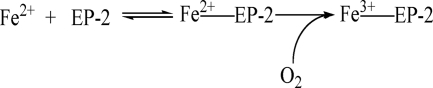Abstract
Naturally occurring phytoferritin is a heteropolymer consisting of two different H-type subunits, H-1 and H-2. Prior to this study, however, the function of the two subunits in oxidative deposition of iron in ferritin was unknown. The data show that, upon aerobic addition of 48–200 Fe2+/shell to apoferritin, iron oxidation occurs only at the diiron ferroxidase center of recombinant H1 (rH-1). In addition to the diiron ferroxidase mechanism, such oxidation is catalyzed by the extension peptide (a specific domain found in phytoferritin) of rH-2, because the H-1 subunit is able to remove Fe3+ from the center to the inner cavity better than the H-2 subunit. These findings support the idea that the H-1 and H-2 subunits play different roles in iron mineralization in protein. Interestingly, at medium iron loading (200 irons/shell), wild-type (WT) soybean seed ferritin (SSF) exhibits a stronger activity in catalyzing iron oxidation (1.10 ± 0.13 μm iron/subunit/s) than rH-1 (0.59 ± 0.07 μm iron/subunit/s) and rH-2 (0.48 ± 0.04 μm iron/subunit/s), demonstrating that a synergistic interaction exists between the H-1 and H-2 subunits in SSF during iron mineralization. Such synergistic interaction becomes considerably stronger at high iron loading (400 irons/shell) as indicated by the observation that the iron oxidation activity of WT SSF is ∼10 times larger than those of rH-1 and rH-2. This helps elucidate the widespread occurrence of heteropolymeric ferritins in plants.
Keywords: Iron, Protein Assembly, Protein Chemistry, Protein Domains, Protein Turnover, H1 and H2 Subunits, Ferritin, Phytoferritin, Protein Association
Introduction
Iron and oxygen chemistry, in a variety of non-heme diiron proteins, has drawn considerable attention because of their various roles in reversible O2 binding for respiration in hemerythrin, oxidation and desaturation of organic substrates in methane monooxygenase, R2 subunit of ribonucleotide reductase, stearoyl-acyl carrier protein Δ-9 desaturase, as well as the detoxification and concentration of iron in Dps and ferritin (1–6). Diiron centers have similar structural motifs containing a combination of carboxylate and histidine ligands that either bind or bridge the two metal ions of the dinuclear active site. Although the dinuclear centers of the protein are very similar in structure, each of the proteins fulfills a different biological function that appears to be mediated by the nature of the first and second coordination sphere of the diiron center.
Ferritins are a special class of diiron proteins that play a role in both iron housekeeping and iron detoxification. They are widely distributed in animals, plants, and bacteria (but not in yeast) and are typically composed of 24 similar or identical subunits assembled into a shell-like structure. In vertebrates, ferritins consist of two types of subunits, H and L, respectively. The two types have about 55% identity in amino acid sequence. The ratio of the two kinds of subunits in the native protein varies according to the nature and the function of the tissue (1, 2). The H-type subunit of vertebrate ferritins contains a dinuclear iron “ferroxidase site” at which the rapid oxidation of Fe2+ to Fe3+ by dioxygen occurs; subsequent hydrolysis of the Fe3+ and its migration away from the ferroxidase site leads to formation of the Fe3+ mineral core. Unlike the H-subunit, the L-subunit lacks a ferroxidase center but has a greater negative charge; in the assembled ferritin, the negative charge presents itself on the interior surface as clusters of acidic residues (Glu and Asp) that comprise the mineral nucleation site (6). Ferritins from lower vertebrates, such as bullfrogs and fish, contain a third subunit type, named the M(H′) chain, which harbors the residues forming both the ferroxidase center and the nucleation site (1, 7). In contrast, two different types of ferritins are found in bacteria, often within the same cell. These are the bacterioferritins and the bacterial ferritins, which are chemically, structurally, and functionally different. All subunits from these two ferritins are of the H chain type; they are not particularly similar to eukaryotic H-chain ferritins in terms of overall sequence similarity (∼20% identity), but conserved within them are some of the key residues that constitute the catalytic ferroxidase center (1, 2).
Compared with animal ferritin, phytoferritin exhibits distinctive structural features. First, only the H-type subunit has been identified in phytoferritin thus far, and it shares ∼40% sequence identity with the animal H-subunit. Second, the H-type subunit in mature phytoferritin contains a specific extension peptide (EP)4 at its N-terminal sequence (8–11). In the case of soybean seed ferritin (SSF), each EP domain is composed of ∼30 amino acid residues. The crystal structure of recombinant SSF shows that the EP is located on the exterior surface of protein and stabilizes the entire oligomeric conformation of phytoferritin by its interaction with a neighboring subunit on the shell surface (12). Recent studies of our group reveal the role of the EP during iron oxidative deposition in phytoferritin as the second binding and ferroxidase center that contributes to mineralization of the iron core at high iron loading of ferritin (>48 irons/shell) (13). On the other hand, the EP exhibits a significant serine protease-like activity (14), which is responsible for protein autodegradation during seed germination (14–16). Associated with the degradation is faster iron release from ferritin to meet the requirements of seedling growth (14). Third, naturally occurring phytoferritin is usually composed of two different H-type subunits. For example, ferritin from soybean and pea seeds consists of H-1 and H-2 subunits sharing ∼80% amino acid sequence identity (8–10, 17). This specific feature of phytoferritin raises the inevitable question of what functions the two subunits serve in phytoferritin. Addressing this question is integral to understanding the relationship between the structure and function of phytoferritin.
The present study is the first to investigate the roles of the H-1 and H-2 subunits in SSF for oxidative deposition of iron in protein. At low iron loading of apoferritin (≤48 irons/protein shell, 2 irons/subunit), no protein association occurs with both recombinant soybean seed H-1 (rH-1) and H-2 (rH-2) ferritins, indicating that iron oxidation occurs only through the diiron ferroxidase mechanism (13). Once the binding capacity of the 24 ferroxidase centers (48 irons/shell) is exceeded, the oxidative addition of Fe2+ to the aporH-2 to form Fe3+ causes protein aggregation, whereas such protein association is not observed in its analog, rH-1, until the amount of added iron is increased to 300 Fe2+/protein shell. This finding suggests that the H-1 subunit has considerably higher regeneration activity (indicative of the ability to turn Fe3+ over from the diiron ferroxidase centers to the inner cavity) compared with H-2. Indeed, regeneration activity measurements show that rH-1 removes Fe3+ from the ferroxidase center to the inner cavity much faster than rH-2. However, rH-2 is able to transfer iron from the outer surface to inside the protein better than rH-1. Thus, the two subunits exhibit different mechanisms to cope with iron oxidative deposition in protein at high iron flux into ferritin: the diiron ferroxidation mechanism for rH-1 versus the EP oxidation mechanism for rH-2. Moreover, a combination of H-1 and H-2 subunits in WT SSF results in stronger catalytic activity in iron mineralization than that of each of the two recombinant proteins because of a synergistic interaction between the two subunits.
EXPERIMENTAL PROCEDURES
Preparation of WT SSF, rH-1, rH-2, and rH-1H-2
SSF was purified as recently described (8, 13). rH-1 was prepared according to reported methods (8, 18). The expression plasmid for rH-2 was constructed by inserting the H-2 cDNA into the NcoI/BamHI site of pET21d using a PCR-based method. The constructs were then introduced into Escherichia coli strain BL21 (DE3). The positive transformants of each construct were grown at 37 °C on LB medium supplemented with 50 mg/liter carbenicillin, and protein expression was induced with 100 μm isopropyl β-d-1-thiogalactopyranoside when the cell density reached an A600 of 0.6 (18). Both rH-1 and rH-2 were purified using the same methods described above for native SSF. Apoferritin was prepared as previously described (19, 20). The concentrations of all types of ferritin were determined according to the Lowry method with BSA as a standard sample. Recombinant heteropolymer rH-1H-2 was constructed by mixing dissociated H-1 and H-2 subunits in a 1:1 ratio and reassembled by slowly raising the pH to 7 according to a reported method (21, 22).
Preparation of rH-1, rH-2, and WT SSF with EP Deleted
The preparations of EP-deleted rH-1, rH-2, and WT SSF were carried out as described recently (13, 14). Purified holophytoferritin (3.0 mg) was added to 0.24 ml of Alcalase 2.4 L (Novozymes, Denmark) and incubated at 60 °C for 5 min, followed by phenylmethanesulfonyl fluoride at a final concentration of 2 mm to stop the proteolysis reaction. The resultant solution was diluted 500-fold with distilled water and then ultrafiltered to its initial concentration using an YM-100 membrane (Millipore Corp., Bedford, MA). Finally, SDS-PAGE was performed to examine the purity of the protein under reducing conditions using 15% minislab gels. The N-terminal amino acid sequences of each subunit treated with the enzyme were also determined.
