Abstract
The origin of Ewing's sarcoma is a subject of much debate. Once thought to be derived from primitive neuroectodermal cells, many now believe it to arise from a mesenchymal stem cell (MSC). Expression of the EWS-FLI1 fusion gene in MSCs changes cell morphology to resemble Ewing's sarcoma and induces expression of neuroectodermal markers. In murine cells, transformation to sarcomas can occur. In knockdown experiments, Ewing's sarcoma cells develop characteristics of MSCs and the ability to differentiate into mesodermal lineages. However, it cannot be concluded that MSCs are the cell of origin. The concept of an MSC still needs to be rigorously defined, and there may be different subpopulations of mesenchymal pluripotential cells. Furthermore, EWS-FLI1 by itself does not transform human cells, and cooperating mutations appear to be necessary. Therefore, while it is possible that Ewing's sarcoma may originate from a primitive mesenchymal cell, the idea needs to be refined further.
1. Introduction
Ewing's sarcoma is a rare malignancy primarily affecting children and adolescents. It arises mainly in bone and less commonly in soft tissues. The poorly differentiated tumors are aggressive and metastasize early to lung, bone marrow, and other tissues [1–4]. In all cases of the disease, there is a characteristic reciprocal chromosomal translocation, which leads to an in-frame fusion between the EWS gene and one of the ETS family gene members [5, 6]. In approximately 85% of cases, the EWS gene is combined with the ETS gene FLI1 in a t(11;22) translocation. In about 10% of cases, EWS is fused to ERG, which has a high degree of homology with FLI1 in the carboxyl terminus. In very rare cases, EWS is fused to other ETS family genes such as FEV and ETV1 [1, 7]. The EWS-ETS fusion gene appears to be critically important for maintaining the tumor phenotype of the disease. Ewing's sarcoma cells with EWS-FLI1 knockdown by siRNA exhibit decreased cell proliferation, and tumor xenografts regress in mice [8, 9].
2. Mechanism for EWS-FLI1 in Tumorigenesis
The mechanism by which EWS-FLI1 contributes to tumorigenesis is complex since the gene affects the cell in many different ways. The best known function of the EWS-FLI1 protein is that of an aberrant transcription factor [10]. Its structure consists of an N-terminal EWS transcriptional activation domain and a C-terminal FLI1 DNA-binding domain. Disruption of either domain impairs the transformation activity of the protein [11, 12].
EWS-FLI1 possesses the winged helix-turn-helix motif of the ETS family of transcription factors, and EWS-FLI1 recognizes the same consensus DNA binding sequence as FLI1. However, because of aberrant protein-protein interactions, in large part due to the EWS portion of the molecule, EWS-FLI1 has an altered profile of gene regulation. It is relevant to note that FLI1 can act as an oncogene in its own right. Fli1 (Friend leukemia virus integration) was originally identified as the gene activated by insertion of the Friend murine leukemia virus (MuLV) [13]. It has the capacity to transform murine cells and is responsible for the development of erythroleukemias in mice [14]. It is presumably the fusion of the N-terminal moiety of EWS that confers the ability to induce Ewing's sarcoma in human cells.
Acting as either a transcriptional activator or repressor, EWS-FLI1 regulates a number of important target genes. EWS-FLI1 has been found to upregulate genes that promote cell survival and proliferation, including IGF1 [15], Myc [16], TOPK [17], NKX2.2 [18], ID2 [19], DAX1 [20], GLI1 [21], EZH2 [22], MK-STYX [23], and PLD2 [24]. EWS-FLI1 concomitantly represses genes that induce cell cycle arrest and apoptosis, including TGFB2 [25], p21 [26], p57kip [27], and IGFBP3 [28]. Interestingly, EWS-FLI1 also upregulates genes that are involved in cell differentiation such as SOX2 [29] and EZH2 [22].
While progress has been made in characterizing the downstream targets of EWS-FLI1, the properties of EWS-FLI1 may not be fully explained on the basis of its activity as a transcription factor. There are other properties of EWS-FLI1 that have not been studied quite so extensively. These include RNA binding, RNA splicing and protein-protein interactions [30–37]. Furthermore, it has become increasingly clear that the cellular context has a large bearing upon the transformation process. Early experiments with EWS-FLI1 in NIH3T3 cells were somewhat artificial in the sense that these cells were immortalized murine fibroblasts. Paradoxically, in normal murine and human fibroblasts, EWS-FLI1 by itself does not transform cells; instead, it results in cell cycle arrest [38, 39]. These findings suggest the possibility that EWS-FLI1 may have complex antagonistic effects on different types of cells and underscore the importance of identifying the correct cell of origin for the disease.
3. Primitive Neuroectodermal Features of Ewing's Sarcoma
The origin of Ewing's sarcoma is a controversial topic [7]. Histologically, Ewing's sarcoma has a certain resemblance to primitive neuroectodermal cells, and it was once widely believed that the tumor arose from such cells [40, 41]. Early neural markers, such as neuron-specific enolase (NSE) and S-100, are present in some tumors [42, 43]. Ultrastructural features, such as neurosecretory granules, can also be observed with electron microscopy in some cases as well [41, 43]. Laboratory experiments have demonstrated that Ewing's sarcoma cells can be induced to differentiate towards the neural lineage in vitro and acquire neuritic processes under appropriate stimulation [44]. Conversely, introduction of EWS-FLI1 into neuroblastoma cell lines has been shown to make the cells less differentiated and acquire characteristics of Ewing's sarcoma [45].
