Abstract
Myocyte enhancer factor 2 (MEF2) regulates specific gene expression in diverse developmental programs and adaptive responses. MEF2 recognizes DNA and interacts with transcription cofactors through a highly conserved N-terminal domain referred to as the MADS-box/MEF2 domain. Here we present the crystal structure of the MADS-box/MEF2 domain of MEF2A bound to DNA. In contrast to previous structural studies showing that the MEF2 domain of MEF2A is partially unstructured, the present study reveals that the MEF2 domain participates with the MADS-box in both dimerization and DNA binding as a single domain. The sequence divergence at and immediately following the C-terminal end of the MEF2 domain may allow different MEF2 dimers to recognize different DNA sequences in the flanking regions. The current structure also suggests that the ligand-binding pocket previously observed in the Cabin1-MEF2B-DNA complex and the HDAC9 (histone deacetylase 9)-MEF2B-DNA complex is not induced by cofactor binding but rather preformed by intrinsic folding. However, the structure of the ligand-binding pocket does undergo subtle but significant conformational changes upon cofactor binding. On the basis of these observations, we generated a homology model of MEF2 bound to a myocardin family protein, MASTR, that acts as a potent coactivator of MEF2-dependent gene expression. The model shows excellent shape and chemical complementarity at the binding interface and is consistent with existing mutagenesis data. The apo structure presented here can also serve as a target for virtual screening and soaking studies of small molecules that can modulate the function of MEF2 as research tools and therapeutic leads.
Keywords: MEF2, myocardin, MASTR, transcription co-regulator recruitment, drug development
Introduction
The myocyte enhancer factor 2 (MEF2) family of proteins was originally identified as a regulator of muscle gene expression in vertebrates;1 its function has now been expanded to diverse cellular processes in eukaryotic cells.2 In yeast Saccharomyces cerevisiae, the MEF2 homolog Rlm1 regulated specific gene expression in response to mitogen-activated protein kinase activation.3 In Drosophila, genetic analyses have demonstrated the central role of MEF2 in myogenesis.4–8 In vertebrates, where four different genes of MEF2 are encoded in the genome (mef2a, b, c, and d), MEF2 has been shown to play critical roles in controlling the differentiation, proliferation, and survival/apoptosis of a wide range of cell types including muscle, T cells, and neurons.2,9–13 In adult tissues, MEF2 also serves as a key regulator of stress responses and adaptive programs in response to environmental signals, including fiber-type switch of skeletal muscle, cardiac hypertrophy, and activity-dependent remodeling of neuronal synapses.14–19
MEF2 is emerging as a potential therapeutic target for a number of human diseases. MEF2 and its associated transcription cofactors such as class IIa histone deacetylases (HDACs) and p300 have been shown to modulate transcription programs involved in muscle metabolism and cardiac growth,16,20–22 suggesting that MEF2 modulators could be used to treat muscle diseases and cardiac hypertrophy. MEF2 regulates cytokine expression and apoptosis in T cells.9,12 Recent studies show that HDAC9 knockout leads to enhanced function of regulatory T cells.23 Since HDAC9 and MEF2 often function as a complex in vivo, these observations raise the possibility that the function of MEF2 might be modulated for treating autoimmune diseases and transplant rejection. A series of studies have established a key role of MEF2 in neuronal survival and synapses.13,17,18,24,25 These findings suggest that MEF2 could be targeted for treating neurodegenerative diseases such as Alzheimer disease, Huntington disease, and other psychiatric disorders such as autism, schizophrenia, and drug addiction. Finally, there is increasing evidence linking MEF2 to cancer development, pointing to a potential role of MEF2 as an oncogene.26,27 MEF2 is highly expressed in certain leukemias and is apparently required for the self-renewal of the leukemia stem cell.28 Moreover, MEF2 activity is elevated through chromosome translocation and gene fusion in some leukemia cell lines.29,30 These results suggest that inhibition of the function of MEF2 in the contexts of tumor cells could be a potential approach to cancer therapy. To develop small molecules that bind MEF2 and modulate its transcription function specifically, it is important to analyze systematically the structure and function of MEF2 and its various complexes in detail.
The MEF2 family of proteins (MEF2A-D) of vertebrates share a highly conserved N-terminal region (residues 1–93) followed by a more divergent C-terminal region. The N-terminal region contains the DNA-binding domain, the MADS-box (residues 1–58), which is named after MCM1, Agamous, Deficiens, and SRF.31 The sequence immediately following the MADS-box (residues 59–93) is unique to the MEF2 family and is thus called the MEF2-specific domain (MEF2 domain). MEF2 recognizes a core consensus DNA sequence of YTA(A/T)4TAR (Y, pyrimidine; R, purine).32–34 MEF2 expressed in different tissues appear to have distinct preferences for sequences flanking the core region, but how such selectivity is achieved is not clear.32 The activity of MEF2 is modulated by posttranslational modifications such as phosphorylation, sumoylation, and acetylation, as well as interactions with a variety of other proteins. These proteins include signaling molecules such as Erk5,35 transcription factors such as MyoD,36 NFAT,14 and Smad3,37 transcriptional corepressors such as Cabin1,9,10 HDAC338,39 and class IIa HDACs,40–42 and transcriptional coactivators such as p300 and myocardin.22,43–47 Most of these MEF2-interacting proteins have been shown to bind to the MADS-box/MEF2 region.
Given the important roles of the MADS-box/MEF2 domain in DNA-binding and protein-protein interactions, substantial effort has been put into characterizing the structure and function of this domain. The structure of the MADS-box/MEF2 domain of MEF2A bound to DNA has been characterized by X-ray crystallography and NMR.48,49 These studies revealed that the MADS-box of MEF2A is similar to that of SRF and MCM1 but also with some key differences. In the crystal structure,48 the C-terminal half of the MEF2 domain (residues 79–86) was truncated off. This region was included in the NMR study but was shown to be disordered.49 The crystal structures of two ternary MEF2 transcription factor complexes, the Cabin1-MEF2B-DNA complex and the HDAC9-MEF2-DNA complex, have also been characterized.50,51 These studies reveal a general corepressor recruiting mechanism by MEF2 wherein an amphipathic helix from Cabin1 and class IIa HDACs bind to a concave hydrophobic groove on the MADS-box/MEF2 domain.
