Abstract
CHARGE syndrome (CS, OMIM #214800) is a rare, autosomal dominant disorder, two-thirds of which are caused by haplo-insufficiency in the Chd7 gene. Here, we show that the Drosophila homolog of Chd7, kismet, is required for proper axonal pruning, guidance and extension in the developing fly's central nervous system. In addition to defects in neuroanatomy, flies with reduced kismet expression show defects in memory and motor function, phenotypes consistent with symptoms observed in CS patients. We suggest that the analysis of this disease model can complement and expand upon the existing studies for this disease, allowing a better understanding of the role of kismet in neural developmental, and Chd7 in CS pathogenesis.
INTRODUCTION
CHARGE Syndrome (CS, OMIM #214800) is a rare disorder, two-thirds of which are caused by haplo-insufficiency in the Chromodomain Helicase DNA Binding Protein 7 (Chd7) gene. Heterozygous loss-of-function mutations in the Chd7 gene were first described as associated with CS in 2004 (1), and since have been estimated to be causative for nearly two-thirds of all CS diagnoses (2), underscoring the importance of analysis of Chd7 function in this disease. CS is characterized by a variety of clinical symptoms and is prevalent in approximately one in every 10 000 live births (3). Although the clinical symptoms of CS vary from case to case, the clinical definition of CS initially relied on six basic symptoms: coloboma, heart malformation, choanal atresia, retardation of growth and/or development, genital anomalies and ear anomalies, making CS a common cause of congenital anomalies (2). Though these clinical symptoms alone can often suggest a diagnosis of CS, the literature stresses the importance of coupling this clinical diagnosis with a molecular diagnosis, through mutational analysis of Chd7 (2–5). Chd7 encodes an evolutionarily conserved protein thought to play a role in the regulation of gene expression through chromatin remodeling (1). Recent studies of Chd7 DNA-binding sites on chromatin have shown that Chd7 binding is correlated to areas of mono- and dimethylated lysine 4 of histone H3 (6), consistent with Chd7 serving a role in mediating transcription through chromatin remodeling.
Attempts to understand the complex etiology of many human diseases have been improved through the study of model organisms. The fruit fly, Drosophila melanogaster, has been tremendously important and influential in furthering our understanding of the molecular and cellular mechanisms of gene function in a variety of human diseases, including neurodegenerative diseases and forms of hereditary mental retardation. We therefore sought to utilize the fly in an attempt to understand the basic functions of the Drosophila homolog of mammalian Chd7 in behavior and neural development. Literature suggests that the Drosophila homolog of Chd7 is the kismet (kis) gene (7–14). Searches of the NCBI Homologene database and information hyperlinked over proteins databases also list kis as the fly homolog of human Chd7. Finally, an NCBI Protein BLAST of the human CHD7 protein (NP_060250.2) lists the Kismet protein as the protein with the highest degree of similarity and identity within Drosophila (E value 0.0).
CHD7 belongs to a subfamily of proteins in mammals that includes CHD6, CHD7, CHD8 and CHD9 (13), collectively referred to as subgroup III (12). Literature has suggested that Kismet protein function in flies is carried out by the proteins within this subfamily III group in mammals (12,13), consistent with the protein sequence homology shared by Kismet and this group of proteins. Taken together, this suggests that the Kismet protein is most closely related to the Bilaterian ancestral protein that fly Kismet and human CHD6, CHD7, CHD8 and CHD9 evolved from ∼670 million years ago (15). Recently, Batsukh et al. (12) have shown that CHD7 interacts with family member CHD8 both directly and indirectly to form a potential CHD7/CHD8 complex. Based on the literature showing that in other diseases (Hereditary Spastic Paraplegia and Cornelia de Lange syndrome), interacting partners are involved in the underlying cause of the disease, these authors suggest that a CHD7/CHD8 containing complex may be involved in the pathogenesis of CS (12). A major advantage of Drosophila as a model system for the study of human disease is the ability to study the functions of genes whose mammalian homologs exist in multi-gene families, in order to gain a deeper understanding of the evolutionarily conserved functions and relationships between these genes. Given that 1) CHD7 is the closest human homolog based on sequence identity to fly Kismet; 2) CHD7 and CHD8 family members are both homologous to the Kismet protein; and 3) CHD7 and CHD8 interact to potentially contribute to CHARGE pathogenesis, determining the functions of kismet in neural development and behavior may significantly contribute to our understanding of the function of both CHD7 and CHD8 in CS pathogenesis.
The Drosophila kismet gene encodes for two protein products (Kis-L and Kis-S) which share a common C terminal stretch of ∼2100 amino acids (16). The common C terminal segment contains a BRK domain of unknown function, whereas the N-terminal domain of Kis-L also contains an ATPase domain similar to those found in other chromatin remodeling enzymes and two chromodomains which can recognize Histone H3 methylation (16). Importantly, the BRK domain, as well as the ATPase and chromodomains found in the Kismet protein are also conserved in human Chd7 (8), suggesting that the Drosophila protein is a good candidate to study in order to better understand human Chd7 protein function.
kis was initially identified in a genetic screen as a suppressor of a dominant homeotic phenotype and as a member of the trithorax group of transcriptional activators (17). Subsequent analysis of kis mutations revealed an essential role for kis in embryonic segmentation and adult and larval cell fate specification (17,18). kis mutations have been identified in genetic screens as modifiers of the Ras and Notch signal transduction pathways (19,20), as modifiers of the proneural gene atonal in retinal development (11) and as a regulator for vein specification in developing wings (21). Kis protein is proposed to facilitate an early step in transcriptional elongation through alterations in chromatin structure (16).
Here, we utilize Drosophila to better understand the role of kis in fly behavior and neural development. We report the first evidence that adult flies with decreased Kis protein exhibit a number of robust and genetically tractable phenotypes that are similar to symptoms observed in CS patients, including defects in gross motor coordination and defective learning and memory. Histological analysis of kis mutant neurons show defects in cell and axonal migration in multiple neuronal populations in the fly central nervous system, suggesting a possible anatomical mechanism for the behavioral defects we observe in adult flies. We suggest that analysis of this disease model can complement and expand upon the studies done in cell culture and vertebrate model organisms for this disease, allowing a better understanding of the role of kis in neural developmental and Chd7 in CS pathogenesis.
RESULTS
Adult flies with decreased kismet expression display numerous phenotypes
Null mutations in kis are embryonic lethal, presumably due to the fact that kis gene function is required to maintain states of homeotic gene transcription in early embryogenesis (18). We hypothesized that if we were to partially knock down kis gene function in specific tissues, we could allow flies to overcome the requirement for kis in the embryo, while sufficiently hindering kis gene function in later stages of development, to generate viable adults with prominent phenotypes. To accomplish this, we utilized the Gal4/UAS system (22). We expressed two different publicly available kis RNAi strains from the Vienna Drosophila RNAi Center (kis RNAi.a, and kis RNAi.b). To examine the extent to which we are able to knock down Kis protein expression with these constructs in vivo, we first expressed them in the posterior compartment of developing wing imaginal discs by using the engrailed-Gal4 driver (Supplementary Material, Fig. S1). engrailed is expressed only in the posterior compartment of tissues (23,24), leaving the anterior compartment as a critical internal control for this assay and allowing us to estimate the amount of knock down these constructs have in vivo. The Kis protein is normally expressed ubiquitously in the developing wing imaginal disc, with roughly equivalent expression levels in the posterior and anterior compartment (Supplementary Material, Fig. S1A and B). We found that the two different kis RNAi constructs knockdown Kis protein expression differentially. kis RNAi.a shows a 60% knockdown (Supplementary Material, Fig. S1C and D), whereas kis RNAi.b shows a very strong 90% knockdown (Supplementary Material, Fig. S1E and F). These results are consistent with recently published data that also show significant knockdown of Kis protein using these constructs (10).