The molecular weights of all ferritin samples were estimated by PAGE using a 4–15% polyacrylamide gradient gel run at 30 V for 15 h at 4 °C and with Tris-HCl (25 mm, pH 8.3) as running buffer. SDS-PAGE was performed under reducing conditions using 15% minislab gels according to a reported method (23). Gels were stained with Coomassie Brilliant Blue R-250.
Sequence of N-terminal Amino Acid
The amino acid sequence of the N terminus was determined on a protein sequencer (Procise® 491, Applied Biosystems) using automated Edman degradation. After SDS-PAGE, the protein was transferred to a polyvinylidene difluoride membrane (Millipore Corp.) and stained with Coomassie Brilliant Blue R-250. The two subunits were eluted from the membrane, and the sequence of the 10 amino acids at the N terminus was determined for each subunit.
Reactions between EP and Fe3+
EP-1 with sequence ASTVPLTGVIFEPFEEVKKSELAVPT (corresponding to the EP from the H-1 subunit) and EP-2 with sequence ASNAPAPLAGVIFEPFQELKKDYLAVPI (corresponding to the EP from the H-2 subunit) were purchased from Scilight Biotechnology (Beijing, China). Purity was more than 95% as indicated by HPLC and mass spectroscopy. A solution of FeCl3 (70 mm) was freshly prepared in pH 2.0 double-distilled H2O. Reaction between the EP and Fe3+ was carried out as follows. 2 μl of FeCl3 was added to 100 μl of EP-1 (140 μm) in 5 mm Mops at pH 7.0, and the resultant solution was incubated overnight at 4 °C. Subsequently, the solution was mixed with 0.1% (v/v) trifluoroacetic acid and α-cyano-4-hydroxycinnamic acid (10 mg/ml) prior to MALDI-TOF-MS analyses. All mass spectra of MALDI-TOF-MS were obtained on a Bruker Ultraflex III TOF/TOF (Bruker Daltonik GmbH, Germany) in positive ion mode at an accelerating voltage of 20 kV with a nitrogen laser set at 337 nm (14, 17).
Stopped-flow Light Scattering and Dynamic Light Scattering Experiments
Stopped-flow light scattering measurement experiments were performed with a Hi-Tech SFA-20 M apparatus in conjunction with a Cary Eclipse spectrofluorimeter (Varian). Both excitation and emission wavelengths were set to 680 nm, and the time-dependent change in scattering light was set to a 90° angle, perpendicular to the beam, and recorded as previously reported (13). All quoted concentrations are final concentrations after mixing the two reagents. All iron solutions were freshly prepared with pH 2.0 water.
The dynamic light scattering (DLS) measurements were performed at 25 °C using a Viscotek model 802 dynamic light scattering instrument (Viscotek Europe Ltd.) as described recently (13). The hydrodynamic radius (RH) was calculated with the regularization histogram method using the spheres model, from which an apparent molecular mass was estimated according to a standard curve calibrated from known globular proteins. OmniSIZE 2.0 software was used to calculate the size distribution of aggregated protein from the addition of iron. Unless stated otherwise, all samples were allowed to stand for 1 h prior to DLS measurements to ensure that the association reactions were complete.
Fluorescence Quenching with Fe2+ Oxidative Deposition in Ferritin
The fluorescence-quenching kinetic experiments were performed with a pneumatic drive Hi-Tech SFA-20 M stopped-flow accessory interfaced to a Cary Eclipse spectrofluorimeter (Varian) as described previously (24). The dead time of the stopped-flow fluorimeter apparatus was determined to be 9.2 ± 0.4 ms using the N-acetyltryptophanamide and N-bromosuccinimide test reaction (25). The dead times consider both the mixing time and software delay of the instrument. The ratio of protein to iron is 1:48, and protein concentration is 0.5 μm. Measurements were carried out at 25 °C in 0.15 m NaCl and 100 mm Mops at pH 7.0. Path lengths for excitation and emission were 1.0 and 0.2 cm at wavelengths of 280 and 325 nm, respectively. A slit width of 5 nm for both excitation and emission was used.
Fe2+ Oxidative Deposition in Ferritin
The fast kinetics experiments were conducted by measuring absorbance at 300 nm using the Hi-Tech SFA-20 M stopped-flow accessory on a Varian Cary 50 spectrophotometer (Varian) at 25 °C. Equal 140-μl volumes of a weakly acidic FeSO4 solution (0.192–1.6 mm) and buffered apoferritin solution (4.0 μm) were mixed at 25 °C in the thermostatted sample compartment containing a 280-μl quartz stopped-flow cuvette with 1-cm optical path length. All quoted concentrations were final concentrations after mixing the two reagents. Fe2+ oxidation was monitored at 300 nm. Data were acquired every 12.5 ms (the shortest acquisition time possible with the Cary 50). The spectrophotometer was operated in single beam mode and zeroed prior to each kinetic run with a cuvette containing apoferritin in buffer. The initial rates of iron oxidation measured as μ-oxo complex formation were obtained from the linear A1 term of a third order polynomial curve fitted to the experimental data as reported previously (13).
Statistical Analysis
All statistical analyses were performed using the software Statistical Program for Social Sciences Analysis System (SPSS Inc.). An analysis of variance and Tukey's test were carried out with a confidence level of 95% (p ≤ 0.05). Each value represents the average of at least three independent measurements.
RESULTS
Protein Association Property in rH-1, rH-2, WT SSF, and rH-1H-2
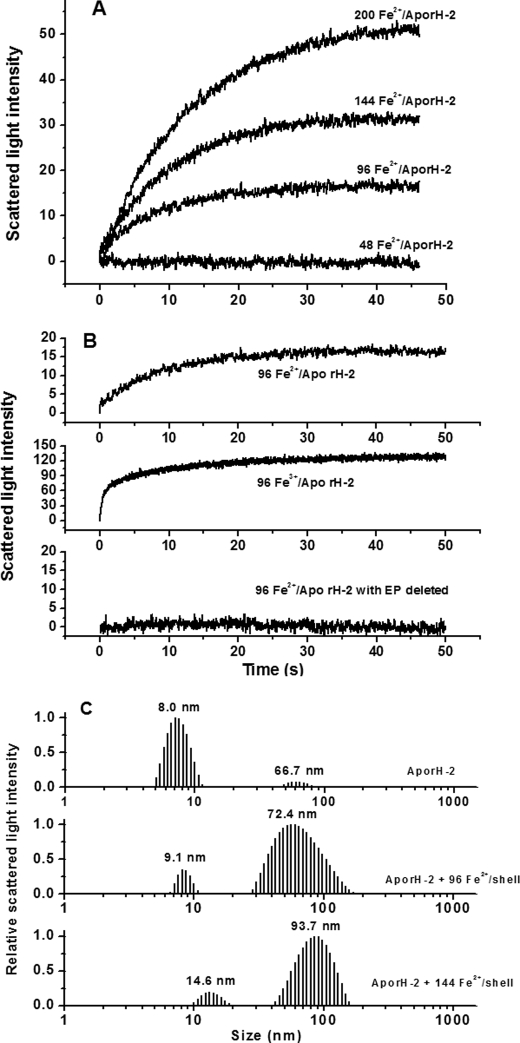
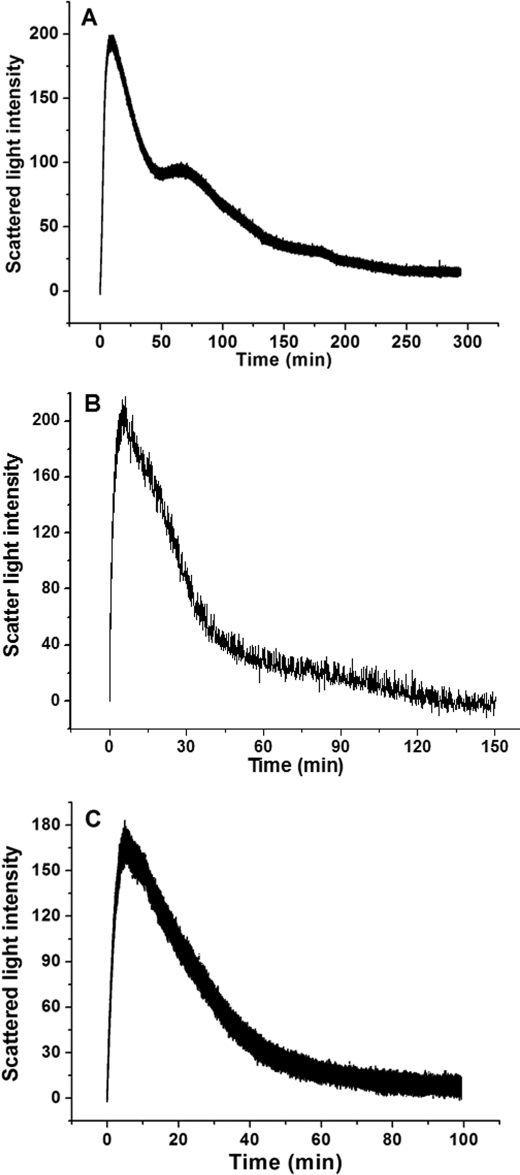
Laser light scattering is a widely applied technique for studying biomolecules in a solution because it can provide information about the size and conformation of proteins and their aggregation state as well as their ability to crystallize (13, 26–28). To shed light on the role of the H-1 and H-2 subunits in SSF for iron oxidative deposition in phytoferritin, we first conducted dynamic and stopped-flow light scattering experiments with H-1 soybean seed ferritin (rH-1) and its analog, rH-2. The stopped-flow experiments were performed with either Fe2+ or Fe3+, rapidly mixed with the aporH-2 and rH-1 under aerobic conditions, respectively (Figs. 1 and 2). Light scattering changed minimally within the 50-s time frame of the experiment when ≤48 iron/shell were mixed with the rH-2 apoproteins (as also found using H2O in place of the iron solution). In contrast, the light scattering intensity increased markedly because of protein aggregation when the aporH-2 was rapidly mixed aerobically with a series of Fe2+ solutions to give ratios from 96 to 200 Fe2+/shell (Fig. 1A), and the rise in the initial rate, ν0, is first order with respect to iron concentration (Table 1). These results suggest that protein-protein association is driven by binding of the additional iron on the exterior surface of the protein shell because the 24 dinuclear ferroxidase centers of rH-2 have a maximal binding capacity of 48 iron ions and are located in the interior of the protein (13).