Based on the systematic structural and biochemical studies of MEF2 complexes,50,51 it is possible to analyze if an MEF2-interacting protein uses a similar binding mechanism described above. This can be achieved by first identifying if the protein of interest has a short amphipathic helix that matches with consensus MEF2-binding motif and then docking the helix into the hydrophobic groove of MEF2. Such analysis will help to interpret existing data and guide functional studies of a variety of MEF2 complexes. However, since the C-terminal half of the MEF2 domain is not observed in the previous crystal structure and is disordered in the NMR study,48,49 it is not clear if the structure of the hydrophobic groove observed in the Cabin1-MEF2B-DNA complex and the HDAC9-MEF2B-DNA complex is induced by the corepressor binding. In the present study, we have solved the crystal structure of the MADS-box/MEF2 domain of human MEF2A bound to DNA in the absence of any cofactor. Our study shows that the entire MADS-box/MEF2 domain of MEF2A is stably folded. The structure of the intact MEF2 domain (residues 59–93) is stabilized by extensive interactions with the MADS-box and DNA. The hydrophobic groove is preformed in the “apo” structure of the MEF2A-DNA complex and is therefore poised to bind transcription factor partners. Using this structure, we analyzed a number of proteins that have been shown to interact with MEF2 physically using the docking approach described. We show that a short amphipathic helix in Myocardin and MASTR, which have been shown to activate MEF2-depdent gene expression and binds to the MADS-box/MEF2 domain,46,47 fits perfectly into the hydrophobic groove on MEF2. The structure of the MEF2-DNA complex presented here will facilitate similar computer-based analyses of other MEF2 interacting proteins. This structure can also be used for virtual screen and soaking studies of small molecules that can bind MEF2 and modulate its transcription function inside cells.
Results
Crystal structure of MEF2A (2–95) bound to DNA
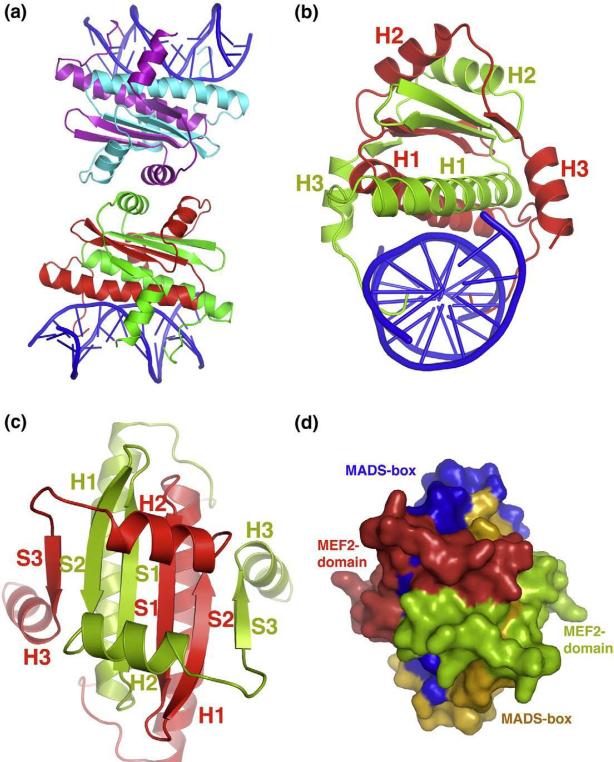
We have crystallized the MADS-box/MEF2 domain of human MEF2A (residues 2–95, the first Met is cleaved off) bound to a double-stranded DNA containing a consensus MEF2-binding site (CTATTTATAA) (Fig. 1). The crystals belong to space group P21212 (a = 77.80 Å, b = 77.96 Å, c = 106.79 Å) and diffract to 2.87 Å. The structure was solved by molecular replacement using the MEF2A (2–78)- DNA complex as a partial search model.48 The final model was refined to an R-factor of 22.06% (Rfree, 27.86%) with excellent geometry (Table 1). The asymmetric unit of the crystals contains two independent MEF2A (2–91; residues 92–95 show no density) complexes bound to DNA that stack head-to-head with each other (Fig. 1a). Interestingly, helix H2 of each complex seems to mimic the amphipathic helix in Cabin1 and HDAC9 to bind the reciprocal complex, but the crystal packing interaction observed here is much more limited than that seen in the Cabin1-MEF2 ad HDAC9- MEF2 complex. Compared with the helix of Cabin1 and HDAC9, helix H2 is further away from its MEF2 binding partner and shows only sparse contacts (data not shown).50,51 We also checked the oligomerization state of MEF2 at various concentrations using multiangle light scattering and found that MEF2 (2–95) exists invariably as a dimer, suggesting that the packing interaction mediated by helix H2 is unstable in solution. Although we cannot completely rule out crystal packing effects on the apo structure of MEF2 observed here, the fact that helix H2 interacts with MEF2 very differently from the corresponding helix of Cabin1 and HDAC9 suggests that the crystal packing interactions had little effect on the structure of the ligand-binding site. The structure of each MEF2 monomer consists of an extended N-terminal tail, three helices (H1, H2, and H3) and three beta strands (β1, β2, and β3). A pair of MEF2 monomers forms an intertwined dimer (Fig. 1b and c). Although the N-terminal tail, helix H1, and strands β1 and β2 have been traditionally described as the MADS-box, our crystal structure reveals that the MADS-box and the MEF2 domain of MEF2A form an intimately folded structure instead of two separated domains (Fig. 1d). The MEF2 dimer has a six-strand beta sheet that is sandwiched by a pair of helices above and below (Fig. 1b and c). The three-layer sandwich structure is clamped from both sides by helix H3 from the MEF2 domain. While the central beta sheet serves as the core for protein folding, the surrounding helices appear to function as the structure elements for DNA binding and protein-protein interactions. The N-terminal tail and helix H1 form the major DNA binding surface; the C-terminal end of helix H3 is also poised to interact with DNA. The two helices (H2) above the beta sheet form the binding site for Cabin1 and HDAC9 and possibly other proteins. As discussed before, helix H3 and strand β3 may also serve as the binding sites of some MEF2-interacting proteins such as MyoD.51 The structure represents the first high-resolution structure of the intact MADS-box/MEF2 domain of MEF2 bound to DNA in the absence of a cofactor. This structure will serve as a key reference for analyzing higher-order MEF2 complexes and for computational docking of potential MEF2-binding proteins and virtual screen of small MEF2 modulators.
Fig. 1.
Overall structure and packing of MEF2 dimer bound to DNA. (a) Asymmetric unit containing two independent MEF2–DNA complexes stacking head-to-head is shown in cartoon diagram. Two monomers in one complex are colored in green and red, respectively, while in the other complex they are colored in cyan and violet, respectively. DNA is colored in blue throughout the illustration. (b) MEF2–DNA complex forming an intertwined dimer is shown in cartoon diagram along the DNA axis. One monomer is colored in red and the other in green. All three α helices for both the monomers are labeled in corresponding colors. (c) Top view of the same complex shown in (b). Here, all three β sheets along with the three α helices of both the monomers are labeled in corresponding colors. DNA is omitted in this view for clarity. (d) Surface representation with the same orientation as shown in (c). Here, MADS-box and MEF2 domain are colored in orange and red in one monomer, respectively. Corresponding colors in the other monomer are blue and green, respectively. This representation shows that the MADS-box and MEF2 domain of the two monomers form an intimately folded domain.
Table 1.
