Abstract
Proteus mirabilis causes complicated urinary tract infections (UTI). While the urinary tract is an iron-limiting environment, iron acquisition remains poorly characterized for this uropathogen. Microarray analysis of P. mirabilis HI4320 cultured under iron limitation identified 45 significantly up-regulated genes (P ≤ 0.05) that represent 21 putative iron-regulated systems. Two gene clusters, PMI0229-0239 and PMI2596–2605, encode putative siderophore systems. PMI0229-0239 encodes a nonribosomal peptide synthetase (NRPS)-independent siderophore (NIS) system for producing a novel siderophore, proteobactin. PMI2596-2605 are contained within the high-pathogenicity island, originally described in Yersinia pestis, and encodes proteins with apparent homology and organization to those involved in yersiniabactin production and uptake. Cross-feeding and biochemical analysis shows that P. mirabilis is unable to utilize or produce yersiniabactin, suggesting that this yersiniabactin-related locus is functionally distinct. Only disruption of both systems resulted in an in vitro iron-chelating defect; demonstrating production and iron-chelating activity for both siderophores. These findings clearly show that proteobactin and the yersiniabactin-related siderophore function as iron acquisition systems. Despite the activity of both siderophores, only mutants lacking the yersiniabactin-related siderophore reduce fitness in vivo. The fitness requirement for the yersiniabactin-related siderophore during UTI shows, for the first time, the importance of siderophore production in vivo for P. mirabilis.
Keywords: Proteus, uropathogenesis, iron acquisition, ferri-siderophore
Introduction
Iron, an essential element, is required for the function of many proteins and enzymes involved in diverse biological processes including oxygen transport, gene regulation, and nitrogen fixation. Under aerobic conditions and neutral pH, iron exists in the insoluble ferric (Fe3+) form, which can be toxic following interaction with oxygen and oxygen-reduced species. During colonization of the host, pathogens must overcome host iron sequestration to establish infections because free iron is essentially unavailable in vivo. To counterbalance iron-limiting conditions and maintain iron homeostasis, bacterial species have evolved various iron transport systems, intracellular iron stores, redox stress resistance systems, and iron responsive regulatory elements to control the expression of genes involved in diverse cellular functions (Andrews et al., 2003).
One way bacteria acquire extracellular ferric iron is by secreting ferric chelators known as siderophores that scavenge iron from the environment during iron-limiting conditions. Over 500 siderophores have been described and can be classified into three groups; catecholates, hydroxamates, and hydroxycarboxylates (Miethke & Marahiel, 2007). Precursors for siderophore biosynthesis include citrate, amino acids, dihydroxybenzoate, and N5-acyl-N5-hydroxyornithine (Winkelmann, 2002). Often, genes that encode the biosynthetic enzymes for siderophores are clustered with genes for ferri-siderophore transport (Wandersman & Delepelaire, 2004, Koster, 2001, Crosa & Walsh, 2002). The mechanism of siderophore synthesis and transport is tightly regulated by the ferric uptake regulator (Fur) protein in response to iron availability (Ernst et al., 1978, Crosa & Walsh, 2002, Hantke, 2001).
Once synthesized and transported outside the bacterial cell, the siderophore chelates ferric iron with high affinity. The ferri-siderophore, in turn, binds with high specificity to a TonB-dependent outer membrane receptor in Gram-negative bacterial envelopes. Transport into the cell is driven by cytosolic membrane potential mediated by the energy-transducing TonB-ExbB-ExbD system which requires the direct contact of TonB and the outer membrane receptor (Braun, 1995, Tuckman & Osburne, 1992). Once in the periplasmic space, the ferri-siderophore is shuttled by a periplasmic binding protein to a cytosolic membrane ATP-binding cassette (ABC) transporter. The ferri-siderophore is delivered to the cytosol of the bacterium where iron is reduced to the ferrous (Fe2+) form and dissociates from the siderophore (Neilands, 1995, Faraldo-Gomez & Sansom, 2003, Wandersman & Delepelaire, 2004, Ratledge & Dover, 2000).
Previous reports have indicated that Proteus mirabilis, a species within the Enterobacteriaceae and an important etiologic agent of complicated urinary tract infection (UTI) (Mobley & Warren, 1987, Warren et al., 1982), lacks detectable siderophore production (Massad et al., 1995, Marcelis et al., 1978, Miles & Khimji, 1975). The Arnow and Csaky tests failed to detect catechol- or hydroxamate-type siderophores, respectively, when testing P. mirabilis P18 (Evanylo et al., 1984). These findings were confirmed with gas chromatography in conjunction with mass spectroscopy (Evanylo et al., 1984). However, analysis of the newly annotated P. mirabilis HI4320 genome (Pearson et al., 2008) revealed at least two gene clusters with genes related to both siderophore biosynthesis and ABC transport. One of these appears to be a novel nonribosomal peptide synthetase (NRPS)-independent siderophore (NIS) system, herein termed proteobactin that has not been previously described in any bacterial species. The other gene cluster contains the nrp operon which has been previously described to be up-regulated during iron limitation in P. mirabilis U6450 (Gaisser & Hughes, 1997). Recently, the nrp operon has been shown to be encoded within the high-pathogenicity island (HPI) in P. mirabils HI4320 that has homology to the HPI of Yersinia spp. (Flannery et al., 2009) In addition to the possibility that nrp has a function related to iron limitation, the metabolite, α-hydroxyisovaleric acid, has been identified as a possible siderophore (Evanylo et al., 1984). α-keto acids produced by amino acid deaminases and α-hydroxycarboxylic acids have also been postulated as possible siderophores in Proteus, Providencia, and Morganella species (Drechsel et al., 1993, Massad et al., 1995). Despite these studies, none of these metabolites nor those produced by the nrp gene products have been shown to possess iron-chelating properties.
Studies on uropathogenic Escherichia coli have shown that the urinary tract is iron-limited and that iron acquisition by outer membrane receptors is important during UTI (Alteri & Mobley, 2007, Torres et al., 2001, Hagan & Mobley, 2009, Snyder et al., 2004). Consistent with this, multiple outer membrane proteins of P. mirabilis are up-regulated in both human urine and iron-limiting medium (Shand et al., 1985) and three outer membrane proteins induced by iron-starvation are involved in heme uptake in P. mirabilis 6515 (Piccini et al., 1998). One of these, was shown to function as a heme receptor that contributes to the uropathogenesis of P. mirabilis 6515 (Lima et al., 2007). Using signature-tagged mutagenesis (STM), we previously identified five genes (PMI3120; putative TonB-dependent receptor, PMI2605; putative 4′-phosphopantetheinyl transferase nrpG, PMI2959; putative iron ABC transporter permease, PMI0842; putative TonB-dependent receptor, PMI0030; biopolymer transport protein exbD) associated with iron acquisition in P. mirabilis HI4320 that when mutated attenuated the bacterium (Burall et al., 2004, Himpsl et al., 2008).
Since iron acquisition remains poorly defined for P. mirabilis, we sought to define the global response to iron limitation by first using microarray analysis. This approach allowed for the identification of 21 putative iron acquisition systems based upon the 45 significantly up-regulated genes during in vitro iron limitation. Molecular and biochemical characterization of two of these iron acquisition systems verified that these genes are transcriptionally up-regulated during iron limitation, repressed by iron, and are responsible for siderophore production in this human pathogen.
Results
Differential expression of P. mirabilis genes during iron limitation
Gene expression of P. mirabilis HI4320 cultured under iron limitation was analyzed by microarray. A concentration of 15 μM Desferal, an iron-chelator, inhibited the growth rate of P. mirabilis in LB medium (Fig. 1). Microarray analysis of cells cultured in LB medium with and without 15 μM Desferal showed that 1017 genes were differentially expressed: 872 were up-regulated and 145 were down-regulated. Fold-change for all 1017 genes was ≥ 2-fold and calculated based upon the ratio of average spot intensity between these conditions. Of these genes, Significance Analysis of Microarray (SAM) identified 45 significantly up-regulated genes (Table S1) and 43 significantly down-regulated genes (Table S2).
Fig. 1. The growth rate of P. mirabilis HI4320 is decreased following culture in LB medium containing Desferal.
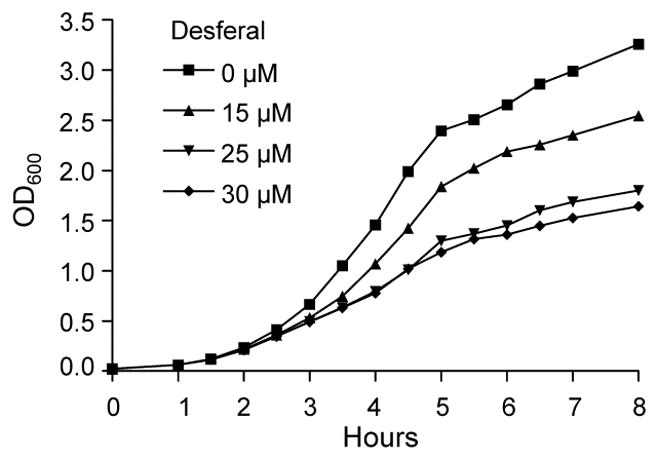
P. mirabilis HI4320 cultured in LB medium containing 0, 15, 25, or 30 μM Desferal, an iron-chelating agent. A concentration of 15 uM Desferal in LB medium decreases the growth rate of wild-type in comparison to wild-type grown in LB medium alone.
The 45 significantly up-regulated genes represent 21 putative iron acquisition systems. These systems include the energy transducing complex TonB-ExbB-ExbD, genes for heme uptake (PMI0409, PMI1424, and hmuR1R2STUV), an aerobic ferrous iron uptake system (sitABCD) (Fisher et al., 2009), two putative ferri-siderophore systems (nrpSUTABG and PMI0229-0239), and three putative ferri-siderophore transporters (ireA, PMI0331, and PMI2957-2960) (Table 1). Other genes induced during iron limitation are a putative iron utilization protein PMI1437, two putative TonB-dependent systems that may be involved in colicin uptake, PMI1551-1548 and PMI0842, and systems with potential iron-sulfur clusters, sufABCDSE, bfd, and PMI0176-0172. As expected, in contrast to sitABCD the P. mirabilis ferrous iron uptake system feoAB was not up-regulated ≥ 2-fold during aerobic iron limitation. Genes repressed during iron limitation include predicted iron storage proteins and iron-metalloproteins.
Table 1.
