Abstract
The human ether-a-go-go-related gene 1 (hERG1) encodes the pore-forming subunit of the rapidly activating delayed rectifier potassium channel. Several hERG1 isoforms with different N- and C-terminal ends have been identified. The hERG1a, hERG1b, and hERG1-3.1 isoforms contain the full-length C terminus, whereas the hERG1USO isoforms, hERG1aUSO and hERG1bUSO, lack most of the C-terminal domain and contain a unique C-terminal end. The mechanisms underlying the generation of hERG1USO isoforms are not understood. We show that hERG1 isoforms with different C-terminal ends are generated by alternative splicing and polyadenylation of hERG1 pre-mRNA. We identified an intrinsically weak, noncanonical poly(A) signal, AGUAAA, within intron 9 of hERG1 that modulates the expression of hERG1a and hERG1aUSO. Replacing AGUAAA with the strong, canonical poly(A) signal AAUAAA resulted in the predominant production of hERG1aUSO and a marked decrease in hERG1 current. In contrast, eliminating the intron 9 poly(A) signal or increasing the strength of 5′ splice site led to the predominant production of hERG1a and a significant increase in hERG1 current. We found significant variation in the relative abundance of hERG1 C-terminal isoforms in different human tissues. Taken together, these findings suggest that post-transcriptional regulation of hERG1 pre-mRNA may represent a novel mechanism to modulate the expression and function of hERG1 channels.
Keywords: Ion channels, Polyadenylation, Potassium Channels, RNA Processing, RNA Splicing
Introduction
The human ether-a-go-go-related gene 1 (hERG1)3 encodes a K+ channel that has properties similar to the rapidly activating delayed rectifier K+ current (IKr) in the heart (1–4). hERG1 channels play an important role in cardiac action potential repolarization, and mutations in hERG1 cause long QT syndrome type 2 (5). Several hERG1 isoforms with different N- and C-terminal ends have been identified (4, 6). The first cloned hERG1 isoform, hERG1a, consists of 1159 amino acids (1). The hERG1b isoform lacks the first 376 amino acids of hERG1a and has an alternate 36-amino acid N-terminal end (7, 8). The hERG1-3.1 isoform lacks the first 102 amino acids of hERG1a, which are replaced by 6 unique amino acids (9). The C-terminal isoforms, hERG1aUSO and hERG1bUSO, contain the hERG1a and hERG1b N-terminal ends, respectively, but lack the last 359 amino acids, which are replaced by an alternate 88-residue C-terminal end (6, 10, 11). For simplicity, we refer to hERG1 isoforms with the full-length C terminus as hERG1FL, and hERG1 isoforms with the USO C terminus as hERG1USO. The mechanisms responsible for the generation of hERG1 N-terminal isoforms have been well characterized (12, 13), however, the molecular determinants that generate hERG1 C-terminal isoforms are not fully understood.
Functional studies have shown that hERG1a, hERG1b, and hERG1-3.1 channels generate K+ currents with distinct gating properties (2, 3, 7–9). Recently, it was suggested that native ventricular IKr channels are heterotetramers containing hERG1a and hERG1b subunits (14). hERG1aUSO has been reported to generate the hERG1 current with markedly reduced amplitude when expressed in Xenopus oocytes (15). However, when expressed in mammalian Ltk− or HEK293 cells, hERG1aUSO and hERG1bUSO fail to form functional channels (10, 11). Coexpression of hERG1a with hERG1aUSO in equimolar concentrations has no obvious effect on the hERG1a-mediated current (10). When an excess of hERG1aUSO or hERG1bUSO is coexpressed with hERG1a or hERG1b, a decrease in hERG1 current is observed (10, 11). Biochemical studies have shown that hERG1USO-containing heterotetrameric channels are retained in the endoplasmic reticulum and degraded by the ubiquitin-proteasome pathway (11). Specific knockdown of hERG1USO isoforms by RNA interference increases the functional expression of hERG1a/b channels. These previous studies suggest that the function and trafficking of hERG1USO isoforms are significantly impaired in comparison to hERG1FL isoforms and that hERG1USO isoforms may post-translationally suppress hERG1a/b channel function (11). Therefore, regulation of hERG1 C-terminal isoform expression has significant functional consequences.
In the present study, we show that the generation of hERG1a and hERG1aUSO isoforms results from the alternative splicing and polyadenylation of hERG1 pre-mRNA. The strengths of the 5′ splice site and poly(A) signal of intron 9 play an important role in determining the relative expression of hERG1a and hERG1aUSO and thus, regulating hERG1 channel function. The alternative processing of hERG1 pre-mRNA may represent a novel post-transcriptional mechanism to modulate the expression and function of hERG1 channels.
EXPERIMENTAL PROCEDURES
Rapid Amplification of cDNA End-Poly(A) Test (RACE-PAT)
RACE-PAT assay was performed as described (16). Human heart mRNA was purchased from Clontech. First-strand cDNA was synthesized by reverse transcription with an oligo(dT) primer linked to a C/G-rich anchor sequence (5′-CGAGCTCCGCGGCCGCG(T)12-3′). Subsequent PCR amplification was performed using a reverse primer with only the anchor sequence without oligo(dT) (R2, 5′-GCGAGCTCCGCGGCCGCG-3′) and the hERG1USO specific forward primer (F1, 5′-GCAGATATAGCAAGCTCTTTCGACCATAG-3′). The control PCR was carried out using the F1 primer and a reverse primer upstream and adjacent to the intron 9 poly(A) signal (R1, 5′-GAAAGAACATACAGTAGTATAGCTTAG-3′). The PCR products were visualized on a polyacrylamide gel stained with ethidium bromide.