Statistics of crystallographic analysis
| Data set | |
|---|---|
| Resolution (Å) | 50−2.87 (last bin 2.97−2.87 Å) |
| R sym a | 0.052 (0.585) |
| Completeness (%)b | 99.6 (99.5) |
| I/σ(I) | 43.7(4.5) |
| Redundancy | 7.0(6.9) |
| Refinement | |
| Resolution (Å) | 44.06−2.90 (last bin 3.12−2.90 Å) |
| R-factor (%)c,d | 0.2207 (0.3017) |
| Rfree (%)c,d | 0.2786 (0.3735) |
| r.m.s.d.'s | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.227 |
| Average B-factor (Å2) | 85.46 |
Rsym=Σ|I−〈I〉|/ΣI where I is the observed intensity and 〈I〉 is the statistically weighted average intensity of multiple observations of symmetry-related reflections.
The number in parentheses is for the outer shell (last bin).
Rwork=Σ||Fo|−|Fc||/Σ| Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
Rfree is calculated for 5% of the data that were withheld from refinement.
Comparison with previous structures
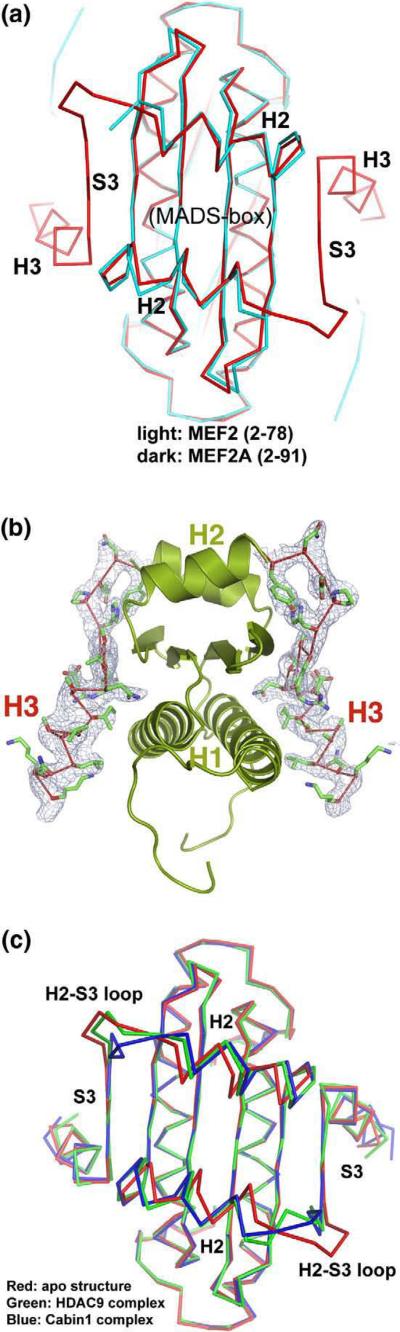
Superposition of the structure of MEF2A (2–91) with that of the MEF2A (2–78) reveals that the MADS-box (residues 2–58) and the first half of the MEF2 domain are very similar between the two complexes (r.m.s.d. of 0.50 Å between 142 Cα atoms) (Fig. 2a). This portion of the MEF2A structure is therefore not affected by the presence or absence of the C-terminal half of the MEF2 domain (strand β3 and helix H3). On the other hand, the MEF2 domain (helix H2, strand β3, and helix H3) is unlikely to form the observed structure in the absence of the MADS-box because these structural elements form an extended structure on the surface of the MADS-box. In the MEF2A (2–91)-DNA complex, the electron density for the entire MEF2 domain is well defined, even though the initial search model only contained the MADS-box (Fig. 2b). In deletion studies to identify the binding region of a given MEF2-interacting protein, loss of binding as a result of disruption of the MADS-box does not necessarily suggest that the MADS-box is the direct binding site. This is because the protein surface of the MADS-box is covered almost entirely by DNA and the MEF2 domain.52 A more likely interpretation is that the MEF2 domain, which depends on the MADS-box for stable folding, is the actual binding site of the MEF2 partners.52
Fig. 2.
Comparison with the previous crystal structures. (a) Cα superposition of the present crystal structure of MEF2A (2–91, red) on that of MEF2A (2–78, cyan) showing the identical core structure of the MADS-box (residues 2–58) and helix H2 and the additional structure of strand S3 and H3 observed here. (b) Side view of the C-terminal region of the MEF2 domain (residues 79–91, stick model) and its corresponding electron density (sigma-A weighted 2Fo – Fc density at 1 e/l̊3). The rest of the MEF2 structure is displayed in cartoon diagram. The DNA is not shown. (c) Superposition of the Cα backbone of MEF2 in the present crystal structure (red) with that in the HDAC9–MEF2B–DNA complex (green) and Cabin1–MEF2B–DNA complex (blue). This view is in the same orientation as Fig. 1c.
The present study suggests that the entire MADS-box/MEF2 domain, including the C-terminal half of the MEF2 domain, can form a stable folded structure upon DNA binding. This finding is in contrast to previous NMR analysis, which showed a disordered region in the C-terminal half of the MEF2 domain (strand β3 and helix H3).49 The four MEF2 molecules in the asymmetric unit assume nearly identical structures despite being in different crystal environments, suggesting that the observed structure of MEF2A (2–91) is not induced by crystal packing. We have previously observed an entirely folded structure of the MADS-box/MEF2 domain of MEF2B in the Cabin1-MEF2B-DNA complex and the HDAC9-MEF2B-DNA complex.50,51 Superposition of the present structure with the corresponding region of MEF2B in the two corepressor complexes reveals that the MADS-box/MEF2 structures are highly similar in the three complexes (r.m.s.d. of 0.984 Å between 180 Cα atoms when compared with the Cabin1 complex and r.m.s.d. of 0.884 Å between 178 Cα atoms when compared with the HDAC9 complex) (Fig. 2c). Thus, the stable structure of the MADS-box/MEF2 domain can form in the absence of corepressors and the overall structure of the MADS-box/MEF2 is largely preserved upon cofactor binding, although some differences in the ligand-binding pocket are notable (discussed further below).