Putative iron acquisition systems up-regulated in P. mirabilis HI4320 during iron-limiting microarraya
| Functionb | PMI no. | Gene | Description | Log2 fold-changec |
|---|---|---|---|---|
| Heme uptake | 0409* | putative TonB-dependent heme receptor | 6.22 | |
| 1424* | putative hemin uptake protein | 8.52 | ||
| 1425* | hmuR1 | hemin receptor | 5.92 | |
| 1426* | hmuR2 | hemin receptor | 7.89 | |
| 1427* | hmuS | hemin transport protein | 6.96 | |
| 1428* | hmuT | hemin binding periplasmic protein | 5.02 | |
| 1429 | hmuU | hemin transport system; permease protein | 3.42 | |
| 1430* | hmuV | hemin transport system; ATP-binding protein | 5.42 | |
| Ferrous iron uptake | 1024 | sitD | iron ABC transporter; membrane protein | 3.41 |
| 1025 | sitC | iron ABC transporter; membrane protein | 4.81 | |
| 1026* | sitB | iron ABC transporter; ATP-binding protein | 5.49 | |
| 1027 | sitA | iron ABC transporter; periplasmic substrate binding protein | 5.93 | |
| Ferric citrate transport | 3704 | exported protease | 1.36 | |
| 3705 | putative ABC transport binding protein | 2.14 | ||
| 3706 | TonB-dependent receptor | 2.45 | ||
| 3707 | FecR-transcriptional regulator | 3.83 | ||
| 3708 | extracytoplasmic function (EFC)-family σ factor | 4.52 | ||
| 3709 | TonB-like protein | 2.92 | ||
| Putative siderophore biosynthesis & ABC transport systems | 0229* | ABC transporter; permease protein | 4.81 | |
| 0230 | ATP-binding protein | 3.94 | ||
| 0231* | pbtI | putative citrate lyase β subunit | 6.45 | |
| 0232* | pbtA | putative siderophore biosynthesis protein | 6.36 | |
| 0233* | pbtB | putative TonB-dependent siderophore receptor | 6.04 | |
| 0234* | pbtC | putative lysine/ornithine decarboxylase | 5.30 | |
| 0235* | pbtD | putative pyridoxal-phosphate dependent enzyme | 4.77 | |
| 0236 | pbtE | putative octopine/opine/tauropine dehydrogenase | 3.23 | |
| 0237 | pbtF | MFS-family transporter | 3.31 | |
| 0238 | pbtG | putative substrate-binding protein | 1.76 | |
| 0239 | pbtH | conserved hypothetical protein | 2.53 | |
| 2596* | putative siderophore TonB-dependent receptor | 7.13 | ||
| 2597 | nrpX | MFS-family transporter | 5.92 | |
| 2598* | nrpY | conserved hypothetical protein | 7.93 | |
| 2599 | nrpR | putative nonribosomal peptide synthase | 5.15 | |
| 2600 | nrpS | putative nonribosomal peptide synthase | 3.62 | |
| 2601 | nrpU | putative siderophore biosynthetic protein | 2.92 | |
| 2602 | nrpT | putative siderophore biosynthetic protein (thioesterase) | 2.90 | |
| 2603 | nrpA | put. sid. ABC transporter; ATP-binding/permease protein | 4.08 | |
| 2604 | nrpB | put. sid. ABC transporter; ATP-binding/permease protein | 3.56 | |
| 2605 | nrpG | putative 4′-phosphopantetheinyl transferase | 1.27 | |
| Additional ferri-siderophore transporters | 0331* | putative ABC transporter; substrate binding protein | 6.49 | |
| 0363 | putative TonB-dependent ferri-siderophore receptor | 3.52 | ||
| 1945* | ireA | putative TonB-dependent ferri-siderophore receptor | 6.65 | |
| 2957* | putative iron ABC transporter; substrate binding protein | 6.20 | ||
| 2958 | putative iron ABC transporter; permease component | 3.93 | ||
| 2959 | putative iron ABC transporter; permease | 2.99 | ||
| 2960 | putative iron ABC transporter; ATP-binding | 3.13 | ||
Putative iron acquisition systems of P. mirabilis HI4320 listed are composed of genes that were up-regulated during in vitro iron limitation with a fold-change ≥ 2-fold.
Genes are grouped by function and listed within putative systems or operons by ascending P. mirabilis gene identification number (PMI no.).
Log2 transformation of fold-change values determined by taking ratio of transcript level of P. mirabilis growth in LB medium and LB medium treated with 15 μM Desferal averaged across all five microarray experiments.
Genes found to be significantly up-regulated as determined by Significance Analysis of Microarray (SAM).
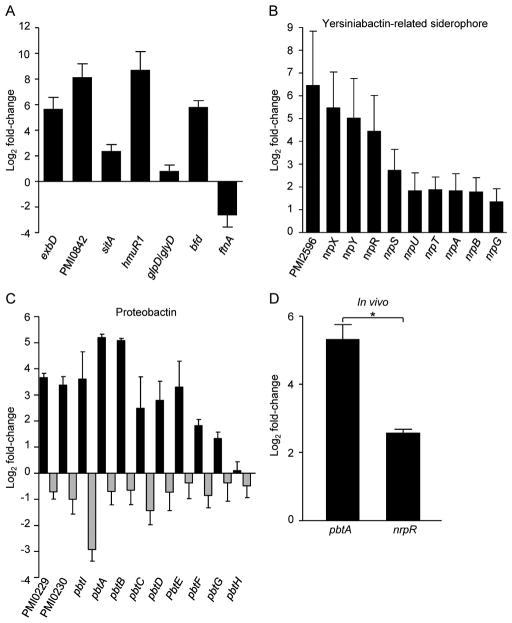
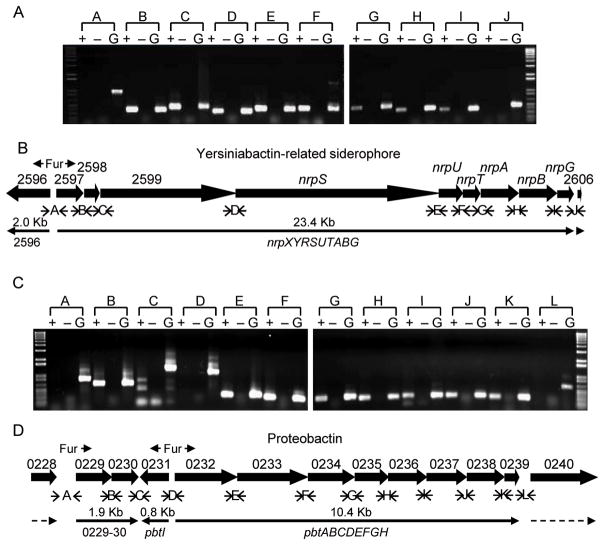
To validate the results of the microarray, quantitative RT-PCR (qPCR) was performed on select genes that were iron responsive in vitro. A total of six individual up-regulated genes (exbD, TonB-dependent receptor PMI0842, sitA, hmuR1, glpD/glyD, bfd) and one significantly down-regulated gene (ftnA) were verified by qPCR (Fig. 2A). As expected, all genes except glpD/glyD were up-regulated > 2-fold and ftnA was down-regulated > 2-fold during culture of P. mirabilis HI4320 in 15 μM Desferal (Fig. 2A). Putative siderophore biosynthesis and ABC transport gene clusters encoding the capacity to synthesize the yersiniabactin-related siderophore, nrp (PMI2596-2605) and proteobactin, pbt (PMI0229-0239) were also tested by qPCR to verify up-regulation (Fig. 2B, C). Putative siderophore TonB-dependent receptor, PMI2596, and all genes in putative nrp operon (PMI2597-2605) were up-regulated > 2-fold (Fig. 2B). Genes of the pbt gene cluster (PMI0229-0239) were up-regulated > 2-fold, except pbtH (PMI0239) (Fig. 2C). Both nrp and pbt gene clusters contain consensus sequences for ferric uptake regulator (Fur) binding (Fig. 3) and, consistent with this, the transcription of pbt genes are repressed by the addition of 25 μM FeCl3·6H2O to bacteria cultured in iron-chelated LB medium (Fig 2C). Both of these putative siderophore systems are organized as operons as demonstrated using RT-PCR; the nrp system consists of two transcripts and the pbt system is transcribed in three units (Fig. 3).
Fig. 2. qPCR of mRNA from P. mirabilis HI4320 cultured in LB medium containing 15 μM Desferal validates microarray results that identified up- and down-regulated genes in response to iron limitation.
Log2 fold-change was determined relative to strain HI4320 cultured in LB medium alone. (A) qPCR validation of select HI4320 genes up- or down-regulated during iron limitation in vitro as determined by microarray. (B) qPCR validation of HI4320 putative yersiniabactin-related siderophore, nrp (PMI2596-2605) that was up-regulated during iron limitation in vitro as determined by microarray. (C) qPCR validation of HI4320 putative siderophore system proteobactin, pbt (PMI0229-0239) that was up-regulated during iron limitation in vitro as determined by microarray (black bars) and down-regulated following the addition of 25 μM FeCl3·6H2O to an iron-chelated LB medium culture of bacteria (grey bars). (D) qPCR confirmation of HI4320 siderophore biosynthesis genes pbtA (PMI0232) and nrpR (PMI2599) expression in vivo. mRNA, isolated from bacteria recovered from the urine of experimentally infected CBA/J mice, was subjected to qPCR. A significant difference in expression levels (*, P = 0.0002) was determined using a two-tailed unpaired t-test.
Fig. 3. Transcriptional organization of putative siderophore system PMI2596-2605 and PMI0229-0239 in P. mirabilis HI4320.
(A, C) mRNA for cDNA synthesis was isolated from wild-type HI4320 following culture in LB medium containing15 μM Desferal. For PCR reactions: +, reverse transcriptase added to cDNA synthesis reaction; −, no reverse transcriptase added, mRNA used as template; G, HI4320 genomic DNA used as template. (B, D) Siderophore biosynthesis and transport gene clusters of HI4320 drawn to scale with primer design for RT-PCR operon mapping. Thick arrows represent open reading frames that indicate direction of transcription. Ferric uptake regulator (Fur) recognition sequences were identified based on homology to those of P. aeruginosa using a promoter analysis program in the Virtual Footprint software suite (Munch et al., 2005). Location and direction of Fur recognition sequences are indicated. Intergenic primer pairs used for RT-PCR are represented by alphabetical letters. Thin solid arrows represent direction and span the length of genes that are required to make a complete transcript. Proposed gene nomenclature for transcripts are indicated below thin solid lines and size of transcript are indicated above the solid lines.
Since the urinary tract is iron-limited (Alteri & Mobley, 2007, Hagan & Mobley, 2009, Torres et al., 2001, Snyder et al., 2004) and the nrp and pbt operons are induced during iron limitation in vitro, the expression of one putative siderophore biosynthesis gene from each of the two systems was examined in vivo. We assessed the transcription of these genes during infection by comparing expression of pbtA (PMI0232) and nrpR (PMI2599) using qPCR to quantify RNA purified from wild-type P. mirabilis cultured in LB medium and from P. mirabilis recovered from the urine of infected mice. Bacteria recovered from urine of infected mice up-regulated both siderophore synthetase genes in vivo as compared to LB medium. Interestingly, pbtA appeared to display a greater level of induction than nrpR under these conditions (P = 0.0002) (Fig. 2D).
