Abstract
Pathogens use specialized secretion systems and targeting signals to translocate effector proteins inside host cells, a process that is essential for promoting disease and parasitism. However, the amino acid sequences that determine host delivery of eukaryotic pathogen effectors remain mostly unknown. The Crinkler (CRN) proteins of oomycete plant pathogens, such as the Irish potato famine organism Phytophthora infestans, are modular proteins with predicted secretion signals and conserved N-terminal sequence motifs. Here, we provide direct evidence that CRN N termini mediate protein transport into plant cells. CRN host translocation requires a conserved motif that is present in all examined plant pathogenic oomycetes, including the phylogenetically divergent species Aphanomyces euteiches that does not form haustoria, specialized infection structures that have been implicated previously in delivery of effectors. Several distinct CRN C termini localized to plant nuclei and, in the case of CRN8, required nuclear accumulation to induce plant cell death. These results reveal a large family of ubiquitous oomycete effector proteins that target the host nucleus. Oomycetes appear to have acquired the ability to translocate effector proteins inside plant cells relatively early in their evolution and before the emergence of haustoria. Finally, this work further implicates the host nucleus as an important cellular compartment where the fate of plant–microbe interactions is determined.
Keywords: plant immunity, Phytophthora, Crinklers, translocation, nuclear localization
Plant and animal pathogens target secreted proteins (cytoplasmic effectors) inside host cells where they directly modify or perturb host cellular processes (1–5). These pathogens use specialized secretion systems for effector delivery, a process that in most cases requires a specific targeting signal. In bacteria, several secretion systems are known, among which the type III secretion system is critical for pathogenicity of several Gram-negative plant pathogenic bacteria (6, 7). In filamentous pathogenic eukaryotes (fungi and oomycetes), the mechanisms and amino acid sequence signals that determine host translocation remain mostly unknown. An exception is the oomycete RXLR family of cytoplasmic effectors (8–10). RXLR effectors are defined by a conserved N-terminal motif similar in sequence, position, and function to a host translocation signal present in malaria parasites that enables delivery of effector proteins inside plant and human cells (11–14). These effectors accumulate in specialized infection structures of Phytophthora, known as haustoria, before their delivery inside plant cells (14). Among the RXLR effectors, the Phytophthora infestans protein AVR3a is well studied and has been shown to confer avirulence on plants expressing the intracellular immune receptor R3a from potato (14–17).
Species of the oomycete genus Phytophthora are arguably the most devastating pathogens of dicotyledonous plants and include notorious pathogens, such as P. infestans and Phytophthora capsici that cause blight on several vegetable crops and related plant species (18, 19). The genomes of all Phytophthora species examined to date harbor large repertoires of RXLR effector genes, as well as another distinct class of candidate effectors termed “Crinklers” (CRNs) (4, 20, 21). CRN1 and CRN2 originally were identified following an in planta functional expression screen of candidate secreted proteins of P. infestans (21). Expression of both genes in Nicotiana spp. and in the host plant tomato results in a leaf-crinkling and cell-death phenotype accompanied by induction of defense-related genes (21). The CRN genes form the most expanded family in P. infestans (196 genes and 255 pseudogenes) relative to Phytophthora sojae (100 and 102, respectively) and Phytophthora ramorum (19 and 42, respectively) (20). Similar to the RXLR effector genes, this dramatic expansion of the CRN genes in P. infestans may be accounted for by their occurrence in gene-sparse genomic regions that are rich in repeats and transposable elements (20). Analogous to RXLR effectors, the CRN proteins have modular architectures that include a signal peptide, conserved N termini, and highly diverse C-terminal domains (20). However, the conserved CRN N termini harbor a distinct LXLFLAK motif, which is followed by the conserved DWL domain. A highly conserved HVLVXXP motif marks the end of the CRN N-terminal domains and forms a junction where diverse C-terminal domains are fused (20). Deletion analyses of the CRN2 protein defined a minimal 234-aa region (173–407, domain DXZ) to be sufficient for induction of cell death when expressed directly inside plant cells (20). Expression of four other C-terminal domains, DC, DBF, D2 and DXW-DXX-DXS, also leads to cell death inside plant cells (20). This modular structure of the CRNs, with conserved N termini and divergent C termini that execute effector activity, is reminiscent of the host-translocated RXLR effectors and has led Haas et al. (20) to propose that the CRNs are a second class of cytoplasmic effectors in the oomycetes. However, direct demonstration that CRN N termini can mediate translocation inside plant cells has not been reported yet.
