extracellular vesicles of relatively strong positive curvature. ATP-consuming processes cannot introduce negative curvature to the virion envelope and virion envelope lipids are not enzymatically remodeled or translocated between leaflets. These characteristics disfavor the formation of the negative curvature required for fusion, and the activation energy required to overcome this limitation is provided solely by the virion glycoproteins (21). In contrast, fusion of cellular vesicles is facilitated by lipid-modifying enzymes, translocases, ATPases, and other proteins that actively modulate changes in curvature (39–43).
Strong amphiphiles of appropriate molecular shapes tend to spontaneously insert in lipid bilayers, when in aqueous environments, to minimize the free energy. Lipids with large hydrophilic heads only inhibit fusion when localized in the outer leaflets (31), as expected from their shape. The RAFI dUY11 is highly unlikely to translocate across leaflets; its polar head is much too large. Therefore, dUY11 most likely localizes to the outer leaflet of extracellular virions. In contrast, any dUY11 that gained access to intracellular vesicles would reside in the inner monolayer (because of the membrane inversion during endocytosis); it would thus not be able to inhibit physiological fusion processes, consistent with the observed lack of cytotoxic effects.
Previously, dUY11 had been evaluated in multiple replication-cycle assays, in which virions produced by one infected cell must infect—and replicate in—the neighboring cells (34). The IC50 reported under such conditions is in close agreement with the IC50 we observed toward the infectivity of virions produced by cells treated after infection (Table S4). The actual target of inhibition was therefore most likely not HSV-1 replication, but the infectivity of the virions produced by the primarily infected cells. Despite extensive attempts, we have been unable to reproduce earlier reports suggesting that dUY11 was specific for HSV-1 (44).
Current entry inhibitors target interactions between viral proteins and their receptors (8). Investigational compounds also nonspecifically crosslink viral glycoproteins (45). All such agents can promptly select for resistance. Previous attempts at targeting lipid envelopes aimed at destabilization with agents such as 9-nonaxonol, or fluidity modulation with cholesterol-like or cholesterol-binding molecules (46), or cholesterol regulating drugs (47). As membrane stability and fluidity are also required for cell integrity, such agents often result in cytotoxicity, mucosal injuries, or inflammation, which enhance infection. In contrast, RAFIs are designed to target lipids to affect the subtle but critical mechanistic differences between viral and cellular fusions.
We have shown that RAFIs inhibit virion fusion as a result of their shapes and amphipathicities. Then, chemically unrelated molecules could be designed to act through similar mechanisms. As this article was undergoing modifications, Wolf et al. reported chemically unrelated inhibitors of viral infectivity (48) with remarkably similar properties to the RAFIs described here. The compounds also inhibit infectivity of a variety of otherwise unrelated enveloped viruses without affecting that of nonenveloped ones. Interestingly, they also have a rigid and planar hydrophobic moiety attached to a hydrophilic head of larger diameter. The most potent compounds, LJ-001, -002, and -003 have a smaller hydrophobic moiety than dUY11 and an ∼200-fold higher IC50 toward enveloped viruses (48). Such high IC50 is consistent with the SAR data (Fig S1), and the mechanism of action herein proposed. However, other antiviral mechanisms are of course also possible for LJ-001, -002, or -003.
In conclusion, we show that synthetic rigid amphiphiles with hydrophilic heads of larger diameter than their hydrophobic tails inhibit viral infectivity by targeting virion envelope lipids. These molecules are active against a variety of otherwise unrelated enveloped viruses, including two important pathogens, HCV and HSV-2.
Methods
Cells and Viruses.
Vero Clone 57 cells express RFP under the control of the HSV-1 ICP0 promoter (35). Huh7.5 cells were obtained from C. Rice (The Rockefeller University, New York, NY). HCV strain JFH-1 was obtained from T. Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan).
Drugs.
The synthesis of dUY1, aUY1, dUY3, dUY11, aUY11, ddUY11, Pv-ddUY11, and Mk-dUY11 is described in the SI Methods (see also Figs. S4 and S5) and that of other compounds, elsewhere (49). Markiewicz's protective group was used to facilitate intermediate purifications. Compounds were prepared in DMSO as 10 mM stocks, and resuspended to the indicated concentrations in DMEM with or without FBS. Equivalent concentrations of DMSO were used in the controls.
Virion Exposure to RAFIs.
Equal volumes of virus and test compounds or vehicle at twice the desired concentration were prewarmed to 37 to 40 °C, mixed, and further incubated for 5 min immediately before infection. Virion infectivity was evaluated by plaquing efficiency.