Characterization of the yersiniabactin-related locus in P. mirabilis
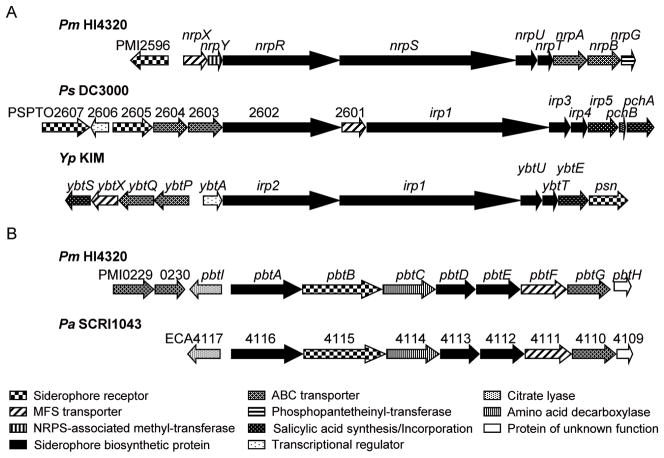
Previously, we have shown that the nrp genes (PMI2597-2605) are located on a pathogenicity island in P. mirabilis HI4320 (Flannery et al., 2009). These genes have homology to genes required for synthesis of yersiniabactin, a nonribosomal peptide synthetase (NRPS) siderophore, which is encoded on the high-pathogenicity island (HPI) of Yersinia spp. (Flannery et al., 2009, Carniel et al., 1996). In P. mirabilis, the putative yersiniabactin receptor, PMI2596, is annotated as a TonB-dependent receptor and shares 27% amino acid sequence identity with the yersiniabactin/pesticin receptor gene psn (Fig. 4A, Table 2). Genes PMI2597-2605 are divergently transcribed from PMI2596 (Fig. 3) and make up an operon encoding a major facilitator superfamily (MFS) transporter, a conserved hypothetical protein, two putative nonribosomal peptide synthases including nrpS, two putative siderophore biosynthesis genes nrpU and nrpT, two putative siderophore ABC transport ATP-binding/permeases nrpA and nrpB, and putative 4′-phosphopantetheinyl transferase nrpG (Fig. 4A). To maintain consistency with the current nomenclature, the three previously uncharacterized genes that are expressed in this operon are designated nrpX (PMI2597), nrpY (PMI2598), and nrpR (PMI2599).
Fig. 4. The yersiniabactin-related siderophore and proteobactin of P. mirabilis are similar to siderophore biosynthesis and transport gene clusters in other Enterobacteriaceae as shown by gene sequence alignments.
(A) P. mirabilis yersiniabactin-related siderophore is homologous to that of P. syringae DC3000 and Y. pestis KIM. The yersiniabactin-related siderophore of P. mirabilis lacks the genes involved in salicyclic acid biosynthesis and incorporation, ybtS and ybtE of Y. pestis and irp5, pchB, and pchA of P. syringae. The nrp locus of P. mirabilis also lacks a transcriptional regulator but encodes a NRPS-associated methyl-transferase and a phosphopantetheinyl-transferase which are not located within the yersiniabactin operons of P. syringae and Y. pestis. (B) The newly identified NIS system of P. mirabilis, proteobactin, is similar to identical gene clusters found in P. carotovora subsp. atroseptica (shown) and P. carotovora subsp. carotovora.
Table 2.
Similarity search between P. mirabilis and Y. pestis, E. coli, P. syringae, and P. luminescens.
| Gene (No. of amino acids) | % Identity (% Similarity) | No. of Amino acids | |
|---|---|---|---|
| PMI2596 (662) | |||
| Yp KIM psn | 27 (48) | 673 | |
| Ec 536 ECP_1947 | 27 (49) | 673 | |
| Ps DC3000 PSPTO2605 | 27 (47) | 685 | |
| Pl TT01 Plu2316 | 30 (46) | 668 | |
| PMI2597 nrpX (409) | |||
| Yp KIM ybtX | 39 (58) | 467 | |
| Ec 536 ybtX | 38 (58) | 462 | |
| Ps DC3000 PSPTO2601 | 25 (46) | 408 | |
| Pl TT01 Plu2317 | 31 (53) | 414 | |
| * PMI2598 nrpY (245) | |||
| Yp KIM y0042 | 34 (51) | 260 | |
| Ec 536 ECP_3773 | 34 (51) | 260 | |
| Ps DC3000 argS | 26 (44) | 578 | |
| Pl TT01 Plu2237 | 40 (59) | 315 | |
| PMI2599 nrpR (2034) | |||
| Yp KIM irp2 | 40 (59) | 2035 | |
| Ec 536 HMWP2 | 40 (59) | 2035 | |
| Ps DC3000 PSPTO2602 | 42 (59) | 2057 | |
| Pl TT01 Plu2320 | 41 (58) | 2049 | |
| PMI2600 nrpS (3071) | |||
| Yp KIM irp1 | 50 (66) | 3163 | |
| Ec 536 HMWP1 | 50 (66) | 3163 | |
| Ps DC3000 irp1 | 52 (67) | 3173 | |
| Pl TT01 Plu2321 | 39 (56) | 3908 | |
| PMI2601 nrpU (362) | |||
| Yp KIM ybtU | 42 (64) | 386 | |
| Ec 536 ybtU | 42 (64) | 366 | |
| Ps DC3000 irp3 | 44 (62) | 360 | |
| Pl TT01 Plu2322 | 38 (57) | 365 | |
| PMI2602 nrpT (257) | |||
| Yp KIM ybtT | 47 (58) | 218 | |
| Ec 536 ybtT | 44 (57) | 262 | |
| Ps DC3000 irp4 | 48 (61) | 271 | |
| Pl TT01 Plu2323 | 47 (57) | 258 | |
| PMI2603 nrpB (588) | |||
| Yp KIM ybtP | 30 (53) | 600 | |
| Ec 536 ybtP | 30 (53) | 570 | |
| Ps DC3000 PSPTO2604 | 45 (67) | 593 | |
| Pl TT01 Plu2318 | 31(52) | 572 | |
| PMI2604 nrpA (575) | |||
| Yp KIM ybtQ | 32 (56) | 600 | |
| Ec 536 ybtQ | 32 (57) | 600 | |
| Ps DC3000 PSPTO2603 | 46 (68) | 581 | |
| Pl TT01 Plu2319 | 31 (52) | 600 | |
| * PMI2605 (249) | |||
| Yp KIM y1938 | 35 (52) | 410 | |
| Ec 536 ECP_1982 | 38 (62) | 62 | |
| Ps DC3000 bphP | 43 (56) | 745 | |
| Pl TT01 Plu4193 | 31 (46) | 237 | |
P. mirabilis gene listed has homology to genes located outside of the yersiniabactin operon of other Enterobacteriaceae.
Despite the homology to yersiniabactin biosynthesis and transport genes in other Enterobacteriaceae (Table 2), the yersiniabactin-related siderophore synthesized by P. mirabilis HI4320 is predicted to lack modification by salicylate. Genes involved in salicylic acid biosynthesis and incorporation, pchB, pchA, and irp5 in Pseudomonas syringae pv. tomato DC3000 and ybtE and ybtS in Y. pestis KIM that are essential for yersiniabactin function in these bacteria are absent in the P. mirabilis nrp locus (Fig. 4A). The P. mirabilis nrp operon also lacks a linked transcriptional regulator analogous to PSPTO2606 and YbtA (Fig. 4A). Another distinctive product of the P. mirabilis nrp operon is the presence of a conserved hypothetical protein encoded by nrpY (PMI2598), which has similarity to methyl-transferases found in NRPS systems (Fig. 4A). Additionally, the 4′-phosphopantetheinyl-transferase located at the end of the P. mirabilis nrp operon, nrpG, is encoded separately from the yersiniabactin operon on the Y. pestis chromosome (ybtD) (Fig. 4A). Likewise, the 4′-phosphopantetheinyl-transferase of P. syringae is absent from the yersiniabactin operon (Fig. 4A).
P. mirabilis is unable to grow on LB agar containing 25 μM Desferal. However, in cross-feeding assays, P. mirabilis growth is restored (Fig. 5A, Table 3) by uropathogenic E. coli (UPEC) strain 536 that produces both enterobactin and yersiniabactin (Brzuszkiewicz et al., 2006). This restoration of P. mirabilis growth was abolished when cross-fed by a UPEC 536 construct that lacks both enterobactin and yersiniabactin (Fig. 5C, Table 3). Unexpectedly, P. mirabilis was incapable of growth on iron chelator when cross-fed by the 536 strain that produces yersiniabactin and lacks only enterobactin (Fig. 5B, Table 3). In addition, UPEC CFT073, a strain incapable of synthesizing yersiniabactin (Bultreys et al., 2006), can also restore the growth defect in P. mirabilis in cross-feeding assays (Fig. 5D, Table 3); in this assay, P. mirabilis swarms around E. coli CFT073. This ability of P. mirabilis to utilize enterobactin is independent of both the nrp and pbt iron acquisition systems (Table 3). These surprising findings show that yersiniabactin is not able to be utilized by P. mirabilis and suggests that despite the clear evolutionary relationship, the nrp locus is not involved in the production and uptake of yersiniabactin per se.
Fig. 5. P. mirabilis is unable to utilize yersiniabactin produced by uropathogenic E. coli 536.
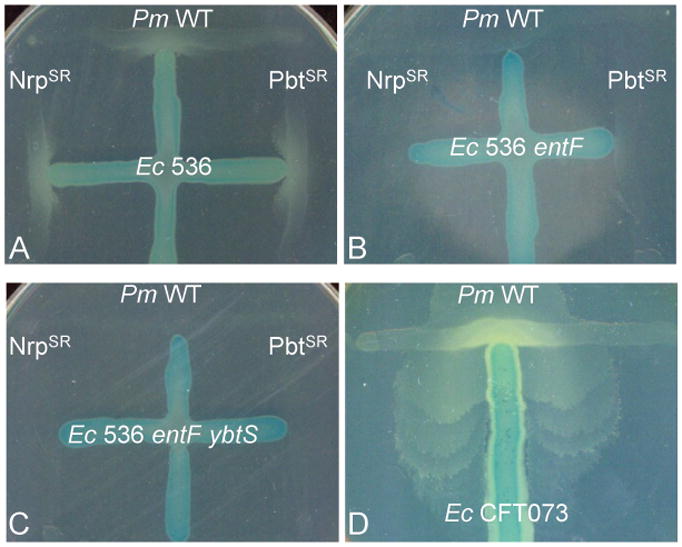
LB medium was supplemented with 25 μM Desferal, the minimum concentration of chelator required to completely suppress growth of P. mirabilis. Lactose-fermenting E. coli are distinguished from P. mirabilis by the using the chromogenic indicator X-gal. (A) Growth of wild-type P. mirabilis, the proteobactin synthesis/receptor mutant (PbtSR), and the yersiniabactin-related siderophore/receptor mutant (NrpSR) can be restored when cross-fed by E. coli 536 that produces both enterobactin and yersiniabactin. (B, C) Restoration of P. mirabilis growth for all strains tested is abolished when cross-fed by the either E. coli 536 enterobactin mutant, entF::kan, or the enterobactin/yersiniabactin double mutant, entF ybtS::kan. (D) P. mirabilis growth can be restored using E. coli CFT073, an enterobactin producing strain incapable of synthesizing yersiniabactin.
Table 3.