Plasmid Constructs and Transfection
The poly(A) reporter construct was generated by introducing tandem poly(A) sites into a modified pGL3 vector (Promega, Madison, WI), in which the synthetic poly(A) signal upstream of the SV40 promoter was removed. The construct contained the SV40 promoter, the firefly luciferase gene, and 308 bp of the hERG1 intron 9 followed by a synthetic poly(A) signal. The 308-bp fragment containing intron 9 poly(A) signal was amplified by PCR and subcloned into pGL3 with XbaI and BamHI, replacing the SV40 poly(A) site of pGL3. A synthetic poly(A) signal fragment was amplified by PCR using the unmodified pGL3 vector as the template and inserted at BamHI and SalI sites of the poly(A) reporter construct. The poly(A) signal mutations were generated by site-directed mutagenesis using the pAlter in vitro mutagenesis system. HEK293 cells were transiently transfected with these constructs as described (17).
The hERG1 minigene composed of hERG1 cDNA (exons 1–6) and hERG1 genomic DNA (intron 6 to poly(A) site) was constructed by replacing the hERG1a cDNA C-terminal intronless fragment with the intron-containing hERG1 genomic DNA fragment obtained from a human PAC clone consisting of hERG1 genomic sequence from intron 6 to the end of hERG1 gene (PR4-548K24). The minigene was subcloned into a modified pcDNA5 vector, in which the BGH poly(A) signal was deleted. Mutations of the hERG1 intron 9 poly(A) signal and 5′ splice site were generated by site-directed mutagenesis using the pAlter in vitro mutagenesis system. The hERG1aUSO cDNA construct was generated by PCR from the human heart Marathon-Ready cDNA (Clontech) using a forward primer (5′-CTGGCCATGAAGTTCAAGACCACAC-3′) and a reverse primer (5′-AGGACTCCCTTTGCTTTGGATGTG-3′). The PCR fragment was inserted into pCRII vector by TA cloning (Invitrogen). The cDNA fragment of hERG1aUSO was sequenced and subcloned into the backbone of hERG1a at SphI and EcoRI sites to obtain the full-length hERG1aUSO cDNA. The hERG1a and hERG1aUSO cDNAs were subcloned into pcDNA5 vector. Flp-In HEK293 cells (Invitrogen) were stably transfected with pcDNA5 cDNA or minigene constructs and selected with 100 μg/ml of hygromycin B. Flp-In HEK293 cells, which contain a single integrated Flp recombination target site, allow stable integration of a single copy of the cDNA or minigene construct at a specific genomic site and, subsequently, similar expression in all cell clones.
Quantitative Analysis of hERG1 Transcripts in Human Tissues by Real Time PCR
The human tissue total RNAs were purchased from Ambion (Austin, TX) and Biochain (Hayward, CA). cDNAs were synthesized with the SuperScript III First Strand Synthesis System for RT-PCR with random hexamers (Invitrogen). PCR was performed on the MX300P real time PCR machine (Stratagene, La Jolla, CA) using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). After denaturing at 95 °C for 10 min, the reaction was run for 40 cycles with denaturation at 95 °C for 15 s, annealing and primer extension at 60 °C for 1 min. The sequences of the primers used to distinguish hERG1 C-terminal isoforms were: hERG1FL forward (5′-CTTTGGGGAGCCTCTGAACC-3′), hERG1FL reverse (5′-GGACCAGAAGTGGTCGGAGAA-3′), hERG1USO forward (5′-CAGTCACTGGGGCTGTGGA-3′), and hERG1USO reverse (5′-GAGAATGTGGGAACCCCAGAG-3′). The primers for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: forward (5′-ACACCATGGGGAAGGTGAAG-3′) and reverse (5′-TGGGTGGAATCATATTGGAACA-3′). To take into account the amplification efficiency of each primer set, we used a plasmid DNA that contains partial cDNAs of hERG1FL, hERG1USO, and GAPDH as template for generating standard curves for each primer set. By using a plasmid in which the hERG1FL and hERG1USO cDNA fragments are at a 1:1 ratio, equal quantities can be assigned to each dilution point of the standard curves (18). The amounts of the hERG1FL and hERG1USO transcripts in the samples were calculated using MxPro Quantitative-PCR software (Stratagene), and normalized to the amount of the GAPDH transcript in the same samples.
RNase Protection Assay
RNA isolation and the RNase protection assay (RPA) were performed as previously described (19). Briefly, antisense RNA riboprobes were transcribed in vitro in the presence of biotin-14-CTP. Yeast RNA was used as a control for the complete digestion of the probes by RNase. The relative intensity of each band was quantified using Scion Image software (Scion Corp., Frederick, MD) and adjusted for the number of biotin-labeled cytidines in each protected fragment. For hERG1 minigene experiments, the expression level of the hygromycin B resistance gene from the pcDNA5 vector was used as a loading control.
Immunoblot Analysis
Immunoblot analysis was performed as previously described (17, 20). The cell lysates were subjected to SDS-polyacrylamide gel electrophoresis and then electrophoretically transferred onto nitrocellulose membranes. The membranes were incubated with an anti-hERG1 antibody against the N terminus of hERG1a and hERG1aUSO proteins (Santa Cruz, Santa Cruz, CA) and visualized with the ECL detection kit (Amersham Biosciences). The expression level of hygromycin B phosphotransferase encoded by the hygromycin B resistant gene in the pcDNA5 vector was used as a loading control (21). The polyclonal anti-hygromycin B phosphotransferase antibody was custom generated by Genscript (Piscataway, NJ) using a peptide antigen corresponding to the last 14 amino acids (CSGNRRPSTRPRAKE) of hygromycin B phosphotransferase. The anti-hygromycin B phosphotransferase antibody was affinity purified and used at a 1:2000 dilution.
Patch Clamp Recordings
Membrane currents were recorded in whole cell configuration using suction pipettes as previously described (17, 20). All patch clamp experiments were performed at 22–23 °C. The patch clamp data are presented as mean ± S.E. and analyzed by Student's t test. p < 0.05 is considered statistically significant.