In this study, we developed a translocation assay in the model oomycete species P. capsici and demonstrated that CRN N termini and, more specifically, the conserved LXLFLAK motif mediate translocation of C-terminal effector domains inside host plant cells. Remarkably, functional CRN-type N termini that can mediate host delivery of AVR3a occur widely in the oomycetes, unlike the more phylogenetically restricted RXLR translocation signals present in some of the species that produce haustoria. Subcellular localization of diverse CRN C termini inside plant cells revealed that they accumulate inside host nuclei in a manner dependent on the host nuclear import factor importin-α, and in the case of CRN8, on a functional nuclear localization signal (NLS). Cell death induced by CRN8 requires its accumulation in host nuclei, suggesting that CRN effectors target and perturb host nuclear processes. We conclude that the oomycetes acquired the ability to translocate effector proteins inside plant cells and target nuclei relatively early in their evolution and before the emergence of haustoria.
Results
AVR3a-Based Translocation Assay in P. capsici.
We devised a translocation assay that relies on recognition of the P. infestans avirulence protein AVR3a (Fig. 1). During P. infestans infection, AVR3a is secreted and subsequently delivered inside host cells in a process that requires its conserved RXLR motif (Fig. 1 B and C) (14). Delivery of AVR3a in the presence of the resistance protein R3a leads to recognition and induction of effector-triggered immunity (ETI) against P. infestans. To implement the assay efficiently, we first developed a rapid DNA transformation protocol for the model oomycete P. capsici that allows faster transgene expression and in vivo characterization than with P. infestans (Fig. S1; ref. 22). Next, we asked whether expression of the Avr3a gene in P. capsici confers avirulence on R3a-transgenic Nicotiana benthamiana leaves. P. capsici strains expressing Avr3a were inoculated onto both wild-type and R3a-transgenic N. benthamiana leaves, and lesion development was monitored on each genotype (Fig. 1 B and C). Strains carrying Avr3a were less virulent than control strains on wild-type N. benthamiana leaves and failed to infect in the presence of R3a, indicating R3a-dependent resistance to P. capsici. Strains lacking Avr3a robustly infected both genotypes, demonstrating that resistance was specified by recognition of AVR3a (Fig. 1 B and C). We further tested translocation requirements in our assays and found that, in contrast to wild-type Avr3a, mutation of its RXLR motif rendered P. capsici virulent on R3a leaves (Fig. 1 B and C). These results are consistent with previous reports demonstrating that the inability to accumulate inside host cells prevents R3a-dependent ETI (14–16). Taken together, these results show that R3a-mediated recognition can be used to infer translocation of the cytoplasmic effector AVR3a in P. capsici and that this assay can be used to test eukaryote translocation signals in vivo.
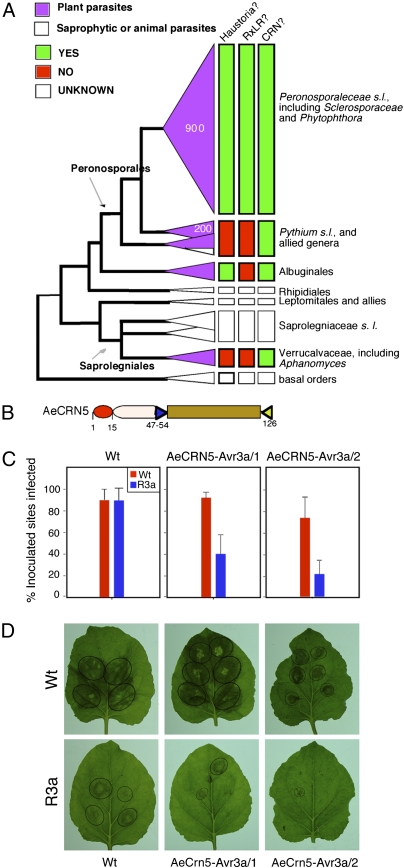
Fig. 1.
Expression of AVR3a in P. capsici results in avirulence on R3a N. benthamiana leaves. (A) Schematic of constructs used in translocation assays. Full-length Avr3a with a functional RXLR-dEER motif as well as its mutated full-length counterpart cloned in pTOR as described in Whisson et al. (14) were used to transform P. capsici. (B) Zoospore suspensions (50,000–100,000 spores/mL) of a P. capsici strain carrying the empty pTOR vector and of strains expressing either wild-type or mutated Avr3a were inoculated onto N. benthamiana wild-type and R3a leaves. Inoculation sites were scored for lesion formation 4 d after inoculation (dpi), and infection frequencies were calculated. Data represent three independent experiments. (C) Images showing representative wild-type and transgenic R3a leaves 4 dpi with strains analyzed in B. Avr3a expression was confirmed by RT-PCR analyses (Fig. S2). Lesions are marked by circles.
CRN N-Termini Mediate Translocation of AVR3a Inside Host Cells.