Interactions that stabilize the structure of the MEF2 domain
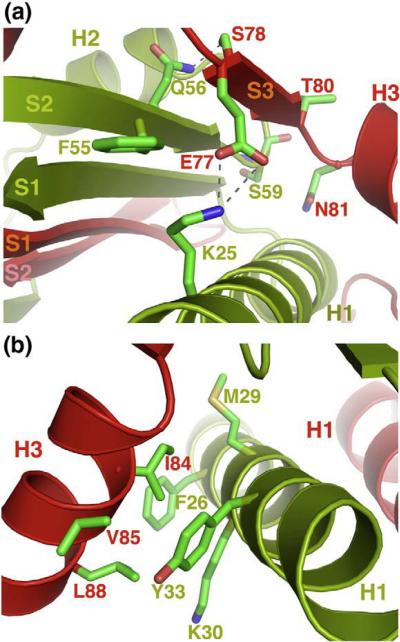
The stable structure of the entire MEF2 domain is likely a result of intrinsic folding interactions conserved in the MEF2 family of proteins. Strand β3 of one MEF2A monomer forms parallel beta-stranded interactions with β2 of its dimer partner. The binding involves extensive main-chain and side-chain contacts (Fig. 3a). For example, Glu77 of monomer A forms a salt bridge with Lys25 of monomer B; the side chain of Glu77 of monomer A also packs against Phe55 of monomer B. Ser78 of monomer A forms a hydrogen bound with Gln56 of monomer B, whereas Thr80 and Asn81 of monomer A pack against the main chain and side chain of Ser59 of monomer B. In addition to the beta sheet interaction, helix H3 of one MEF2A monomer packs perpendicularly on helix H1 of its dimer partner (Fig. 3b). Here, a patch of hydrophobic residues on helix H3 of monomer A, including Ile84, Val85, Leu88, interact with a hydrophobic surface on monomer B that is composed of Met29, Phe26, the aliphatic side chain of Lys30, and Tyr33. Most of the residues involved in the interactions are also conserved in the MEF2 family, suggesting that the folded structure of the intact MADS-box/MEF2 domain is a conserved structural feature of homo-and heterodimers of the MEF2 family of proteins.
Fig. 3.
Interactions that stabilize the structure of the MEF2 domain. (a) Interactions between β sheet S3 of one monomer (red) and β sheet S2 and helix H1 of the other (green). Similar interactions (not shown) are also observed at the other end of the hydrophobic groove. (b) Hydrophobic interactions between helix H3 of one monomer (red) and helix H1 of the other (green). Similar interactions (not shown) are also observed between helix H1 and helix H3 on the other side of the MEF2 dimer.
DNA binding by the intact MADS-box/MEF2 domain
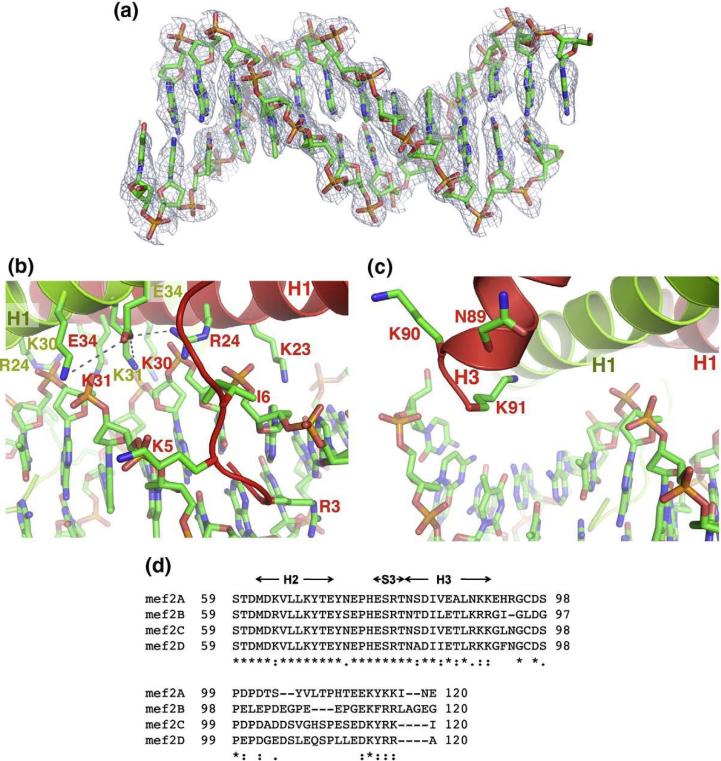
The present structure provides new insights into the DNA-binding mechanisms of MEF2. In previous structures of the MEF2A (2–78)-DNA complex, Cabin1-MEF2B-DNA complex, and the HDAC9- MEF2-DNA complex,48,50,51 the density of DNA is scrambled in certain regions because the asymmetric DNA adopts two equal orientations in the crystal lattice. While the structures could be refined using alternative conformations for DNA, the poor electron density of DNA made it difficult to interpret the detailed protein-DNA interactions. In the present crystal, the orientation of DNA in one of the two complexes in the asymmetric unit is fortuitously fixed due to asymmetric crystal packing environment. The electron density of this DNA is well defined (Fig. 4a), allowing detailed analyses of the DNA binding by the intact MADS-box/MEF2 domain of MEF2A.
Fig. 4.
DNA binding by the intact MADS-box/MEF2 domain. (a) DNA in stick model shown in sigma-A weighted 2Fo – Fc map at a contour level of 2.5 e/Å3. (b) Interactions of the N-terminal tail and helix H1 of the MADS-box with the minor groove of the DNA. (c) Interactions between the C-terminal end of the MEF2 domain of one monomer (red) with DNA. (d) Sequence alignment of MEF2A-D showing the conserved basic residues at the C-terminal of helix H3 and the divergent sequences immediately following the C-terminal end of the MEF2 domain.
The DNA is in a straight B-form conformation. MEF2A (2–95) binds DNA largely through the N-terminal tail and helix H1 of the MADS-box, which forms a clamp that grips the DNA by the minor groove (Fig. 4b). A number of residues from the N-terminus, Gly2, Arg3, Lys4, Lys5, and Ile6, insert deeply into the minor groove to interact with DNA bases and backbone sugars and phosphates. A network of positively charged residues from helix H1, including Arg24, Lys30, and Lys31 from both monomers, form extensive electrostatic interactions with the phosphate backbones bordering the minor groove at the center of the MEF2-binding site. Many of these positively charged residues are linked together by bridging interactions. For example, Glu34 of monomer A interacts with Lys30, Lys31 of monomer A, and Arg24 of monomer B. Such bridging interactions may contribute to DNA binding by restricting the flexibility of positively charged residues that contact DNA directly. A predominant feature of DNA binding by MEF2 is the recognition of the narrowed minor groove, which is consistent with the fact that MEF2-binding sites are rich in A and T in the middle region.8,24,53 The only base-specific recognition in the major grove is mediated by Lys23, which specifies the two base pairs at both ends of the MEF2-binding site [YTA(A/T)4TAR, in bold].