RESULTS
Polyadenylation of hERG1aUSO mRNA
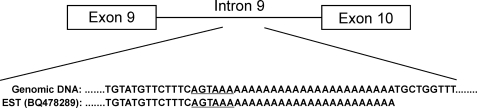
When hERG1aUSO was originally identified, it was unclear whether hERG1aUSO mRNA was polyadenylated (10). A BLAST search of the human EST data base revealed that many USO-containing ESTs have a short poly(A) tail. However, these EST cDNA sequences lack the canonical poly(A) signal (AAUAAA) upstream of the 3′-poly(A) tail. Interestingly, hERG1 genomic DNA contains a 25-bp poly(A) stretch within intron 9. An alignment of the EST sequence BQ478289 with human genomic DNA revealed that the poly(A) tail of the EST matched the intronic sequence of hERG1 (Fig. 1). It is noted that none of the USO-containing ESTs have more than 25 adenosines in the poly(A) tails, suggesting that they may result from internal priming (i.e. oligo(dT) primers hybridizing to internal poly(A) stretches instead of the expected 3′ end poly(A) sequence). Because of this uncertainty, the poly(A) data base did not include hERG1USO transcripts as polyadenylated hERG1 variants (22). Therefore, it is important to confirm that hERG1USO transcripts are generated by polyadenylation at the 3′ end rather than internal priming.
FIGURE 1.
Sequence comparison of an EST clone (BQ478289) with hERG1 intron 9 of human genomic DNA. The poly(A) tail in the EST matched the 25-nt poly(A) stretch in hERG1 intron 9. The noncanonical poly(A) signal (AGTAAA) is underlined.
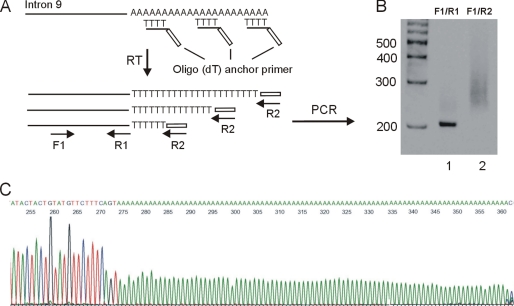
To examine the polyadenylation status of hERG1USO transcripts we performed the RACE-PAT assay outlined in Fig. 2A. Annealing of the oligo(dT) portion of the primer to the poly(A) tail can occur anywhere along the entire length of the poly(A) tail. Therefore, a heterogeneous pool of first strand cDNA, primed at all possible positions along the poly(A) tail, is synthesized after reverse transcription. Subsequent PCR amplification using a reverse primer containing the anchor sequence without oligo(dT) and a forward primer specific to hERG1USO isoforms yielded a smeared band between 250 and 350 bp (Fig. 2B, lane 2). To demonstrate that the smeared band represents hERG1USO transcripts with different lengths of the poly(A) tail, the PCR products from lane 2 were cloned into the pCRII vector and 6 clones were sequenced. All 6 clones matched the hERG1USO sequences and contained poly(A) tails with 36, 38, 75, 85, 88, and 97 adenosines (Fig. 2C). This suggests that hERG1USO transcripts are polyadenylated due to cleavage and the addition of a poly(A) tail. Although it is not known from this experiment where the cleavage site is located, the cleavage should occur somewhere along the 25-nt poly(A) stretch. Thus, up to 25 adenosines of the poly(A) tail of hERG1USO transcripts are from the genomic DNA.
FIGURE 2.
Polyadenylation analysis of hERG1USO transcripts by RACE-PAT. A, schematic outlining the strategy for analyzing poly(A) tail status by RACE-PAT. Reverse transcription using an oligo(dT) anchor primer can occur anywhere along the stretch of poly(A) tail, generating cDNA products of variable length. Primers: F1, hERG1USO specific forward primer; R1, reverse primer before the intron 9 poly(A) signal; R2, reverse primer of the anchor sequence without oligo(dT). B, polyacrylamide gel electrophoresis of PCR products. Lane 1, PCR with F1 and R1 primers. Lane 2, PCR with F1 and R2. C, sequence analysis of a representative clone that contains an 88-adenosine poly(A) tail.
Identification of an Intrinsically Weak, Noncanonical Polyadenylation Signal in hERG1 Intron 9
Although the RACE-PAT assay demonstrated that hERG1USO isoforms arise from alternative polyadenylation, the poly(A) tail of hERG1USO transcripts lacks the upstream canonical poly(A) signal AAUAAA. Examination of the hERG1 genomic intron 9 sequence reveals the presence of a noncanonical poly(A) signal (AGUAAA) at the beginning of the 25-nt poly(A) stretch and several putative U/GU-rich elements downstream of the poly(A) stretch (supplemental Fig. S1). The AGUAAA sequence has been previously shown to initiate polyadenylation in other genes (23, 24). To determine whether this intronic poly(A) signal and downstream U/GU elements can elicit polyadenylation, we performed a well described competition assay using a set of constructs with tandem poly(A) signals (25, 26). The hERG1 intron 9 poly(A) signal AGUAAA and flanking sequences (−130/+172 bp) were positioned upstream of a relatively strong synthetic poly(A) signal (Fig. 3A and supplemental Fig. S1). The tandem poly(A) signals are in competition with each other and the utilization of the intron 9 poly(A) signal relative to the synthetic poly(A) signal reflects the relative strength of the intron 9 poly(A) signal. This tandem poly(A) construct allows the experiments to be independent of transfection efficiency, as the two products are derived from the same plasmid (27).
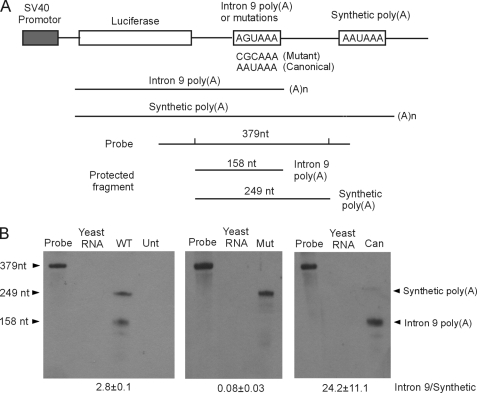
FIGURE 3.