Cytotoxicity Assays.
Vero cells were treated with the indicated concentrations of each RAFI in phenol red-free DMEM supplemented with 5% FBS. For incubations longer than 48 h, medium (with fresh drug) was replaced at 24, 48, and 72 h. Relative cell number was analyzed by MTT assays.
Infectivity of GFP-Expressing Viruses.
Recombinant HSV-1 or adenovirus expressing GFP were exposed to dUY11 before infecting HEK293 cells. GFP expression was monitored at 18 (HSV-1) or 24 (adenovirus) h later.
Infectivity of Progeny Virions.
Vero cells infected with 3 pfu/cell of HSV-1 or SIN were treated with dUY11 from 1 to 2 or 1 to 24 h after infection. Infectivity of progeny virions was evaluated in equal volumes of cell lysates or supernatants harvested at 24 h. To analyze budded virions, extracellular virions were purified from the supernatant by high-speed centrifugation followed by a sucrose cushion.
Binding and Entry Assays.
[35S]methionine labeled HSV-1 virions (∼1.2 cpm/pfu) were exposed to 7 μM dUY11 (140-fold above IC50) or 100 μg/mL heparin, and diluted in cold DMEM with dUY11, heparin, or no drug. Next, 1.6 × 105 cpm of virions was adsorbed onto Vero cells at 4 °C for 1 h before washing three times. Binding was calculated by cpm bound to cells divided by total cpm, adjusted by background. For secondary gD binding, radiolabeled virions exposed to dUY11 were adsorbed for 15 min at 4 °C. Cells were washed twice and either lysed (total binding) or washed for 1 h with heparin. Cell-bound cpm before and after heparin washes were used to calculate percentage of gD-mediated binding. For entry, HSV-1 stocks UV-inactivated by at least 10,000-fold, and which still induced RFP expression in clone 57 cells, were exposed to 0.050 to 2 μM dUY11 (3.5-fold below the concentrations that did not inhibit binding). RFP expression was evaluated 24 h after infection.
Temperature-Dependence Assays.
HSV-1 virions were mixed with an equal volume of 4 μM dUY11 (final concentration, 2 μM, 40-fold above IC50) for 5 min at 4 or 37 °C. Virions were then serially diluted 10-fold in cold DMEM before adsorption onto cells at 4 °C.
Cellular Localization of dUY11.
Near-confluent Vero cells were treated with 600 nM dUY11 (sixfold above IC50) for 5 min at 37 °C, washed, and incubated with 250 nM PKH26 membrane dye (PKH26). Fixed cells were evaluated by confocal microscopy.
HCV Infectivity Assays.
For HCV infectivity assays, 1.5 × 106 HCV genome equivalent copies (JFH-1 strain) were exposed to dUY11 in 500 μL at 37 °C for 5 min before infecting Huh7.5 cells. Infectivity of the exposed virions was evaluated by Western blot with mouse monoclonal anti-HCV NS3 (a nonstructural protein) helicase domain antibody (Chemicon International) 96 h after infection.
Membrane Curvature.
DEPE (Avanti Polar Lipids)-dUY11 mixes were dried under nitrogen gas and placed in a vacuum desiccator for 3 h. The dried films were hydrated with 0.8 mL of 20 mM Pipes, 0.14 M NaCl, 1 mM EDTA pH 7.4 by extensive vortexing, degassed, and placed in the calorimeter cell. The bilayer to hexagonal phase transition temperature was evaluated at a scan rate of 1°/min. Results were plotted in ORIGIN 7.0 and analyzed with DA-2 (Microcal, Inc.).
Lipid Fusion.
Lipid fusion was evaluated by octadecyl rhodamine B chloride (R18) dequenching. R18-labeled VSV virions were mixed with Vero cells on ice at pH 7.4. Fusion was triggered by decreasing the pH to 5.5.
Supplementary Material
Acknowledgments
We thank Dr. G. Eitzen for sharing his fluorometer and his advice on fusion assays and Mr. Le for providing technical assistance. This work was supported by the Burroughs Wellcome Fund and the Canadian Institutes of Health Research. L.M.S. is a Burroughs Wellcome Fund investigator. D.L.J.T. is a Canadian Institutes of Health Research/GlaxoSmithKline senior scientist. V.A.K. and A.V.U. were supported by the Molecular and Cellular Biology Program of the presidium of RAS and Dynasty Foundation.
Footnotes
Conflict of interest statement: M.R.S., A.V.U., and L.M.S. are co-inventors in a series of patent applications describing these discoveries.