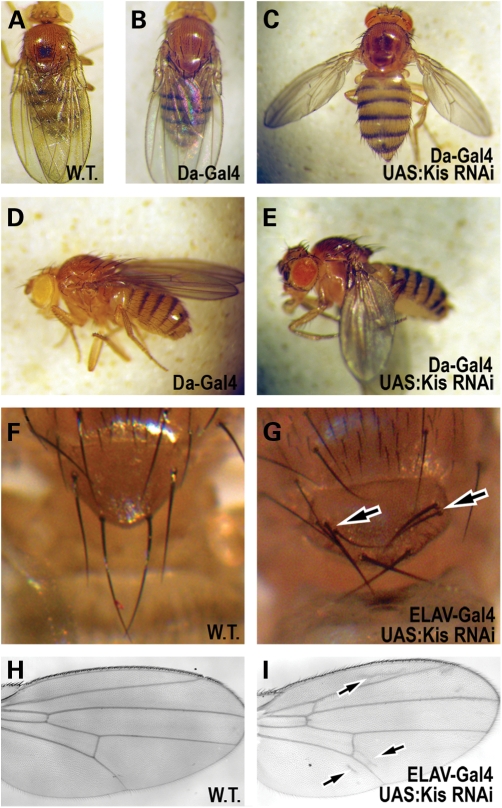
In order to generate viable adult flies with decreased Kis expression, we first expressed both of these kis RNAi constructs ubiquitously with the daughterless-Gal4 driver (da-Gal4). Expression of kis RNAi.b with da-Gal4 at 25°C leads to late pupal lethality, suggesting that Kis is required for some critical function during pupal development. However, expression of kis RNAi.a with da-Gal4 at 25°C produces viable adults with obvious morphological defects (Fig. 1). These flies are unable to fly, and exhibit a prominent postural defect where they hold their wings apart and below their bodies with 100% penetrance (Fig. 1C and E). This postural defect is reminiscent of defects associated in muscle cells, and is also observed in mutants for other chromatin remodeling factors in flies (25,26). Further, this phenotype is consistent with hypotonia-related posture problems observed in CHARGE patients (27).
Figure 1.
Adult phenotypes in Kismet knockdown flies. (A–E) Adult female flies. (A–C) Dorsal view. (D–E) Lateral view. (A) Wild-type (Canton S) fly. Note the position of the adult wings. (B and D) A fly expressing only the Daughterless-Gal4 reagent (Da-Gal4/ +) holds its wings normally. (C and E) A fly where Kis protein is ubiquitously knocked down (Da-Gal4/UAS:kis RNAi.a) holds its wings abnormally apart from its body. (F and G) Dorsal view of bristles on scutellum in (F) wild-type flies and (G) flies where Kis protein is knocked down in nervous tissue (Elav-Gal4/UAS:kis RNAi.a). Arrow notes duplication of bristles in Kis knockdown flies. (H and I) Adult wings from (H) wild-type and (I) Kis knockdown flies (Elav-Gal4/UAS:kis RNAi.a). Arrows denote presence of extra vein tissue near wing vein L2 (top arrow), the posterior crossvein (middle arrow) and wing vein L5 (bottom arrow). In cases not shown, Da-Gal4/+ and ELAV/+ flies show wild type phenotypes.
To initially assess kis function in the developing nervous system, we expressed both of the RNAi constructs using ELAV-Gal4, a pan-neural driver. Expression of kis RNAi.b at 25°C leads to late pupal lethality, suggesting that the cause of the lethality observed at this stage with the da-Gal4 driver may be due to loss of Kis protein within the nervous system. Expression of kis RNAi.a with ELAV-Gal4 produces viable flies with various morphological defects (Fig. 1). These flies exhibit duplicated bristles on the adult thorax [Fig. 1G, 24% penetrance (n = 50) compared with 0% in sibling controls (n = 50)]. This is a defect in the development of peripheral nervous system structures and has been previously observed in other regulators of chromatin structure in Drosophila (28,29). These flies also display extra vein differentiation in adult wings (Fig. 1I). These defects were associated mostly near wing veins L2 [45% (n = 50) compared with 0% in sibling controls (n = 70)] and L5 [24% (n = 50) compared with 0% in sibling controls (n = 70)]. This is also consistent with previously reported loss-of-function kis effects in wing vein differentiation (21).
Kismet function is required in muscles for early climbing behavior
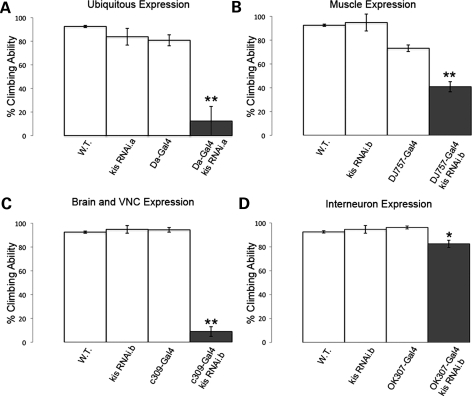
Patients with CHARGE display delayed or impaired motor coordination and adaptive motor skills, musculoskeletal anomalies and hypotonia (3,27). To specifically test motor control of our CS flies, we utilized a simple, yet powerful behavioral assay based on climbing ability (30). Because we are interested in looking for early defects that may be associated with developmental problems due to loss of kis function, we restricted our analysis of climbing ability to the first 10 days after eclosion (early climbing behavior, days 1–10). Flies with ubiquitous expression of UAS:kis RNAi.a by Da-Gal4 exhibit a significant decrease in climbing behavior within 10 days after eclosion (Fig. 2A) compared with controls. To more precisely determine specific tissue(s) in which loss of kis function affects this behavior, we expressed the UAS:kis RNAi constructs with a number of different tissue specific Gal4 drivers (Fig. 2, Table 1). Although the expression of kis RNAi.b leads to late pupal lethality with the pan-neural ELAV-Gal4 (see above), the expression of kis RNAi.a with this driver has no significant effect on early climbing behavior (Table 1). Further, specific knockdown of Kis protein within motor neurons, dopaminergic neurons, glutamatergic neurons, cholinergic neurons, mushroom body neurons and glial cells all have no significant effect on early climbing behavior (Table 1). However, knockdown of Kis within muscles shows a strong effect (Fig. 2B, Table 1), reducing climbing frequency by nearly half compared with control flies. These data suggest that kis function is critically required in Drosophila muscles for coordinated motor function.
Figure 2.
Kismet knockdown in muscles causes defective climbing behavior. (A–D) shows compiled climbing ability of wild-type, control and Kis knockdown flies at days 2–10. (A) Ubiquitous expression of UAS:Kis RNAi.a (Da-Gal4/UAS:Kis RNAi.a) shows severely reduced climbing ability compared with wild-type, Da-Gal4/+ and UAS:Kis RNAi.a/+ outcrossed control flies. (B) Kis knockdown restricted to muscles (DJ757-Gal4/UAS:Kis RNAi.b) also shows a strong reduction in climbing ability compared with wild-type, DJ757-Gal4/+ and UAS:Kis RNAi.b/+ outcrossed controls. (C) Kis knockdown in mushroom body neurons and the ventral nerve cord (c309-Gal4/UAS:Kis RNAi.b) shows a strong reduction in climbing ability compared to wild-type, c309-Gal4/+ and UAS:Kis RNAi.b/+ outcrossed controls. (D) Kis knockdown in the giant fiber inter-neurons (OK307-Gal4/UAS:Kis RNAi.b) shows a small but significant effect compared with wild-type, OK307-Gal4/+ and UAS:Kis RNAi.b/+ outcrossed controls. Error bars represent ± SEM. In all cases, double asterisk indicates P < 0.001 and asterisk indicates P < 0.05 compared with controls.
Table 1.