Analysis of hERG1 intron 9 poly(A) signal. A, diagram of the tandem poly(A) signal construct including the SV40 promoter, firefly luciferase gene, hERG1 intron 9 poly(A) signal, and the synthetic poly(A) signal. The RPA probe and RNase-protected fragments corresponding to RNA terminated at the intron 9 poly(A) and the synthetic poly(A) sites are indicated. B, RPA analysis of wild-type (WT), mutant (Mut), and canonical (Can) hERG1 intron 9 poly(A) signals. Unt, untransfected control. Quantifications of the ratio of the intron 9 and synthetic poly(A) signals are shown as the mean ± S.E. from three independent experiments.
The RNA derived from the tandem poly(A) construct was studied by RPA using a probe specific to 249 nt of hERG1 intron 9. Two possible fragments, 158 and 249 nt, can be generated after RNase digestion depending on which poly(A) signal is utilized (Fig. 3A). The total length of the probe was 379 nt and contained sequences from the pCRII vector at both ends. RPA analysis of the tandem poly(A) construct showed that both the hERG1 intron 9 poly(A) signal and the synthetic poly(A) signal were used (Fig. 3B, left panel). The ratio of intron 9 to the synthetic (intron 9/synthetic) poly(A) signal usage was 2.8 ± 0.1 (n = 3). To further define a functional role of this noncanonical poly(A) signal we mutated AGUAAA to CGCAAA and performed RPA analysis using a probe containing the same mutation. The results showed that transcription was terminated predominantly at the synthetic poly(A) signal (Fig. 3B, middle panel). These results suggest that the poly(A) signal AGUAAA in hERG1 intron 9 contributes to the formation of poly(A) tail of hERG1USO transcripts.
Previous studies have shown that the noncanonical poly(A) signal AGUAAA is less efficient than canonical poly(A) signal AAUAAA (23). To test whether the hERG1 intron 9 noncanonical poly(A) signal is weak, we mutated AGUAAA to the canonical poly(A) sequence AAUAAA. We performed RPA analysis using a probe containing the AAUAAA mutation and found that the 158-nt protected fragment was predominantly generated from the intron 9 poly(A) signal (Fig. 3B, right panel). These results strongly suggest that the poly(A) signal in hERG1 intron 9 is intrinsically weak due to the presence of the noncanonical hexamer AGUAAA.
Design of a Minigene System to Study the Alternative Processing of hERG1
The presence of a poly(A) signal in hERG1 intron 9 suggests that this intron is alternatively processed during hERG1 pre-mRNA maturation. To study the alternative processing of hERG1 pre-mRNA to hERG1a and hERG1aUSO isoforms we constructed a minigene that contained the hERG1a cDNA from exon 1 to exon 6 and the genomic sequence spanning intron 6 to the poly(A) signal of the hERG1 gene (Fig. 4A). If intron 9 is spliced out and the poly(A) site in exon 15 is used, the full-length hERG1a transcript will be produced. If the minigene transcript is polyadenylated at the intron 9 poly(A) site, the hERG1aUSO transcript will be generated. We transfected the minigene into Flp-In HEK293 cells and analyzed the expressed mRNA by RPA. We designed a probe to span regions of exons 9 and 10. This probe protects a 309-nt fragment of the hERG1a transcript and a 210-nt fragment of the hERG1aUSO transcript. The length of the probe was 440 nt and contained sequences from the pCRII vector at both ends. As shown in Fig. 4B, both the 309- and 210-nt fragments were observed in the minigene RPA experiments. The hERG1aUSO to hERG1a (hERG1aUSO/hERG1a) expression ratio was 1.9 ± 0.1 (n = 3). To confirm the validity of this result, a second probe was designed, which contains hERG1aUSO sequences from exon 9 to intron 9. A similar hERG1aUSO/hERG1a expression ratio (2.1 ± 0.1, n = 3) was obtained in RPA analysis (supplemental Fig. S2). These results demonstrate that the hERG1 minigene expressed in HEK293 cells can be alternatively processed to form hERG1a and hERG1aUSO transcripts.
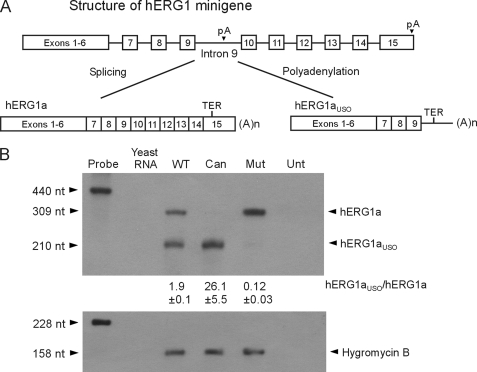
FIGURE 4.
Effect of poly(A) signal mutations on the alternative processing of hERG1 mRNA. A, diagram of the hERG1 minigene structure. Splicing intron 9 generates the full-length hERG1a isoform. Polyadenylation at the noncanonical poly(A) site in intron 9 generates the truncated hERG1aUSO isoform. B, RPA analysis of the minigenes carrying the wild-type (WT), canonical (Can), and mutant (Mut) intron 9 poly(A) signals. Unt, untransfected control. The expression level of the hygromycin B resistance gene served as the loading control. Quantifications of the hERG1aUSO to hERG1a ratio are shown as the mean ± S.E. from three independent experiments.
To determine whether the minigene can produce hERG1a and hERG1aUSO proteins, we performed immunoblot analysis using an antibody that recognizes both hERG1a and hERG1aUSO isoforms. As shown in supplemental Fig. S3A, the hERG1 minigene expressed three protein bands at about 155, 135, and 100 kDa, consistent with the sizes of hERG1a and hERG1aUSO proteins expressed from cDNA constructs (supplemental Fig. S3A, lanes 1, 4, and 5). The 155-kDa band represents the complex-glycosylated mature form of the hERG1a protein, the 135-kDa band represents the core-glycosylated immature form of the hERG1a protein (20), and the 100-kDa band represents the core-glycosylated form of the hERG1aUSO protein (11). The protein expression ratio of hERG1aUSO to hERG1a in the minigene was 1.2 ± 0.2 (n = 4), which is lower than the expression ratio of hERG1aUSO/hERG1a transcripts. This is likely due to degradation of the hERG1aUSO protein by the proteasome pathway (11).