FIGURE 1.
A, time course of rH-2 aggregation during Fe2+ oxidation. B, time course of rH-2 or EP-deleted rH-2 aggregation upon the addition of Fe2+ or Fe3+. The curve represents an average of six experimental measurements. C, relative scattered light intensity distribution curves for aporH-2 and aporH-2 plus Fe2+ ion. Conditions were as follows: [aporH-2] = 0.5 μm in 100 mm Mops (pH 7.0), 24–100 μm FeSO4, 25 °C.
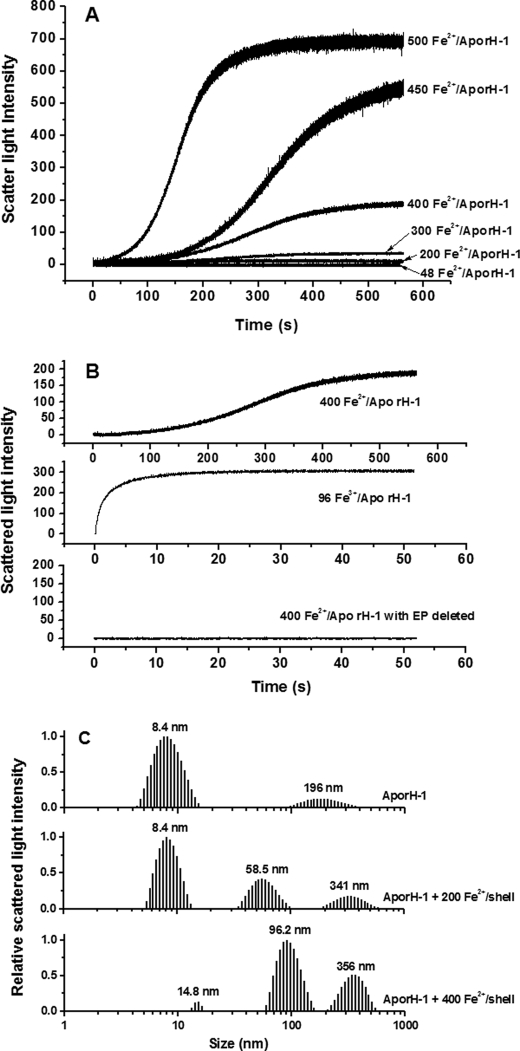
FIGURE 2.
A, time course of rH-1 aggregation during Fe2+ oxidation. B, time course of rH-1 or EP-deleted rH-1 aggregation upon the addition of Fe2+ or Fe3+. The curve represents an average of six experimental measurements. C, relative scattered light intensity distribution curves for aporH-1 and aporH-1 plus Fe2+ ion. Conditions were as follows: [aporH-1] = 0.5 μm in 100 mm Mops (pH 7.0), 24–250 μm FeSO4, 25 °C.
TABLE 1.
Initial rates of protein association obtained from Figs 1, 2, and 4 and supplemental Fig. S4
| Fe2+/shell | Initial rate |
|||
|---|---|---|---|---|
| rH-1 | rH-2 | SSF | rH-1H-2 | |
| ΔY/s | ||||
| 48 | 0.03 ± 0.01a | 0.02 ± 0.01a | 0.02 ± 0.01a | 0.04 ± 0.02a |
| 96 | 0.02 ± 0.01b | 1.65 ± 0.07a | 0.80 ± 0.03c | 0.91 ± 0.07c |
| 144 | 0.03 ± 0.01b | 2.66 ± 0.13a | 1.59 ± 0.06c | 1.21 ± 0.04c |
| 200 | 0.04 ± 0.01b | 4.69 ± 0.11a | 3.84 ± 0.08c | 3.57 ± 0.14c |
| 300 | 0.12 ± 0.02 | |||
| 400 | 0.23 ± 0.02 | |||
| 450 | 0.30 ± 0.03 | |||
| 500 | 0.35 ± 0.03 | |||
a–c Values followed by the same letter are not significantly different according to the least significant difference test (p < 0.05).
Stopped-flow light scattering experiments were also carried out to determine whether Fe2+ or Fe3+ is responsible for the observed protein aggregation. A rapid increase in intensity (ν0 = 12.31 ± 0.34 ΔY/s) is observed when Fe3+ is directly added to aporH-2 (96 Fe3+/shell), whereas light scattering intensity slowly increases under the same conditions (ν0 = 1.65 ± 0.07 ΔY/s) when Fe2+ is mixed aerobically with aporH-2 to produce Fe3+. The protein association reaction induced by Fe3+ is completed within ∼0.5 s, whereas the reaction requires ∼20 s to complete when Fe2+ is used instead of Fe3+. By contrast, the anaerobic addition of Fe2+ causes no change in scattering light intensity (data not shown). Thus, Fe3+ facilitates protein-protein association, not Fe2+. A similar observation was obtained with PSF (13).
Protein association facilitated by iron was also investigated by DLS under the same conditions used in the stopped-flow light scattering experiment. Two populations with RH values of 8.0 and 66.7 nm are evident in the scattered light intensity distribution curve of the aporH-2 sample (Fig. 1C). The corresponding mass distribution curve (supplemental Fig. S1) shows that the population centered at 8.0 nm is rich in monomers, accounting for about 99.9% of the total (supplemental Table S1), whereas the second population having RH = 66.7 nm represents a very small amount (0.1%) of aggregates composed of ∼134 monomers (supplemental Table S1). This result indicates that at pH 7.0, essentially all of the aporH-2 molecules exist in a dissociated state. By contrast, the size distribution is markedly altered toward large aggregates upon aerobic introduction of 96 Fe2+/shell to aporH-2 (Fig. 1C). The population with RH = 72.4 nm represents ∼36.0% of the total and corresponds to a large aggregate of ∼164 monomers (supplemental Table S1). As the added iron is increased to 144 Fe2+/shell, the population with RH = 93.7 nm accounts for about 98.3% of the total, indicating that most of the rH-2 molecules participate in protein association upon treatment with iron. These findings are in accord with the stopped-flow results shown in Fig. 1, A and B.
The EP domains located on the exterior surface of PSF molecules serve as a second ferroxidase center and are involved in iron mineralization, which contains initial Fe2+ oxidation and Fe3+ translocation from the outer protein surface to the inner cavity through EP-regulated protein association (13). To elucidate whether the EP domains in the H-2 subunit (EP-2) play the same role, EP-deleted rH-2 was mixed aerobically with different solutions of Fe2+ or Fe3+ under the same experimental conditions used for rH-2. Results show that no protein association occurs in all tested samples (Fig. 1B), confirming the idea that EP-2 acts in a manner similar to that of the EP from PSF (13).
Similarly, light scattering intensity is essentially unchanged within 550 s, when ≤48 Fe2+/shell were shot against aporH-1 (Fig. 2A). Surprisingly, when the amount of iron added ranged from 48 to 200 Fe2+/shell, the light scattering intensity exhibits minimal change within the 550-s time frame of the experiment, indicating that no iron is bound to the outer surface. Thus, rH-1 appears to have the larger regeneration activity of its ferroxidase centers, which corresponds to the ability of the protein to remove iron from the ferroxidase centers in the inner cavity (see below). By contrast, light scattering intensity significantly increases as a result of protein aggregation when aporH-1 was rapidly mixed aerobically with a series of Fe2+ solutions in ratios from 300 to 500 Fe2+/shell, with the rate and magnitude of the increase being a strong function of the Fe2+/shell ratio (Fig. 2A and Table 1). Such protein association was also confirmed by DLS results (Fig. 2C). At 200 Fe2+/shell loading of the protein, the mass populations with a large size center at both 58.5 and 341 nm, representing only 3.8 and 0.5%, respectively, of the total (supplemental Fig. S2 and Table S2). This indicates that no significant protein association under this experimental condition is observed. However, at high iron flux into the protein (400 Fe2+/shell), the mass population at both 96.2 and 356 nm accounts for ∼59% of the total, demonstrating that protein association occurs in rH-2 to a great extent, confirming the observations from the stopped-flow light scattering experiments (Fig. 2A).