We used the AVR3a translocation assay to show that CRN N termini mediate host delivery (Fig. 2 B and C and Fig. S3). We exploited the fact that both CRN and RXLR effectors are modular proteins by fusing AVR3a C-terminal domains with several CRN N termini and transforming P. capsici with resultant CRN–AVR3a fusion constructs (Fig. 2A). Zoospore suspensions of P. capsici transformants expressing fusions between the N termini (SigP, LFLAK, and DWL domains) of P. infestans CRN2 [GenBank accession no. (GB): AF424677.1], CRN16 (GB: AY961464.1), and CRN8 (GB: AY961456.1) to the effector domain of AVR3a were inoculated onto both wild-type and transgenic R3a leaves (Fig. 2 and Fig. S3). Multiple independent transgenic P. capsici strains infected N. benthamiana wild-type plants (lacking R3a) but were unable to infect R3a plants, indicating recognition of the AVR3a effector domain by intracellular R3a (Fig. 2 B and C and Fig. S3). These results show that the CRN N termini are functionally equivalent to the N termini of RXLR effector proteins and that they can mediate the delivery of effector proteins inside host cells.
Fig. 2.
The CRN protein family forms a class of modular proteins that are translocated inside host cells. (A) Schematic representation of CRN2, CRN8, and CRN16 N-terminal domains identified in P. infestans and used for translocation assays presented. (B) Virulence assessment of transgenic P. capsici strains on wild-type and transgenic R3a N. benthamiana leaves. Infection rates of transgenic strains carrying empty vector (pTOR), CRN2:AVR3a, CRN16:AVR3a, and their mutated counterparts (CRN2mut:AVR3a, CRN16mut:AVR3a) were determined on wild-type and R3a leaves 4 dpi. Values represent the mean percentage of successful infections over three independent experiments. (C) Representative pictures showing wild-type and transgenic R3a leaves inoculated with strains carrying (Left to Right) the empty vector pTOR, CRN2:AVR3a, and CRN16:AVR3a as well as their mutated counterparts CRN2mut:AVR3a and CRN16mut:AVR3a. Pictures were taken 4 dpi. Crn-Avr3a expression was confirmed by RT-PCR analyses (Fig. S2). Lesions are marked by circles.
LXLFLAK Motif Is Required for Host Translocation.
The LXLFLAK motif is highly conserved in the N termini of CRN proteins, and in the case of several proteins of the downy mildew pathogen Hyaloperonospora arabidopsidis the RXLR motif overlaps with LXLFLAK, resulting in RXLRLFLAK (20, 23). This overlap suggested that, similar to the RXLR sequence, the LXLFLAK motif might be required for host targeting. To test this hypothesis, we constructed CRN2:AVR3a and CRN16:AVR3a derivatives with mutations in the LXLFLAK motif (to LXAAAA) and introduced these constructs into P. capsici (Fig. 2). P. capsici transformants expressing the mutated CRN–AVR3a fusions were unaltered in their virulence on R3a N. benthamiana leaves, in contrast to the wild-type CRN–AVR3a constructs (Fig. 2 B and C). Development of disease lesions was evident on R3a leaves inoculated with the mutated CRN–AVR3a strains, indicating absence of R3a-mediated resistance (Fig. 2 B and C). These results indicate that the LXLFLAK motif is required for effector targeting and translocation. Interestingly, this finding suggests that genome-wide searches for LXLFLAK-type motifs can be used to identify cytoplasmic effectors in oomycetes as routinely performed for RXLR effectors.
Unlike the RXLR Domain, the LXLFLAK Domain Occurs Widely in Oomycetes.
Within the Oomycota, plant parasitism has emerged at least three times in distinct phylogenetic lineages (24). Given the pivotal functions of effectors in pathogenicity, we assessed the occurrence of cytoplasmic effector classes across oomycete species for which genome sequences are available. Previous examinations of the genome of the Peronosporaleceae species P. sojae, P. ramorum, P. infestans, and H. arabidopsidis revealed large and diverse repertoires of RXLR effectors ranging from ∼150 in H. arabidopsidis to >500 in P. infestans (Fig. 3A) (20, 23, 25). In contrast, no or only a few classical RXLR genes are present in the phylogenetically diverse oomycetes Pythium ultimum, Albugo candida, and Aphanomyces euteiches (SI Text) (26, 27). These observations suggest that RXLR effectors have emerged and/or diversified following the evolution of haustoria in the Peronosporaleceae lineage (Fig. 3A). Surveys of genome and transcriptome sequences for the occurrence of the LXLFLAK domain typical of the CRNs (SI Text) revealed that every examined species, including P. ultimum, A. candida, and A. euteiches, have candidate CRN genes (26–28) and indicate that, unlike the RXLR effectors, the CRN family is ubiquitous in plant pathogenic oomycetes (26–28). These findings suggest that the CRNs form an ancient effector family that arose early in oomycete evolution before the emergence of haustoria.
Fig. 3.