Cross-feeding of P. mirabilis wild-type and siderophore mutants by E.coli 536, 536 siderophore mutants, and CFT073.
| Ability to cross-feed P. mirabilis strains | ||||
|---|---|---|---|---|
| E. coli 536 | E. coli 536 entF::kan | E. coli 536 entF ybtS::kan | E. coli CFT073 | |
| P. mirabilis | + | − | − | + |
| P. mirabilis PbtSR | + | − | − | + |
| P. mirabilis NrpSR | + | − | − | + |
Strain noted as synthetaseS and receptorR mutant in the proteobactin (Pbt) and/or yersiniabactin-related (Nrp) siderophore system.
+, growth
−, no growth
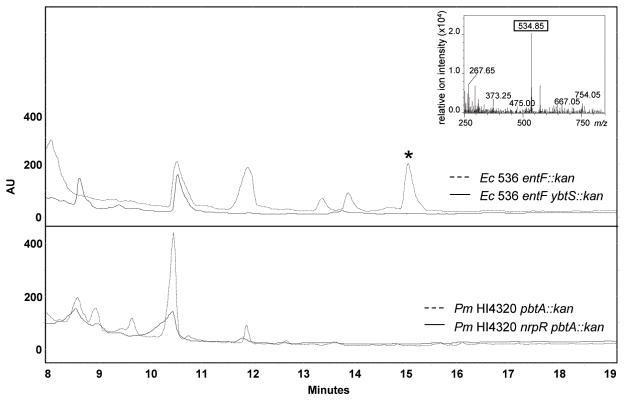
The lack of yersiniabactin production is also supported by an HPLC analysis showing the absence of UV absorbance maxima (310 and 385 nm) characteristic of yersiniabactin (Jones et al., 2007) when concentrated supernatants from P. mirabilis cultured under iron limitation were analyzed by HPLC and compared directly to concentrated supernatants of E. coli 536 mutants entF::kan and entF ybtS::kan (Fig. 6). Furthermore, extracts of the same experimental P. mirabilis HI4320 and control E. coli 536 cultures were prepared using previously reported methods (Bultreys et al., 2006, Jones et al., 2007) and analyzed by ESI-LC-MS. Monitoring in positive ion mode, m/z matching that of a ferri-yersiniabactin species described by Jones and colleagues (2007) could be detected in E. coli 536 entF::kan (expected: 535.1, observed: 534.8); the same signal could not be detected from E. coli entF ybtS::kan negative control or P. mirabilis HI4320 samples (Fig. 6). These data support the assertion that P. mirabilis nrp, while highly similar to the yersiniabactin operon of other species, is incapable of producing a molecule identical to yersiniabactin.
Fig. 6. Preparative HPLC and LC-MS analysis demonstrates P. mirabilis HI4230 is incapable of producing yersiniabactin.
Preparative HPLC chromatogram (upper traces) performed on crude supernatant extracts from yersiniabactin-producing E. coli 536 entF::kan (dashed line) and yersiniabactin-deficient entF ybtS::kan (solid line) cultures to demonstrate production/lack of production of yersiniabactin. The lower traces represent chromatograms using the same HPLC conditions from P. mirabilis HI4230 pbtA::kan (dashed line) and nrpR pbtA::kan (solid line). The y-axis represents arbitrary units (AU) to offset sample traces. The inset indicates the LC-MS (positive ion mode) analysis of E. coli yersiniabactin present in the dominant peak (*) isolated at 15 min by HPLC. Iron-bound yersiniabactin is known to have an ion intensity at m/z of 535.1.
Identification of a novel NRPS-independent siderophore system that synthesizes proteobactin
The second siderophore biosynthesis operon of P. mirabilis HI4320 encodes a novel NRPS-independent siderophore (NIS) pathway that is predicted to produce a hydroxycarboxylate siderophore. The first two genes, PMI0229-0230, are predicted to encode an ABC transport permease protein and an ABC transport ATP-binding subunit, are expressed as a two gene transcript (Fig. 3C, D). PbtI (PMI0231) encodes a putative citrate lyase β subunit and is transcribed as a single mRNA from the opposite coding strand (Fig. 3C, D). Citrate lyase β activity converts citric acid to oxaloacetate, which is a key biosynthetic precursor and could be the preferred substrate of the putative siderophore biosynthesis protein encoded by pbtA (PMI0232). Because oxaloacetate is a closely related metabolic intermediate to α-ketoglutarate, a well known substrate for NIS type B synthetases (Challis, 2005), it is probable that pbtA also belongs to the type B subfamily of NIS synthetases. The polycistron that contains the putative NIS synthetase also codes for a putative TonB-dependent siderophore receptor, a putative lysine/ornithine decarboxylase, a putative pyridoxal-phosphate dependent enzyme, a putative octopine/opine/tauropine dehydrogenase, a MFS-family transporter, a substrate-binding protein, and a hypothetical protein (Fig. 3C, D).
All known NIS biosynthetic pathways utilize at least one siderophore synthetase with homology to hydroxamate synthesis enzymes IucA (NIS type A) and IucC (NIS type C) (Challis, 2005). In line with this, the siderophore synthetase from this operon in P. mirabilis has 21% and 25% identity to IucA and IucC of uropathogenic E. coli, respectively. Upon further examination, the closest homolog to this P. mirabilis hydroxamate synthetase is found in an uncharacterized siderophore system of the phytopathogens Pectobacterium (formerly Erwinia) carotovora subsp. atroseptica and P. carotovora subsp. carotovora (54% identity to both). The primary siderophore biosynthetic enzyme used by these systems and the P. mirabilis synthetase encoded by PbtA have only 27–28 % identity to the achromobactin biosynthetic enzyme AcsD, which has recently been classified as the prototype member of a new family of enzymes, NIS type B, that synthesize a hydroxycarboxylate NIS in P. chrysanthemi (Munzinger et al., 2000, Franza et al., 2005, Schmelz et al., 2009).
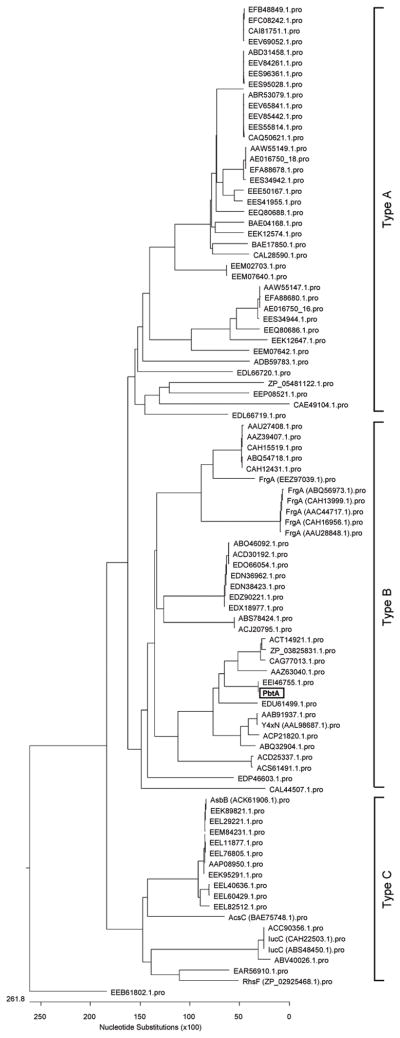
To determine which NIS synthetase subfamily the synthetase of proteobactin represents, 92 protein sequences homologous to siderophore synthetase PbtA, identified by BLASTP, were aligned using Clustal W (Thompson et al., 1994) and used for phylogenetic analysis of the related NIS enzymes (Fig. 7). The resulting phylogeny clearly identified the three known NIS subfamilies as distinct clades and placed PbtA within the same clade as Y4xN (Fig. 7), a siderophore synthetase of Sinorhizobium fredii described to be a type B NIS synthetase (Challis, 2005). This phylogenetic analysis suggests that the NIS of P. mirabilis is a type B NIS synthetase and the genes that encode for the capacity to produce this NIS, proteobactin, are referred to as pbtABCDEFGHI, for proteobactin synthesis (Fig. 4B).
Fig. 7. Phylogenetic analysis of 92 homologous protein sequences to NIS synthetase PbtA involved in the assembly of proteobactin.
The NIS synthetase enzymes are split into three subfamilies A, B, and C. Proteobactin synthetase, PbtA, (boxed) is placed with the type B subfamily clade of NIS synthetases which includes Y4xN, a siderophore synthetase previously found to be homologous to other NIS synthetases of this group.
Siderophore production by P. mirabilis HI4320
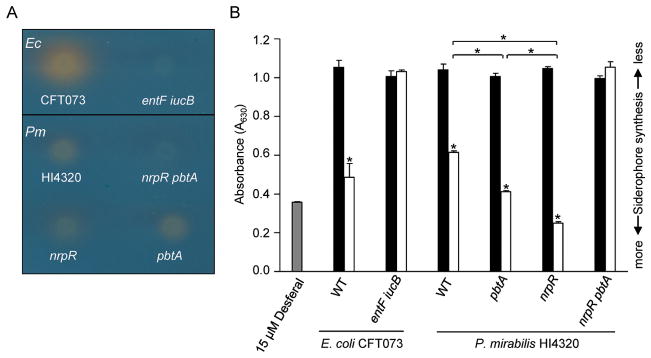
To detect siderophore production by both the yersiniabactin-related and proteobactin systems, we examined P. mirabilis on CAS agar and by the CAS assay. Following growth of HI4320 on CAS agar, a color change of CAS from blue to orange, indicating iron chelation, was observed as a halo surrounding the bacteria. This effect was less pronounced than that caused by uropathogenic E. coli CFT073 (Fig. 8A). The iron chelation effect produced by wild-type HI4320 on CAS agar is similar to that of either proteobactin (pbtA) or yersiniabactin-related (nrpR) biosynthesis mutants (Fig. 8A). However, a double mutant in both siderophore systems (nrpR pbtA) is unable to chelate iron from the dye (Fig. 8A). This iron-chelating defect in the double mutant is similar to E. coli CFT073 entF::kan iucB::cam, which does not produce siderophores (Torres et al., 2001) (Fig. 8A).
Fig. 8. Siderophore production by P. mirabilis HI4320.
(A) Detectable siderophore production on chrome azurol S (CAS) agar of overnight LB bacterial cultures spotted in 5 μl volumes. Blue to orange color change of agar indicates siderophore production. (B) CAS assay depicting siderophore production from filtered bacterial supernatants following culture in MOPS defined medium with (black bars) and without (white bars) 0.1 mM FeCl3·6H2O. Only supernatants of P. mirabilis samples were concentrated 50-fold. Supernatant of E. coli CFT073 and double mutant, entF::kan iucB::cam, were not concentrated. Positive control of phosphate buffered saline containing 15 μM Desferal (grey bar). A lower absorbance (A630) indicates greater siderophore production. Significant differences in absorbance (*, P ≤ 0.0022) was determined using a two-tailed unpaired t-test.