In parallel experiments, Fe3+ was added to aporH-1 with two different ratios (48 and 96 Fe3+/shell). There is no increase in the intensity of scattered light upon the addition of 48 Fe3+/shell (not shown), indicating that the diiron ferroxidase sites likewise accommodate 48 ferric ions, one ferric ion per site. By contrast, light scattering intensity considerably and rapidly increases upon 96 Fe3+/shell addition (Fig. 2B). Thus, the diiron ferroxidase sites in the H-1 subunit are not able to remove these monoferric complexes from the cavity as effectively as they do with the diferric complexes formed by Fe2+ oxidation, reflecting the discrepancy between the absorption of Fe2+ and Fe3+ by the protein.
Similarly, upon removal of the EP of H-1 (EP-1), protein association disappears with rH-1 under the same experimental conditions (Fig. 2B), indicating that EP-1 is an essential structural component for protein association caused by either Fe3+ or Fe2+ added aerobically. Notably, the kinetic curve for the aerobic addition of Fe2+ to aporH-1 is sigmoidal, the initial lag phase of which is characteristic of slow iron oxidation by oxygen on the outer surface of protein (13), a finding indicative of the lower catalyzing activity of EP-1 compared with the EP-2.
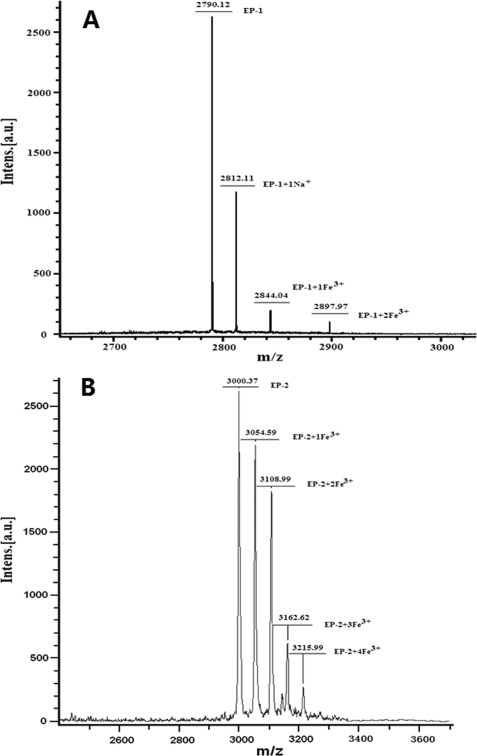
To gain insight into why EP-2 exhibits higher activity than EP-1, the interaction of the two EPs with iron was studied by MALDI-TOF-MS. The MS profile of the intact EP-1 shows a single peak at m/z 2790.12 Da (data not shown), indicating a singly charged peptide monomer [M + H]+, which is in good agreement with the predicted molecular mass, 2790.24 Da. Upon treatment with FeCl3, three new peaks appear at m/z 2812.11, 2844.04, and 2897.97 Da, which correspond to EP-1 + Na+, EP-1 + Fe3+, and EP-1 + 2 Fe3+, respectively (Fig. 3A). Thus, EP-1 can bind up to 2 ferric ions/molecule. Except for a peak at m/z 3000.37 Da corresponding to the EP-2 + H+, the MS profile of EP-2 exhibits four peaks at m/z 3054.59 Da (EP-2 plus Fe3+), 3108.99 Da (EP-2 plus 2 Fe3+), 3162.62 (EP-2 plus 3 Fe3+), and 3215.99 (EP-2 plus 4 Fe3+) after treatment with FeCl3 under the same experimental conditions. This indicates that EP-2 is able to bind a maximum of 4 ferric ions/molecule (Fig. 3B). Therefore, the observed difference in the catalyzing activity between the two EPs may be derived from their different binding capacities with ferric ions.
FIGURE 3.
MS profiles of EP-1 (A) and EP-2 (B) treated with FeCl3 (Fe3+/EP ratio = 10:1) acquired by MALDI-TOF-MS. Conditions were as follows: [EP-1 or EP-2] = 140 μm in 5 mm Mops, pH 7.0.
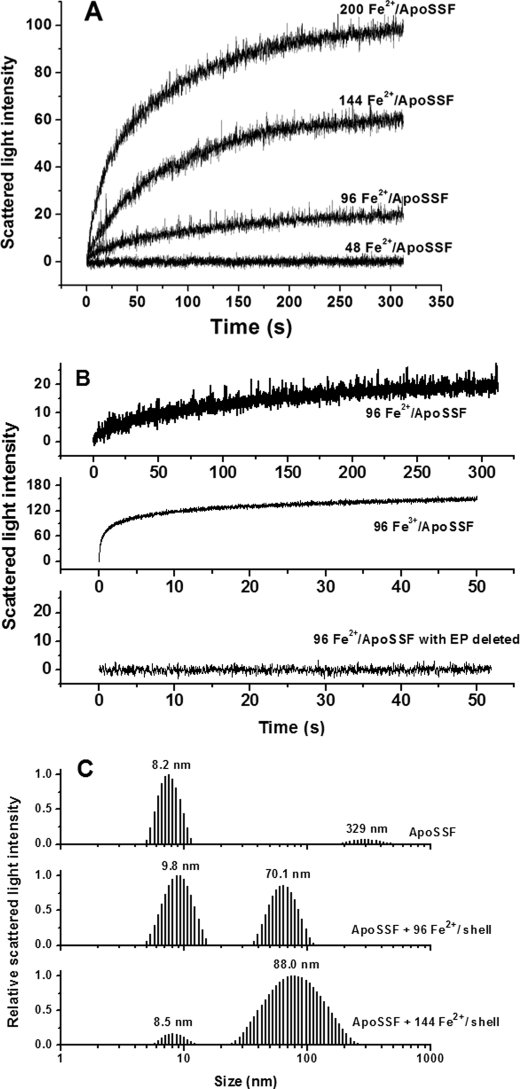
To better understand the roles of the H-1 and H-2 subunits in protein association, the same stopped-flow light scattering and DLS experiments were carried out with WT SSF, and results are shown in Fig. 4. Similar to rH-1 and rH-2, no protein association occurs with WT SSF at low iron loading of ferritin, whereas a flux of intermediate iron into SSF causes protein association (Fig. 4A). The protein association induced by the direct addition of Fe3+ is considerably faster than that induced by the aerobic addition of Fe2+ (Fig. 4B), again demonstrating that Fe3+, not Fe2+, triggers the protein aggregation. Such a process was likewise confirmed by DLS results (Fig. 4C), and the corresponding mass distribution curve and parameters are given in supplemental Fig. S3 and Table S3, respectively. Similar results were obtained with the recombinant H-1 and H-2 soybean seed ferritin (rH-1H-2) (supplemental Fig. S4, A and B), consistent with the above observation. Thus, both WT SSF and rH-1H-2 exhibit assembly behavior similar to that of rH-2 rather than rH-1. However, upon the addition of 144 Fe2+/shell, the mass population at 88.0 nm in SSF is only ∼38.2% of the total (supplemental Fig. S3 and Table S3), a value significantly smaller than that in rH-2 (98.3%), indicating that the degree of protein association in SSF is lower than that in rH-2. Taken together, a combination of the H-1 and H-2 subunits in either SSF or rH-1H-2 appears to result in an increase in the regeneration activity of the diiron ferroxidase centers in SSF but does not cause an activity as large as that in rH-1.
FIGURE 4.
A, time course of WT SSF aggregation during Fe2+ oxidation. B, time course of WT SSF or EP-deleted SSF aggregation upon the addition of Fe2+ or Fe3+. The curve represents an average of six experimental measurements. C, relative scattered light intensity distribution curves for apoSSF and apoSSF plus Fe2+ ion. Conditions were as follows: [WT SSF] = 0.5 μm in 100 mm Mops (pH 7.0), 24–100 μm FeSO4, 25 °C.