The Relative Strength of the Intron 9 Poly(A) Signal Alters the Ratio of hERG1a to hERG1aUSO Transcripts and Proteins
To study how the strength of the poly(A) signal affects the alternative processing of hERG1 mRNA, we performed mutagenesis in the context of the hERG1 minigene. When the intron 9 poly(A) signal AGUAAA was changed to the canonical poly(A) signal, AAUAAA, the expression of hERG1aUSO was increased at both mRNA and protein levels, whereas the expression of hERG1a was almost completely inhibited (Fig. 4B and supplemental Fig. S3A, lane 2). In contrast, mutation of the intron 9 poly(A) signal from AGUAAA to CGCAAA resulted in a significant decrease in the levels of hERG1aUSO mRNA and protein, which was accompanied by a reciprocal increase in the expression of hERG1a mRNA and protein (Fig. 4B and supplemental Fig. S3A, lane 3). These results indicate that the strength of the intron 9 poly(A) signal plays an important role in determining the relative expression of hERG1a and hERG1aUSO. A strong poly(A) signal results in the predominant production of hERG1aUSO, whereas the elimination of the poly(A) signal increases the efficiency of splicing from exon 9 to exon 10, leading to the predominant expression of hERG1a.
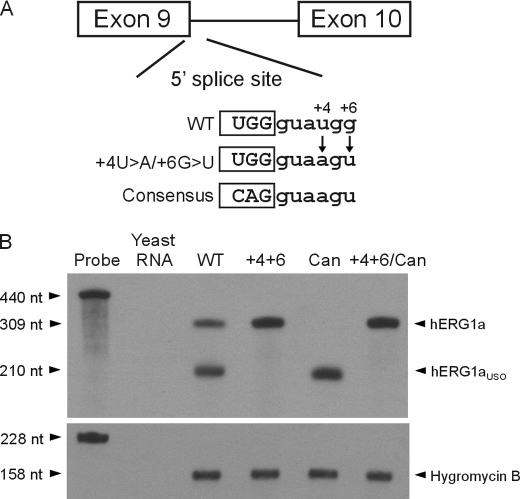
Weak 5′ Splice Site in Intron 9 Contributes to the Regulation of Alternative Processing of hERG1 Pre-mRNA
To investigate the role of intron 9 splicing in the alternative processing of hERG1 pre-mRNA, we examined the 5′ splice site of intron 9. Compared with the consensus 5′ splice site sequence (CAG/gtaagt), the 5′ splice site of hERG1 intron 9 (TGG/gtatgg) deviates at four positions (Fig. 5A). We hypothesized that the deviation of the 5′ splice site from the consensus accounts for the low efficiency of intron 9 splicing. To test this possibility, we increased the strength of this site by mutating the nucleotides at the highly conserved +4 and +6 positions toward the consensus sequence in the hERG1 minigene. When the +4U>A and +6G>U mutations were introduced into the hERG1 minigene, hERG1a was exclusively produced at the mRNA and protein levels (Fig. 5B and supplemental Fig. S3B, lane 2). This result indicates that the 5′ splice site of hERG1 intron 9 is intrinsically weak, and suggests that the presence of a strong 5′ splice site would preclude the formation of hERG1aUSO.
FIGURE 5.
Effect of 5′ splice site mutations on the alternative processing of hERG1 mRNA. A, schematic representation of the 5′ splice site sequence and mutations. B, RPA analysis of the minigenes carrying the wild-type (WT), 5′ splice site mutation +4U>A/+6G>U (+4+6), canonical (Can) intron 9 poly(A) signal, and +4U>A/+6G>U plus canonical poly(A) signal mutation (+4+6/Can). The expression level of the hygromycin B resistance gene served as the loading control. Results shown are representative of three independent experiments.
The above experiments demonstrate that both the 5′ splicing and poly(A) signals of hERG1 intron 9 are weak. Increasing the strength of the poly(A) signal results in the predominant production of hERG1aUSO, whereas increasing the strength of the 5′ splicing signal leads to the predominant production of hERG1a. To further study the competition between these two reactions, we introduced the +4U>A and +6G>U mutations into the hERG1 minigene containing the canonical poly(A) signal. This change resulted in the reversal of the predominant production of hERG1aUSO to the predominant production of hERG1a at the mRNA and protein levels (Fig. 5B and supplemental Fig. S3B, lanes 4 and 5). These results indicate that a strong 5′ splicing signal can outcompete not only the noncanonical poly(A) signal in intron 9 but also a poly(A) signal with the canonical sequence. Thus, the relative strengths of 5′ splice site and the poly(A) signal in intron 9 are important for alternative processing of hERG1 pre-mRNA.
Alternative Processing of hERG1 Pre-mRNA Regulates hERG1 Channel Function
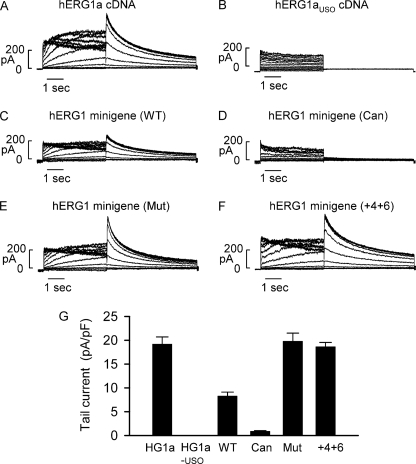
To study functional properties of hERG1 channels expressed from the minigene constructs, we performed patch clamp experiments. As controls, we first studied the hERG1 currents generated by hERG1a and hERG1aUSO cDNA. hERG1 current was activated by depolarizing steps between −70 and 50 mV from a holding potential of −80 mV. The hERG1 tail current was recorded following repolarization to −50 mV. Similar to previous reports (10, 11, 20), hERG1a cDNA produced a typical hERG1 current (Fig. 6A), whereas cells transfected with hERG1aUSO cDNA did not express the hERG1 current (Fig. 6B). The hERG1 minigene produced the hERG1 current, but the current amplitude was reduced compared with the hERG1a cDNA (Fig. 6C). The tail current amplitudes for hERG1a cDNA and the minigene were 19.2 ± 1.5 (n = 8) and 8.3 ± 0.8 pA/pF (n = 13, p < 0.05), respectively (Fig. 6G). The reduced current amplitude in the hERG1 minigene is consistent with the RPA experiments, in which only about one-third of minigene transcripts were processed to the functional hERG1a isoform.