Gal4 lines tested for climbing behavior
| Name | BL# | Organ/tissue | Climbing affected |
|---|---|---|---|
| ELAV-Gal4 | 458 | Pan-neuronal | No |
| Ple-Gal4 | 8845 | Dopaminergic neurons | No |
| Cha-Gal4 | 6798/6793 | Cholinergic neurons | No |
| OK371-Gal4 | 26160 | Glutamatergic neurons | No |
| OK107-Gal4 | 854 | entire MB | No |
| 1471-Gal4 | 9465 | Gamma lobes of MB | No |
| D42-Gal4 | 8816 | Motor neurons | No |
| Pdf-Gal4 | 6900 | Ventrolateral neurons | No |
| Repo-Gal4 | 7415 | Glial cells | No |
| C309-Gal4 | 6906 | MB, thoracic ganglion, eye | Yes |
| OK307-Gal4 | 6488 | Giant fiber interneuron | Yes |
| A51-Gal4 | 8764 | Muscles and motor neurons | Yes |
| DJ757-Gal4 | 8184 | Muscles | Yes |
The effect of different Gal4 lines tested for climbing behavior with UAS:kis RNAi constructs. BL, Bloomington Stock number; MB, mushroom body. Climbing is affected when compared with wild-type flies, as well as Gal4 and UAS controls outcrossed to w−.
In addition to identifying a requirement for kis function in muscles, we also observe a strong decrease in early climbing ability by simultaneously reducing kis function in the ventral nerve cord, Kenyon neurons (neurons involved in learning and memory) and pars intercerebralis (the major neuroendocrine gland in flies) (Fig. 2C, Table 1). We also observe a small but significant decrease in early climbing ability when we knock down Kis in the giant fiber circuit with the OK307-Gal4 reagent (Fig. 2D, Table 1). The giant fiber system in Drosophila is a pair of symmetrical inter-neurons that mediate the flight escape response by relaying sensory visual information to the thoracic flight muscles and leg extensor muscles of the thorax (31–33). We also observe that roughly half of these flies exhibit abnormal postural defects where they hold their wings out to the side of their bodies (53%, n = 62). Taken together, these data show that Kis function is also required outside of muscles in these inter-neuronal populations within the brain and ventral nerve cord to control climbing behavior and wing posture. However, because we restricted our analysis of kis function to the first 10 days post-eclosion, we cannot rule out the possibility that kis is required within the tissues we tested at later times during the fly's life cycle.
Kismet protein is broadly expressed in the developing fly brain
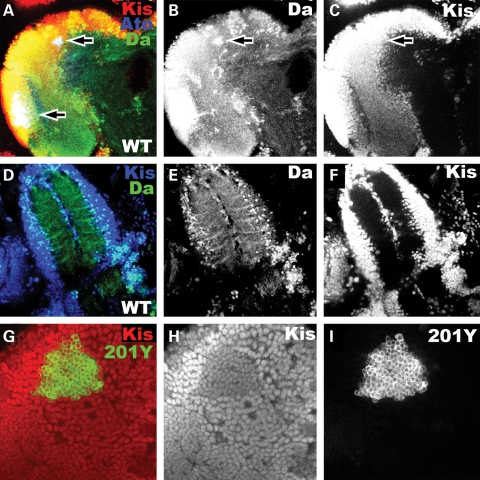
To begin to explore Kis protein function in the fly brain, we first analyzed Kis protein expression in the developing central nervous system. Because we had identified kis as a regulator of atonal and daughterless proneural gene expression in the retina (11), we compared Kis protein expression in the brain with the expression pattern of both these proneural proteins. We find that Kis protein is strongly expressed in many areas of the larval brain, including the cortex, the brain lobes and the ventral nerve cord (Fig. 3), and strongly co-localized with both Atonal and Daughterless protein expression in this tissue. Our analysis of Ato protein expression is consistent with what has been previously reported in this tissue (34). Ato is expressed in the inner proliferation center of the optic lobe (lower arrow in Fig. 3A), as well as in a group of 20–30 cells in the dorso-lateral region of the central brain (upper arrow in Fig. 3A). This group of 20–30 cells is known as the dorsal cluster neurons (DCNs), which send out bundles of axons ventrally and then contralaterally toward the opposite optic lobe (34). We find that the Da protein is strongly co-localized to both the Ato expressing neurons in the optic lobe as well as in the central brain (Fig. 3A–F). Da protein is also significantly expressed in neurons outside of the Ato-specific neurons, within regions of higher expression in both the cortex and central brain (Fig. 3B). Similarly, Kis protein expression is also co-localized to sites of Ato expression (Fig. 3A). Like Da protein expression, Kis is also significantly expressed outside of the Ato-specific expression domains, but unlike Da, Kis expression is more uniform in the brain (Fig. 3C). Kis protein is also expressed within the neurons of the Kenyon cells (Fig. 3G–I), which are required for learning and memory in the fly.
Figure 3.
Kismet is widely expressed in the developing larval central nervous system. (A–C and G–I) Developing brain lobe of wild-type late third instar central nervous system. (D–F) Developing ventral nerve cord of wild-type late third instar larval central nervous system. (A–F) are whole brain reconstructions from individual confocal image slices. (A) Kis expression (red), Atonal expression (blue) and Daughterless expression (green) in wild-type brain lobe. Upper arrow denotes the Atonal-expressing DCNs. Lower arrow denotes the developing optic lobe. (B) Daughterless expression (white) from (A). Arrow denotes Daughterless expression in DCNs. (C) Kis expression (white) from (A). Arrow denotes Kis expression in DCNs. (D) Kis expression (blue) and Daughterless expression (green) in developing ventral nerve cord. (E) Daughterless expression (white) from (D). (F) Kis expression (white) from (D). (G) GFP expression (green) in the membrane as driven from the 201Y-Gal4 driver (201Y-Gal4/UAS:CD8-GFP) to denote the location of the γ-Kenyon neurons. Kis expression (red). Note strong co-localization. (H) Kis expression (white) from (G). (I) GFP expression (white) from (G).
Kismet function is required for immediate recall memory
Based on the expression pattern of Kis in the developing brain, and the fact that patients with CS often suffer from intellectual disability, we tested the learning and memory ability of adult flies with reduced kismet function. To test for deficits in learning and memory in these flies, we performed the conditioned courtship suppression assay (35). This assay is an associative conditioning procedure that is ethologically based and capable of measuring both learning and memory in individual flies (36). The conditioning aspect of this assay is based on the observation that male courtship behavior is modified by exposure to a previously mated female that is unreceptive to courting (35,37). Thus, after 1 h of unsuccessful courting of a mated female, wild-type males suppress their courtship behavior, even toward subsequent receptive virgin females, for 1–3 h (35,38–40).
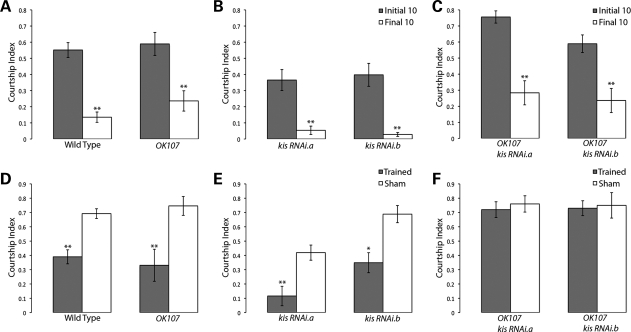
In order to knock down Kis protein in this assay, we expressed each of the UAS:kis RNAi constructs continuously during development with the OK107-Gal4 driver. This driver is expressed in discrete populations of neurons during development in the fly brain, including the Kenyon cells (41), neurons of the Ventral Ganglion (42), and early in the optic lobes, central brain regions and protocerebrum (43). To determine the effects on learning, male flies were placed in a courtship chamber with a previously mated (unreceptive) wild-type female for 60 min. The amount of time the male spent performing courtship behavior was assessed during the first 10 min of this training and compared with the last 10 min of the training period. Wild-type (Canton S) control flies show a significant drop in courtship behavior in the last 10 min of training when compared with the first 10 min (Fig. 4A), indicative of an appropriate learning response. Similarly, outcrossed control flies (Fig. 4A and B) and Kis knockdown flies (Fig. 4C) also show a significant decrease in courtship behavior in the last 10 min of the training period compared with the first 10. Importantly, this indicates that our Kis knockdown flies are capable of successful perception and interpretation of the sensory stimuli required in this assay and that these flies are able to alter their behavior appropriately (learn) in response to this training.