CRN N-terminal domains are widespread within the oomycete lineage and function as translocation domains. Previously, examinations of A. euteiches sequence collections identified the CRN5 homolog AeCRN5 (26). (A) The CRN protein family arose before acquisition of haustorium formation. Genome-wide surveys for effector genes (described in SI Text) were superimposed on a phylogenetic tree describing oomycete evolutionary relationships (adapted from ref. 24). (B) Comparisons between the N terminus of AeCRN5 and known P. infestans CRNs highlight similarity in domain architecture. An LXLFLAK-like motif (LXLYLALK) is present 47 residues from the N terminus, and the conserved HVLVXXP motif is present. (C) The AeCRN5 N terminus fused to C-terminal AVR3a conditions avirulence on R3a but not on wild-type N. benthamiana leaves. Quantification of infection rates across three independent experiments (4 dpi). (D) The wild-type and transgenic R3a leaves inoculated with strains analyzed in C. Pictures were taken 4 dpi with zoospore suspensions. Transgene expression was confirmed by RT-PCR for each strain tested (Fig. S2). Lesions are marked by circles.
Functional CRN N Terminus in the Phylogenetically Distinct Oomycete A. euteiches.
A. euteiches is a root pathogen of legume plants that belongs to the Saprolegniales and is distantly related to Phytophthora and the downy mildews (24). A. euteiches CRN-like sequences carry a conserved N-terminal LQLYLALK motif similar to the Phytophthora LXLFLAK sequence (26). To determine if this LXLFLAK-like domain in A. euteiches also enables host translocation, we tested the AeCRN5 N terminus using the AVR3a translocation assay (Fig. 3 B–D). For this purpose, we made translational fusions between the AeCRN5 (Ae_1AL4462 in AphanoDB) (26) N terminus and the AVR3a C-terminal effector domain and transformed the resulting construct into P. capsici. Multiple independent strains expressing AeCrn5:Avr3a gene fusions were avirulent only in the presence of R3a (Fig. 3 C and D). Transformants were tested in three independent experiments, and R3a-mediated resistance was observed in all cases. Our results show that the LXLFLAK-like N-terminal domains found in A. euteiches can mediate translocation. These results show that CRN proteins form a widely distributed class of cytoplasmic effectors in the oomycetes. In summary, these observations point to the emergence of cytoplasmic effectors early in oomycete evolution and before the acquisition of haustoria.
Diverse CRN Effectors Localize to Plant Cell Nuclei.
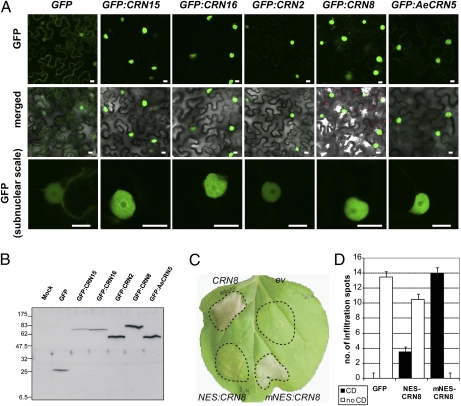
Because we have obtained significant evidence that CRN proteins are translocated effectors, we investigated their subcellular localization inside plant cells. To assess localization, we created N-terminal GFP fusions to the effector domains of CRN2 (DXZ), CRN8 (D2), CRN15 (DXW-DXX-DXV), CRN16 (DXW-DXX-DXV), and AeCRN5 (DN17) [effector domains after the HVLVXXP motif as defined by Haas et al. (20)]. We used agroinfiltration to express ectopically GFP:CRN fusions as well as free (cytosolic) GFP in N. benthamiana leaves and performed confocal microscopy 24 h after infiltration (Fig. 4A). Localization studies revealed that all five CRN fusion proteins accumulated in plant nuclei, indicating that distinct CRN C-terminal domains target host nuclear compartments (Fig. 4A). This nuclear localization contrasted sharply with the GFP control, which was distributed equally in the cytosol and nucleus, as reported previously (Fig. 4A) (29). We assessed GFP:CRN steady-state levels 24 h after infiltration by immunoblot analyses. Specific bands corresponding to the expected sizes were detected for all five GFP:CRN proteins, excluding the possibility of proteolytic degradation driving nuclear localization (Fig. 4B). Consistent with previous reports (20, 21), ectopic expression of CRN2 and CRN8 fusion proteins resulted in cell death, as did the AeCRN5 C-terminal domain (Fig. S4), indicating that GFP fusions did not perturb CRN function. In summary, these results indicate that, even though the tested CRN C-terminal domains are unrelated in sequence and originate from two divergent species, they specifically target the host nuclear compartment.
Fig. 4.
