Abstract
miR-103 and miR-107, microRNAs hosted by pantothenate kinase genes, are proposed to regulate cellular lipid metabolism. microRNA-mediated regulation is complex, potentially affecting expression of the host gene, related enzymes within the same pathway, or apparently distinct targets. Using qRT-PCR, we demonstrate that miR-103 and miR-107 expression does not correlate with expression of host pantothenate kinase genes in mouse tissues. The miR-103/7 family thus provides an intriguing model for dissecting microRNA transcription, processing and coordinated function within host genes.
Keywords: intronic microRNA, pantothenate kinase
1. Introduction
Mammalian microRNAs (miRNAs) are post-transcriptional regulators that bind to complementary regions generally within 3′ untranslated regions (UTRs) of messenger RNA transcripts (mRNAs), forming double-stranded RNA complexes that ultimately result in gene silencing. Approximately 37% of mammalian miRNAs are located within the introns of protein-coding genes [1]. Initially, intronic miRNAs were predicted to be co-transcribed with their host transcript because many miRNAs and their host genes have been shown to have similar expression patterns [2]. However, not all miRNA expression correlates with host gene expression [3, 4]. miRNAs thus likely represent an integral part of a complex regulatory network controlling many key cellular pathways, including metabolism. Differential miRNA expression may be governed by post-transcriptional regulation [5], independent promoters [6] or disparate transcript stabilities [7].
miR-103 and miR-107 are highly conserved miRNAs that map to intron 5 of three distinct pantothenate kinase (Pank) genes. Pank2 and Pank3 host the precursor miRNA (pre-miRNA) sequences miR-103-2 and miR-103-1, respectively, which are processed into miR-103. Pank1 encodes miR-107, which differs from miR-103 by a single nucleotide. Pantothenate kinase is the rate limiting enzyme in the biosynthesis of coenzyme A, a cofactor that is involved in over 100 metabolic reactions [8]. In addition, PANK2 is the causative gene in pantothenate kinase-associated neurodegeneration (PKAN), which is characterized by dystonia, brain iron accumulation and neuroaxonal spheroid deposition [9].
The conservation of miR-103/7 within pantothenate kinase genes suggests a role for these miRNAs in metabolism and neurodegeneration. miR-103 and miR-107 are both ubiquitously expressed, with relative abundance in the brain [3, 10]. However, differential expression of miR-103 and miR-107 has been reported in adipogenesis [11], models of diabetes [12] and following glucose treatment in a pancreatic β cell line [13], as well as in neurological disease [14, 15] and cancer [16-20]. Functionally, miR-103/7 are predicted to target many enzymes that are important in regulating metabolism [21]. Moreover, experimentally validated miR-103/7 targets include β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) [15], granulin (GRN) [22] and hypoxia inducible factor-1β (HIF-1β) [16], which have all been implicated in neurodegeneration.
Microarray analyses of miRNA and mRNA profiles in human tissues previously revealed a low correlation between the ubiquitously expressed miR-103/7 family and their host genes [3, 4]. However, in cell culture, correlated expression has been observed. For example, there is similar induction of miR-103/7, Pank1, Pank2 and Pank3 expression during 3T3-L1 cell adipogenesis [11]. Also, p53 can induce expression of PANK1 and miR-107; presumably, through a p53 element located ~1kb upstream of the PANK1 transcriptional start site [16]. Finally, studies in a pancreatic cancer cell line suggest that miR-107 expression can be regulated through epigenetic silencing of the PANK1 promoter [20].
However, evidence is also surfacing for independent transcription of miR-107 and Pank1. In particular, Monteys et al. recently described an independent miR-107 promoter that is sufficient to drive miR-107 expression in a promoterless plasmid [6]. In another study, Corcoran et al. also predicted miR-107 to have an independent promoter, based on RNA polymerase II chromatin immunoprecipitation experiments [23]. Interestingly, however, miR-103-1 and miR-103-2 are not predicted to have promoters independent of Pank3 and Pank2, respectively [23].