Regeneration Activities of rH-1, rH-2, and WT SSF
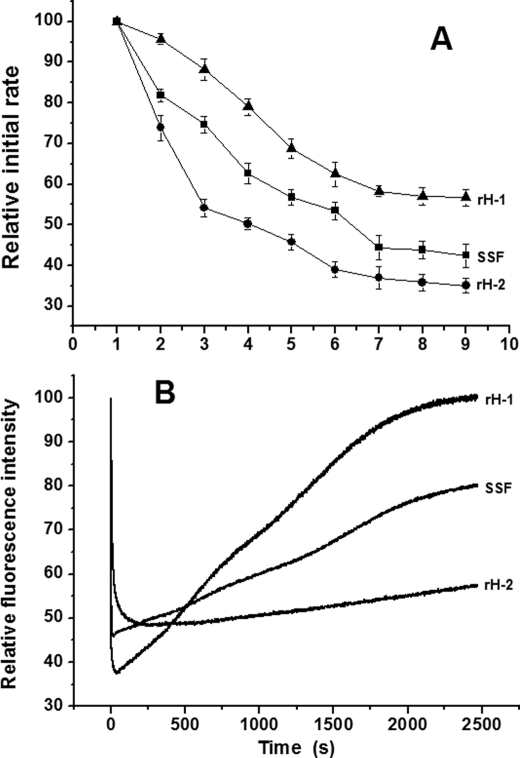
No protein association occurred when aporH-1 was mixed with a series of Fe2+ solutions with ratios between 48 and 200 Fe2+/shell, indicating that all added ferrous ions enter into the protein shell, probably because of the stronger regeneration activity of its diiron ferroxidase centers than that of rH-2 and SSF (the regeneration activity reflecting the ability of protein to remove Fe3+ from the diiron ferroxidase centers to the inner cavity). To confirm this idea, the regeneration activities of rH-1, rH-2, and SSF were measured, and results are shown in Fig. 5A. A single Fe2+ addition corresponds to half-saturation of the ferroxidase centers (i.e. 24 Fe2+/protein for each ferritin). The initial rates were normalized to the initial rate observed for the first injection. Generally, rH-1 exhibits the strongest activity among the three proteins, whereas the activity of rH-2 is weakest. For example, there is a sharp drop in initial rate on the third injection into rH-2 right after an iron/protein stoichiometry corresponding to the saturation of the diiron ferroxidase centers. By contrast, after the third injection, the regeneration activity of rH-1 still retains ∼90% of its original value. The ninth injection leads to only a 43% decrease in the regeneration activity for rH-2, whereas the same injection induces a 65% decrease for rH-1. The regeneration activity of SSF is midway between that of rH-1 and rH-2, which decreases by ∼57% after the ninth injection. These results provide a satisfactory explanation on why protein association occurs in rH-2 and WT SSF but not in rH-1 at moderate iron flux.
FIGURE 5.
A, regeneration of oxidation activities in rH-1, rH-2, and WT SSF as normalized initial rates. Each injection is half-saturation of the ferroxidase sites, 1 Fe2+/site. Conditions were as follows: [aporH-1, aporH-2, or WT apoSSF] = 2.0 μm in 0.15 m NaCl and 100 mm Mops, pH 7.0, 25 °C, time intervals between injections 3 min. Error bars, S.E. B, change of relative fluorescence intensity of apoferritin after the aerobic addition of 48 Fe2+ atoms/shell. Conditions were as follows: λex = 280 nm, λem = 325 nm, [aporH-1, aporH-2, or WT apoSSF] = 0.5 μm in 0.15 m NaCl and 0.1 m Mops, pH 7.0, 25 °C.
Fluorescence-quenching kinetic experiments were carried out according to a reported method (24, 29), in which iron solutions (48 added per shell) were shot against the three apoferritins (Fig. 5B). The intrinsic fluorescence of aporH-1, rH-2, or WT SSF was not quenched when the protein was rapidly mixed with an Fe2+ solution (48 irons/shell) anaerobically (data not shown). By contrast, the fluorescence is considerably quenched when each of the proteins was aerobically shot against Fe2+, indicating that the observed fluorescence quenching stems from iron oxidation but not ferrous ion binding. However, the three proteins exhibit different kinetic quenching behaviors. The intrinsic fluorescence in rH-2 is quenched rapidly, reaches a constant value of about half of the original signal after 15–20 s, and then increases more slowly until it attains about 55% of the initial intensity at around 2950 s. In a different manner, the fluorescence in rH-1 recovers markedly faster than that in rH-2 and retains about 100% of the initial intensity at 2000 s. The first fast fluorescence-quenching phase corresponds to Fe2+ oxidation at the ferroxidase centers, whereas the second phase of slow fluorescence recovery represents the transfer of Fe3+ from the diiron ferroxidase centers to the cavity (24, 29). Thus, the fluorescence results again support the conclusion that the H-1 subunit is able to turn over Fe3+ from the diiron ferroxidase centers to the cavity better than H-2. The rate of the fluorescence recovery in SSF is slower than that in rH-1 but faster than in rH-2, a result also in good agreement with the above mentioned observation (Fig. 5A).
Reversibility of rH-1, rH-2, and WT SSF Association/Dissociation
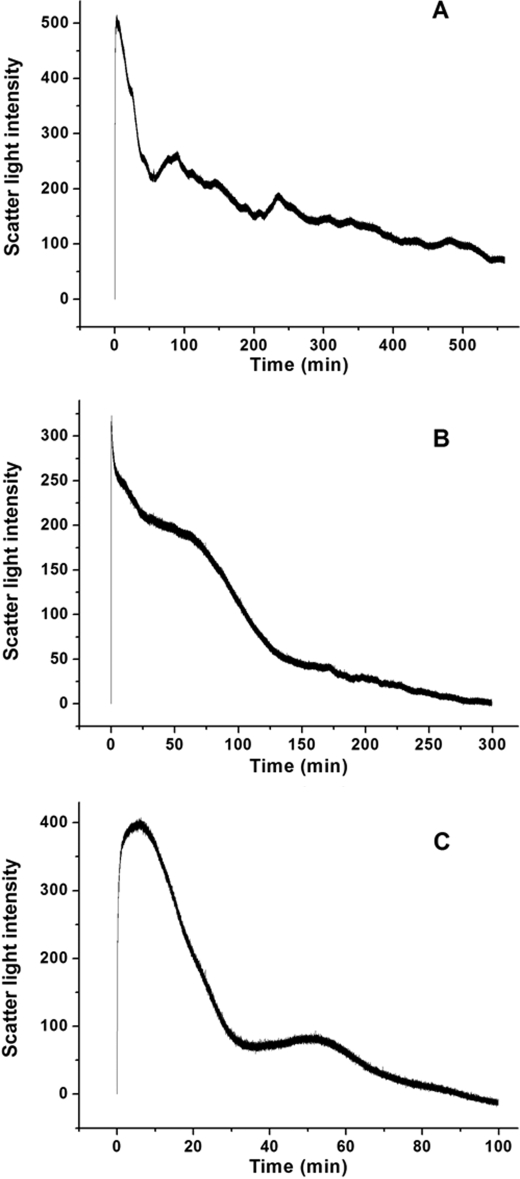
To determine whether ferritin association during oxidative deposition of iron is reversible, a stopped-flow experiment was carried out in which 400 Fe2+/shell were rapidly mixed aerobically with aporH-1, aporH-2, and apoSSF. Results are shown in Fig. 6. Generally, all kinetic curves consist of two phases, namely the rapid initial rise in light scattering intensity, followed by a slow decrease to its original value. The first phase corresponds to protein association, whereas the second is due to protein dissociation as described recently (13). Therefore, protein association induced by the aerobic addition of Fe2+ is a reversible process in the three proteins. DLS measurements confirm the reversibility of protein association/dissociation induced by iron (supplemental Fig. S5).
FIGURE 6.
Time course of the formation and dissociation of rH-1 (A), rH-2 (B), and WT SSF (C) aggregates during the oxidative deposition of iron in the protein shell. The curve represents an average of six independent experimental measurements. Conditions were as follows: [aporH-1, aporH-2, or WT apoSSF] = 0.5 μm in 100 mm Mops (pH 7.0), 200 μm Fe2+, 25 °C.
Based on a recent report (13), the rate of protein dissociation positively correlates with the ability of phytoferritin to move Fe3+ from the outer surface of the protein to its inner cavity. Protein dissociation occurs with aporH-2 over a period of 150 min (Fig. 6A), whereas this process is completed within 300 min for aporH-1 (Fig. 6B), indicating that rH-2 is able to transfer Fe3+ from the protein outer surface to the cavity better than rH-1. Interestingly, protein aggregates dissociate completely within 100 min for SSF (Fig. 6C), much faster than for rH-2, indicating that there is a positively synergistic interaction between the H-1 and H-2 subunits in iron translocation from the outer surface of the protein to the cavity.