FIGURE 6.
Voltage clamp recordings of hERG1 currents from minigenes and cDNAs expressed in HEK293 cells. A and B, representative currents recorded from cells transfected with hERG1a or hERG1aUSO cDNA. C–F, representative currents recorded from cells transfected with the minigenes carrying the wild-type (WT), canonical (Can), and mutant (Mut) intron 9 poly(A) signals, and 5′ splice site mutation +4U>A/+6G>U (+4+6). G, histogram showing the averaged tail current density measured at −50 mV following a 4-s depolarizing pulse to 20 mV for hERG1a cDNA (n = 8), hERG1aUSO cDNA (n = 9), wild-type minigene (n = 13), minigene with canonical intron 9 poly(A) signal (n = 11), minigene with mutant intron 9 poly(A) signal (n = 12), and minigene with 5′ splice site mutation +4U>A/+6G>U (n = 8).
To study if hERG1 channel function is modulated by the relative expression of hERG1a and hERG1aUSO we recorded current from cells transfected with minigenes containing mutations that alter the strength of the 5′ splice site and intron 9 poly(A) signal. hERG1 current was nearly abolished by the AGUAAA to AAUAAA poly(A) signal mutation (Fig. 6D), whereas the hERG1 current was significantly increased in the AGUAAA to CGCAAA poly(A) signal mutation (Fig. 6E). The tail current amplitudes for AAUAAA and CGCAAA mutations were 0.9 ± 0.1 pA/pF (n = 11, p < 0.05 compared with the wild-type minigene) and 19.8 ± 1.7 pA/pF (n = 12, p < 0.05 compared with the wild-type minigene), respectively (Fig. 6G). The 5′ splice site mutations +4U>A/+6G>U increased hERG1 tail current amplitude (18.6 ± 0.9 pA/pF, n = 8, p < 0.05 compared with the wild-type minigene) (Fig. 6F, G). The expression levels of hERG1 current generated from the minigene with the AGUAAA to CGCAAA and the +4U>A/+6G>U mutations were comparable with that of the hERG1a cDNA. These findings are consistent with the immunoblot experiments and suggest that changes in the relative expression of hERG1a and hERG1aUSO can modulate functional expression of hERG1 channels.
Differential Expression of hERG1 C-terminal Isoforms in Human Tissues
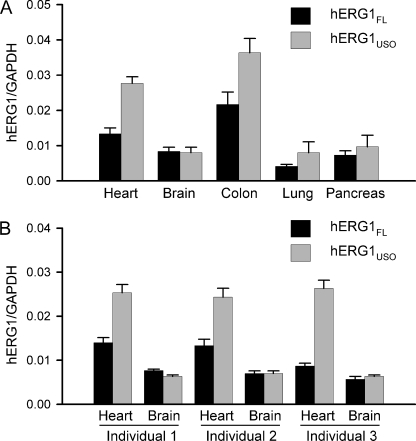
Many genes that undergo alternative RNA processing exhibit tissue-specific regulation (28–30). To determine whether alternative processing of hERG1 pre-mRNA is regulated in a tissue-specific context, we analyzed the relative expression levels of hERG1FL and hERG1USO in different tissues by real time RT-PCR. Quantitative RT-PCR revealed that the relative abundance of hERG1 C-terminal isoforms exhibited significant variation in different human tissues (Fig. 7A). The hERG1USO/hERG1FL expression ratio was highest in the heart (2.08) and lowest in the brain (0.96). To further study the hERG1USO/hERG1FL expression ratio in heart and brain, we analyzed both heart and brain RNA samples from the same individuals. In each individual the hERG1USO/hERG1FL expression ratio was higher in the heart than in the brain (Fig. 7B). The averaged hERG1USO/hERG1FL expression ratios in the heart and brain were 2.22 ± 0.41 and 0.98 ± 0.08, respectively (n = 3 individuals). These results indicate that alternative processing of hERG1 pre-mRNA is regulated in a tissue-specific manner.
FIGURE 7.
Real time RT-PCR analysis of relative expression of hERG1 C-terminal isoforms in different human tissues. Quantitative expression of hERG1FL and hERG1USO transcripts in human tissues was analyzed by real time PCR. The levels of hERG1 transcripts are normalized to the level of the GAPDH. Data are shown as the mean ± S.E. from three independent experiments. A, expression level of hERG1FL and hERG1USO in different tissues. The hERG1USO/hERG1FL expression ratios for heart, brain, colon, lung, and pancreas were 2.08, 0.96, 1.68, 1.94, and 1.31, respectively. B, expression level of hERG1FL and hERG1USO in the hearts and brains from three individuals.