In this short report, we present the relative expression of the pantothenate kinase genes and their intronic miRNAs in various mouse tissues and discuss the relationship of these expression levels to the regulation and function of these miRNAs.
2. Material and Methods
qRT-PCR
Total RNA was harvested from three 100 day old C.D2 Es-Hba wildtype mice using RNAqueous-Micro kit (Ambion, Inc.) for retinal tissue and RNA-STAT60 (Tel-Test, Inc.) for all other tissue.
To quantify Pank1, Pank2 and Pank3 mRNA levels, cDNA was synthesized from 50ng total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Inc.) according to the manufacturer’s instructions. For PCR, cDNA was diluted using RNase free water, mixed with primer/probe sets for Pank1 (Mm00458408_m1), Pank2 (Mm00463258_m1), Pank3 (Mm00461298_m1) or Gusb (Mm03003537_s1) and 2× PCR Universal Master Mix (Applied Biosystems, Inc.). PCR reactions were performed in triplicate on an ABI Prism 7000 Sequence Detections System following the manufacturer’s directions.
To quantify miRNAs, cDNA was reverse transcribed from 4ng total RNA using primers from miR-103 (assay ID 439), miR-107 (assay ID 443) or snoRNA234 (assay ID 1234) TaqMan MicroRNA Assays and reagents from the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Inc.). The resulting cDNA was amplified by PCR using primer/probe sets supplied with the TaqMan MicroRNA Assays and 2x TaqMan Universal PCR Master Mix (Applied Biosystems, Inc.). PCR reactions were performed in triplicate on an ABI Prism 7000 Sequence Detections System following the manufacturer’s directions.
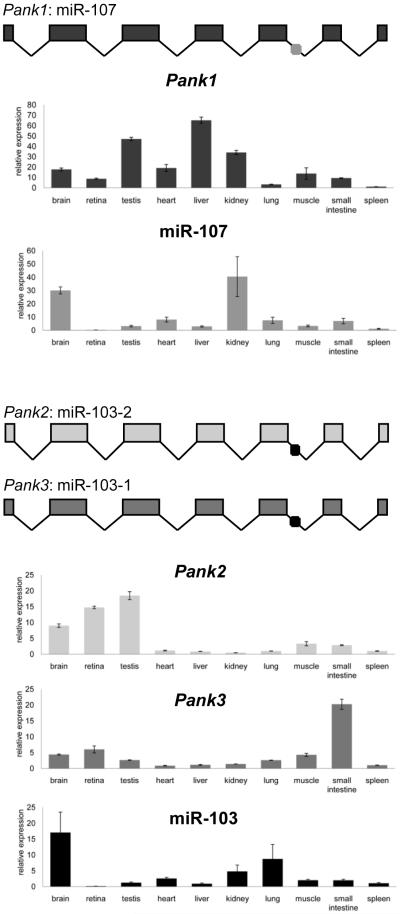
Gusb and snoRNA234 were endogenous controls for the analysis of Pank and miRNA expression, respectively. For each tissue, the mean Ctcontrol value was subtracted from the mean Ctexperimental value (ΔCt). Spleen was chosen as a reference tissue and the mean ΔCt value for spleen was subtracted from each tissue ΔCt value (ΔΔCt). Relative expression (RE) in each tissue was calculated as RE = 2−ΔΔCt. The mean of the RE for three biological replicates is plotted in Figure 1 and error bars represent the standard error of the mean.
Figure 1.
Relative expression of miR-103, miR-107 and their host genes in mouse tissues. RNA was harvested from mouse tissues (n=3) and analyzed by qRT-PCR. Error bars represent standard error.