To elucidate whether the same synergistic interaction also occurs in protein dissociation upon direct treatment with Fe3+, a stopped-flow experiment was undertaken in which 144 Fe3+/shell were rapidly shot against aporH-1, aporH-2, and apoSSF (Fig. 7). Similarly, complete dissociation is observed with all samples prepared by the direct addition of Fe3+, although it is markedly slower than that induced by the aerobic addition of Fe2+ (Fig. 6). Dissociation in rH-2 is completed within 300 min (Fig. 7A), which is significantly faster than that in rH-1 (500 min) (Fig. 7B), indicating that the H-2 subunit exhibits a stronger activity in transferring Fe3+ to the cavity than H-1. This result is in accordance with that obtained by the aerobic addition of Fe2+ to apoprotein. By contrast, complete protein dissociation in SSF occurs within 100 min (Fig. 7C), a process again much faster than that in rH-1 and rH-2, confirming the above mentioned synergistic interaction between the two subunits in SSF.
FIGURE 7.
Kinetic curves of the formation and dissociation of rH-1 (A), rH-2 (B), and WT SSF (C) aggregates during the oxidative deposition of iron in the protein shell. The curve represents an average of six independent experimental measurements. Conditions were as follows: 0.5 μm apoferritin in 100 mm Mops (pH 7.0), 72 μm Fe3+, 25 °C.
Iron Oxidative Deposition in rH-1, rH-2, WT SSF, and rH-1H-2
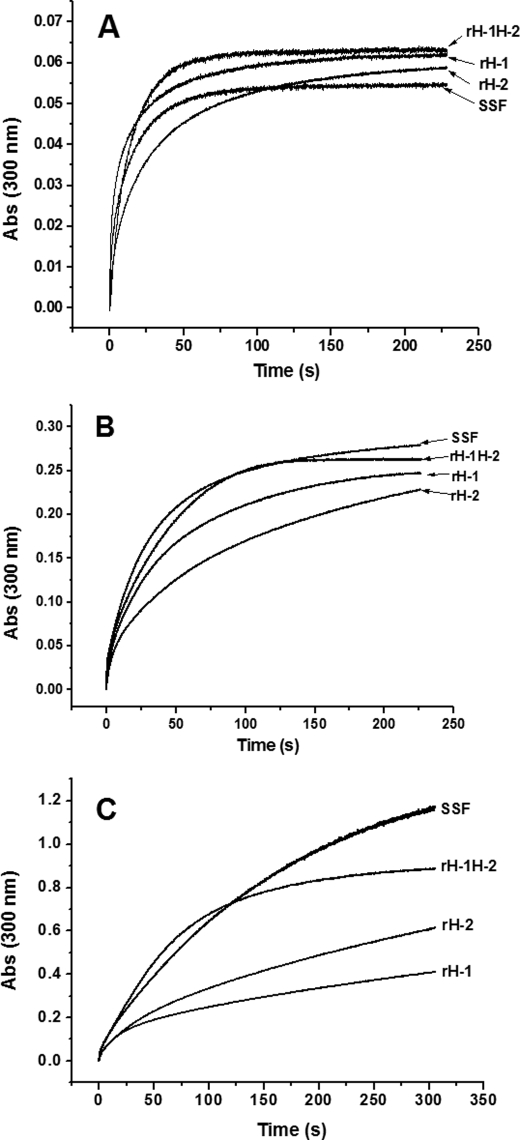
UV absorption in the 300–330 nm spectral region has been traditionally used to monitor the formation of Fe3+ species during oxidative deposition of iron in the ferritins (13, 17, 20, 30, 31). Spectrophotometric kinetic measurements of iron deposition in rH-1, rH-2, WT SSF, and rH-1H-2 were conducted to evaluate the role of the H-1 and H-2 subunits in the formation of the mineral core of ferritin at different iron loadings (48, 200, and 400 irons/protein). The stopped-flow kinetic traces were obtained at 300 nm upon the addition of 48, 200, or 400 Fe2+/shell to the four protein samples under the same conditions as those in the light scattering experiments shown in Fig. 8. Unlike the above kinetic curves shown in Figs. 1, 2, 3, 6, and 7, the stopped-flow UV-visible kinetic traces are very smooth, suggesting that no insoluble protein aggregates form during iron oxidative deposition in protein. Otherwise, the noise in the curves caused by light scattering from the formation of protein or FeOOH particles would be observed as described previously (32). Additionally, all curves are hyperbolic, consistent with protein catalysis of iron oxidation (33, 34) and with these proteins having fully intact H-chain ferroxidase sites.
FIGURE 8.
Kinetic curves of Fe2+ oxidation by O2 in aporH-1, aporH-2, WT apoSSF, and aporH-1H-2 at different Fe2+ concentrations, 48 (A), 200 (B), and 400 Fe2+/shell (C). Conditions were as follows: final [aporH-1, aporH-2, aporH-1H-2, or WT apoSSF] = 0.5 μm in 100 mm Mops (pH 7.0), 24–200 μm FeSO4, 25 °C.
At low iron loading (48 Fe2+/shell), the initial rate of iron oxidation catalyzed by rH-1 (1.02 ± 0.05 μm iron/subunit/s) is ∼3.5-fold larger than that by rH-2 (0.29 ± 0.02 μm iron/subunit/s) when compared on a subunit basis (2 Fe2+/subunit) under the present conditions (Fig. 8A). This indicates that the ferroxidase center in rH-1 has markedly stronger catalyzing activity than that in rH-2. The iron initial rate of SSF is 0.47 ± 0.04 μm iron/subunit/s, a value midway between the values of rH-1 and rH-2, suggesting that the relative contribution of the two subunits in SSF to iron mineralization is essentially equal. Consistent with this conclusion, the value of the initial rate of rH-1H-2 (0.42 ± 0.02 μm iron/subunit/s) is also between the values of rH-1 and rH-2 (Fig. 8A).
At intermediate iron loading (200 Fe2+/shell), the four ferritins exhibit a markedly different kinetic behavior (Fig. 8B) (i.e. biphasic kinetic curves are observed, which are characterized by an initial fast phase followed by a slow phase). The initial rapid rise in absorbance is attributed to fast Fe2+ oxidation at the diiron ferroxidase site. For rH-2 and SSF, the EP is responsible, at least partially, for iron mineralization in the second phase because of low regeneration activity of their diiron ferroxidase sites. The specific domain in rH-1 is not believed to be involved in this process, as suggested by protein association results (Fig. 1). Similarly, the initial rate of iron oxidation catalyzed by rH-1 (0.59 ± 0.07 μm iron/subunit/s) is slightly larger than that by rH-2 (0.48 ± 0.04 μm iron/subunit/s), suggesting that the catalyzing activity of the diiron ferroxidase site in rH-1 is stronger than both the activity of the diiron ferroxidase site in rH-2 and in EP-2. Different from the situation at low iron flux into ferritin, the iron oxidation rates of both SSF (1.10 ± 0.13 μm iron/subunit/s) and rH-1H-2 (0.91 ± 0.03 μm iron/subunit/s) are considerably larger than those of rH-1 and rH-2, again indicating that a positive interaction exists between the H-1 and H-2 subunits in SSF or rH-1H-2 in iron oxidation at medium iron loading.
At high iron loading of the protein (400 Fe2+/protein shell), a situation where added iron (400 Fe2+/shell) exceeds the regeneration activity of the diiron ferroxidase center in rH-1, the order of the kinetic activity of rH-2 and rH-1 is reversed, and consequently, rH-2 (ν0 = 1.42 ± 0.04 μm iron/subunit/s) exhibits stronger catalyzing activity than rH-1 (ν0 = 0.99 ± 0.06 μm iron/subunit/s) (Fig. 8C). This implies that iron mineralization processed by EP-2 is more effective than that by EP-1. More importantly, under the present condition, SSF (ν0 = 10.84 ± 0.36 μm iron/subunit/s) is the most kinetically active of the three proteins (rH-1, rH-2, and SSF), again indicating that the H-1 and H-2 subunits in SSF have a synergistic effect on iron oxidative deposition in the protein. Support for this conclusion comes from the observation that rH-1H-2 (ν0 = 11.77 ± 0.87 μm iron/subunit/s) exhibits a much stronger catalyzing iron oxidation activity compared with those of rH-1 and rH-2. This effect becomes more pronounced when iron loading increases from 200 to 400 Fe2+/protein shell (Fig. 8C). Additionally, it is evident that the initial rate of iron oxidation catalyzed by ferritin is generally proportional to iron concentration, a result that accords with previous observations (30, 32).
DISCUSSION
To date, all known phytoferritins naturally occurring in seeds (except Arabidopsis) are heteropolymers composed of H-1 and H-2 subunits with high sequence identity (8, 17, 35). This property is reminiscent of heteropolymeric H/M(H′) ferritins found in the liver of Antarctic teleosts (36). The M subunit displays the features of both H and L subunits (7, 36), indicating that it is an H and L hybrid subunit. Similarly, all subunits in phytoferritin characterized thus far likewise contain both a typical H-type ferroxidase center and all of the amino acid residues characteristic of an L-type subunit for efficient iron nucleation with the cavity (11). Based on these structural features, phytoferritin is considered an H/L hybrid ferritin (11). Therefore, it is of special interest to elucidate the role of H-1 and H-2 in SSF for oxidative deposition of iron in protein.