DISCUSSION
Polyadenylation is essential for 3′-end formation of mRNA in eukaryotes. It involves two tightly coupled steps: the cleavage of the nascent pre-mRNA and the addition of a 3′ poly(A) tail (31, 32). The poly(A) signal is defined by the hexamer AAUAAA, or a one-base variant, found 11–23 nt upstream from the cleavage site. A downstream U- or GU-rich element, located <30 nt from the cleavage site, is also required for the formation of the poly(A) tail. Bioinformatic analyses of poly(A) signals indicate that as many as 50% of human genes contain multiple poly(A) signals and ∼20% human genes have at least one intronic poly(A) signal (30, 33, 34). The use of intronic poly(A) sites is often coupled with alternative splicing. The present findings demonstrate that hERG1 C-terminal isoforms are generated by alternative processing of a single pre-mRNA precursor by use of alternative splice or polyadenylation sites. hERG1a is produced by splicing from exon 9 to 10 and use of a poly(A) signal in exon 15, whereas hERG1aUSO is generated by the silencing of the intron 9 5′ splice site and the activation of a poly(A) site in intron 9. The poly(A) signal within intron 9 of the hERG1 gene consists of a noncanonical hexamer, AGUAAA. Although the canonical hexamer AAUAAA is the most widely used poly(A) signal, the AGUAAA sequence has been reported to occur with a frequency of 5–8% in human genes (24). Furthermore, several putative U/GU-rich elements are present downstream of the AGUAAA poly(A) signal (supplemental Fig. S1). The poly(A) competition assay shows that the AGUAAA and surrounding sequences in hERG1 intron 9 are capable of eliciting polyadenylation in a tandem poly(A) signal construct. Mutational analysis confirmed the identity of the AGUAAA hexamer as a relatively weak, poly(A) signal in intron 9 of the hERG1 gene.
Although the intron 9 poly(A) signal is weak, approximately two-thirds of hERG1 pre-mRNA is processed to hERG1USO in the heart. This is due to the presence of a weak 5′ splice site in intron 9. When the 5′ splice site of intron 9 is mutated toward the consensus sequence, the splicing reaction dominates over polyadenylation, leading to the predominant expression of hERG1a. In contrast, when the noncanonical intron 9 poly(A) signal is changed to the strong, canonical poly(A) signal, polyadenylation becomes the dominant reaction, leading to predominant expression of hERG1aUSO. These results suggest that the relative strengths of the 5′ splice site and poly(A) signal of intron 9 contributes to the alternative processing of hERG1 pre-mRNA. This notion is also supported by the finding that elimination of the intron 9 poly(A) signal by the AGUAAA to CGCAAA mutation results in predominant expression of hERG1a. Thus, the inefficient splicing observed in hERG1 pre-mRNA in the minigene experiment is due to the presence of the competing poly(A) signal in intron 9. These findings suggest that the proper balance between splicing and polyadenylation efficiencies, mediated by intrinsically weak splicing and poly(A) signals, is important for the alternative processing of hERG1 mRNA and the relative expression of hERG1a and hERG1aUSO isoforms.
Our results show that the relative abundance of hERG1 C-terminal isoforms varies in different tissues, suggesting that alternative processing of hERG1 pre-mRNA is regulated in a tissue-specific manner. Regulation of gene expression by alternative processing of pre-mRNA has been reported in many genes (28–30, 35, 36). A well studied example is the regulation of the IgM heavy chain gene during B cell differentiation (28, 36). In developing B cells, a membrane-bound form of IgM heavy chain is produced through splicing of intron 4 and the utilization of the poly(A) signal in exon 6. In mature plasma cells, a relatively weak intronic polyadenylation site within intron 4 is used, leading to the predominant production of the secreted form of IgM heavy chain. This regulation depends on the competition between splicing and polyadenylation. The ratio of membrane-bound to secreted forms of IgM mRNA is regulated by the relative efficiencies of these two reactions (36). Our findings suggest that a similar mechanism may contribute to the generation of hERG1 C-terminal isoforms. Altering splicing and polyadenylation efficiencies by mutating the splicing and poly(A) signals in hERG1 intron 9 is sufficient to shift the alternative processing of hERG1 pre-mRNA toward the hERG1a or hERG1aUSO pathway. It is conceivable that the efficiencies of splicing and polyadenylation of hERG1 intron 9 vary in different tissues, leading to the observed differential expression of hERG1 C-terminal isoforms.
Functional studies show that changes in the relative expression ratio of hERG1a and hERG1aUSO modulate hERG1 channel function. Although the hERG1aUSO isoform does not form functional channels, changes in the expression level of hERG1aUSO may regulate hERG1 channel function through post-transcriptional and post-translational mechanisms. Because hERG1a and hERG1aUSO are produced from alternative processing of a single pre-mRNA precursor, an increase in hERG1aUSO would limit the production of full-length hERG1a, whereas a decrease in hERG1aUSO would allow a given pre-mRNA transcript to be processed to the functional hERG1a isoform. In our minigene experiments, a decrease or increase in hERG1aUSO expression induced by 5′ splice site and poly(A) signal mutations is always accompanied by a reciprocal increase or decrease in hERG1a. These findings suggest that alternative processing of hERG1 pre-mRNA represents a novel post-transcriptional mechanism that regulates the expression level of the hERG1a isoform, and thereby hERG1 channel function. Alternative processing of hERG1 pre-mRNA may also regulate hERG1 channel function through a post-translational mechanism as recently proposed by Guasti et al. (11), who showed that the hERG1USO isoforms coassemble with hERG1a/b and inhibit hERG1 channel function. Thus, differential expression of hERG1 C-terminal isoforms mediated by the alternative processing of hERG1 pre-mRNA may play an important role in modulating hERG1 channel function and electrophysiological properties in different tissues.
In summary, our results demonstrate that hERG1 C-terminal isoforms are generated by alternative splicing and polyadenylation of hERG1 intron 9. The alternative processing of hERG1 pre-mRNA is regulated in a tissue-specific manner. This regulation depends on the relative efficiencies of RNA splicing and polyadenylation events. The alternative processing of hERG1 pre-mRNA may represent a novel post-transcriptional mechanism to modulate the expression and function of hERG1 channels.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL-68854 (to Z. Z.) and a Collins Trust Medical grant.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- hERG1
- human ether-a-go-go related gene 1
- RACE-PAT
- rapid amplification of cDNA end-poly(A) test
- EST
- expression sequence tag
- RPA
- RNase protection assay
- nt
- nucleotide(s).