Figure 4.
Kismet knockdown flies can learn, but are deficient in their immediate recall memory. (A–C) denote learning during the first 10 (gray columns) and last 10 minutes (white columns) of the training phase during the courtship suppression assay. Genotypes are indicated. Note normal response of Kis knockdown mutants. (D–F) Panels denote immediate recall memory (0–2 min post-training) of trained flies (gray columns) when compared with sham-trained matching genotypes (white columns). Kis knockdown showed no significant difference between trained and sham-trained flies, indicative of no immediate recall memory of training. Error bars represent ± SEM. In all cases, double asterisk indicates P < 0.001 and asterisk indicates P < 0.05 compared with controls.
There have been five phases of memory defined in Drosophila, immediate recall (0–2 min post-training), short-term memory (out to 1 h post-training), medium-term memory (out to 6h), anesthesia-resistant memory (out to 2 days) and long-term memory (out to 9 days) (44,45). In order to test the earliest phase of memory first, we assayed Kis knockdown flies for their immediate recall memory by transferring trained male flies to clean mating chambers with a receptive virgin female within 2 min of training and assaying their courtship behavior for 10 min. Trained wild-type males show a clear decrease in courtship activity when compared with parallel sham-trained wild-type flies (Fig. 4D), indicating a change in behavior consistent with normal immediate recall memory of training. Similarly, outcrossed Gal4 and UAS control flies also show a significant decrease in their courtship activity compared with genotype equivalent outcrossed sham-trained flies (Fig. 4D and E). However, both Kis knockdown flies show no significant decrease in their courtship activity within 2 min of training (Fig. 4F). Because these flies are capable of successful perception, and experience-dependent alteration of their behavior in the learning component of this assay, their inability to suppress their courtship frequency in the second component of this assay indicates that these flies are defective in their immediate recall memory of this learning.
Kismet mutants display abnormal axonal pruning and migration during development of Kenyon cells
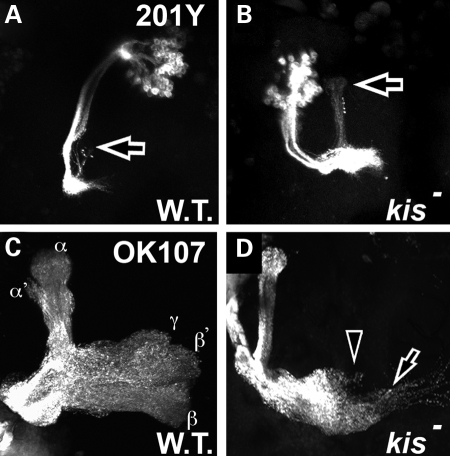
Because the kis gene encodes for a transcription factor, these results suggest that kis may be regulating the gene expression of some critical factor(s) important for immediate recall memory formation during development. This could either be through the control of neurogenesis of appropriate neurons in this memory circuit, control of circuit connectivity once these neurons have already differentiated and/or control of the development and/or morphology of these neuronal target tissues. To begin to address these questions, we first analyzed the effect of loss of function in kis on neuronal development and morphology of the Kenyon neurons, as these neurons are associated with learning and memory in multiple experimental paradigms in Drosophila (46–50). We utilized the MARCM technique (51) to create clones of neurons that are homozygous mutant for kisLM27, a protein null mutant (11). The population of Kenyon cells consists of three distinct groups of neurons, each with its own characteristic developmental morphology (41). During larval development, the γ subset of Kenyon neurons are differentiated first, and these neurons undergo highly stereotyped dendritic and axonal pruning during pupal development (41,52). Pruning of these neurons occurs by localized degeneration (53) and is evident by 18 h after puparium formation in wild-type neurons (Fig. 5A, arrow shows remnants of γ axonal projects into the α lobes). Although γ neurons mutant for kis undergo proper dendritic pruning, they display defects in their axonal pruning (arrow in Fig. 5B) and continue to display unpruned γ axons projecting into α lobes 40% of the time (Table 2).
Figure 5.
Kismet is required for proper axon pruning and axon migration in developing mushroom bodies. All panels show GFP in Kenyon neurons by MARCM analysis. (A and B) Pupal brains 18–20 h after puparium formation (apf). (C and D) Adult brains 48 h after eclosion. (A) Wild-type MARCM clones (FRT 40A) in pupal brains driven by 201Y-Gal4 show remnants of proper axonal pruning of γ neurons that were previously populating the α lobes (arrow). (B) kisLM27 homozygous mutant MARCM clones display unpruned γ axons that continue to populate the α lobes (arrow). (C) Wild-type MARCM clones (FRT 40A) in adult brains driven by OK107-Gal4 show normal pattern of α, α′, β, β′ and γ axons innervating the mushroom body lobes, as labeled. (D) kisLM27 homozygous mutant MARCM clones in these adult brains often display abnormal axon migration beyond the midline (demarcated by the arrowhead).
Table 2.
Neuronal defects observed in kismet MARCM mutants
| Neuron | Gal4-line | n | Defects observed | % | P-value |
|---|---|---|---|---|---|
| γ neurons, MB | 201Y | ||||
| control (pupal) | 8 | Unpruned axons | 0% | — | |
| kis mutant (pupal) | 10 | Unpruned axons | 40% | <0.05 | |
| α, β, γ neurons, MB | OK107 | ||||
| Control (adult) | 9 | Lobe structure | 0% | — | |
| Cell migration | 0% | — | |||
| Axon migration | 11% | — | |||
| kis mutant (adult) | 11 | Lobe structure | 55% | <0.05 | |
| Cell migration | 36% | <0.05 | |||
| Axon migration | 55% | <0.05 | |||
| DCNs | Atonal | ||||
| Control (larval) | 9 | Cell position | 0% | — | |
| Axon migration | 0% | — | |||
| Cells out of cluster | 0% | — | |||
| Control (adult) | 6 | Axon extension (13.5 axons in medulla) | 0% | — | |
| kis mutant (larval) | 19 | Cell position | 53% | <0.001 | |
| Axon migration | 32% | <0.05 | |||
| Cells out of cluster | 32% | <0.05 | |||
| kis mutant (adult) | 6 | Axon extension (5.5 axons in medulla) | 100% | <0.001 |
Quantification of defects observed in different kismet mutant neurons. MB, mushroom body. Unpruned axons refers to the number of clones with unpruned γ axons extending into the α lobe at 18–20 h APF. Lobe structure indicates missing or altered lobe morphology in α, β or γ lobes. Cell position indicates the number of soma that appear lateral to their appropriate locations by the developmental time indicated. Axon migration indicates either axons crossing the midline for adult Kenyon cells or commissural axons failing to cross the midline or failing to migrate to their proper contralateral targets in larval DCNs. Axon extension indicates the presence of axons failing to extend from lobulla into the medulla in adult brains. P-values are indicated from a two-tailed Student's t-test comparing kis mutant with control.
The final adult structure of the mushroom body axonal projections consist of five distinct projections to the α, α′, β, β′ and γ lobes (Fig. 5C). To determine whether structures other than γ lobes are affected by mutations in kis, we created MARCM clones mutant for kisLM27 using the OK107-Gal4 driver, which is expressed in the α, β and γ neurons. While Kenyon cell dendrite morphology was normal in these kis mutant neurons, we did observe axonal migration defects in these cells where axons cross the midline of the brain to migrate into the opposite brain lobe (arrow in Fig. 5D, 55%, Table 2).