To quantify the observed iron-binding properties of both P. mirabilis proteobactin and the yersiniabactin-related siderophore, a soluble CAS assay was performed using 50-fold concentrated P. mirabilis supernatants from log-phase cells cultured in MOPS defined medium (Neidhardt et al., 1974) with and without 0.1 mM FeCl3·6H2O. Results of this quantitative CAS assay clearly demonstrate that when cultured under iron limitation, P. mirabilis produces and releases one or more molecules that chelate iron from the ferri-CAS complex (P = 0.0001) (Fig. 8B). This same result was seen for each P. mirabilis single siderophore biosynthesis mutant; presence of either proteobactin or the yersiniabactin-related siderophore was sufficient to chelate iron during iron limitation (indicated by a decrease in A630) (P < 0.0001) (Fig. 8B). In contrast, significant iron-chelation by the P. mirabilis double siderophore biosynthesis mutant was not observed during iron limitation (Fig. 8B). This is consistent with the iron-chelating defect seen with the E. coli CFT073 enterobactin/aerobactin double siderophore mutant entF::kan iucB::cam. Interestingly, iron chelation by either single synthetase mutant was modestly increased as compared to wild-type HI4320 (P < 0.0001) (Fig. 8B). Despite our best efforts to standardize bacterial cultures by optical density, it is possible that variation and siderophore stability during sample preparation could be responsible for this result. qPCR analysis determined this phenotype is not due to the up-regulation of one synthetase gene in the absence of the other synthetase gene (data not shown).
To determine whether the yersiniabactin-related siderophore and proteobactin are common in other P. mirabilis isolates, ten urine isolates and ten non-UTI isolates were screened by PCR for the presence of siderophore biosynthesis genes pbtA and nrpR. As expected, 100% (10/10) of the P. mirabilis urine isolates contain both pbtA and nrpR while only 50% (5/10) of the P. mirabilis non-UTI isolates contain both genes. Therefore, urine isolates are more likely to have both pbtA and nrpR, (P = 0.0325) compared to P. mirabilis non-UTI isolates.
In vivo contribution of siderophore systems during ascending UTI
To determine the role of siderophores during murine ascending UTI, siderophore synthetase and TonB-dependent receptor mutants of proteobactin or the yersiniabactin-related siderophore were transurethrally inoculated into the bladders of CBA/J mice in independent and cochallenge competition experiments with wild-type HI4320. In independent infections, the loss of the proteobactin siderophore system alone or a double mutant in both siderophore synthetase genes did not attenuate HI4320 (Table 4). During independent infection, the yersiniabactin-related synthetase/receptor mutant (NrpSR) appeared to colonize better than wild-type HI4320 (P < 0.05) (Table 4) where the median CFU/g was less than one-half log greater for NrpSR than HI4320 (Fig. S2). In cochallenge competition experiments, the strain lacking only proteobactin did not have a fitness defect and loss of PbtSR may slightly increase fitness in the kidneys (P = 0.034) (Table 5). Interestingly, the yersiniabactin-related synthetase/receptor mutant (NrpSR) was significantly out-competed by wild-type in the bladder (P = 0.004) and kidneys (P = 0.027) (Table 5). Similarly, the yersinibactin-related synthetase and proteobactin synthetase/receptor mutant (NrpSPbtSR) was significantly out-competed by wild-type in the bladder (P = 0.019) (Table 5). These data, from both independent and cochallenge infections, consistently indicate that the yersiniabactin-related siderophore had a greater contribution for P. mirabilis fitness in vivo.
Table 4.
In vivo virulence of siderophore biosynthesis mutants following independent challenge experiments.
| Strain defecta | Bladder | Kidneys | ||
|---|---|---|---|---|
| Median CFU | P-valueb | Median CFU | P-valueb | |
| Wild-type | 3.77×106 | 4.01×105 | ||
| PbtSR | 5.17×107 | 0.208 | 2.72×106 | 0.068 |
| Wild-type | 2.26×107 | 6.29×105 | ||
| NrpSR | 6.25×107 | 0.045 | 2.28×106 | 0.036 |
| Wild-type | 3.04×107 | 2.15×106 | ||
| NrpSPbtSR | 2.70×107 | 0.741 | 3.00×105 | 0.106 |
Strain defect noted as synthetaseS and/or receptorR mutant in the proteobactin (Pbt) and/or yersiniabactin-related (Nrp) siderophore system.
P-value determined by Mann Whitney test. Significant P-values are bolded.
Table 5.
In vivo fitness of siderophore biosynthesis mutants following cochallenge competition experiments with P. mirabilis HI4320.
| Strain defecta | Bladder | Kidneys | ||
|---|---|---|---|---|
| CIb | P-valuec | CIb | P-valuec | |
| PbtSR | 0.195 | 0.850 | 0.040 | 0.034 |
| NrpSR | 7.900 | 0.004 | 30.95 | 0.027 |
| NrpSPbtSR | 2.069 | 0.019 | 1.470 | 0.051 |
Strain defect noted as synthetaseS and/or receptorR mutant in the proteobactin (Pbt) and/or yersiniabactin-related (Nrp) siderophore system.
Competitive index, determined by dividing the ratio of the CFU of wild-type to mutant at 7 days post-infection by the ratio present in the input inoculum. Significant CI>1 indicates mutant has a fitness defect.
CI P-value as determined by Wilcoxon signed-rank test. Significant P-values are bolded.
Discussion
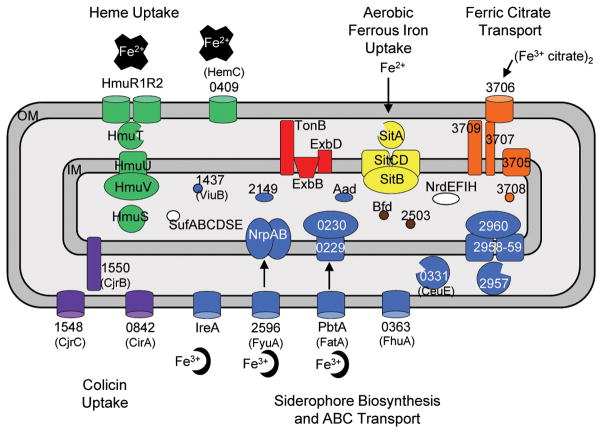
It is well known that the urinary tract is iron-limited. Despite this, little is known about iron acquisition by the common uropathogen P. mirabilis. To improve our understanding of iron acquisition and identify potential siderophore systems utilized by this pathogen; we examined the genome-wide transcriptome from bacteria cultured under iron-replete and iron-deficient conditions. As expected, many genes predicted to encode iron acquisition systems or proteins were up-regulated during iron limitation including exbD and three additional genes that have been previously identified by our signature-tagged mutagenesis (STM) screens (Burall et al., 2004, Himpsl et al., 2008). The iron acquisition systems identified in the present study are grouped into functional classes that include genes required for heme uptake, aerobic ferrous iron uptake (Fisher et al., 2009), ferric citrate transport, and siderophore biosynthesis and transport. These microarray findings may not comprehensively demonstrate expression of every iron acquisition system employed by P. mirabilis HI4320; however, this study has been useful to propose a model of iron acquisition and transport strategies utilized by this human pathogen (Fig. 9).
Fig. 9. A proposed model for iron acquisition systems in P. mirabilis.
Hypothetical functions of the proteins listed are based on up-regulation of their genes under iron limitation, homology with genes from other bacterial species, and functional studies conducted in this report and related bacterial species. Outer membrane (OM) receptors are dependent on the energy transducing complex TonB-ExbB-ExbD (shown in red) for intracellular transport of iron. Heme uptake systems are displayed in green. Heme uptake may also be carried out by the outer membrane receptor, HemC. Import of periplasmic ferrous iron is performed by the ABC transporter SitABCD (shown in yellow) located on the inner membrane (IM). A putative ferric citrate system (shown in orange) encodes a TonB-dependent receptor PMI3706, and accessory proteins. Proteins involved in siderophore biosynthesis and transport of ferri-siderophores are shown in blue. Ferri-siderophores are transported by outer membrane receptors IreA, FyuA, FatA, and FhuA. Ferri-siderophores transported to the periplasm are shuttled to inner membrane transport proteins, NrpAB and PMI0229-0230. Additional putative ferri-siderophore transport proteins include the periplasmic binding protein, CeuE, and inner membrane transport proteins, PMI2957-2960. Cellular proteins, Aad, PMI2149, ViuB and, may be involved in siderophore synthesis and the regulation and utilization of iron of ferri-siderophore. Putative TonB-dependent receptors possibly involved in colicin uptake are shown in purple. Cytoplasmic iron-containing proteins include nrdEFIH and sufABCDSE (shown in white). Bfd, an iron removal protein, and PMI2503 involved in iron metabolism are shown in brown.
Numerous unsuccessful attempts to detect siderophores in P. mirabilis have led to speculation that P. mirabilis is a poor siderophore producer (Marcelis et al., 1978, Massad et al., 1995, Miles & Khimji, 1975, Evanylo et al., 1984). However the results of the microarray indicated the up-regulation of a number of genes related to siderophore synthesis during iron limitation including a known and unknown amino acid deaminase aad and PMI2149 both of which could possibly chelate iron (Drechsel et al., 1993, Massad et al., 1995). Also, genes homologous to the transport and metabolism of ferri-siderophores, such as putative ABC substrate-binding protein PMI0331, ABC transport system PMI2957-2960, putative iron utilization protein PMI1437, and putative esterase PMI2503 were up-regulated during iron limitation. A total of four putative TonB-dependent ferri-siderophore receptors were up-regulated during iron limitation: ireA, PMI0363, PMI2596, and PMI0233. Two of these ferri-siderophore receptors, PMI0233 and PMI2596, are located in regions that encode for both siderophore biosynthesis and ABC transport, which is a common feature of known ferri-siderophore systems (Wandersman & Delepelaire, 2004, Koster, 2001, Crosa & Walsh, 2002).
One of these P. mirabilis siderophore systems encodes the nrp operon and is closely related to the synthesis, uptake, and ABC transport of yersiniabactin, a nonribosomal peptide synthetase (NRPS) siderophore encoded on the HPI of Yersinia spp. (Flannery et al., 2009). Despite apparent homology and organization to yersiniabactin, cross-feeding assays indicate that P. mirabilis is unable to utilize yersiniabactin produced by UPEC strain 536 (Table 3, Fig. 5). This finding suggests that the nrp locus of P. mirabilis is functionally distinct from yersiniabactin. Indeed, the nrp synthesis and ABC transport genes of P. mirabilis differ equally from the known yersiniabactin synthesis and ABC transport genes of Y. pestis KIM, E. coli 536, P. syringae pv. tomato DC3000, and Photorhabdus luminescens TT01 (Table 2). Previously, three yersiniabactin loci evolutionary groups have been described in Y. pestis, P. syringae, and P. luminescens (Bultreys et al., 2006). The nrp locus of P. mirabilis may likely encode a fourth yersiniabactin evolutionary group. Interestingly, the genes for salicylic acid biosynthesis and incorporation required for yersiniabactin synthesis in these Enterobacteriaceae are absent from the P. mirabilis nrp operon. Despite the absence of these genes, we have shown that the nrp system of P. mirabilis is able to chelate iron suggesting the salicylic acid biosynthesis genes may be located elsewhere on the chromosome or perhaps the nrp locus has evolved to synthesize a novel yersiniabactin-related siderophore that does not require this modification. Furthermore, efforts to chemically detect yersiniabactin from P. mirabilis were unsuccessful (Fig. 6), supporting the possibility of an evolutionary divergence from producing traditional yersiniabactin.