The present work demonstrates that the H-1 and H-2 subunits have markedly distinctive functions in oxidative deposition of iron in SSF (i.e. the H-1 subunit is mainly responsible for rapid Fe2+ oxidation followed by mineralization at low iron loading of ferritin, whereas the H-2 subunit plays a more important role at high iron flux into protein or upon direct addition of Fe3+ to ferritin). Consequently, the H-1 and H-2 subunits have complementary functions in iron uptake into ferritin, a situation similar to that of the H and L subunits in animal ferritin (1–3). More significantly, they have a synergistic effect on the oxidative deposition of iron in SSF, resulting in stronger catalyzing activity of both WT SSF and rH-1H-2. This property has not been previously recognized. Although the amino acids involved in the definition of the ferroxidase center are strictly conserved in the two subunits (8, 10–12), their ferroxidase centers exhibit markedly different regeneration and catalyzing activities. For example, at low iron flux into apoferritin (48 Fe2+/shell), the rate of iron oxidation by rH-1 is markedly larger than that of rH-2 (Fig. 8A). On the other hand, when the amount of iron added to apoferritin is in the range of 48–200 Fe2+/shell, no protein aggregation occurs with rH-1, whereas such an addition considerably facilitates protein association in rH-2 (Fig. 1). The disparity in protein aggregation between the two proteins stems from the difference in regeneration activity of the diiron ferroxidase centers. As shown in Fig. 5A, the regeneration activity of the diiron ferroxidase center in the H-1 subunit is much greater than that in H-2, resulting in the absence of ferric iron bound to the exterior surface of the protein. Indeed, measurement of the regeneration activity by fluorescence spectrophotometry reveals that rH-1 exhibits the stronger ability to remove Fe3+ from the ferroxidase center to the inner cavity compared with rH-2 (Fig. 5B), confirming the above mentioned idea. These results demonstrate that other amino acid residues near or far away from the ferroxidase center likewise have significant effects on the regeneration activity. Support for this idea comes from the recent observation that, unlike human H-chain ferritin, mitochondrial ferritin does not regenerate its ferroxidase activity after oxidation of its initial complement of Fe(II) and generally has considerably slower ferroxidation and mineralization activities as well, although mitochondrial ferritin has the same ferroxidase center residues and overall structure as human H-chain ferritin (37, 38).
With the addition of more than 200 Fe2+/shell, protein association also takes place in rH-1 because such addition exceeds the turnover capability of the protein. Interestingly, the kinetic curves of protein association of rH-1 are sigmoidal in contrast to rH-2, in which the hyperbolic kinetic curves are observed, indicating that the catalyzing activity of EP-2 is stronger than that of EP-1 (Figs. 1 and 2) (13). This may be because EP-2 has a higher capacity for iron than its analog, EP-1 (Fig. 3). Moreover, when 400 Fe2+/shell were aerobically mixed with apoferritin, protein dissociation is completed within ∼300 min with rH-2 and is faster than that of rH-1 (∼500 min) (Fig. 6), demonstrating that EP-2 is able to translocate Fe3+ from the outer surface of the protein to the inner shell better than EP-1. As expected, upon the direct addition of Fe3+/shell, the same result was obtained with the two proteins (Fig. 7), again supporting the above mentioned conclusion. Thus, with increasing iron concentration from 48 to 200 irons/shell or higher, the mechanism of iron oxidative deposition in deposition in both rH-1 and rH-2 is changing from the diiron ferroxidase mechanism to the EP-involved catalyzing mechanism (13); the EP domains play a more important role in iron mineralization than the diiron ferroxidase center. The difference in the catalyzing activity between EP-1 and EP-2 is consistent with their difference in structure, namely sequence ASTVPLTGVIFEPFEEVKKSELAVPT for EP-1 versus sequence ASNAPAPLAGVIFEPFQELKKDYLAVPI for EP-2. These findings support the idea that the EP domains might regulate the iron oxidation activity of plant ferritin, especially at moderate and high iron loadings. Consistent with this idea, two other soybean ferritin subunits, H-3 and H-4, have distinct EP domains from their analogues H-1 and H-2, and homopolymer rH-3 exhibits lower iron incorporation activity as compared with rH-1 and rH-2 (18). Further support for this proposal comes from previous studies showing that cowpea leaf ferritin (39), maize ferritin (40), and pea seed ferritin (17) consist of two or more different subunits, which contain specific EP domains at the N-terminal extremity.
REACTION 1.
Although the iron oxidation activity of rH-1 is markedly larger than that of rH-2 at low iron flux into the protein (≤48 iron/shell), the rate of iron oxidation catalyzed by rH-1 is comparable with that by rH-2 at both moderate (200 iron/shell) and high (400 iron/shell) iron fluxes into the protein (Table 2). Agreeing with the above observation, the H and M subunits likewise exhibit a similar iron oxidation activity at high iron loading (41). In contrast, there exists a 600–1000-fold disparity in the iron oxidation activity between H and L subunits under identical conditions (41). Therefore, the H-2 subunit is much closer to the M subunit in promoting iron mineralization but not to the L chain.
TABLE 2.
Initial rates of Fe2+ oxidative deposition obtained from Fig. 8
| Fe2+/shell | Initial rate |
|||
|---|---|---|---|---|
| rH-1 | rH-2 | SSF | rH-1H-2 | |
| μm iron/subunit/s | ||||
| 48 | 1.02 ± 0.05a | 0.29 ± 0.02b | 0.47 ± 0.04c | 0.42 ± 0.02c |
| 200 | 0.59 ± 0.07c | 0.48 ± 0.04c | 1.10 ± 0.13a | 0.91 ± 0.03a |
| 400 | 0.99 ± 0.06c | 1.42 ± 0.04c | 10.84 ± 0.36a | 11.77 ± 0.87a |
a–c Values followed by the same letter are not significantly different according to the least significant difference test (p < 0.05).
Notably, the rate and magnitude of protein association is likewise a function of Fe2+ aerobically added to apoferritin (Fig. 1). These results suggest that binding of Fe2+ to EP-2 is an equilibrium reaction followed by iron oxidation by O2 as shown in Reaction 1. Resultant ferric ions ultimately induce protein aggregation (Fig. 1), as recently observed with PSF (13).
What advantage might the assembly of the H-1 and H-2 subunits into the protein render to phytoferritin in oxidative deposition of iron? The answer to this question may lie in the experimental results of iron mineralization in protein (Fig. 8). Among the four proteins, rH-1 is the most kinetically active at low iron loading of protein (48 Fe2+/shell) (Fig. 8A). However, as the amount of Fe2+ added to protein aerobically increases to about 200 Fe2+/shell, either SSF or rH-1H-2 is generally superior to rH-2 and rH-1 in promoting iron mineralization, suggesting that there is a synergistic interaction between the H-1 and H-2 subunits in this process. Consistent with this conclusion, both WT SSF and rH-1H-2 have a stronger ability to catalyze this process than rH-1 and rH-2 upon the addition of 400 Fe2+/shell. In parallel experiments in which 400 Fe2+/shell were shot against apoferritin followed by stopped-flow light scattering measurement, protein aggregates clearly dissociate more rapidly in SSF than in rH-2 and rH-1 (Fig. 6), again demonstrating that the H-1 and H-2 subunits have a positively cooperative effect in facilitating the transfer of Fe3+ from its exterior surface to the cavity of the protein. This effect becomes more evident when Fe3+ is directly used instead of Fe2+ to mix with the apoferritins (Fig. 7). Thus, the combination of the H-1 and H-2 subunits in SSF considerably enhances the efficiency of iron oxidative deposition in the protein. This view is supported by previous studies indicating that the L-chain facilitates mineral formation in horse spleen ferritin, presumably because of increased iron turnover at the ferroxidase site of the H-chain in the presence of the L-chain (42). In contrast, such cooperative interaction seems to be lacking in the H/M heteropolymer found in fish as indicated by the fact that the co-assembly of H- and M-chains in Trematomus newnesi and Trematomus bernacchii liver ferritin results in a decreased catalytic and mineralization efficiency relative to the M homopolymer, which favors adaptation to low temperatures (36, 43). These comparative results imply that the specific EP domains of phytoferritin might be responsible, at least partly, for the above observed synergistic interaction between the H-1 and H-2 subunits during oxidative deposition of iron in protein.