Kismet mutants display abnormal axon extension and migration in DCNs and photoreceptor axons
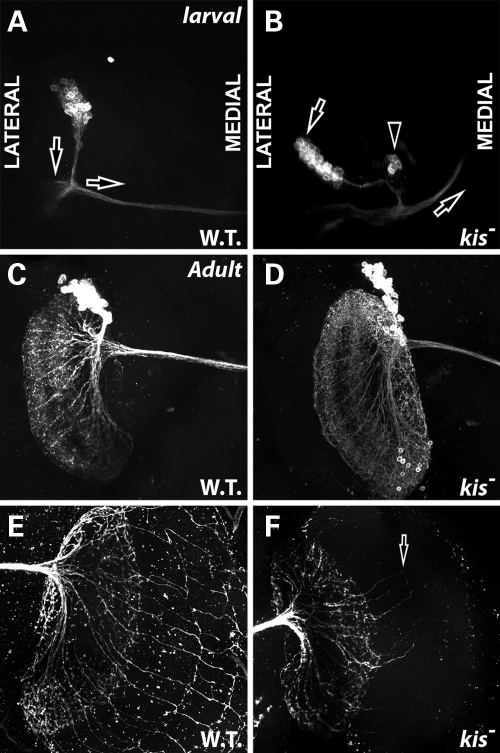
The Kis protein is widely expressed in the fly brain (Fig. 3). Given that mutations in kis affect axonal pruning and axonal migration in developing Kenyon neurons, we analyzed the effect of kis loss of function in other neuron populations in the developing and adult brain to determine whether the Kis protein may play a broader role in regulating neuronal morphology. Based on the co-localization of Kis and Atonal proteins in the developing larval brain and that kis function is known to regulate atonal transcription in the developing retina (11), we created MARCM clones mutant for kis function in the DCNs, a subset of Atonal-expressing neurons in the larval and adult brain (34). These neurons form a highly stereotypical pattern of connections in the fly brain that innervate the optic lobes (34,54). The regular array of these neuronal connections has been extremely useful to determine how genes function to regulate neurite extension, outgrowth, neuronal morphology and arborization patterns (54,55). During larval development, DCN soma forms a distinct cluster of roughly 20–30 cells in the dorso-lateral region of the central brain (34). These neurons then extend a bundle of neurites posteriorly, and then form a bundle of commissural axons that migrate contralaterally to innervate targets of the opposite brain lobe (Fig. 6A). Developing larval DCNs mutant for kisLM27 exhibit several morphological abnormalities indicative of abnormal axon development (Table 2, Supplementary Material, Table S1). These neurons display abnormal commissural axon projections (arrow in Fig. 6B, Table 2), consistent with the defects in axonal migration observed in kis mutant Kenyon neurons. kis mutant cells also display abnormal positioning within the brain, appearing laterally in more than half of the clones observed (left arrow in Fig. 6B, Table 2). Interestingly, these neurons appear to send their initial neurite bundles to the proper location for initial branching however (Fig. 6B). Additionally, we occasionally observe groups of DCN soma that are not part of the larger soma cluster (arrowhead in Fig. 6B, Table 2). We observe similar morphological defects in larval DCNs where kis is knocked down with both UAS:kis RNAi constructs (Supplementary Material, Fig. S1), providing further validation of the specificity of these defects for kis function in these cells.
Figure 6.
Kismet is required for proper DC position and axon migration in developing DCNs. All panels show GFP in DCNs by MARCM analysis. (A and B) Late third instar larval brains. Both images show the left hemisphere. Anterior is up. Medial is right. Lateral is left. (C–F) Adult brains 48 h after eclosion. (A) Wild-type MARCM clones (FRT 40A) in larval DCNs. Note position of the soma cluster axon bundles (parallel to vertical arrow) compared with commissural axon bundles (below horizontal arrow). (B) kisLM27 homozygous mutant MARCM clones display abnormal positioning of soma cluster compared with commissural axon bundles (left arrow), soma that have developed outside of the normal cluster (arrowhead) and disrupted commissural axon migration (right arrow). (C) Wild-type MARCM clones (FRT 40A) in adult DCNs displaying normal dendritic morphology on ipsilateral brain hemisphere. (D) kisLM27 homozygous mutant MARCM clones in adult brains. Overall morphology of dendritic field appears normal. (E) Wild-type MARCM clones (FRT 40A) in adult DCNs displaying normal axonal morphology on contralateral brain hemisphere. (F) kisLM27 homozygous mutant MARCM clones in adult brains. Arrow displays abnormal and reduced number of axonal extensions from lobulla into the medulla.
In adults, the atonal expressing neurons form highly complex dendritic and axonal fields (Fig. 6C and E) (34,55). Adult DCNs project neurite bundles that branch ipsilaterally to form the dendritic field (Fig. 6C), and also project a large bundle of commissural axons contralaterally to innervate the lobula and medulla in a stereotypical fan-like pattern (Fig. 6E) (34,54,55). Adult DCNs homozygous mutant for kis do not display abnormal dendrite formation (Fig. 6D). However, kis mutant DCNs show a severe reduction in the number of axons extending into the lobulla, and subsequently extending from the lobulla into the medulla (arrow Fig. 6F, Table 2). Wild-type MARCM clones exhibit 13.5 ± 1.51 neurites extending from the lobulla into the medulla, whereas kis mutant MARCM clones exhibit 5.5 ± 2.59 neurites extending from the lobulla to the medulla, a significantly lower number (P = 0.00018). These results are similar to those observed in Kenyon cells, in that the dendrites develop normally, whereas the axons do not.
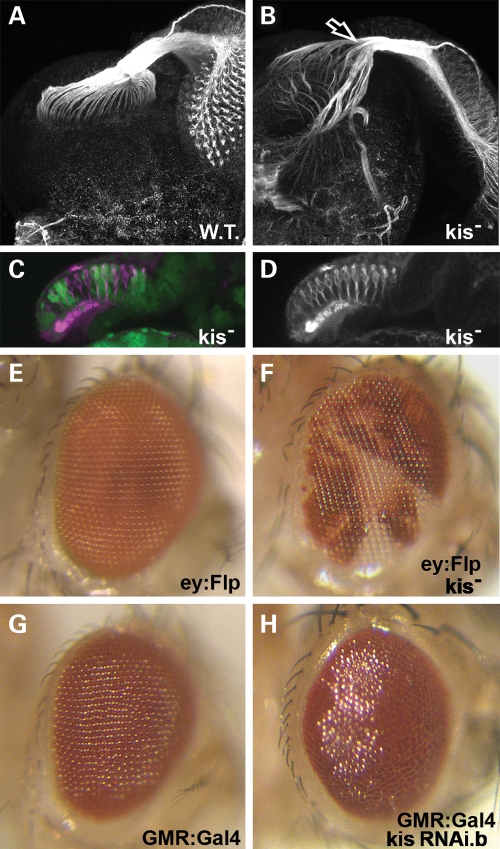
To further validate the axonal defects we observe in kis mutant neurons, we analyzed the axonal projections of photoreceptor cells into the optic lobe of developing larval brains, a standard technique for analyzing photoreceptor axonal guidance (56,57). We dissected third instar retina–brain complexes from developing larvae and stained these tissues for the MAP1B-like protein Futsch (58). Wild-type photoreceptor cells extend their axons from retinal cells away from retinal tissue toward the optic stalk where the axons then fasciculate into a bundle. These axons then migrate to innervate the lamina and medulla of the larval brain in a highly stereotypical fashion (Fig. 7A). We then created clones of kisLM27 homozygous mutant cells in the developing retina and analyzed migration of photoreceptor axons into the brain. In these retinas, photoreceptor axons exhibit a severe defect in axonal migration into the optic lobe (Fig. 7B), with photoreceptor axons extending beyond their normal targets and radiating out into the surrounding brain areas. These axons also show abnormal defasciculation of axonal bundles in the optic stalk (arrow in Fig. 7B). Interestingly, the photoreceptor cells migrate normally into their proper positions in the ommatidial clusters in kis mutant retinal tissue (Fig. 7C, non-green tissue) and initially send their axons to properly bundle in the optic stalk. Further, these clones in the adult eye do not show any abnormal external development of mutant ommatidia (Fig. 7E and F).
Figure 7.