CRN effector domains traffic to and function in the host nucleus. (A) Confocal imaging of GFP, GFP:CRN15, GFP:CRN16, GFP:CRN2, GFP:CRN8, and GFP:AeCRN5 in N. benthamiana epidermal cells 24 h after infiltration. (Scale bars: 10 μm.) (B) Immunoblot analyses of GFP fusion protein accumulation in planta 24 h after infiltration. Blots were probed with α-GFP antibody. Sizes in kDa are indicated on the left. (C) Nuclear localization is required for effector-induced cell death. N. benthamiana leaves infiltrated with Agrobacterium strains carrying PVX CRN8, PVX NES:CRN8, or PVX mNES:CRN8 and an empty vector (ev) control. A representative picture was taken 5 d after infiltration. (D) Quantification and direct comparisons of cell death (CD) induced by nuclear-localized and nuclear-excluded CRN8. For each experiment, 14 leaves were infiltrated with strains as shown in C. Leaf panels were scored 3 d after infiltration, and the number of infiltration sites showing cell death was counted.
Nuclear Localization of the CRNs Requires Host Importin-α.
In eukaryotic cells, protein import and subsequent accumulation of nuclear proteins requires functional importin-α, a component of the nuclear import machinery that binds to basic NLS motifs (30, 31). Because diverse CRN effector domains accumulate in nuclei, we asked whether these proteins harbor functional NLS motifs. Analyses of the five CRN effector domains unveiled predicted NLS motifs in CRN8, CRN16, and AeCRN5 (Fig. S5), indicating possible reliance on the host nuclear import machinery for nuclear accumulation. To test this notion, we used virus-induced gene silencing (VIGS) to knock down the expression of the N. benthamiana importin-α homologs NbImpα1 and NbImpα2 and expressed the GFP:CRN proteins as well as a fusion to the bacterial nuclear effector AvrHah1 in silenced plants using the methods described by Kanneganti et al. (30). GFP:CRN2, GFP:CRN8, GFP:CRN16, and GFP:AeCRN5 showed marked localization shifts toward the cytosol, similar to shifts seen for GFP:AvrHah1 in NbImpα-silenced leaves, compared with negative controls (Fig. S6). These results indicate that the CRN proteins recruit the host nuclear import machinery to achieve nuclear accumulation. This observation further underlines our view of CRN proteins as host-translocated effectors that target the nucleus during infection.
CRN8 Requires Nuclear Localization to Trigger Plant Cell Death.
Ectopic expression of several CRN C termini inside plant cells results in cell death (20, 21). Given the observation that CRNs accumulate inside plant nuclei, we hypothesized that nuclear localization is required for cell-death induction. To test this hypothesis, we fused the C-terminal domain of CRN8 to a nuclear export signal (NES) or its mutated (mNES) counterpart (32) and ectopically expressed the resulting constructs in N. benthamiana leaves using agroinfiltration (Fig. 4C). NES:CRN8 consistently failed to elicit cell death in planta, whereas CRN8 fused to the mutated (inactive) NES domain robustly induced cell death (Fig. 4 C and D). NES prevented nuclear accumulation of CRN8, as inferred from their accumulation in the cytosol in our localization experiments. (Fig. S7). To test further the involvement of nuclear import in CRN function, we created an N-terminal GFP fusion to a C-terminal CRN8 effector domain carrying a mutated NLS. Ectopic expression of GFP:CRN8 fusion proteins harboring either the mutated or wild-type NLS resulted in a significant shift toward cytosolic GFP accumulation in the mutant and abolishment of cell-death induction in planta (Fig. S8 A and B). Western blot analyses demonstrated that mutating the CRN8 NLS did not affect protein stability (Fig. S8D). These results are consistent with the observed reduction in cell death upon nuclear exclusion of CRN8. We therefore conclude that inhibition of CRN8 nuclear accumulation prevents cell death and thus that CRN8 nuclear localization forms an important requirement for cell-death induction. These results indicate that CRN8 requires nuclear accumulation to carry out its effector activities and, by extension, suggest that CRN proteins are targeted to the host nucleus during infection.
Discussion
The oomycete RXLR and RXLR-dEER motifs are the only conserved sequence determinants of host translocation that have been reported so far in filamentous plant pathogens (8–10, 33). Analyses of the recently published genome of the Irish potato famine pathogen P. infestans unveiled the CRNs as a large and complex family of candidate cytoplasmic effectors (20). Here we provide direct evidence of host translocation mediated by CRN N termini. We developed a translocation assay that relies on accumulation and recognition of the C-terminal effector domain of AVR3a. Delivery of AVR3a leads to R3a-dependent ETI, evidenced by avirulence in the presence of R3a (Fig. 1). We demonstrate that CRN N termini are functionally interchangeable with those of RXLR effectors and can deliver the C-terminal half of AVR3a inside plant cells. Host translocation requires the conserved LXLFLAK motif, which is distributed ubiquitously in plant pathogenic oomycetes (26, 27). Remarkably, the N terminus of AeCRN5, a CRN-like protein from the unrelated oomycete pathogen A. euteiches, also can mediate translocation of AVR3a inside plant cells. Translocation activity specified by CRN N termini present in divergent species implies that the CRNs are an ancient family of cytoplasmic effectors that arose early in oomycete evolution.