Overall, protein-DNA interactions observed in the MEF2A (2–95)-DNA complex are similar to those seen in the MEF2A (2–78)-DNA complex,48 suggesting that the DNA-binding interactions by the MADS-box are conserved in complexes formed by protein fragments of different lengths. However, longer fragments of MEF2 may engage in additional DNA-binding interactions through regions outside the MADS-box. In the present structure of the intact MADS-box/MEF2 domain, helix H3 of the MEF2 domain is projected toward DNA. Its C-terminus, which is rich in positively charged residues, is poised to interact with DNA (Fig. 4c). Based on the trajectory of helix H3, it is possible that sequences immediately following the MEF2 domain may also interact with DNA flanking the core MEF2 site. Interestingly, MEF2 from different tissues have been shown to bind the same core MEF2 site but display distinct preferences for particular sequences in the flanking regions.32 It is possible that different MEF2 homo- or heterodimers expressed in different tissues is at least partly responsible for such binding-site selectivity. Consistent with this hypothesis, the sequences at and immediately following the C-terminal end of the MEF2 domain are highly divergent in the MEF2 family (Fig. 4d). However, using electrophoretic mobility shift assay, we found that MEF2A (2–78) and MEF2A (2–95) bound short DNA fragments (14mer and 20mer) with similar affinities (Y.W. et al., data not shown), although we previous showed that these two fragments of MEF2 bounbinding by helix H3 may need to be examined on longer DNA substrates or in the context of chromatin. Further studies will be needed to address these questions.
Cofactor-binding site
Our previous studies of the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex identified a hydrophobic groove on the MADS-box/MEF2 domain that serves as a cofactor-binding site. This hydrophobic groove is bordered by the central beta sheet (the floor) and helix H2 from each monomer (the two rims). The present structure indicates that this cofactor-binding site is preformed in the apo state of the MEF2A–DNA complex (Fig. 2). Detailed analyses reveal that the cofactor-binding site undergoes notable structural changes upon ligand binding (Fig. 2c). Superposition of the present structure with that of the Cabin1–MEF2–DNA complex and the HDAC9–MEF2B–DNA complex reveals that helix H2, which is the major structural element interacting with cofactors, displays significant shift along the axis of the alpha helix, whereas the underlying central beta sheet is nearly identical in the three structures. While the positions of helix H2 in the Cabin1 and HDAC9 complexes are very similar, its position in the apo structure shows a large offset. In other words, upon the binding of cofactor Cabin1 and HDAC9, helices H2 from the two monomers shift toward each other along the helical axis. Such shift of helix alignment apparently allows for optimal binding of the cofactor. We have previously shown that the loop connecting helix H2 and strand β3 can undergo conformational changes to accommodate the binding of different corepressors (Cabin1 and HDAC9).50 As shown in Fig. 2c, this region of the apo structure also shows a conformation different from that seen in the Cabin1 and HDAC9 complexes. Thus, helix H2 and the flexible linker between H2 and β3 can undergo structural changes to mediate corepressor binding through induced fit.
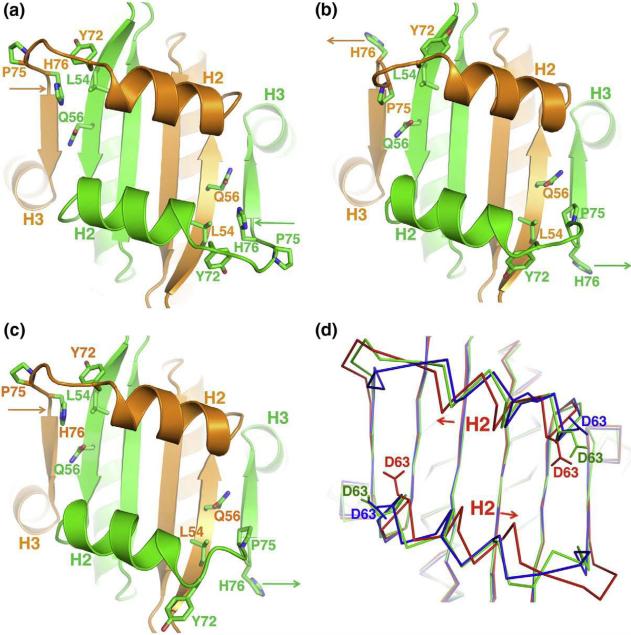
In addition to the conformational changes of protein backbone, residues in the hydrophobic groove also display different side-chain orientations in the apo structure when compared with the repressor-bound complexes. The most significant difference can be seen on His76. In the apo structure, His76 of both monomers is tucked inside the hydrophobic groove and packs against a number of residues that line the interior of the cofactor binding site (Tyr72 of the same monomer, Leu54 and Gln56 of the dimer partner) (Fig. 5a). In the Cabin1–MEF2B–DNA complex, His76 of both monomers swings outside (Fig. 5b). Although Pro75 seems to take its position in the groove, the backbone trajectory of this region is very different in the two structures. In the HDAC9–MEF2B–DNA complex, His76 of one monomer adopts the tucked-in conformation as in the apo structure, while the other adopts the outward conformation seen in the Cabin1 complex (Fig. 5c). A similar histidine-associated structural change is also observed in glucocorticoid receptors binding to different DNA sequences. Such a histidine switch is proposed to play a role in transmitting the allosteric effect of DNA binding to transcriptional activity of glucocorticoid receptors.54 Here, His76 seems to play a key role in the binding of different cofactors by MEF2. Asp63 at the N-terminus of helix H2 also displays different side-chain orientations in the apo structure and in the two-corepressor complex (Fig. 5d), suggesting that this residue also plays an important role in the induced-fit binding of the cofactors. The detailed analyses of the ligand-binding pocket described above are important for structure-based virtual screen of small molecules that bind MEF2 and modulate its function. In work to be published elsewhere, we have indeed identified such small molecules and verified that these compounds can bind MEF2 competitively with HDAC4 in vitro and modulate MEF2's function in vivo (N.J. et al., unpublished results).
Fig. 5.
Cofactor binding site. (a) Cofactor binding site of the present structure in apo MEF2A (2–91) displaying the key residues involved in binding with the cofactor. Two monomers in the intertwined dimer are colored in green and orange. The orientation of the cartoon representation is same as in Fig. 1c. (b) and (c) are similar representations for the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex, respectively. Different side-chain orientations of His76 are marked by arrows in corresponding colors of the monomers in the three representations (a), (b) and (c). (d) Superposition of the Cα of the three MEF2 structures (apo in red, HDAC9–MEF2B complex in green, and Cabin1–MEF2B complex in blue) showing different side-chain orientations for Asp63. A pair of arrows also indicates the shift of helix H2 in the apo structure with respect to the HDAC9 and Cabin1 complexes.