Kismet is required for proper photoreceptor axon migration and eye development. (A–D) Late third instar larval retinas with attached brains. (E–H) Adult eyes, anterior right. (A) Wild-type retina with attached larval brain stained for the Futsch protein (white) display normal axonal bundles in the optic stalk that innervate the developing optic lobes. (B) kisLM27 homozygous mutant clones in the developing retina stained for Futsch (white) display abnormal axonal migration into the developing brain. Arrow indicates defasciculation of axon bundles in the optic stalk. (C) Cross-section of kisLM27 homozygous mutant clones in the developing retina posterior to the morphogenetic furrow. Heterozygous (control) tissue is denoted by the presence of GFP (green). Homozygous clones are denoted by lack of GFP. Retinas are stained with Futsch (magenta). Note normal positioning of ommatidia clusters near the apical (top) portion of the retina. (D) Futsch stain (white) from (C). (E) Normal eye expressing only eyeless:Flip. (F) Adult eye showing kisLM27 homozygous mutant clones (in white). (G) GMR-Gal4 adult eye raised at 25°C displays normal morphology. (H) GMR-Gal4 / UAS:kis RNAi.b adult eye raised at 25°C displays abnormal morphology and glassy appearance.
Greater than 80% of CHD7-positive CHARGE patients display coloboma of the eye, retina or both tissues (3,59). Though external adult eye morphology appears normal when discrete patches of eye tissue are mutant for kis, we tested whether loss of function in kis throughout the whole eye would show abnormal external morphology. We expressed UAS:kis RNAi.b in all cells posterior to the morphogenetic furrow using the GMR:Gal4 driver at 25°C. At this temperature, GMR:Gal4 eyes alone display normal external eye morphology (Fig. 7G), as do UAS:kis RNAi.b flies (data not shown). However, when the Kis protein is significantly reduced throughout development of the eye, adult morphology is disrupted (Fig. 7H). These eyes have a slightly rough and significantly glassy appearance (Fig. 7H), although eye size is not affected. Taken together, our results suggest that proper kis function is required both for normal eye development, as well as normal photoreceptor axon guidance in the developing fly eye.
DISCUSSION
In summary, the data presented here describe a requirement for kis gene function in adult Drosophila early climbing behavior, memory, eye development and neural development. Our results suggest that the reduced early climbing ability observed in kis mutants can at least partially be attributed to a requirement for kis gene function in muscle cells. This is particularly of interest when we consider that knockdown of kis function in motor neurons, as well as pan-neural knockdown of kis function has no effect on the early climbing behavior we analyzed. Thus, kis may either be regulating the expression of critical post-synaptic target gene(s) in muscle cells required to facilitate synaptic transmission and muscle coordination and/or may be required for the morphology and/or development of the muscle cells themselves. Further, the postural defect we observe in adult flies with reduced Kismet protein is also reminiscent of defects associated with muscle cells, and both phenotypes are consistent with hypotonia, impaired motor coordination and muscle-related posture problems observed in CHARGE patients (27). Based on these similarities, our results may suggest that there is a similar requirement for Chd7 function in muscles of vertebrates and may help to elucidate a possible mechanism for these symptoms often observed in CHARGE patients.
Our data suggest that decreased kis function does not alter the fly's ability to learn, although it does have an effect on immediate recall memory. Our data have also shown that kis function is required for the proper development of the Kenyon cells, as kis mutants show defective axonal pruning and axonal migration in these neuronal populations. The Kenyon neurons are associated with learning and memory in multiple experimental paradigms in Drosophila, including the conditioned courtship suppression behavior (46–50,60). Thus, it may be that the defects we observe in Kenyon cell development are responsible for the impaired learning behavior we observe in adult flies with decreased kis function. kis-mediated transcriptional regulation of genes involved in the pruning and/or migration of axons in this cell population would therefore make attractive targets for further investigation into this memory defect.
In analyzing the morphology of neuronal populations mutant for kis, we consistently observe defects in axon morphology and positioning. These defects may be due to abnormal axonal pruning observed in some kis mutant neurons or may be due to defective axonal migration or defects in axonal extension and/or retraction. Interestingly, in each of the neuronal populations studied, dendritic development was normal. When taken together, these data suggest that kis may function to regulate the expression of target gene(s) normally required for axon morphology and connectivity, as opposed to dendritic morphology, at least in the neuronal populations and developmental times studied here. The specific target genes that Kismet regulates most likely differ between different neuronal populations. However, the defects observed in kis mutant DCNs suggest that kis functions in DCN positioning as well as axonal morphology. Abnormal positioning of larval DC soma is also observed in larval brains mutant for ato gene function (34). As kis regulates ato transcription in the larval retina (11), this neuron migration defect may be due to kis mediated regulation of ato transcription in these cells as well.
Taken together, we suggest that the analyses presented here can complement and expand upon the studies done in cell culture and vertebrate model organisms toward a better understanding of the role of kis in neural developmental and Chd7 in CS pathogenesis.
MATERIALS AND METHODS
Drosophila stocks and genetics
All flies were maintained at 25°C in a 12:12 light:dark cycle at 60% humidity. All crosses were carried out at 25°C. Normal food consisted of a standard cornmeal, yeast, molasses recipe. BL# refers to Bloomington Stock Center stock number (http://flystocks.bio.indiana.edu/bloomhome.htm). VDRC# refers to the Vienna Drosophila Resource Center stock number (http://stockcenter.vdrc.at/control/main). Wild-type flies used were Canton S. Stocks used are described: kisLM27 (11), UAS:kis RNAi.a (VDRC #10762) and UAS:kis RNAi.b (VDRC #46685).
All Gal4 stocks were obtained from the Drosophila Bloomington Stock Center. The tissues affected and Bloomington Stock number are listed in Table 1.
Clones in developing retinas to analyze axonal guidance were generated using ey:FLP as previously described (56). MARCM mosaic analysis was performed as previously described (51,61). Genotypes for MARCM analysis were:
y−,w−,hs:Flp, UAS:CD8-GFP; Gal80,Frt40A/Cyo; +/+; OK107-Gal4
y−,w−,hs:Flp, UAS:CD8-GFP; Gal80,Frt40A, 201Y-Gal4
y−,w−,hs:Flp, UAS:CD8-GFP; Gal80,Frt40A/Cyo; Ato-Gal4(14a)/TM6B
w−; kisLM27,Frt40A/Cyo
w−; Frt40A
Immunohistochemistry and antibodies
Kis antibody was a kind gift from J. Tamkun and is described in Srinivasan et al. (16). Daughterless antibody was a kind gift from C. Cronmiller and is described in Cronmiller et al. (62). Futsch antibody was obtained from the Iowa Developmental Hybridoma Bank (22C10). Secondary antibodies for immunohistochemistry used were goat anti-mouse TRITC (# 115-116-072, 1:150), goat anti-rabbit TRITC (# 111-116-144, 1:250), goat anti-rabbit Cy5 (#111-176-144, 1:1000) and goat anti-mouse Cy5 (# 115-176-072, 1:500). All secondary antibodies were from Jackson ImmunoResearch.
Adult and larval brains were dissected, fixed and prepared essentially as described (63). Adult and larval brains were dissected directly in fix. Brains were mounted in vectashield (Vector Laboratories, H-1000). All fluorescent imaging was done using an Olympus FluoView FV1000 laser scanning confocal microscope.
Behavioral testing and training
For climbing assays, a modified version of Le Bourg and Lints (30) was used. Flies were collected between 0 and 8 h after eclosion and assayed every 2 days. Groups of 10 or fewer flies were transferred to a clean, empty vial and given 18 s to climb 5 cm. The number of flies that successfully reach the 5 cm line were recorded.
For courtship behavioral training, virgin male flies of the appropriate genotype were collected between 0 and 6 h after eclosion and transferred to individual food vials. All flies were maintained at 25°C in a 12:12 light:dark cycle at 60% humidity. All behavioral tests were performed in a separate room maintained at 25°C and 60% humidity and illuminated under a constant 130 V white light Kodak Adjustable Safelight Lamp mounted above the courtship chambers. All behavior was digitally recorded using a Sony DCR-SR47 Handycam with Carl Zeiss optics. Subsequent digital video analysis of time spent performing courtship behavior was quantified using iMovie software (Apple). The total time that a male performed courtship activity was measured and scored. The courtship index was calculated as the total time observed performing courting behavior divided by the total time assayed, as described (35).