Although the CRNs are ubiquitous in plant pathogenic oomycetes, the RXLR effectors appear to be restricted phylogenetically to Phytophthora and downy mildews within the Peronosporales and to be missing in P. ultimum and the more distantly related oomycete A. euteiches (26, 27) (Fig. 3A). Levesque et al. (27) suggested that RXLR effectors may have evolved relatively recently in the Phytophthora/downy mildew lineage, coinciding with the emergence of haustoria. Indeed, RXLR effectors such as AVR3a were shown to accumulate in haustoria (14) (Fig. S1), and the expression of many RXLR effector genes peaks during the biotrophic phase when the maximal number of haustoria are produced (20). Interestingly, P. ultimum and A. euteiches, which do not form haustoria, have relatively few RXLR effectors but have a number of CRN-like effectors (26, 27). These observations point to a possible conserved translocation pathway that preceded the emergence of RXLR effectors during oomycete evolution and raise important questions regarding the evolution of oomycete pathogenesis.
Future work will determine the extent to which effector trafficking is confined to the haustorial interface, as suggested for RXLR effectors (14, 34). The identification of cytoplasmic effectors in pathogenic oomycetes that lack haustoria suggests that host translocation can occur during various infectious and developmental stages. These findings also highlight the importance of studying phylogenetically diverse pathogenic oomycetes, such as Pythium, Albugo, and Aphanomyces species. Comparative analyses of a broader spectrum of oomycete species probably will expand our knowledge of pathogenicity mechanisms (24).
What are the mechanisms of CRN effector entry into host cells? Contrasting models of effector entry have emerged following the identification of host-translocated RXLR and PEXEL proteins in oomycetes and Plasmodium spp., respectively (2, 11, 12, 35). Although the RXLR and PEXEL domains are known to be functionally interchangeable, different mechanisms of translocation have been proposed. Dou et al. (12) reported that soybean cells internalize purified AVR1b fusion proteins in the absence of any pathogen, thereby ruling out the involvement of pathogen-encoded machinery. However, the recent identification of proteins that form part of a translocon in Plasmodium is inconsistent with passive uptake and argues in favor of pathogen-encoded machineries (35, 36). Future studies addressing CRN translocation requirements may provide insights into effector translocation mechanisms.
Here we show that a large class of translocated effectors target host nuclei by co-opting the host nuclear import machinery (37, 38). The identification and characterization of nuclear effectors in bacterial pathogens such as Xanthomonas spp. and Phytoplasma asteris (39–42) support the view that effector function is contingent upon fulfilling spatial requirements (nuclear localization) during infection. Exclusion of CRN8 from the plant nucleus, either by fusion to an NES or by mutation of its NLS, reduced cell-death induction in our experiments (Fig. 4 C and D and Fig. S8), indicating that CRN8 effector activity requires nuclear accumulation. Therefore, we propose that during infection oomycete plant pathogens traffic CRN effectors to the host nucleus to promote virulence. Identification of CRN host targets and signaling events therefore will help delineate effector function and the nuclear processes perturbed during host–microbe interactions.
Specialized secretion systems and their effector substrates are principal determinants of pathogenicity in both plant and animal parasites. In this study, we validated the CRN family as a class of host-translocated oomycete effectors that exclusively target the host nucleus. Similar to the well-known RXLR effector class, CRN proteins are modular, with the N termini functioning in host delivery and the C termini encoding the effector activity. Our findings further emphasize the importance of host translocation processes in pathogenic microbes. They also imply that the cytoplasmic effector repertoire of oomycete pathogens, such as P. infestans, is remarkably large and complex (20). We conclude that CRN effectors arose early in oomycete evolution and then diversified across plant pathogenic species. Given that such a large and ancestral class of cytoplasmic effectors specifically targets the host nucleus, our work confirms this compartment as a key interface where the outcomes of host–microbe interactions are determined.
Materials and Methods
Plant and Bacterial Growth Procedures and P. capsici Culturing.
All N. benthamiana plant genotypes used in this study were grown from seeds in growth chambers at 22 °C with 16-h photoperiod and 55% humidity. Fully expanded leaves from 3- to 4-wk-old plants were used for all experiments in this study. Escherichia coli and Agrobacterium tumefaciens bacterial strains were routinely grown in LB at 37 °C and 28 °C, respectively, using appropriate selectable markers. P. capsici strain LT1534 was used in this study, and cultures were grown on solid (1.5% agar) 10% (vol/vol) unclarified V8 medium supplemented with 50 mg/L β-sitosterol and 1.0 g/L CaCO3. P. capsici transformants were generated as described in SI Text and subsequently maintained on solid V8 medium, supplemented with 50 μg/mL carbenicillin, 25 μg/mL vancomycin, and 50 μg/mL G418 (Geneticin).
Plasmid Construction and Preparation.