In summary, the present work defined the key features of the H-1 and H-2 subunits in SSF for oxidative deposition of iron in ferritin and showed that their function in the above mentioned process depends on the amount of iron added to the protein. At low iron flux into the protein (≤48 Fe2+/shell), both the H-1 and H-2 subunits catalyze iron oxidation via the diiron ferroxidase center through a ferroxidase mechanism as proposed previously (42). Under conditions of moderate iron flux, the above mechanism likewise dominates iron oxidation in the H-1 subunit because of its stronger ability to remove Fe3+ from the outer surface of protein to the inner cavity than that of the H-2 subunit. However, in the H-2 subunit, this mechanism is gradually replaced by another mechanism by which the iron oxidation is processed by EP-2. At high iron loading of the protein, EP-1 also appears and participates in the iron oxidation, but its ability to catalyze iron oxidation and transfer Fe3+ to the cavity is pronouncedly weaker than that of EP-2. More interestingly, when more than 48 Fe2+ or Fe3+ per protein are loaded, there is a positively synergistic interaction between the H-1 and H-2 chains in SSF in iron oxidation and translocation. Thus, heteropolymeric ferritin may facilitate plant cell absorption of both ferrous and ferric ions from soil more effectively than homopolymeric ferritin.
Supplementary Material
Acknowledgment
We thank Chris Janus-Chandler for corrections to the manuscript.
This project was supported by National Natural Science Foundation of China Grant 30972045, Chinese Universities Scientific Fund Grant 2009-3-10, and Innovation Fund for Graduate Students Grant 15059203.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
- EP
- extension peptide
- EP-1
- extension peptide of 26.5-kDa subunit of soybean seed ferritin
- EP-2
- extension peptide of 28.0-kDa subunit of soybean seed ferritin
- rH-2
- recombinant soybean seed H-2 chain ferritin
- rH-1
- recombinant soybean seed H-1 chain ferritin
- rH-3
- recombinant soybean seed H-3 chain ferritin
- rH-1H-2
- recombinant H-1 and H-2 soybean seed ferritin
- PSF
- pea seed ferritin
- SSF
- soybean seed ferritin
- apoSSF
- apo-soybean seed ferritin
- DLS
- dynamic light scattering.
REFERENCES
- 1.Harrison P. M., Arosio P. (1996) Biochim. Biophys. Acta 1275, 161–203 [DOI] [PubMed] [Google Scholar]
- 2.Le Brun N. E., Crow A., Murphy M. E., Mauk A. G., Moore G. R. (2010) Biochim. Biophys. Acta 1800, 732–744 [DOI] [PubMed] [Google Scholar]
- 3.Zhao G., Ceci P., Ilari A., Giangiacomo L., Laue T. M., Chiancone E., Chasteen N. D. (2002) J. Biol. Chem. 277, 27689–27696 [DOI] [PubMed] [Google Scholar]
- 4.Solomon E. I., Brunold T. C., Davis M. I., Kemsley J. N., Lee S. K., Lehnert N., Neese F., Skulan A. J., Yang Y. S., Zhou J. (2000) Chem. Rev. 100, 235–350 [DOI] [PubMed] [Google Scholar]
- 5.Kopp D. A., Lippard S. J. (2002) Curr. Opin. Chem. Biol. 6, 568–576 [DOI] [PubMed] [Google Scholar]
- 6.Crichton R. R., Herbas A., Chavez-Alba O., Roland F. (1996) J. Biol. Inorg. Chem. 1, 567–574 [Google Scholar]
- 7.Dickey L. F., Sreedharan S., Theil E. C., Didsbury J. R., Wang Y. H., Kaufman R. E. (1987) J. Biol. Chem. 262, 7901–7907 [PubMed] [Google Scholar]
- 8.Masuda T., Goto F., Yoshihara T. (2001) J. Biol. Chem. 276, 19575–19579 [DOI] [PubMed] [Google Scholar]
- 9.Zhao G. (2010) Biochim. Biophys. Acta 1800, 815–823 [DOI] [PubMed] [Google Scholar]
- 10.Theil E. C. (2004) Annu. Rev. Nutr. 24, 327–343 [DOI] [PubMed] [Google Scholar]
- 11.Lobreaux S., Yewdall S. J., Briat J. F., Harrison P. M. (1992) Biochem. J. 288, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masuda T., Goto F., Yoshihara T., Mikami B. (2010) J. Biol. Chem. 285, 4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li C., Fu X., Qi X., Hu X., Chasteen N. D., Zhao G. (2009) J. Biol. Chem. 284, 16743–16751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X., Deng J., Yang H., Masuda T., Goto F., Yoshihara T., Zhao G. (2010) Biochem. J. 427, 313–321 [DOI] [PubMed] [Google Scholar]
- 15.Van Wuytswinkel O., Savino G., Briat J. F. (1995) Biochem. J. 305, 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobreaux S., Briat J. F. (1991) Biochem. J. 274, 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C., Hu X., Zhao G. (2009) Biochimie 91, 230–239 [DOI] [PubMed] [Google Scholar]
- 18.Masuda T., Goto F., Yoshihara T., Ezure T., Suzuki T., Kobayashi S., Shikata M., Utsumi S. (2007) Protein Expr. Purif. 56, 237–246 [DOI] [PubMed] [Google Scholar]
- 19.Bauminger E. R., Harrison P. M., Hechel D., Nowik I., Treffry A. (1991) Biochim. Biophys. Acta 1118, 48–58 [DOI] [PubMed] [Google Scholar]
- 20.Treffry A., Hirzmann J., Yewdall S. J., Harrison P. M. (1992) FEBS Lett. 302, 108–112 [DOI] [PubMed] [Google Scholar]
- 21.Chiaraluce R., Consalvi V., Cavallo S., Ilari A., Stefanini S., Chiancone E. (2000) Eur. J. Biochem. 267, 5733–5741 [DOI] [PubMed] [Google Scholar]
- 22.Kang S., Oltrogge L. M., Broomell C. C., Liepold L. O., Prevelige P. E., Young M., Douglas T. (2008) J. Am. Chem. Soc. 130, 16527–16529 [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 24.Cavallo S., Mei G., Stefanini S., Rosato N., Finazzi-Agrò A., Chiancone E. (1998) Protein Sci. 7, 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterman B. F. (1979) Anal. Biochem. 93, 442–444 [DOI] [PubMed] [Google Scholar]
- 26.Zhelev N., Barudov S. (2005) Biotechnol. Biotec. Eq. 19, 3–8 [Google Scholar]
- 27.Janmey P. A., Hvidt S., Käs J., Lerche D., Maggs A., Sackmann E., Schliwa M., Stossel T. P. (1994) J. Biol. Chem. 269, 32503–32513 [PubMed] [Google Scholar]
- 28.Pal G. P., Elce J. S., Jia Z. (2001) J. Biol. Chem. 276, 47233–47238 [DOI] [PubMed] [Google Scholar]
- 29.Li C., Qi X., Li M., Zhao G., Hu X. (2009) Biochimie 91, 1475–1481 [DOI] [PubMed] [Google Scholar]
- 30.Bou-Abdallah F., Zhao G., Mayne H. R., Arosio P., Chasteen N. D. (2005) J. Am. Chem. Soc. 127, 3885–3893 [DOI] [PubMed] [Google Scholar]
- 31.Wallar B. J., Lipscomb J. D. (1996) Chem. Rev. 96, 2625–2658 [DOI] [PubMed] [Google Scholar]
- 32.Zhao G., Bou-Abdallah F., Arosio P., Levi S., Janus-Chandler C., Chasteen N. D. (2003) Biochemistry 42, 3142–3150 [DOI] [PubMed] [Google Scholar]
- 33.Crichton R. R., Roman F. (1978) J. Mol. Catal. 4, 75–82 [Google Scholar]
- 34.Macara I. G., Hoy T. G., Harrison P. M. (1972) Biochem. J. 126, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravet K., Touraine B., Boucherez J., Briat J. F., Gaymard F., Cellier F. (2009) Plant J. 57, 400–412 [DOI] [PubMed] [Google Scholar]
- 36.Giorgi A., Mignogna G., Bellapadrona G., Gattoni M., Chiaraluce R., Consalvi V., Chiancone E., Stefanini S. (2008) Arch. Biochem. Biophys. 478, 69–74 [DOI] [PubMed] [Google Scholar]
- 37.Bou-Abdallah F., Santambrogio P., Levi S., Arosio P., Chasteen N. D. (2005) J. Mol. Biol. 347, 543–554 [DOI] [PubMed] [Google Scholar]
- 38.Arosio P., Ingrassia R., Cavadini P. (2009) Biochim. Biophys. Acta 1790, 589–599 [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Theil E. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8557–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wicks R. E., Entsch B. (1993) Biochem. Biophys. Res. Commun. 192, 813–819 [DOI] [PubMed] [Google Scholar]
- 41.Lobreaux S., Massenet O., Briat J. F. (1992) Plant Mol. Biol. 19, 563–575 [DOI] [PubMed] [Google Scholar]
- 42.Yang X., Chen-Barrett Y., Arosio P., Chasteen N. D. (1998) Biochemistry 37, 9743–9750 [DOI] [PubMed] [Google Scholar]
- 43.Mignogna G., Chiaraluce R., Consalvi V., Cavallo S., Stefanini S., Chiancone E. (2002) Eur. J. Biochem. 269, 1600–1606 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.