A model of the recruitment of myocardin and MASTR by MEF2
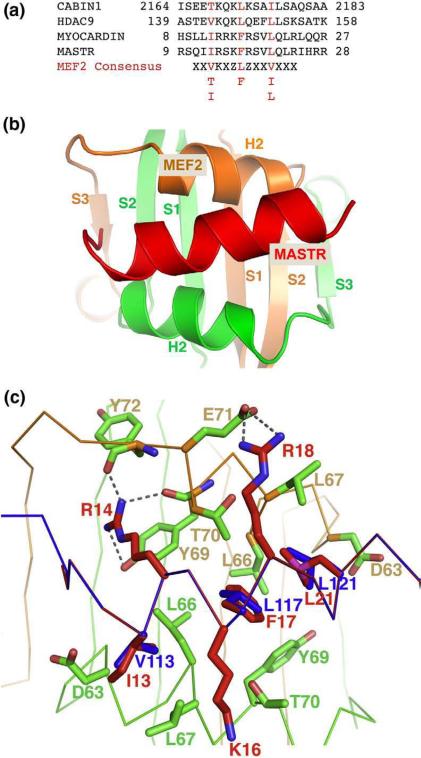
Myocardin and myocardin-related transcription factors (MRTFs) are potent activators of muscle-specific genes in cardiac and smooth muscle cells.46,47 These proteins generally do not bind DNA directly but associate with other sequence-specific transcription factors to regulate specific gene expression. One such sequence-specific transcription factor is the serum response factor (SRF) that binds myocardin through its MADS-box. Several members of the MRTF family, such as the longer splicing variant of myocardin (myocardin-935) enriched in cardiac muscle and the newly identified MASTR, have been shown to activate MEF2-dependent gene expression.46 This subset of myocardin family members shares a short peptide motif that is necessary and sufficient to bind the MADS-box/MEF2 domain. Although the MADS-box of MEF2 is highly homologous to that of SRF, its binding mechanism to myocardin seems to be distinct from that of SRF.46 As discussed above, proteins that interact with MEF2 through the MADS-box/MEF2 region most likely bind the MEF2 domain directly, since the MADS-box is largely buried, whereas the structure elements of the MEF2 domain (helix H2, strand β3, and helix H3) are exposed. The sequences of the MEF2-binding peptide in myocardin-935 and MASTR match very well with the consensus MEF2-binding motif defined by the crystal structures of the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex (Fig. 6a).50 Secondary-structure prediction also suggests that the MEF2-binding region of myocardin-935 and MASTR has a high propensity to form an amphipathic alpha helix. These observations suggest that myocardin-935 and MASTR may bind MEF2 through mechanisms similar to that of Cabin1 and HDAC9 complexes. To address this question, we used two different methods to model the interactions between myocardin-935/MASTR and MEF2. The first method is homology modeling using the program MODDLER.55 The sequence of the MEF2-binding motif of myocardin-935 or MASTR is threaded on that of Cabin1 and HDAC9 in their respective complexes (Fig. 6a). The generated models have well-matched binding interfaces; only minor adjustments of local side-chain orientations are needed to remove a few clashes. The second method is protein docking using the program ZDOCK.56 Here we first generate the helix structure of the MEF2-binding motif of myocardin-935 and MASTR and then perform flexible docking to the cofactor-binding site on MEF2. In the flexible docking approach, we take into consideration localized structural changes induced by Cabin1 and HDAC9 (see above). Models generated by different methods are similar in overall structure as well as residue-to-residue contacts, although some differences are observed. In the following, we will describe the details of the homology model of the MASTR–MEF2–DNA complex (Fig. 6b). The homology model of myocardin-935 bound to MEF2 on DNA is very similar and will be discussed briefly in comparison with that of the MASTR–MEF2–DNA complex.
Fig. 6.
Binding of MASTR to MEF2. (a) Sequence alignment of different MEF2-binding motifs from HDAC9, Cabin1, myocardin, and MASTR along with the consensus MEF2-binding motif. Shown in red are conserved hydrophobic residues that bind nonpolar pockets on MEF2 (degenerated residues are also listed below the consensus sequence). (b) The homology model of MASTR (red) bound to MEF2 (orange and green). The MEF2-binding motif of MASTR adopts a helical structure to bind the hydrophobic groove of MEF2 diagonally. (c) Detailed interactions at the modeled MASTR–MEF2 interface. Residues from MASTR are colored red and residues from MEF2 in green. The Cα backbones of MEF2 (green) and MASTR (blue) are also shown as spatial references. The key hydrophobic residues from HDAC9 that bind to MEF2 (Val143, Leu 147, and Leu151, colored in blue) are also shown for comparison. These residues and their corresponding residues in MASTR (Ile13, Phe17, and Leu21, respectively) occupy the same nonpolar pockets on the surface of MEF2.
The amphipathic helix of MASTR binds the hydrophobic groove on the MADS-box/MEF2 domain in a diagonal manner (Fig. 6b). A number of hydrophobic residues, which define the consensus features of the MEF2-binding motif (Fig. 6a), bind discrete hydrophobic pockets on MEF2. For example, Ile13 of MASTR inserts into a pocket surrounded by Leu66, Leu67, and the side chain of Asp63, whereas Leu21 of MASTR binds a similar pocket on the other side of the symmetric MEF2 dimer. At the center of the binding interface is a hydrophobic pocket formed by Leu66, Tyr69, and Thr70 from the two MEF2 monomers. In the Cabin1 and HDAC9 complexes, a leucine residue highly conserved in class IIa HDACs and Cabin1 inserts into this hydrophobic pocket. Mutation of this leucine to alanine in Cabin1, HDCA4 and HDAC9 completely abolished the binding of MEF2, demonstrating its critical role in MEF2-dependent recruitment.50,51 Here in the MASTR complex, a highly conserved phenylalanine (Phe17) occupies the equivalent position. In addition to the hydrophobic contacts, a number of hydrogen-bond and electrostatic interactions also form at the MASTR–MEF2 binding interface. Most notably, Arg14 forms a hydrogen bond with the main-chain carbonyl of Tyr72; its guanidinium group stacks on the aromatic ring of Tyr69 to form a charged pi-stacking interaction. The long aliphatic side chain of Arg14 also packs against Thr70. Similarly, the aliphatic side chains of Lys16 and Arg18 make a number of van der Waals contacts to Leu67 and Thr70 on both sides of the amphipathic helix. Arg18 also forms a salt bridge with Glu71 (Fig. 6c).
Mutations of a number of residues in myocardin-935 have been shown to disrupt the binding and activation of MEF2.46 These mutations include a triple mutant wherein Arg13, and Arg14 and Lys15, were substituted with Gln (RRK (13–15)/QQQ), a double mutant wherein Phe16 and Arg17 were replaced with Asp (FR (16–17)/DD), and another double mutant wherein Leu22 and Leu24 were replaced with Ser (LRL (22–24)/SRS). Residues Arg13, Lys15, Phe16, and Arg17 are identical between myocardin-935 and MASTR (note that residue numbering between myocardin-935 and MASTR differs by one). As shown by the homology model (Fig. 6c), these residues are located at the center of the binding interface and engage in extensive interactions with the cofactor-binding pocket of MEF2.
Discussion
The diverse roles of MEF2 in development and adaptive responses have now been well recognized, but the mechanisms by which MEF2 regulate distinct transcriptional programs in different cellular contexts remain enigmatic. A plausible model is the combinatorial mechanism wherein MEF2 interacts with different transcription factors and cofactors to regulate specific gene expression. Indeed, a variety of protein factors binds MEF2 and modulates its transcriptional activity in a tissue-specific manner. Interestingly, most of these proteins bind MEF2 at the highly conserved MADS-box/MEF2 domain located at the N-terminus. Thus, the MADS-box/MEF2 domain of MEF2 serves not only as the major DNA recognition motif but also as the binding site of a variety of transcription factors and cofactors. Despite its functional importance, the structure of the MADS-box/MEF2 domain has not been fully resolved by previous studies.48,49 In the present study, we have solved the crystal structure of the intact MADS-box/MEF2 domain of MFE2A (2–95) bound to DNA. The crystal structure reveals new insights into DNA recognition and cofactor recruitment by MEF2.