All primers used in this study are listed in Table S1. Cloning methods are described in SI Text. Phytophthora transformation constructs CRN2:AVR3a, CRN8:AVR3a, and CRN16:AVR3a were synthesized and cloned into pTOR by Genscript Corp. Fusion genes were flanked by ClaI (5′) and SacII (3′) sites, and internal AscI sites were inserted between the CRN and AVR3a coding regions to facilitate subsequent cloning applications.
Phytophthora Infection Assays.
Phytophthora infection assays were performed on detached leaves that were placed on prewetted paper in large square (22 × 22 cm) incubator plates at room temperature. P. capsici translocation assays were performed by drop inoculation of zoospore suspensions (50,000–100,000 spores/mL) onto N. benthamiana wild-type and transgenic R3a leaves. In each experiment, three leaves of each genotype were placed in a single plastic tray (six total), and each leaf was inoculated by placing six 20-μL droplets of zoospore suspension on the lower side of the leaf. For each strain and plant genotype, infection rate as determined by the formation of spreading and/or sporulating lesions was recorded. Each transgenic P. capsici strain used in this assay was tested in at least three independent experiments.
Protein Extractions and Analyses.
Immunoblot analyses were performed on protein extracts prepared by grinding an ≈100-mg leaf sample in liquid nitrogen, after which the powder was suspended into GTEN extraction buffer [10% (vol/vol) glycerol, 25 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl] supplemented with 10 mM DTT, 2% (wt/vol) polyvinylpolypyrrolidone (PVPP), and 1× protease inhibitor mixture (Sigma). Resultant suspensions were mixed and spun at full speed (16,000 × g) at 4 °C, and supernatants were separated from pellets. Samples were loaded onto and separated in standard SDS/PAGE gels (12.5% acryl amide) and blotted using standard procedures. Blots were blocked using Tris-buffered saline-Tween 20 (TBST)-milk (5% wt/vol) for 3 h before α-GFP antibody (Invitrogen) was incubated in TBST-milk (1:2,000, 5%) at 4 °C for 48 h. Blots were washed four times in TBST for 10 min before being probed with HRP-conjugated α-rabbit polyclonal antibody (Sigma). Blots were washed four times in TBST before being developed using the Pierce Horse Radish Peroxidase detection kit (Thermo Scientific) and exposed for 2 min.
Transient Agro-Infiltration Assays.
For in planta expression of all relevant 35S constructs, A. tumefaciens strain GV3101 was used to deliver respective transgenes in N. benthamiana leaves, using methods described elsewhere (43, 44). A. tumefaciens cultures were grown to late-log phase, spun down by centrifugation, and resuspended in infiltration medium [10 mM MgCl2, 5 mM MES (pH 5.3), and 150 μM acetosyringone] to an OD600 = 0.3 before infiltration.
Confocal Microscopy.
Cut leaf patches were mounted in water and analyzed on a Leica DM6000B/TCS SP5 confocal microscope (Leica Microsystems) with the following excitation wavelengths: CFP, 458 nm; GFP, 488 nm; RFP, 561 nm. When required, scanning was performed in sequential mode to prevent signal bleed-through. Identical microscope power settings were applied for all individual images of a figure to allow comparison of fluorescence intensities between samples. Fluorescence intensity transects were generated using the Leica analysis software LAS AF1.8.2.
Supplementary Material
Acknowledgments
We thank S. Whisson (Scottish Crop Research Institute, Dundee, UK) for the pTOR constructs, D. Baulcombe (University of Cambridge, Cambridge, UK) for the PVX vector, and E. Kemen and J. D. G. Jones (The Sainsbury Laboratory, Norwich, UK) for access to A. candida sequence data. S.S. was supported by Grant SCHO1347/1-1 from the Deutsche Forschungsgemeinschaft, T.O.B. by Marie Curie Grant FP7-PEOPLE-2007-2-1-IEF, and S.K.’s laboratory was supported by the Gatsby Charitable Foundation and the Biotechnology and Biological Sciences Research Council. E.H. is supported by the Royal Society of Edinburgh.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008491107/-/DCSupplemental.