REFERENCES
- 1.Warmke J. W., Ganetzky B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 3.Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A. (1996) Science 272, 1087c. [DOI] [PubMed] [Google Scholar]
- 4.Tseng G. N. (2001) J. Mol. Cell Cardiol. 33, 835–849 [DOI] [PubMed] [Google Scholar]
- 5.Curran M. E., Splawski I., Timothy K. W., Vincent G. M., Green E. D., Keating M. T. (1995) Cell 80, 795–803 [DOI] [PubMed] [Google Scholar]
- 6.Larsen A. P. (2010) Pflugers Arch. 460, 803–812 [DOI] [PubMed] [Google Scholar]
- 7.Lees-Miller J. P., Kondo C., Wang L., Duff H. J. (1997) Circ. Res. 81, 719–726 [DOI] [PubMed] [Google Scholar]
- 8.London B., Trudeau M. C., Newton K. P., Beyer A. K., Copeland N. G., Gilbert D. J., Jenkins N. A., Satler C. A., Robertson G. A. (1997) Circ. Res. 81, 870–878 [DOI] [PubMed] [Google Scholar]
- 9.Huffaker S. J., Chen J., Nicodemus K. K., Sambataro F., Yang F., Mattay V., Lipska B. K., Hyde T. M., Song J., Rujescu D., Giegling I., Mayilyan K., Proust M. J., Soghoyan A., Caforio G., Callicott J. H., Bertolino A., Meyer-Lindenberg A., Chang J., Ji Y., Egan M. F., Goldberg T. E., Kleinman J. E., Lu B., Weinberger D. R. (2009) Nat. Med. 15, 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupershmidt S., Snyders D. J., Raes A., Roden D. M. (1998) J. Biol. Chem. 273, 27231–27235 [DOI] [PubMed] [Google Scholar]
- 11.Guasti L., Crociani O., Redaelli E., Pillozzi S., Polvani S., Masselli M., Mello T., Galli A., Amedei A., Wymore R. S., Wanke E., Arcangeli A. (2008) Mol. Cell. Biol. 28, 5043–5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crociani O., Guasti L., Balzi M., Becchetti A., Wanke E., Olivotto M., Wymore R. S., Arcangeli A. (2003) J. Biol. Chem. 278, 2947–2955 [DOI] [PubMed] [Google Scholar]
- 13.Luo X., Xiao J., Lin H., Lu Y., Yang B., Wang Z. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1371–1380 [DOI] [PubMed] [Google Scholar]
- 14.Jones E. M., Roti Roti E. C., Wang J., Delfosse S. A., Robertson G. A. (2004) J. Biol. Chem. 279, 44690–44694 [DOI] [PubMed] [Google Scholar]
- 15.Aydar E., Palmer C. (2006) J. Membr. Biol. 211, 115–126 [DOI] [PubMed] [Google Scholar]
- 16.Sallés F. J., Strickland S. (1999) Methods Mol. Biol. 118, 441–448 [DOI] [PubMed] [Google Scholar]
- 17.Gong Q., Zhang L., Moss A. J., Vincent G. M., Ackerman M. J., Robinson J. C., Jones M. A., Tester D. J., Zhou Z. (2008) J. Mol. Cell Cardiol. 44, 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenbroucke I. I., Vandesompele J., Paepe A. D., Messiaen L. (2001) Nucleic Acids Res. 29, E68-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong Q., Zhang L., Vincent G. M., Horne B. D., Zhou Z. (2007) Circulation 116, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z., Gong Q., Epstein M. L., January C. T. (1998) J. Biol. Chem. 273, 21061–21066 [DOI] [PubMed] [Google Scholar]
- 21.Gritz L., Davies J. (1983) Gene 25, 179–188 [DOI] [PubMed] [Google Scholar]
- 22.Lee J. Y., Yeh I., Park J. Y., Tian B. (2007) Nucleic Acids Res. 35, D165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheets M. D., Ogg S. C., Wickens M. P. (1990) Nucleic Acids Res. 18, 5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caron H., van Schaik B., van der Mee M., Baas F., Riggins G., van Sluis P., Hermus M. C., van Asperen R., Boon K., Voûte P. A., Heisterkamp S., van Kampen A., Versteeg R. (2001) Science 291, 1289–1292 [DOI] [PubMed] [Google Scholar]
- 25.Brackenridge S., Ashe H. L., Giacca M., Proudfoot N. J. (1997) Nucleic Acids Res. 25, 2326–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall-Pogar T., Zhang H., Tian B., Lutz C. S. (2005) Nucleic Acids Res. 33, 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Putten H. H., Spaargaren-van Riel C. C., Bertina R. M., Vos H. L. (2006) J. Thromb. Haemost. 4, 2285–2287; author reply 2288–2289 [DOI] [PubMed] [Google Scholar]
- 28.Edwalds-Gilbert G., Veraldi K. L., Milcarek C. (1997) Nucleic Acids Res. 25, 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Z., Zhang H., Hague L. K., Lee J. Y., Lutz C. S., Tian B. (2006) Gene 366, 325–334 [DOI] [PubMed] [Google Scholar]
- 30.Tian B., Pan Z., Lee J. Y. (2007) Genome Res. 17, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colgan D. F., Manley J. L. (1997) Genes Dev. 11, 2755–2766 [DOI] [PubMed] [Google Scholar]
- 32.Zhao J., Hyman L., Moore C. (1999) Microbiol. Mol. Biol. Rev. 63, 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iseli C., Stevenson B. J., de Souza S. J., Samaia H. B., Camargo A. A., Buetow K. H., Strausberg R. L., Simpson A. J., Bucher P., Jongeneel C. V. (2002) Genome Res. 12, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian B., Hu J., Zhang H., Lutz C. S. (2005) Nucleic Acids Res. 33, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas C. P., Andrews J. I., Liu K. Z. (2007) FASEB J. 21, 3885–3895 [DOI] [PubMed] [Google Scholar]
- 36.Peterson M. L., Perry R. P. (1989) Mol. Cell. Biol. 9, 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.