The C-terminal half of the MEF2 domain was found to be disordered in the NMR analysis of the MEF2A (2–86)–DNA complex.49 Yet we observed a fully folded MADS-box/MEF2 domain in the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex.50,51 It is possible that the folded structure of the MEF2 domain observed in the Cabin1 and HDAC9 complexes was induced by the corepressor binding and/or due to the sequence difference between MEF2A and MEF2B. Our present study ruled out these two possibilities by showing that the MEF2 domain in the MEF2A (2–95)–DNA complex can form a stably folded structure through interactions with the MADS-box. The interactions that stabilize the folded structure of the MEF2 domain are conserved in the MEF2 family, suggesting that the structure of the MADS-box/MEF2 domain observed here in the MEF2A (2–95)–DNA complex and in previous Cabin1 and HDAC9 complexes is an intrinsic property of MEF2 and is likely conserved in homo- and heterodimers formed by various MEF2 family members.
Why is the MEF2 domain folded here but partially disordered in the previous study? Different experimental techniques (crystallography versus NMR) could contribute to the difference. However, as discussed above, crystal packing is unlikely the cause of the difference, since the four copies of MEF2 in the asymmetric unit adopt the same structure despite being in different packing environments. A more likely possibility is the slightly different protein constructs used in the two studies. In the present study, we used a longer MEF2 fragment MEF2A (2–95) than that used in the NMR study, MEF2A (2–86). The extra seven residues (87–93) at the C-terminus of the MEF2 domain show well-defined electron density as an alpha helix in the MEF2A (2–95)–DNA complex (Fig. 2b). These residues could stabilize the structure of the MEF2 domain through a number of different mechanisms that are not mutually exclusive. First, these residues could stabilize helix H3 by extending the helix with two additional turns. Second, as described above (Fig. 3b), the C-terminal end of helix H3 engages in extensive interactions with helix H1 of the MADS-box. These interactions could in turn stabilize helix H3. Finally, the C-terminal end of helix H3, which is rich in positively charged residues, is positioned to interact with the negatively charged DNA backbone (Fig. 4c). These interactions could also enhance the stability of helix H3. The stable folding of helix H3 can lead to the ordering of strand β3 through cooperative folding of the entire MEF2 domain. The observation that helix H3 interacts with DNA also has important implications for DNA recognition by MEF2. Based on the structure, we speculate that helix H3 and its immediately C-terminal sequences could interact with DNA flanking the core MEF2 site. These binding interactions may allow MEF2 homo- or heterodimers expressed in different tissues to bind MEF2 sites with different flanking sequences in vivo (Fig. 4d).32
Our studies demonstrated that the ligand-binding pocket previously observed in the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex is not induced by the particular corepressors, but rather an intrinsic structural property of MEF2. The fact that the MEF2–DNA complex has a preformed ligand-binding pocket allows us to analyze the binding of other cofactors to MEF2 using homology modeling and protein docking. Since the MADS-box is covered by DNA and the MEF2 domain in the MEF2–DNA complex, many MEF2-interacting proteins that are known to bind the MADS-box/MEF2 domain most likely interact with the MEF2 domain directly. Some of these proteins may bind the hydrophobic groove on MEF2 through a mechanism similar to that of Cabin1 and HDAC9. Based on this hypothesis, we have analyzed the interactions between MEF2 and several members of the myocardin family that are potent coactivators of MEF2-dependent gene expression. These myocardin family members, including the cardiac-specific splicing isoform myocardin-935 and MASTR, are known to bind MEF2 at the MADS-box/MEF2 domain through a short amphipathic helix. Our modeling analyses suggest that these myocardin proteins may indeed bind MEF2 at the hydrophobic groove in a manner similar to that of Cabin1 and HDAC9. The generated models are supported by several observations. First, the MEF2-binding motif of myocardin-935 and MASTR share a high level of similarity with the consensus motif we previously derived from the crystal structure of the Cabin1–MEF2B–DNA complex and the HDAC9–MEF2B–DNA complex. Second, the binding interfaces in the generated models have excellent shape and chemical complementarity. Finally, the modeled structure is consistent with existing mutagenesis data. The homology models could guide further mutagenesis studies of the myocardin–MEF2 complex and the MASTR–MEF2 complex in vitro and in vivo. Our studies also establish a general computation approach for studying the interaction between MEF2 and other transcription factors and cofactors.
Deregulation of MEF2 activity, either through mutations of the MEF2 gene or by aberrant cellular signals affecting the activity of cofactors,16,21,22,57 has been implicated in a number of human diseases such as coronary artery disease and cardiac hypertrophy. An attractive idea to develop MEF2-based therapeutics is to find small molecules that can specifically block the recruitment of MEF2 cofactors that are aberrantly overexpressed under pathological conditions. The apo structure of the MEF2A–DNA complex presented here will facilitate the virtual screening of such small molecules as research tools and as leads for drug development.
Materials and Methods
Protein expression and purification
Human MEF2A (residues 1–95) was cloned in pET30b. The MEF2A protein was expressed in Escherichia coli Rosetta BL21(DE3) pLysS and purified in a Sepharose column (GE) and a Superdex 200 column (GE) as described previously.50,51 The final concentration of MEF2A is 32 mg/ml in storage buffer [10 mM, Hepes (pH 7.6), 250 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM DTT].
DNA preparation
DNA (5′-AAC TAT TTA TAA GA-3′) and its complimentary one (5′-TTC TTA TAA ATA GT-3′) were purchased from Integrated DNA Technologies (Coralville, IA) at 1-μmol scale in the crude but desalted form. The crude DNA was dissolved in a buffer (100 mM NaCl, 10 mM NaOH, pH 12.0) and purified by a Mono Q cation-exchange column on FPLC (Amersham Biosciences, Piscataway, NJ). The peak fractions were pooled and neutralized to pH 7.0 by Hepes prior to overnight dialysis against water. The desalted DNA sample was lyophilized to powder, resuspended in water, and quantified at 260 nm. Complementary DNA strands were annealed at 95 °C in the annealing buffer (100 mM NaCl, 5 mM Hepes, pH 7.6) at a concentration of 1.36 mM.