In addition to the nrp/yersiniabactin-related system, P. mirabilis also has a previously uncharacterized NRPS-independent siderophore (NIS) system. This newly identified NIS system, proteobactin, has similarity to NIS group A (IucA) and NIS group C (IucC) synthetases. Closer scrutiny reveals the proteobactin synthetase, PbtA, is most closely related to a predicted synthetase that is part of an uncharacterized siderophore system found in the phytopathogens, Pectobacterium carotovora subsp. atroseptica and subsp. carotovora. Proteobactin and the molecules produced by these putative systems are distinct from achromobactin, a recently identified NIS system in P. chrysanthemi (Franza et al., 2005, Munzinger et al., 2000, Schmelz et al., 2009). Furthermore, both these subspecies of P. carotovora and P. mirabilis do not possess the acs genes required for achromobactin synthesis suggesting proteobactin represent a new NIS system (Franza et al., 2005). Phylogenetic analysis (Fig. 7) places PbtA among the type B subfamily clade of NIS synthetases. Other genes in the NIS operon potentially involved in the synthesis of proteobactin include pbtD and pbtE. PbtD is a putative pyridoxal-phosphate dependent enzyme and PbtE is a putative octopine/opine/tauropine dehydrogenase. The pyridoxal-phosphate dependent enzyme could be responsible for the incorporation or tailoring of thiol-containing moieties into the proteobactin hydroxycarboxylate siderophore (Kessler, 2006). The octopine/opine/tauropine dehydrogenase could be involved in modification of the primary amine of a siderophore precursor by condensing it to an enzymatically-reduced pyruvate or similar small ketone. This may function in incorporating an additional chelating carboxylate to siderophore biosynthesis intermediates (Mihara et al., 2005). Future biochemical studies will be useful to discern the molecular basis and biochemical function for the NIS genes and enzymes (Oves-Costales et al., 2009).
Initial studies verified that proteobactin and the nrp genes were up-regulated during in vitro iron limitation of P. mirablis HI4320. Likewise, proteobactin and the yersiniabactin-related siderophore biosynthesis genes pbtA and nrpR were up-regulated following experimental urinary tract infection of mice with HI4320, not only validating our in vitro findings but supporting prior evidence that the urinary tract is an iron-limiting environment and confirming that both siderophores are expressed in vivo. Interestingly, P. mirabilis urine isolates were significantly more likely to have both pbtA and nrpR, (P = 0.0325) compared to P. mirabilis non-UTI isolates. This finding suggests that both siderophore systems are significantly more prevalent in P. mirabilis strains that infect the urinary tract than those that colonize other body sites.
The universal chemical assay to detect siderophore production independent of structure (Schwyn & Neilands, 1987) clearly shows that the yersiniabactin-related siderophore and proteobactin of P. mirabilis HI4320 chelate iron in vitro (Fig. 8). However, we speculate that iron chelation in this assay is inefficient compared to other species, such as E. coli CFT073. Unlike the robust conversion from blue to orange observed for E. coli, P. mirabilis generates a less intense, but nevertheless significant, color change from blue to orange on CAS agar (Fig. 8A). Similarly, in quantitative liquid CAS assays, comparable levels of siderophore activity to E. coli CFT073 could only be detected when supernatant from P. mirabilis was concentrated 50-fold. A mutant with defects in both proteobactin and yersiniabactin-related biosynthesis (but not single mutants) abolished iron-chelation, suggesting that both systems are producing functional molecules that chelate iron in vitro (Fig. 8A, B). Additional experiments determined that this phenotype was not due to a growth defect, as inactivation of both siderophore synthesis systems did not affect the in vitro growth of P. mirabilis in LB medium alone or in LB medium with 15 μM Desferal (Fig. S1). During growth, the presence of other iron transport systems as identified by microarray could compensate for the loss of one or both siderophore biosynthesis systems (Table 1).
It is likely that the compensatory action of other iron acquisition and uptake systems in P. mirabilis which were identified in the iron-limiting microarray play a role during uropathogenesis and will be the focus of future studies. In support of this, results of the cross-feeding assays depict E. coli CFT073, an enterobactin producing, yersiniabactin-negative strain, able to cross-feed P. mirabilis (Fig. 5D, Table 3). This suggests that P. mirabilis, while incapable of producing enterobactin, there is a putative TonB-dependent receptor capable of importing enterobactin. Both the P. mirabilis NIS type B proteobactin synthesis/receptor mutant (PbtSR) and the yersiniabactin-related synthesis/receptor mutant (NrpSR) could utilize enterobactin (Fig. 5A, Table 3). These results show that P. mirabilis does possess a TonB-dependent receptor or other mechanism capable of importing enterobactin. Microarray findings support this conclusion, as at least two other putative TonB-dependent receptors, PMI0842 and IreA, were up-regulated during the iron limitation. The TonB-dependent receptor Iha has been shown to function as a catecholate siderophore receptor that mediates uptake of enterobactin (Leveille et al., 2006). Both PMI0842 and IreA of P. mirabilis have 33% and 34% identity to Iha of E. coli, respectively. It is likely that enterobactin is being taken up by these putative TonB-dependent receptors in P. mirabilis.
To investigate the role of these siderophore systems in vivo, independent and cochallenge competition experiments were performed with the mutant strains and wild-type HI4320. Independent challenge experiments showed that loss of both siderophore synthesis genes did not attenuate virulence. During independent challenge the yersiniabactin-related synthetase/receptor mutant (NrpSR) was recovered at modestly higher levels than wild-type HI4320 in the bladder (P = 0.045) and kidneys (P = 0.036) (Table 4). This small difference in the median CFU/g may not be biologically relevant as the ability of both NrpSR and wild-type to colonize the urinary tract was nearly indistinguishable (Fig. S2). Despite the lack of attenuation during independent infections we found that cochallenge competition experiments between mutant and wild-type strains revealed key differences in fitness. In these experiments the proteobactin sythetase/receptor mutant (PbtSR) was able to slightly out-perform wild-type HI4320 (P = 0.034) where a small difference in CFUs was observed. However, these findings may also suggest the yersiniabactin-related system is predominant in vivo as this strain only produces the yersiniabactin-related siderophore. Consistent with this, only mutants lacking the yersiniabactin-related siderophore reduce P. mirabilis fitness in vivo (Table 5). This also agrees with a previous study from our lab, where a STM screen identified another yersiniabactin-related gene, nrpG, as being outcompeted by wild-type HI4320 (Himpsl et al., 2008).
This study is the first genome-wide view of the P. mirabilis HI4320 response to iron limitation. These iron-limiting microarray findings identified systems important in ferri-siderophore uptake, heme uptake, aerobic ferrous iron uptake (Fisher et al., 2009) and ferric citrate transport (Fig. 9). This study is also the first report to demonstrate siderophore production both in vitro and in vivo by two distinct iron acquisition systems in P. mirabilis HI4320. Together, proteobactin and the yersiniabactin-related siderophore system, along with the numerous potential iron acquisition and transport systems identified by our microarray analysis shows that P. mirabilis employs a general strategy of encoding multiple systems with similar function like other extraintestinal pathogens, including uropathogenic E. coli.
Experimental procedures
Bacterial strains and culture conditions
Proteus mirabilis HI4230 and ten additional P. mirabilis caUTI isolates were isolated from urine samples from patients presenting with bacteriuria during long-term catheterization (Mobley & Warren, 1987, Warren et al., 1982). P. mirabilis colonizing strains isolated from the anterior nares, oropharynx, groin, or wound sources (non-UTI isolates) have been previously described (Mody et al., 2007). Escherichia coli CFT073 was isolated from the blood and urine of a patient with acute pyelonephritis (Mobley et al., 1990). The E. coli CFT073 enterobactin and aerobactin mutant, entF::kan iucB::cam, has been previously described (Torres et al., 2001). E.coli 536 has been previously described (Berger et al., 1982). E. coli 536 entF::kan and entF ybtS::kan deletion mutants (Hagan & Mobley, unpublished) were constructed using the lambda Red recombinase system (Datsenko & Wanner, 2000). Bacteria were routinely cultured in Luria-Bertani (LB) medium (per liter, 0.5 g NaCl, 5.0 g yeast extract, 10.0 g tryptone) or on LB agar (15.0 g/L agar). Iron-limiting medium was prepared by adding 15 μM deferoxamine mesylate (Desferal) (Sigma) to LB medium. Desferal was chosen for this study based upon previous findings in which P. mirabilis urinary tract isolates were found to be highly susceptible to this agent (Lowy et al., 1984). We determined a concentration of 15 uM Desferal in LB medium that decreased the growth rate of wild-type in comparison to wild-type grown in LB medium alone (Fig. 1). Growth curves were performed using a Bioscreen growth curve analyzer (Growth Curves) following culture of bacterial samples in triplicate. Error bars were calculated by the standard error of the mean. Iron-limited LB agar supplemented with 25 μM Desferal (Sigma) restricted P. mirabilis growth for siderophore cross-feeding assays. Neidhardt MOPS Minimal Medium (Neidhardt et al., 1974) with and without 0.1 mM FeCl3·6H2O was used for culture of bacterial strains. For growth of P. mirabilis, Neidhardt MOPS Minimal Medium was supplemented with 1 ml 1% nicotinic acid, 1 ml 1 M MgSO4·7H20, 10 ml 20% glycerol, and 1 ml 20% chelex-treated casamino acids per liter. Chrome azurol S (CAS) agar and CAS shuttle solution was prepared as previously described (Schwyn & Neilands, 1987). Antibiotics were added as necessary at the following concentrations: kanamycin (25 μg/ml), tetracycline (15 μg/ml), ampicillin (50 μg/ml), and chloramphenicol (20 μg/ml).
Microarray analysis
70-mer oligonucleotides representing open reading frames within the genome of P. mirabilis HI4320 were spotted (Microarrays, Inc.) in triplicate onto Ultra GAPSll slides (Corning)(Pearson et al., 2010, Flannery et al., 2009). Overnight LB medium cultures of HI4320 were diluted 1:100 in fresh LB medium or LB medium containing 15 μM of Desferal and incubated for three hours at 37°C, or until the culture reached an OD600 of 1.0. Cultures were treated with 2 volumes of RNA protect (Qiagen), incubated at room temperature for five minutes to stabilize RNA, and centrifuged for (10 min, 5000 × g, 25°C). RNA (4 μg) was purified (Qiagen) and DNase-treated (Ambion) as described above. First strand cDNA synthesis reactions, labeling of cDNA with CyDyes, and hybridization to the microarray slides were performed following the protocols; ftp://ftp.jcvi.org/pub/data/PFGRC/pdf_files/protocols/M007.pdf and ftp://ftp.jcvi.org/pub/data/PFGRC/pdf_files/protocols/M008.pdf, found on The Institute for Genomic Research (TIGR) website. The microarray was scanned by a ScanArray Express Microarray Scanner (Perkin Elmer) at 10 μm resolution.