Virgin female wild-type (Canton S) flies were collected and kept in normal food vials in groups of 10. Male flies were aged for 5 days before behavioral training and testing. All tests were performed during the relative light phase. Mated Canton S females used for training were 5 days old and observed to have mated with a Canton S male the evening prior to training. Virgin female Canton S targets used were 4 days old. Male flies were assigned to random groups the day of training, and assays were set up and scored blind. Male flies were transferred without anesthesia to one half of a partitioned mating chambers from Aktogen (http://www.aktogen.com) that contained a previously mated Canton S female in the other partitioned half. Males were allowed to acclimate for 1 min, then the partition between the male and female was removed. Male flies were then trained for 60 min. After 60 min, male flies were transferred within 2 min without anesthesia to one half of a clean partitioned mating chamber that contained a virgin Canton S female in the other partitioned half. The partition was removed and the flies were recorded for 10 min.
Statistical analysis
All statistical analyses were performed on SPSS version 17. To determine the significance between multiple different genotypes, a one-way ANOVA analysis was performed with Tukey post hoc analysis. Genotype is the independent variable. To determine the significance between different measures of the same genotype, a two-tailed paired Student's t-test was performed. An unpaired Student's t-test was performed between two groups of different genotypes. Significance was determined at the 95% confidence interval.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Pennsylvania Department of Health (CURE Award to D.R.M.), Drexel University and the National Institutes of Health, (NCCR) (grant no. R21RR026074 to D.R.M.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank L. Luo for the MARCM stocks, J. Tamkun for Kis antibodies, C. Cronmiller for Da antibodies, the Bloomington Stock center for stocks and the Iowa Developmental Hybridoma Bank for antibodies. We would like to thank K. Siwicki for assistance and advice on the courtship conditioning assays. We would like to thank the Marenda laboratory members and J. Twiss for helpful comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C., et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. doi:10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 2.Sanlaville D., Verloes A. CHARGE syndrome: an update. Eur. J. Hum. Genet. 2007;15:389–399. doi: 10.1038/sj.ejhg.5201778. doi:10.1038/sj.ejhg.5201778. [DOI] [PubMed] [Google Scholar]
- 3.Blake K.D., Prasad C. CHARGE syndrome. Orphanet. J. Rare Dis. 2006;1:34. doi: 10.1186/1750-1172-1-34. doi:10.1186/1750-1172-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jongmans M.C., Hoefsloot L.H., van der Donk K.P., Admiraal R.J., Magee A., van de Laar I., Hendriks Y., Verheij J.B., Walpole I., Brunner H.G., et al. Familial CHARGE syndrome and the CHD7 gene: A recurrent missense mutation, intrafamilial recurrence and variability. Am. J. Med. Genet. A. 2007;146A:43–50. doi: 10.1002/ajmg.a.31921. [DOI] [PubMed] [Google Scholar]
- 5.Van de Laar I., Dooijes D., Hoefsloot L., Simon M., Hoogeboom J., Devriendt K. Limb anomalies in patients with CHARGE syndrome: an expansion of the phenotype. Am. J. Med. Genet. A. 2007;143:2712–2715. doi: 10.1002/ajmg.a.32008. [DOI] [PubMed] [Google Scholar]
- 6.Schnetz M.P., Bartels C.F., Shastri K., Balasubramanian D., Zentner G.E., Balaji R., Zhang X., Song L., Wang Z., Laframboise T., et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. doi:10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasan S., Dorighi K.M., Tamkun J.W. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. doi:10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen M.D., Religa T.L., Freund S.M., Bycroft M. Solution structure of the BRK domains from CHD7. J. Mol. Biol. 2007;371:1135–1140. doi: 10.1016/j.jmb.2007.06.007. doi:10.1016/j.jmb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Bajpai R., Chen D.A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C.P., Zhao Y., Swigut T., Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. doi:10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubruille R., Murad A., Rosbash M., Emery P. A constant light-genetic screen identifies KISMET as a regulator of circadian photoresponses. PLoS Genet. 2009;5:e1000787. doi: 10.1371/journal.pgen.1000787. doi:10.1371/journal.pgen.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melicharek D., Shah A., DiStefano G., Gangemi A.J., Orapallo A., Vrailas-Mortimer A.D., Marenda D.R. Identification of novel regulators of atonal expression in the developing Drosophila retina. Genetics. 2008;180:2095–2110. doi: 10.1534/genetics.108.093302. doi:10.1534/genetics.108.093302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batsukh T., Pieper L., Koszucka A.M., von Velsen N., Hoyer-Fender S., Elbracht M., Bergman J.E., Hoefsloot L.H., Pauli S. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum. Mol. Genet. 2010;19:2858–2866. doi: 10.1093/hmg/ddq189. doi:10.1093/hmg/ddq189. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Paredes M., Ceballos-Chavez M., Esteller M., Garcia-Dominguez M., Reyes J.C. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009;37:2449–2460. doi: 10.1093/nar/gkp101. doi:10.1093/nar/gkp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Layman W.S., Hurd E.A., Martin D.M. Chromodomain proteins in development: lessons from CHARGE syndrome. Clin. Genet. 2010;78:11–20. doi: 10.1111/j.1399-0004.2010.01446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn C.W., Hejnol A., Matus D.Q., Pang K., Browne W.E., Smith S.A., Seaver E., Rouse G.W., Obst M., Edgecombe G.D., et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. doi:10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan S., Armstrong J.A., Deuring R., Dahlsveen I.K., McNeill H., Tamkun J.W. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–1635. doi: 10.1242/dev.01713. doi:10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- 17.Kennison J.A., Tamkun J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl Acad. Sci. USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. doi:10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daubresse G., Deuring R., Moore L., Papoulas O., Zakrajsek I., Waldrip W.R., Scott M.P., Kennison J.A., Tamkun J.W. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- 19.Go M.J., Artavanis-Tsakonas S. A genetic screen for novel components of the Notch signaling pathway during Drosophila bristle development. Genetics. 1998;150:211–220. doi: 10.1093/genetics/150.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Therrien M., Morrison D.K., Wong A.M., Rubin G.M. A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila. Genetics. 2000;156:1231–1242. doi: 10.1093/genetics/156.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marenda D.R., Zraly C.B., Dingwall A.K. The Drosophila Brahma (SWI/SNF) chromatin remodeling complex exhibits cell-type specific activation and repression functions. Dev. Biol. 2004;267:279–293. doi: 10.1016/j.ydbio.2003.10.040. doi:10.1016/j.ydbio.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg T. engrailed: a gene controlling compartment and segment formation in Drosophila. Proc. Natl Acad. Sci. USA. 1981;78:1095–1099. doi: 10.1073/pnas.78.2.1095. doi:10.1073/pnas.78.2.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg T., Siden I., O'Farrell P., Simon M. The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment specific expression. Cell. 1985;40:45–53. doi: 10.1016/0092-8674(85)90307-1. doi:10.1016/0092-8674(85)90307-1. [DOI] [PubMed] [Google Scholar]
- 25.Vazquez M., Moore L., Kennison J.A. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development. 1999;126:733–742. doi: 10.1242/dev.126.4.733. [DOI] [PubMed] [Google Scholar]
- 26.Marenda D.R., Zraly C.B., Feng Y., Egan S., Dingwall A.K. The Drosophila SNR1 (SNF5/INI1) subunit directs essential developmental functions of the Brahma chromatin remodeling complex. Mol. Cell. Biol. 2003;23:289–305. doi: 10.1128/MCB.23.1.289-305.2003. doi:10.1128/MCB.23.1.289-305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartshorne T.S., Hefner M.A., Davenport S.L. Behavior in CHARGE syndrome: introduction to the special topic. Am. J. Med. Genet. A. 2005;133A:228–231. doi: 10.1002/ajmg.a.30541. [DOI] [PubMed] [Google Scholar]