This article is a PNAS Direct Submission. L.C. is a guest editor invited by the Editorial Board.
See Commentary on page 17069.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010026107/-/DCSupplemental.
References
- 1.Pillay D. The priorities for antiviral drug resistance surveillance and research. J Antimicrob Chemother. 2007;60(Suppl 1):i57–i58. doi: 10.1093/jac/dkm159. [DOI] [PubMed] [Google Scholar]
- 2.Phillips AN, et al. UK Collaborative HIV Cohort (CHIC) Study Risk of extensive virological failure to the three original antiretroviral drug classes over long-term follow-up from the start of therapy in patients with HIV infection: An observational cohort study. Lancet. 2007;370:1923–1928. doi: 10.1016/S0140-6736(07)61815-7. [DOI] [PubMed] [Google Scholar]
- 3.De Clercq E, Brancale A, Hodge AV, Field HJ. Antiviral chemistry and chemotherapy's current antiviral agents FactFile 2006 (1st edition) Antivir Chem Chemother. 2006;17:113–166. doi: 10.1177/095632020601700302. [DOI] [PubMed] [Google Scholar]
- 4.De Clercq E. Three decades of antiviral drugs. Nat Rev Drug Discov. 2007;6:941. [Google Scholar]
- 5.Gao M, et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96–100. doi: 10.1038/nature08960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schinazi RF, Bassit L, Gavegnano C. HCV drug discovery aimed at viral eradication. J Viral Hepat. 2010;17:77–90. doi: 10.1111/j.1365-2893.2009.01246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soares MM, King SW, Thorpe PE. Targeting inside-out phosphatidylserine as a therapeutic strategy for viral diseases. Nat Med. 2008;14:1357–1362. doi: 10.1038/nm.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwyer JJ, et al. Design of helical, oligomeric HIV-1 fusion inhibitor peptides with potent activity against enfuvirtide-resistant virus. Proc Natl Acad Sci USA. 2007;104:12772–12777. doi: 10.1073/pnas.0701478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münch J, et al. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129:263–275. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 10.Tan S-L, Ganji G, Paeper B, Proll S, Katze MG. Systems biology and the host response to viral infection. Nat Biotechnol. 2007;25:1383–1389. doi: 10.1038/nbt1207-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schang LM, St Vincent MR, Lacasse JJ. Five years of progress on cyclin-dependent kinases and other cellular proteins as potential targets for antiviral drugs. Antivir Chem Chemother. 2006;17:293–320. doi: 10.1177/095632020601700601. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Alvarez L, et al. Long-term monitoring of genotypic and phenotypic resistance to T20 in treated patients infected with HIV-1. J Med Virol. 2006;78:141–147. doi: 10.1002/jmv.20520. [DOI] [PubMed] [Google Scholar]
- 13.Baba M, Miyake H, Wang X, Okamoto M, Takashima K. Isolation and characterization of human immunodeficiency virus type 1 resistant to the small-molecule CCR5 antagonist TAK-652. Antimicrob Agents Chemother. 2007;51:707–715. doi: 10.1128/AAC.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schang LM. Herpes simplex viruses in antiviral drug discovery. Curr Pharm Des. 2006;12:1357–1370. doi: 10.2174/138161206776361174. [DOI] [PubMed] [Google Scholar]
- 15.Hao L, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlov MM, Leikin SL, Chernomordik LV, Markin VS, Chizmadzhev YA. Stalk mechanism of vesicle fusion. Intermixing of aqueous contents. Eur Biophys J. 1989;17:121–129. doi: 10.1007/BF00254765. [DOI] [PubMed] [Google Scholar]
- 17.Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J. 2006;91:487–495. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markin VS, Albanesi JP. Membrane fusion: Stalk model revisited. Biophys J. 2002;82:693–712. doi: 10.1016/S0006-3495(02)75432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chernomordik LV, Kozlov MM. Membrane hemifusion: Crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Huang HW. Observation of a membrane fusion intermediate structure. Science. 2002;297:1877–1879. doi: 10.1126/science.1074354. [DOI] [PubMed] [Google Scholar]
- 21.Epand RM. Fusion peptides and the mechanism of viral fusion. Biochim Biophys Acta. 2003;1614:116–121. doi: 10.1016/s0005-2736(03)00169-x. [DOI] [PubMed] [Google Scholar]
- 22.Chizmadzhev YA. The mechanisms of lipid-protein rearrangements during viral infection. Bioelectrochemistry. 2004;63:129–136. doi: 10.1016/j.bioelechem.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Rothman JE, Lenard J. Membrane asymmetry. Science. 1977;195:743–753. doi: 10.1126/science.402030. [DOI] [PubMed] [Google Scholar]