Five independent RNA samples were subjected to microarray analysis. The cyanine 3 or cyanine 5 dye was swapped for each condition for two runs. Spot intensities were extracted and LOWESS normalized with ScanArray Express v 4.0 (Perkin Elmer) software, spot replicates were merged with Microarray Data Analysis System (MIDAS) software, and statistically analyzed using Multiexperiment Viewer (MeV) software (Saeed et al., 2003). Using MeV software, a one-class unpaired test was performed by Significance Analysis of Microarray (SAM) to identify genes significantly up-regulated or down-regulated in response to in vitro iron limitation (Tusher et al., 2001, Saeed et al., 2003). The False Discovery Rate was manually set to zero. Fold-change of gene expression is presented as the log2 transformation of the ratio of the transcript level of P. mirabilis growth in LB medium and LB medium with Desferal, averaged across the five experiments. Microarray data are accessible through the Gene Expression Omnibus (GEO) database at with accession number GSE18051.
RNA preparation and quantitative RT-PCR
Overnight bacterial cultures were diluted 1:100 in 5 ml of fresh medium and incubated at 37°C until an OD600 of 1.0 was reached. A 500 μl volume of cells was treated with 62.5 μl of 5% phenol-ethanol stop solution and incubated at room temperature for 10 minutes. For RNA from in vivo urine samples, urine was collected into RNAprotect (Qiagen) following manufacturer’s recommendation. Bacterial suspensions were centrifuged (5 min, 5000 × g, 4°C) and RNA was purified using an RNeasy (Qiagen) column following the manufacturer’s protocol. Contaminating genomic DNA was removed with TURBO DNA-free (Ambion) following the manufacturer’s protocol. Lack of genomic DNA contamination was determined by PCR analysis using primers to amplify the housekeeping gene rpoA prior to cDNA synthesis.
cDNA synthesis of total RNA was performed using SuperScript II first strand synthesis kit (Invitrogen) using 50 ng of random hexamers per 5 μg total RNA following the manufacturer’s protocol. cDNA was purified on a QIAquick column (Qiagen) according to the manufacturer’s protocol and eluted in 30 μl of the supplied elution buffer. Purified cDNA was quantified using a NanoDrop spectrophotometer. Transcripts were quantified on a MX3000P real-time PCR machine (Stratagene) using Brilliant SYBR green QPCR master mix (Stratagene) in 25 μl reactions containing up to 30 ng of total cDNA. Real-time oligonucleotide sequences of genes were designed using the PrimerQuest program (Steve Rozen, Helen J. Skaletsky (1996, 1997, 1998)) on the Integrated DNA Technologies website and are listed in Table S3. Transcript levels were normalized to the housekeeping gene rpoA and log2 fold-change was determined relative to an experiment-specific calibrator using the MXPro v 4.1 software package (Stratagene). Error bars were calculated by the standard error of the mean and statistical analysis was performed using an unpaired t-test.
Reverse transcriptase PCR
Transcriptional organization of siderophore biosynthesis and ABC transport gene clusters, PMI0229-0239 and PMI2596-2605, was examined by reverse transcriptase PCR (RT-PCR) from mRNA isolated following culture of P. mirabilis HI4320 in LB medium with 15 μM Desferal. RNA purification, Dnase-treatment and cDNA synthesis was performed as previously described. A total of 30 ng of cDNA or genomic DNA was used in 25 μl reactions. The intergenic regions spanning genes within these systems were amplified to identify contiguous genes on the same transcript.
Mutant construction
To test siderophore production and colonization in the murine model of ascending UTI, single mutations and a double mutation of PMI0232 and PMI2599, a putative siderophore biosynthesis protein and a putative non-ribosomal peptide synthase, respectively, were constructed using the TargeTronGene Knockout System (Sigma) following a modified protocol (Pearson & Mobley, 2007). Oligonucleotides for mutant design were created using the TargeTron Design Site (Sigma) and are listed in Table S1. PCR confirmation of mutants was performed using oligonuceotides designed with the PrimerQuest program as previously described and are listed in Table S1. Mutants constructed are identified as pbtA::kan, nrpR::kan, nrpR pbtA::kan, and nrpR 2596::kan; the last two designations for these mutants refer to the nrpR insertion mutant with the kanamycin marker deleted and the pbtA or PMI2596 insertion mutant with the kanamycin marker retained. The kanamycin resistance cassette was removed from the intron by IPTG induction of the cre recombinase in pQL123 (Liu et al., 1998).
Detection of siderophore production
Bacteria, cultured in LB medium with aeration at 37°C to stationary phase, were spotted in 5 μl volumes onto chrome azurol S (CAS) agar plates (Schwyn & Neilands, 1987) and incubated at 30°C for 18 hours. A visible color change of agar from blue to orange around bacterial growth indicates siderophore production. E. coli CFT073 and E. coli CFT073 entF::kan iucB::cam, an enterobactin aerobactin double siderophore biosynthesis mutant, were used as positive and negative controls, respectively.
For the CAS assay, overnight bacterial cultures in LB medium were diluted 1:100 into MOPS defined medium with 0.1 mM FeCl3·6H2O. Cells were grown to stationary phase, harvested by centrifugation (5 min, 8000 × g, 4°C), and washed before diluting 1:100 into fresh MOPS buffer with and without 0.1mM FeCl3·6H2O. Following growth to OD600 = 1.0, cells were centrifuged (20 min, 10,000 × g, 4°C), supernatant was collected and passed through a 0.22 μm filter (Millipore). Wild-type and mutant samples of P. mirabilis were lyophilized in 50 ml volumes and subsequently resuspended in 1 ml of phosphate buffered saline. A total of 150 μl of siderophore-containing supernatant was mixed with 150 μl of CAS shuttle solution as previously described (Schwyn & Neilands, 1987). Phosphate buffered saline was used for resuspension of lyophilized supernatant and phosphate buffered saline containing 15 μM Desferal served as a reference for P. mirabilis samples and a positive control, respectively. Following one hour incubation at room temperature, samples were measured at an absorbance of 630 nm using a MicroQuant Spectrophotometer (Bio-Tek). Each sample was measured in triplicate. Absorbance readings were calculated by dividing the samples’ averaged absorbance by the absorbance of the reference. Error bars were calculated by the standard error of the mean and statistical analysis was performed using an unpaired t-test.
Presence of pbt and nrp genes in other Enterobacteriaceae
P. mirabilis isolates were screened by PCR for the presence or absence of genes, PMI0232 and PMI2599, a putative siderophore biosynthesis protein and a putative non-ribosomal peptide synthase, respectively, using genomic DNA (Flannery et al., 2009). Forward and reverse qPCR primers, pbtA and 2599 (Table S3), were used to amplify within the open reading frame of both genes using 60 ng of gDNA as template. Primers for amplification of PMI3255 were used as a positive DNA control as previously described (Flannery et al., 2009). Associations were determined by using the Fisher exact test (GraphPad Prism).
For sequence analysis of the proteobactin synthesis protein PbtA, protein sequences homologous to PbtA were identified by BLAST (Altschul et al., 1990). The resulting 92 sequences were input into MegAlign (DNASTAR) and subjected to multiple alignment using ClustalW (Thompson et al., 1994) using a BLOSUM protein weight matrix and a gap penalty of 10.00. The results of the multiple alignment were used to generate a phylogenetic cladogram where the branch distances correspond to sequence divergence. The cladogram was visualized by bootstrap analysis comparing tree constructs using the neighbor-joining methodology performed with 10,000 trials initialized with a random seed.
Cross-feeding assays
Cross-feeding assays were performed to determine whether yersiniabactin produced by other bacterial species could cross-feed P. mirabilis HI4320. LB agar was supplemented with 25 μM Desferal, an iron-limited condition in which P. mirabilis was unable to grow. For cross-feeding assays, wild-type P. mirabilis, the proteobactin synthesis/receptor and mutant (PbtSR), and the yersiniabactin-related synthesis/receptor mutant (NrpSR) were inoculated from overnight LB medium cultures onto the surface of the iron-limited plate using a sterile 1 μl loop. E. coli CFT073, E. coli 536, and E. coli 536 mutant strains entF::kan and entF ybtS::kan were cross-struck from LB medium perpendicular to P. mirabilis strains. X-gal (40 μg/ml) was added to the iron-limited plates to distinguish growth of the lacZ positive, E. coli 536 and CFT073, and lacZ negative, P. mirabilis, strains. Following incubation at 37°C for 18 hours plates were scored for promotion of growth of the P. mirabilis strains.
Chromatographic detection of yersiniabactin
A final concentration of 100 μM FeCl3·6H2O was added to supernatants of previously described MOPS iron-depleted cultures of E. coli 536 mutants entF::kan and entF ybtS::kan and P. mirabilis mutants pbtA::kan and nrpR pbtA::kan. 50 ml of each sample were then evaporated to dryness and redissolved in 2 ml of methanol. 500 μl of this concentrated preparation was injected onto a Beckman-Coulter System Gold HPLC in line with a Waters XBridge C18 (4 10 × 250 mm) column running the following method: 5% acetonitrile (MeCN) in ddH2O for 1 minute followed by a linear increase to 90% MeCN over 20 minutes and a final 3 minute 100% MeCN wash step. Peaks not shared between all samples were collected for further investigation. Prior to LC-MS analysis, concentrated supernatant samples were diluted with an equal volume of additional methanol and all insoluble material was centrifuged out of solution. 2 μl of the remaining supernatant was injected onto a Shimadzu electospray ionization (ESI) LCMS-2010A mass spectrometer in line with an HPLC system using a Phenomenex Luna C18 (1.5 × 250 mm) column and the following elution method: 5% (MeCN) in ddH2O for 10 minutes (over which time material was diverted from the detector) followed by a jump to 20% MeCN and increasing by a linear gradient to 95% MeCN over 30 minutes. Detection occurred in positive scanning and selected ion monitoring (SIM) modes. Collected peaks from previous HPLC analysis were evaporated under N2, redissolved in methanol, and analyzed by LC-MS in an identical manner.
CBA/J mouse model of ascending UTI
The CBA mouse model of ascending UTI (Johnson et al., 1987, Hagberg et al., 1983) was used to assess the contribution to fitness and virulence of the siderophore biosynthesis and ABC transport systems. For cochallenge competition (to determine fitness) experiments, 6–8 week old female CBA/J mice (20 to 22 g; Jackson Laboratories) were transurethrally infected with a 50 μl bacterial suspension of 5 × 107 CFU containing a 1:1 ratio of P. mirabilis HI4320 and P. mirabilis antibiotic-resistant mutant. For independent challenge (to determine virulence) mice were infected with wild-type HI4320 alone or mutant alone. Urine was collected and pooled from mice infected with wild-type P. mirabilis HI4320 at 24 hours, 48 hours, and 5 days post-infection for transcriptional analysis. Mice were euthanized 7 days post infection. Bladder and kidneys were collected, homogenized, and plated on plain LB agar and/or LB agar with antibiotic (Autoplate 4000, SpiralBiotech). Viable colony counts were enumerated using a Q Count (Spiral Biotech) and expressed as CFU/g of tissue. For cochallenge competition experiments, wild-type infection was determined by subtracting the number of colonies on LB agar containing antibiotic from the number of colonies on plain LB agar. Competitive indices (CI) were calculated by dividing the ratio of the CFU of wild-type to mutant recovered from mice following infection by the ratio of the CFU of wild-type HI4320 to the CFU of mutant present in the input. A CI > 1 indicates the wild-type out-competes the mutant strain and a CI < 1 indicates the wild-type is out-competed by the mutant. The Mann-Whitney test was used to determine the statistical significance of data obtained in independent challenge experiments and cochallenge competition experiments were analyzed using the Wilcoxon signed-rank test.