Crystallization, data collection, and structure determination
The MEF2A–DNA complex was prepared by mixing protein and DNA at 2:1 molar ratio in storage buffer at a final concentration of 10 mg/ml. Crystals were grown by the hanging-drop method at 18 °C using a reservoir buffer of 50 mM acetic acid buffer (pH 4.7), 142 mM NaCl, 5 mM MgCl2, 10 mM CaCl2, 3.3% glycerol, and 22.5% polyethylene glycol (PEG) 3000. The cubic crystals grew to 200 μm in 1 week. Crystals were stabilized in harvest/cryoprotectant buffer [20 mM Mg(OAc)2, 50 mM acetic acid buffer (pH 4.7), 30% PEG 3000, and 25% (w/v) glycerol] and flash-frozen with liquid nitrogen for cryocrystallography. The data were collected at a home X-ray source (Micromax) and Raxis IV++ detector. The crystal diffracted to only 2.8 Å. The data were processed with DENZO and SCALEPACK.58 The structure of MEF2–DNA complex was solved by molecular replacement method using MOLREP (version 9.4.09) from the CCP4 program suite (version 6.0).59 The crystal structure of MEF2A–DNA complex [Protein Data Bank (PDB) code 1EGW] was taken as a partial search model in this molecular replacement.48 The C-terminal half of the MEF2 domain was built in O and the structure was refined with CNS.60,61 Noncrystallographic symmetry (NCS) restraints were applied to two copies of MEF2 homodimer throughout the refinement. A DNA molecule in one MEF2 dimer was modeled as two equally probable alternate conformations and therefore has very high B-factors.
Modeling and docking of the myocardin–MASTR complexes bound to MEF2
For homology modeling, we used the crystal structure of HDAC9 bound to MEF2 on DNA (PDB code 1TQE) as a template to build the homology models using the program MODELLER.50,55 The sequence alignment is shown in Fig. 6a. The myocardin–MEF2–DNA and MASTR–MEF2–DNA complexes were built by superposing the homology models of myocardin and MASTR on HDAC9 in HDAC9–MEF2–DNA complex (PDB code 1TQE). Adjustments of a few side chains at the interface were required to remove the clashes. We also performed a protein–protein docking between myocardin–MASTR and MEF2 dimer (1TQE). Protein–protein docking overall gives an interaction interface similar to that generated by homology modeling (data not shown).
Acknowledgements
The authors thank Dr. Xiaojiang Chen, Dr. Yongheng Chen, and Reza Kalhor for helpful discussions and Dr. Dahai Gai and Dr. Ganggang Wang for help with data collection. This research is supported by grants from NIH (L.C.).
Abbreviations used
- MEF2
myocyte enhancer factor 2
- HDAC
histone deacetylase
- SRF
serum response factor
Footnotes
PDB accession numbers Coordinates and structure factors have been deposited in the PDB with accession number 3KOV.
References
- 1.Gossett LA, Kelvin DJ, Sternberg EA, Olson EN. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol. Cell. Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 3.Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, et al. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 5.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 6.Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev. Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 7.Lovato TL, Adams MM, Baker PW, Cripps RM. A molecular mechanism of temperature sensitivity for mutations affecting the Drosophila muscle regulator Myocyte enhancer factor-2. Genetics. 2009;183:107–117. doi: 10.1534/genetics.109.105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandmann T, Jensen LJ, Jakobsen JS, Karzynski MM, Eichenlaub MP, Bork P, Furlong EE. A temporal map of transcription factor activity: Mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Youn HD, Sun L, Prywes R, Liu JO. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science. 1999;286:790–793. doi: 10.1126/science.286.5440.790. [DOI] [PubMed] [Google Scholar]
- 10.Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity. 2000;13:85–94. doi: 10.1016/s1074-7613(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 11.Youn HD, Grozinger CM, Liu JO. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem. 2000;275:22563–22567. doi: 10.1074/jbc.C000304200. [DOI] [PubMed] [Google Scholar]
- 12.Pan F, Ye Z, Cheng L, Liu JO. Myocyte enhancer factor 2 mediates calcium-dependent transcription of the interleukin-2 gene in T lymphocytes: a calcium signaling module that is distinct from but collaborates with the nuclear factor of activated T cells (NFAT) J. Biol. Chem. 2004;279:14477–14480. doi: 10.1074/jbc.C300487200. [DOI] [PubMed] [Google Scholar]
- 13.Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- 14.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, et al. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, Phan D, van Rooij E, Wang DZ, McAnally J, Qi X, et al. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J. Clin. Invest. 2008;118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 18.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, Mao Z. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei JQ, Shehadeh LA, Mitrani JM, Pessanha M, Slepak TI, Webster KA, Bishopric NH. Quantitative control of adaptive cardiac hypertrophy by acetyltransferase p300. Circulation. 2008;118:934–946. doi: 10.1161/CIRCULATIONAHA.107.760488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 24.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D, et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, et al. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 28.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 29.Yuki Y, Imoto I, Imaizumi M, Hibi S, Kaneko Y, Amagasa T, Inazawa J. Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13) Cancer Sci. 2004;95:503–507. doi: 10.1111/j.1349-7006.2004.tb03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prima V, Gore L, Caires A, Boomer T, Yoshinari M, Imaizumi M, et al. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19) (q23;p13.3) in acute lymphoblastic leukemia. Leukemia. 2005;19:806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- 31.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 32.Andres V, Cervera M, Mahdavi V. Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissue-specific sequence constraints. J. Biol. Chem. 1995;270:23246–23249. doi: 10.1074/jbc.270.40.23246. [DOI] [PubMed] [Google Scholar]
- 33.Fickett JW. Quantitative discrimination of MEF2 sites. Mol. Cell. Biol. 1996;16:437–441. doi: 10.1128/mcb.16.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 35.Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl Acad. Sci. USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinn ZA, Yang CC, Wrana JL, McDermott JC. Smad proteins function as co-modulators for MEF2 transcriptional regulatory proteins. Nucleic Acids Res. 2001;29:732–742. doi: 10.1093/nar/29.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 39.Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol. Cell. Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sparrow DB, Miska EA, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, et al. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 43.Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youn HD, Chatila TA, Liu JO. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 2000;19:4323–4331. doi: 10.1093/emboj/19.16.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Luca A, Severino A, De Paolis P, Cottone G, De Luca L, De Falco M, et al. p300/cAMP-response-element-binding-protein (`CREB')-binding protein (CBP) modulates co-operation between myocyte enhancer factor 2A (MEF2A) and thyroid hormone receptor-retinoid X receptor. Biochem. J. 2003;369:477–484. doi: 10.1042/BJ20020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creemers EE, Sutherland LB, Oh J, Barbosa AC, Olson EN. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol. Cell. 2006;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 47.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 48.Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 49.Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 2000;19:2615–2628. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 51.Han A, Pan F, Stroud JC, Youn HD, Liu JO, Chen L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- 52.Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 53.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 56.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Fan C, Topol SE, Topol EJ, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578–1581. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Minor W, Tomchick D, Otwinowski Z. Strategies for macromolecular synchrotron crystallography. Structure. 2000;8:R105–R110. doi: 10.1016/s0969-2126(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 59.Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1994;50:760–776. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 60.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 61.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]