3. Results & Discussion
To determine whether miR-103 and miR-107 are co-expressed with their host genes in mouse, we measured relative miR-103, miR-107, Pank1, Pank2 and Pank3 expression by qRT-PCR (Figure 1). We detected widespread expression of all pantothenate kinase genes. Pank2 has highest expression in brain, retina and testis, three tissues that are affected by PKAN [24]. Pank3, in contrast, is most abundantly expressed in the small intestine, and Pank1 has relative abundance in the liver. Notably, the relative expression patterns of Pank1, Pank2 and Pank3 in testis, brain and liver differ from a previously published report [25]. miR-103 is broadly expressed across mouse tissues with highest relative expression in brain and lung, consistent with previous findings using microarrays [10]. In contrast, miR-107 was most abundant in brain and kidney.
Despite the shared characteristic of ubiquitous expression, miR-103 and miR-107 expression profiles poorly correlate with that of their host genes. For miR-107, this result supports the recent model of transcription from an independent promoter [6]. For miR-103, the discordant expression is likely attributed, at least in part, to its redundant expression from Pank2 and Pank3 transcripts. However, post-transcriptional factors may also play a role in controlling the relative abundance of the mature transcript. In support of this concept, Lee et al have demonstrated that regulation at the level of miRNA processing is common and often controls tissue-specific regulation of ubiquitously expressed miRNA precursor transcripts [26].
The miR-103/7 family represents an intriguing model of disease-associated intronic miRNA. They are highly conserved within pantothenate kinase genes; one of which, PANK2, is associated with a neurodegenerative disease. Moreover, miR-103/7 likely regulates proteins involved in acetyl-coA metabolism [21], as well as neurodegeneration. However, expression of the mature miRNAs is not synchronized with that of the host genes. These distinct expression patterns are likely due to multiple factors, including the existence of alternate promoters, variable stabilities of the RNA transcripts and regulation of post transcriptional processing. Therefore, the miR-103/7 family provides an intriguing model for dissecting miRNA transcription, processing and coordinated function with host genes.
ACKNOWLEDGEMENTS
This work was supported by the Huebner Family Pediatric Neurobiology of Disease Fellowship (to B.P.).
Abbreviations
- miRNA
microRNA
- PANK
pantothenate kinase
- PKAN
pantothenate kinase-associated neurodegeneration
- precursor microRNA
pre-miRNA
- UTRs
untranslated regions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNAs promoters. RNA. 2010 doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- [8].Robishaw JD, Neely JR. Coenzyme A metabolism. Am J Physiol. 1985;248:E1–9. doi: 10.1152/ajpendo.1985.248.1.E1. [DOI] [PubMed] [Google Scholar]
- [9].Zhou B, Westaway SK, Levinson B, Johnson MA, Gitschier J, Hayflick SJ. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden- Spatz syndrome. Nat Genet. 2001;28:345–349. doi: 10.1038/ng572. [DOI] [PubMed] [Google Scholar]
- [10].Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S, Kaisaki P, Argoud K, Fernandez C, Travers ME, Grew JP, Randall JC, Gloyn AL, Gauguier D, McCarthy MI, Lindgren CM. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53:1099–1109. doi: 10.1007/s00125-010-1667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nelson PT, Wang WX. MiR-107 is Reduced in Alzheimer’s Disease Brain Neocortex: Validation Study. J Alzheimers Dis. 2010 doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- [19].Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. MiR-107 Regulates Granulin/Progranulin with Implications for Traumatic Brain Injury and Neurodegenerative Disease. Am J Pathol. 2010 doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hayflick SJ, Westaway SK, Levinson B, Zhou B, Johnson MA, Ching KH, Gitschier J. Genetic, clinical, and radiographic delineation of Hallervorden-Spatz syndrome. N Engl J Med. 2003;348:33–40. doi: 10.1056/NEJMoa020817. [DOI] [PubMed] [Google Scholar]
- [25].Leonardi R, Zhang YM, Lykidis A, Rock CO, Jackowski S. Localization and regulation of mouse pantothenate kinase 2. FEBS Lett. 2007;581:4639–4644. doi: 10.1016/j.febslet.2007.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]