Supplementary Material
Acknowledgments
The authors would like to thank Shelley Payne for helpful procedural advice regarding CAS agar, Alfredo Torres for generously providing the CFT073 entF::kan iucB::cam strain, Erin Hagan for construction of E. coli 536 mutants entF::kan and entF ybtS::kan and helpful insight to the study, Lona Mody for generously providing P. mirabilis non-UTI colonizing isolates, Sara Smith for assistance with our animal studies, and Chris Alteri for critical reading of the manuscript.
This study was supported in part by the Public Health Service Grants AI043360 and AI059722 from the National Institutes of Health.
References
- Alteri CJ, Mobley HL. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infection and immunity. 2007;75:2679–2688. doi: 10.1128/IAI.00076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS microbiology reviews. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Berger H, Hacker J, Juarez A, Hughes C, Goebel W. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. Journal of bacteriology. 1982;152:1241–1247. doi: 10.1128/jb.152.3.1241-1247.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS microbiology reviews. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultreys A, Gheysen I, de Hoffmann E. Yersiniabactin production by Pseudomonas syringae and Escherichia coli, and description of a second yersiniabactin locus evolutionary group. Appl Environ Microbiol. 2006;72:3814–3825. doi: 10.1128/AEM.00119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burall LS, Harro JM, Li X, Lockatell CV, Himpsl SD, Hebel JR, Johnson DE, Mobley HL. Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infection and immunity. 2004;72:2922–2938. doi: 10.1128/IAI.72.5.2922-2938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. Journal of bacteriology. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel H, Thieken A, Reissbrodt R, Jung G, Winkelmann G. Alpha-keto acids are novel siderophores in the genera Proteus, Providencia, and Morganella and are produced by amino acid deaminases. Journal of bacteriology. 1993;175:2727–2733. doi: 10.1128/jb.175.9.2727-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF, Bennett RL, Rothfield LI. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. Journal of bacteriology. 1978;135:928–934. doi: 10.1128/jb.135.3.928-934.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanylo LP, Kadis S, Maudsley JR. Siderophore production by Proteus mirabilis. Canadian journal of microbiology. 1984;30:1046–1051. doi: 10.1139/m84-163. [DOI] [PubMed] [Google Scholar]
- Faraldo-Gomez JD, Sansom MS. Acquisition of siderophores in gram-negative bacteria. Nat Rev Mol Cell Biol. 2003;4:105–116. doi: 10.1038/nrm1015. [DOI] [PubMed] [Google Scholar]
- Fisher CR, Davies NM, Wyckoff EE, Feng Z, Oaks EV, Payne SM. Genetics and virulence association of the Shigella flexneri sit iron transport system. Infection and immunity. 2009;77:1992–1999. doi: 10.1128/IAI.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery EL, Mody L, Mobley HL. Identification of a modular pathogenicity island that is widespread among urease-producing uropathogens and shares features with a diverse group of mobile elements. Infection and immunity. 2009;77:4887–4894. doi: 10.1128/IAI.00705-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franza T, Mahe B, Expert D. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Molecular microbiology. 2005;55:261–275. doi: 10.1111/j.1365-2958.2004.04383.x. [DOI] [PubMed] [Google Scholar]
- Gaisser S, Hughes C. A locus coding for putative non-ribosomal peptide/polyketide synthase functions is mutated in a swarming-defective Proteus mirabilis strain. Mol Gen Genet. 1997;253:415–427. doi: 10.1007/s004380050339. [DOI] [PubMed] [Google Scholar]
- Hagan EC, Mobley HL. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Molecular microbiology. 2009;71:79–91. doi: 10.1111/j.1365-2958.2008.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infection and immunity. 1983;40:273–283. doi: 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Current opinion in microbiology. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Himpsl SD, Lockatell CV, Hebel JR, Johnson DE, Mobley HL. Identification of virulence determinants in uropathogenic Proteus mirabilis using signature-tagged mutagenesis. Journal of medical microbiology. 2008;57:1068–1078. doi: 10.1099/jmm.0.2008/002071-0. [DOI] [PubMed] [Google Scholar]
- Johnson DE, Lockatell CV, Hall-Craigs M, Mobley HL, Warren JW. Uropathogenicity in rats and mice of Providencia stuartii from long-term catheterized patients. The Journal of urology. 1987;138:632–635. doi: 10.1016/s0022-5347(17)43287-3. [DOI] [PubMed] [Google Scholar]
- Jones AM, Lindow SE, Wildermuth MC. Salicylic acid, yersiniabactin, and pyoverdin production by the model phytopathogen Pseudomonas syringae pv. tomato DC3000: synthesis, regulation, and impact on tomato and Arabidopsis host plants. Journal of bacteriology. 2007;189:6773–6786. doi: 10.1128/JB.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D. Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS microbiology reviews. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- Koster W. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Research in microbiology. 2001;152:291–301. doi: 10.1016/s0923-2508(01)01200-1. [DOI] [PubMed] [Google Scholar]
- Leveille S, Caza M, Johnson JR, Clabots C, Sabri M, Dozois CM. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infection and immunity. 2006;74:3427–3436. doi: 10.1128/IAI.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima A, Zunino P, D’Alessandro B, Piccini C. An iron-regulated outer-membrane protein of Proteus mirabilis is a haem receptor that plays an important role in urinary tract infection and in in vivo growth. Journal of medical microbiology. 2007;56:1600–1607. doi: 10.1099/jmm.0.47320-0. [DOI] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Lowy FD, Pollack S, Fadl-Allah N, Steigbigel NH. Susceptibilities of bacterial and fungal urinary tract isolates to desferrioxamine. Antimicrobial agents and chemotherapy. 1984;25:375–376. doi: 10.1128/aac.25.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelis JH, den Daas-Slagt HJ, Hoogkamp-Korstanje JA. Iron requirement and chelator production of staphylococci, Streptococcus faecalis and enterobacteriaceae. Antonie van Leeuwenhoek. 1978;44:257–267. doi: 10.1007/BF00394304. [DOI] [PubMed] [Google Scholar]
- Massad G, Zhao H, Mobley HL. Proteus mirabilis amino acid deaminase: cloning, nucleotide sequence, and characterization of aad. Journal of bacteriology. 1995;177:5878–5883. doi: 10.1128/jb.177.20.5878-5883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara H, Muramatsu H, Kakutani R, Yasuda M, Ueda M, Kurihara T, Esaki N. N-methyl-L-amino acid dehydrogenase from Pseudomonas putida. A novel member of an unusual NAD(P)-dependent oxidoreductase superfamily. The FEBS journal. 2005;272:1117–1123. doi: 10.1111/j.1742-4658.2004.04541.x. [DOI] [PubMed] [Google Scholar]
- Miles AA, Khimji PL. Enterobacterial chelators of iron: their occurrence, detection, and relation to pathogenicity. Journal of medical microbiology. 1975;8:477–490. doi: 10.1099/00222615-8-4-477. [DOI] [PubMed] [Google Scholar]
- Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infection and immunity. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Warren JW. Urease-positive bacteriuria and obstruction of long-term urinary catheters. Journal of clinical microbiology. 1987;25:2216–2217. doi: 10.1128/jcm.25.11.2216-2217.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody L, Maheshwari S, Galecki A, Kauffman CA, Bradley SF. Indwelling device use and antibiotic resistance in nursing homes: identifying a high-risk group. Journal of the American Geriatrics Society. 2007;55:1921–1926. doi: 10.1111/j.1532-5415.2007.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics (Oxford, England) 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Munzinger M, Budzikiewicz H, Expert D, Enard C, Meyer JM. Achromobactin, a new citrate siderophore of Erwinia chrysanthemi. Zeitschrift fur Naturforschung. 2000;55:328–332. doi: 10.1515/znc-2000-5-605. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. Journal of bacteriology. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. Siderophores: structure and function of microbial iron transport compounds. The Journal of biological chemistry. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Oves-Costales D, Kadi N, Challis GL. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chemical communications (Cambridge, England) 2009:6530–6541. doi: 10.1039/b913092f. [DOI] [PubMed] [Google Scholar]
- Pearson MM, Mobley HL. The type III secretion system of Proteus mirabilis HI4320 does not contribute to virulence in the mouse model of ascending urinary tract infection. Journal of medical microbiology. 2007;56:1277–1283. doi: 10.1099/jmm.0.47314-0. [DOI] [PubMed] [Google Scholar]
- Pearson MM, Rasko DA, Smith SN, Mobley HL. Transcriptome of swarming Proteus mirabilis. Infection and immunity. 2010;78:2834–2845. doi: 10.1128/IAI.01222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MM, Sebaihia M, Churcher C, Quail MA, Seshasayee AS, Luscombe NM, Abdellah Z, Arrosmith C, Atkin B, Chillingworth T, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Walker D, Whithead S, Thomson NR, Rather PN, Parkhill J, Mobley HL. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. Journal of bacteriology. 2008;190:4027–4037. doi: 10.1128/JB.01981-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini CD, Barbe FM, Legnani-Fajardo CL. Identification of iron-regulated outer membrane proteins in uropathogenic Proteus mirabilis and its relationship with heme uptake. FEMS microbiology letters. 1998;166:243–248. doi: 10.1111/j.1574-6968.1998.tb13897.x. [DOI] [PubMed] [Google Scholar]
- Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annual review of microbiology. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schmelz S, Kadi N, McMahon SA, Song L, Oves-Costales D, Oke M, Liu H, Johnson KA, Carter LG, Botting CH, White MF, Challis GL, Naismith JH. AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis. Nature chemical biology. 2009;5:174–182. doi: 10.1038/nchembio.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Analytical biochemistry. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shand GH, Anwar H, Kadurugamuwa J, Brown MR, Silverman SH, Melling J. In vivo evidence that bacteria in urinary tract infection grow under iron-restricted conditions. Infection and immunity. 1985;48:35–39. doi: 10.1128/iai.48.1.35-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infection and immunity. 2004;72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Redford P, Welch RA, Payne SM. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infection and immunity. 2001;69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckman M, Osburne MS. In vivo inhibition of TonB-dependent processes by a TonB box consensus pentapeptide. Journal of bacteriology. 1992;174:320–323. doi: 10.1128/jb.174.1.320-323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annual review of microbiology. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Warren JW, Tenney JH, Hoopes JM, Muncie HL, Anthony WC. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. The Journal of infectious diseases. 1982;146:719–723. doi: 10.1093/infdis/146.6.719. [DOI] [PubMed] [Google Scholar]
- Winkelmann G. Microbial siderophore-mediated transport. Biochemical Society transactions. 2002;30:691–696. doi: 10.1042/bst0300691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.