- 28.Elfring L.K., Daniel C., Papoulas O., Deuring R., Sarte M., Moseley S., Beek S.J., Waldrip W.R., Daubresse G., DePace A., et al. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zraly C.B., Marenda D.R., Nanchal R., Cavalli G., Muchardt C., Dingwall A.K. SNR1 is an essential subunit in a subset of Drosophila brm complexes, targeting specific functions during development. Dev. Biol. 2003;253:291–308. doi: 10.1016/s0012-1606(02)00011-8. doi:10.1016/S0012-1606(02)00011-8. [DOI] [PubMed] [Google Scholar]
- 30.Le Bourg E., Lints F.A. Hypergravity and aging in Drosophila melanogaster. 6. Spontaneous locomotor activity. Gerontology. 1992;38:71–79. doi: 10.1159/000213309. [DOI] [PubMed] [Google Scholar]
- 31.Thomas J.B., Wyman R.J. A mutation in Drosophila alters normal connectivity between two identified neurones. Nature. 1982;298:650–651. doi: 10.1038/298650a0. doi:10.1038/298650a0. [DOI] [PubMed] [Google Scholar]
- 32.Tanouye M.A., Wyman R.J. Motor outputs of giant nerve fiber in Drosophila. J. Neurophysiol. 1980;44:405–421. doi: 10.1152/jn.1980.44.2.405. [DOI] [PubMed] [Google Scholar]
- 33.Koto M., Tanouye M.A., Ferrus A., Thomas J.B., Wyman R.J. The morphology of the cervical giant fiber neuron of Drosophila. Brain Res. 1981;221:213–217. doi: 10.1016/0006-8993(81)90772-1. doi:10.1016/0006-8993(81)90772-1. [DOI] [PubMed] [Google Scholar]
- 34.Hassan B.A., Bermingham N.A., He Y., Sun Y., Jan Y.N., Zoghbi H.Y., Bellen H.J. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25:549–561. doi: 10.1016/s0896-6273(00)81059-4. doi:10.1016/S0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 35.Siegel R.W., Hall J.C. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl Acad. Sci. USA. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. doi:10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broughton S.J., Tully T., Greenspan R.J. Conditioning deficits of CaM-kinase transgenic Drosophila melanogaster in a new excitatory courtship assay. J. Neurogenet. 2003;17:91–102. [PubMed] [Google Scholar]
- 37.Siwicki K.K., Riccio P., Ladewski L., Marcillac F., Dartevelle L., Cross S.A., Ferveur J.F. The role of cuticular pheromones in courtship conditioning of Drosophila males. Learn. Mem. 2005;12:636–645. doi: 10.1101/lm.85605. doi:10.1101/lm.85605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kane N.S., Robichon A., Dickinson J.A., Greenspan R.J. Learning without performance in PKC-deficient Drosophila. Neuron. 1997;18:307–314. doi: 10.1016/s0896-6273(00)80270-6. doi:10.1016/S0896-6273(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 39.Joiner Ml.A., Griffith L.C. CaM kinase II and visual input modulate memory formation in the neuronal circuit controlling courtship conditioning. J. Neurosci. 1997;17:9384–9391. doi: 10.1523/JNEUROSCI.17-23-09384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamyshev N.G., Iliadi K.G., Bragina J.V. Drosophila conditioned courtship: two ways of testing memory. Learn. Mem. 1999;6:1–20. [PMC free article] [PubMed] [Google Scholar]
- 41.Lee T., Lee A., Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 42.Branco J., Al-Ramahi I., Ukani L., Perez A.M., Fernandez-Funez P., Rincon-Limas D., Botas J. Comparative analysis of genetic modifiers in Drosophila points to common and distinct mechanisms of pathogenesis among polyglutamine diseases. Hum. Mol. Genet. 2008;17:376–390. doi: 10.1093/hmg/ddm315. doi:10.1093/hmg/ddm315. [DOI] [PubMed] [Google Scholar]
- 43.Evans C.J., Olson J.M., Ngo K.T., Kim E., Lee N.E., Kuoy E., Patananan A.N., Sitz D., Tran P., Do M.T., et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. doi:10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McBride S.M., Choi C.H., Wang Y., Liebelt D., Braunstein E., Ferreiro D., Sehgal A., Siwicki K.K., Dockendorff T.C., Nguyen H.T., et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. doi:10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 45.Greenspan R.J. Flies, genes, learning, and memory. Neuron. 1995;15:747–750. doi: 10.1016/0896-6273(95)90165-5. doi:10.1016/0896-6273(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 46.Heisenberg M., Borst A., Wagner S., Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. doi:10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 47.Connolly J.B., Roberts I.J., Armstrong J.D., Kaiser K., Forte M., Tully T., O'Kane C.J. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. doi:10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 48.McBride S.M., Giuliani G., Choi C., Krause P., Correale D., Watson K., Baker G., Siwicki K.K. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron. 1999;24:967–977. doi: 10.1016/s0896-6273(00)81043-0. doi:10.1016/S0896-6273(00)81043-0. [DOI] [PubMed] [Google Scholar]
- 49.Dubnau J., Tully T. Functional anatomy: from molecule to memory. Curr. Biol. 2001;11:R240–R243. doi: 10.1016/s0960-9822(01)00115-4. doi:10.1016/S0960-9822(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 50.Zars T., Fischer M., Schulz R., Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. doi:10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 51.Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. doi:10.1016/S0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 52.Schuldiner O., Berdnik D., Levy J.M., Wu J.S., Luginbuhl D., Gontang A.C., Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. doi:10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watts R.J., Hoopfer E.D., Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin–proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. doi:10.1016/S0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 54.Srahna M., Leyssen M., Choi C.M., Fradkin L.G., Noordermeer J.N., Hassan B.A. A signaling network for patterning of neuronal connectivity in the Drosophila brain. PLoS Biol. 2006;4:e348. doi: 10.1371/journal.pbio.0040348. doi:10.1371/journal.pbio.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng X., Zugates C.T., Lu Z., Shi L., Bai J.M., Lee T. Baboon/dSmad2 TGF-beta signaling is required during late larval stage for development of adult-specific neurons. EMBO J. 2006;25:615–627. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newsome T.P., Asling B., Dickson B.J. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 57.Fan Y., Soller M., Flister S., Hollmann M., Muller M., Bello B., Egger B., White K., Schafer M.A., Reichert H. The egghead gene is required for compartmentalization in Drosophila optic lobe development. Dev. Biol. 2005;287:61–73. doi: 10.1016/j.ydbio.2005.08.031. doi:10.1016/j.ydbio.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 58.Hummel T., Krukkert K., Roos J., Davis G., Klambt C. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 2000;26:357–370. doi: 10.1016/s0896-6273(00)81169-1. doi:10.1016/S0896-6273(00)81169-1. [DOI] [PubMed] [Google Scholar]
- 59.Lalani S.R., Safiullah A.M., Fernbach S.D., Harutyunyan K.G., Thaller C., Peterson L.E., McPherson J.D., Gibbs R.A., White L.D., Hefner M., et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype–phenotype correlation. Am. J. Hum. Genet. 2006;78:303–314. doi: 10.1086/500273. doi:10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joiner M.A., Griffith L.C. Mapping of the anatomical circuit of CaM kinase-dependent courtship conditioning in Drosophila. Learn. Mem. 1999;6:177–192. [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J.S., Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat. Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. doi:10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- 62.Cronmiller C., Cummings C.A. The daughterless gene product in Drosophila is a nuclear protein that is broadly expressed throughout the organism during development. Mech. Dev. 1993;42:159–169. doi: 10.1016/0925-4773(93)90005-i. doi:10.1016/0925-4773(93)90005-I. [DOI] [PubMed] [Google Scholar]
- 63.Tio M., Moses K. The Drosophila TGF alpha homolog Spitz acts in photoreceptor recruitment in the developing retina. Development. 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.