References
- 1.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Crabb BS, de Koning-Ward TF, Gilson PR. Protein export in Plasmodium parasites: From the endoplasmic reticulum to the vacuolar export machine. Int J Parasitol. 2010;40:509–513. doi: 10.1016/j.ijpara.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Hogenhout SA, Van der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact. 2009;22:115–122. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- 4.Kamoun S. Groovy times: Filamentous pathogen effectors revealed. Curr Opin Plant Biol. 2007;10:358–365. doi: 10.1016/j.pbi.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Haase S, de Koning-Ward TF. New insights into protein export in malaria parasites. Cell Microbiol. 2010;12:580–587. doi: 10.1111/j.1462-5822.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 6.Alfano JR. Roadmap for future research on plant pathogen effectors. Mol Plant Pathol. 2009;10:805–813. doi: 10.1111/j.1364-3703.2009.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block A, Li G, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birch PR, et al. Oomycete RXLR effectors: Delivery, functional redundancy and durable disease resistance. Curr Opin Plant Biol. 2008;11(4):373–279. doi: 10.1016/j.pbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Morgan W, Kamoun S. RXLR effectors of plant pathogenic oomycetes. Curr Opin Microbiol. 2007;10:332–338. doi: 10.1016/j.mib.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Panstruga R, Dodds PN. Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science. 2009;324:748–750. doi: 10.1126/science.1171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhattacharjee S, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathog. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou D, et al. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grouffaud S, van West P, Avrova AO, Birch PR, Whisson SC. Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology. 2008;154:3743–3751. doi: 10.1099/mic.0.2008/021964-0. [DOI] [PubMed] [Google Scholar]
- 14.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong MR, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos JI, et al. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 2006;48:165–176. doi: 10.1111/j.1365-313X.2006.02866.x. [DOI] [PubMed] [Google Scholar]
- 17.Bos JIB, Chaparro-Garcia A, Quesada-Ocampo LM, McSpadden Gardener BB, Kamoun S. Distinct amino acids of the Phytophthora infestans effector AVR3a condition activation of R3a hypersensitivity and suppression of cell death. Mol Plant Microbe Interact. 2009;22:269–281. doi: 10.1094/MPMI-22-3-0269. [DOI] [PubMed] [Google Scholar]
- 18.Lamour K, Kamoun S. Oomycete Genetics and Genomics: Diversity, Interactions and Research Tools. Hoboken, NJ: Wiley-Blackwell; 2009. [Google Scholar]
- 19.Lamour KH, Win J, Kamoun S. Oomycete genomics: New insights and future directions. FEMS Microbiol Lett. 2007;274:1–8. doi: 10.1111/j.1574-6968.2007.00786.x. [DOI] [PubMed] [Google Scholar]
- 20.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 21.Torto TA, et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 2003;13:1675–1685. doi: 10.1101/gr.910003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huitema E, Smoker M, Kamoun S. A straightforward protocol for electro-transformation of Phytophthora capsici zoospores. In: McDowell JM, editor. Methods in Molecular Biology: Plant Immunity. Humana, Clifton, NJ; 2010. in press. [DOI] [PubMed] [Google Scholar]
- 23.Win J, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thines M, Kamoun S. Oomycete-plant coevolution: Recent advances and future prospects. Curr Opin Plant Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Jiang RH, Tripathy S, Govers F, Tyler BM. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci USA. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaulin E, et al. Transcriptome of Aphanomyces euteiches: New oomycete putative pathogenicity factors and metabolic pathways. PloS One. 2008;3(3):e1723. doi: 10.1371/journal.pone.0001723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lévesque CA, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biology. 2010;11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung F, et al. Analysis of the Pythium ultimum transcriptome using Sanger and Pyrosequencing approaches. BMC Genomics. 2008;9:542. doi: 10.1186/1471-2164-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grebenok RJ, et al. Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 1997;11:573–586. doi: 10.1046/j.1365-313x.1997.11030573.x. [DOI] [PubMed] [Google Scholar]
- 30.Kanneganti T-D, et al. A functional genetic assay for nuclear trafficking in plants. Plant J. 2007;50:149–158. doi: 10.1111/j.1365-313X.2007.03029.x. [DOI] [PubMed] [Google Scholar]
- 31.Lange A, et al. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 33.Oliva R, et al. Recent developments in effector biology of filamentous plant pathogens. Cell Microbiol. 2010;12:705–715. doi: 10.1111/j.1462-5822.2010.01471.x. [DOI] [PubMed] [Google Scholar]
- 34.Avrova AO, et al. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol. 2008;10:2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- 35.de Koning-Ward TF, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schornack S, et al. Ten things to know about oomycete effectors. Mol Plant Pathol. 2009;10:795–803. doi: 10.1111/j.1364-3703.2009.00593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 38.Kanneganti TD, Huitema E, Kamoun S. In planta expression of oomycete and fungal genes. Methods Mol Biol. 2007;354:35–43. doi: 10.1385/1-59259-966-4:35. [DOI] [PubMed] [Google Scholar]
- 39.Bai X, et al. AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol Plant Microbe Interact. 2009;22:18–30. doi: 10.1094/MPMI-22-1-0018. [DOI] [PubMed] [Google Scholar]
- 40.Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Römer P, et al. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 2008;179:546–556. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 43.Huitema E, et al. Linking sequence to phenotype in Phytophthora-plant interactions. Trends Microbiol. 2004;12:193–200. doi: 10.1016/j.tim.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Lindbo JA. High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors. BMC Biotechnol. 2007;7:52. doi: 10.1186/1472-6750-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.