- 24.Chan R, et al. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Til NP, et al. Alteration of viral lipid composition by expression of the phospholipid floppase ABCB4 reduces HIV vector infectivity. Retrovirology. 2008;5:14. doi: 10.1186/1742-4690-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teissier É, Pécheur E-I. Lipids as modulators of membrane fusion mediated by viral fusion proteins. Eur Biophys J. 2007;36:887–899. doi: 10.1007/s00249-007-0201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheetham JJ, Nir S, Johnson E, Flanagan TD, Epand RM. The effects of membrane physical properties on the fusion of Sendai virus with human erythrocyte ghosts and liposomes. Analysis of kinetics and extent of fusion. J Biol Chem. 1994;269:5467–5472. [PubMed] [Google Scholar]
- 28.Yeagle PL, Young J, Hui SW, Epand RM. On the mechanism of inhibition of viral and vesicle membrane fusion by carbobenzoxy-D-phenylalanyl-L-phenylalanylglycine. Biochemistry. 1992;31:3177–3183. doi: 10.1021/bi00127a019. [DOI] [PubMed] [Google Scholar]
- 29.Miao L, et al. Potent inhibition of viral fusion by the lipophosphoglycan of Leishmania donovani. Biochemistry. 1995;34:4676–4683. doi: 10.1021/bi00014a022. [DOI] [PubMed] [Google Scholar]
- 30.Kooijman EE, Chupin V, de Kruijff B, Burger KN. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 2003;4:162–174. doi: 10.1034/j.1600-0854.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 31.Gaudin Y. Rabies virus-induced membrane fusion pathway. J Cell Biol. 2000;150:601–612. doi: 10.1083/jcb.150.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chernomordik LV, Leikina E, Kozlov MM, Frolov VA, Zimmerberg J. Structural intermediates in influenza haemagglutinin-mediated fusion. Mol Membr Biol. 1999;16:33–42. doi: 10.1080/096876899294733. [DOI] [PubMed] [Google Scholar]
- 33.WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skorobogatyi MV, et al. 5-Arylethynyl-2′-deoxyuridines, compounds active against HSV-1. Org Biomol Chem. 2006;4:1091–1096. doi: 10.1039/b516804j. [DOI] [PubMed] [Google Scholar]
- 35.Diwan P, Lacasse JJ, Schang LM. Roscovitine inhibits activation of promoters in herpes simplex virus type 1 genomes independently of promoter-specific factors. J Virol. 2004;78:9352–9365. doi: 10.1128/JVI.78.17.9352-9365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandenburg B, et al. Imaging poliovirus entry in live cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skorobogatyi MV, et al. Fluorescent 5-alkynyl-2′-deoxyuridines: High emission efficiency of a conjugated perylene nucleoside in a DNA duplex. Chem Bio Chem. 2006;7:810–816. doi: 10.1002/cbic.200600040. [DOI] [PubMed] [Google Scholar]
- 38.Polozov IV, Bezrukov L, Gawrisch K, Zimmerberg J. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat Chem Biol. 2008;4:248–255. doi: 10.1038/nchembio.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattila PK, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 41.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 42.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 43.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 44.Andronova VL, et al. Antiviral activity of some 5-arylethynyl 2′-deoxyuridine derivatives. Bioorg Chem. 2003;29:290–295. doi: 10.1023/a:1023936516589. (Translated from Russian) [DOI] [PubMed] [Google Scholar]
- 45.Balzarini J. Targeting the glycans of glycoproteins: A novel paradigm for antiviral therapy. Nat Rev Microbiol. 2007;5:583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harada S. The broad anti-viral agent glycyrrhizin directly modulates the fluidity of plasma membrane and HIV-1 envelope. Biochem J. 2005;392:191–199. doi: 10.1042/BJ20051069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabatov AA, Pollakis G, Linnemann T, Paxton WA, de Baar MP. Statins disrupt CCR5 and RANTES expression levels in CD4(+) T lymphocytes in vitro and preferentially decrease infection of R5 versus X4 HIV-1. PLoS ONE. 2007;2:e470. doi: 10.1371/journal.pone.0000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf MC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci USA. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skorobogatyi MV, et al. 5-Alkynyl-2′-deoxyuridines, containing bulky aryl groups: Evaluation of structure-anti-HSV-1 activity relationship. Tetrahedron. 2006;62:1279–1287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.