In spite of considerable data pointing to a neuroectodermal origin, some doubt has been cast upon this idea. The neural markers seen in Ewing's sarcoma are present only in a minority of cases, and even in such tumors, they occur sporadically in cells. More importantly, EWS-FLI1 has been shown to induce neuroectodermal differentiation and upregulate a number of genes associated with early neural differentiation [46, 47]. The findings raise the possibility that the neuroectodermal characteristics of Ewing's sarcoma might be a direct result of EWS-FLI1 expression and not necessarily the cell of origin.
4. Mesenchymal Stem Cells
An alternative hypothesis is that Ewing's sarcoma derives from a mesenchymal stem cell. This addresses one simple but nagging question regarding the neuroectodermal theory of histogenesis, namely, whether bone normally contains primitive neuroectodermal cells. While cranial bones develop from mesenchymal condensation of neuroectoderm [48], the long bones of the limbs originate from mesoderm [49], and there may not normally be primitive neuroectodermal cells in bone. Since most cases of Ewing's sarcoma arise in bone, it seems plausible that the cell of origin ought to be a normal resident of bone. It may be significant to note that mesenchymal cells in bone marrow can exhibit some characteristics of neuroectodermal cells [48, 50]. The cells sometimes express neural markers spontaneously, including S-100 and neurofilament M [50]. They can also be induced in vitro to differentiate towards the neural lineage [51, 52].
A fundamental problem with the mesenchymal hypothesis is that one must define what exactly constitutes a “mesenchymal stem cell.” The term is widely used in scientific literature, and yet the existence of the cell has not been rigorously proven or defined [53]. The term MSC usually implies a bone marrow-derived cell that has the capacity to differentiate towards various mesodermal lineages, such as osteoblasts, chondrocytes, and adipocytes. In order to satisfy the criteria for being a stem cell, it must be shown to be (1) self-renewing and (2) able to generate different tissue types in vivo from a single cell. This has proven to be a daunting task, and perhaps as a result, the putative MSC has yet to be defined in terms of a reliable set of cell surface markers that would aid greatly in consistent extraction of the cells [54]. Cells considered to be MSCs have been reported to be positive for a number of markers, including Sca-1, Stro-1, SH2, SH3, SH4, CD29, CD44, CD90, and CD105, but none of these are entirely specific for MSCs [53–58].
Most preparations of MSCs are heterogeneous [59]. Bone marrow extracts are typically plated on plastic dishes, and the adherent cells are enriched for cells that have some of the desired properties of MSCs. However, the cultures contain a mixture of cells, including hematopoietic stem cells, endothelial cells, osteoblasts, and other cells. The cells can be stimulated to undergo differentiation towards osteoblastic, chondroblastic, and adipocytic lineages in vitro under appropriate culture conditions. One notes though that unless cultures are derived from single cell colonies, it is possible that different cell subpopulations differentiate into the various lineages, and it is therefore not accurate to call the cells MSCs. Furthermore, expression of in vitro markers of differentiation does not equate to the capacity for in vivo differentiation [53]. For these reasons, some authors prefer to call these cell preparations by less presumptive terms, such as “bone marrow stromal cells” as opposed to MSCs.
There are data to support the idea that there is more than one type of pluripotential nonhematopoietic cell in bone marrow. Colter et al. showed that within the population of plastic adherent cells, there are at least two distinct subgroups which could be separated on the basis of size and cell surface markers [60]. A small, round cell with rapid doubling time seems to predominate at early passage whereas a larger spindle cell tends to divide more rapidly at later times. The latter cell type may be an osteoblast precursor and perhaps less pluripotential since it spontaneously expresses alkaline phosphatase, an osteoblast marker.
Jiang et al. showed that bone marrow contains a small population of pluripotential cells, which they termed “marrow-associated progenitor cells” (MAPCs) [61]. The cells represent a subset of the bone marrow cells that adhere to plastic. The cells were found to have clonogenic self-renewal capabilities. Furthermore, when injected into blastocysts, the cells developed into many different tissues, including those derived from ectoderm, endoderm, and mesoderm. These cells may be distinct from the majority of cells typically termed MSCs since their cell surface markers appear to be different from most of the plastic-adherent cells extracted from bone marrow.
At the present time, it should be recognized that our understanding of the “mesenchymal stem cell” is far from complete. The exact nature and composition of the majority of plastic-adherent bone marrow cells is still in question. The very primitive stem cell described by Jiang et al. [61] may be similar to the stem cells found in other tissues [62] and may not represent the more prevalent bone marrow stromal cell. Within the latter group of cells, there may be multiple sub-populations of cell types with varying pluripotential capabilities for in vivo differentiation, analogous to hematopoietic cells [54].
5. MSC Characteristics of Ewing's Sarcoma
There are several lines of evidence that support the notion that Ewing's sarcoma is derived from an MSC-like cell (accepting for the moment the concept of the MSC). Using Ewing's sarcoma cell lines, researchers have knocked down the expression of EWS-FLI1 and shown that the cells have some capacity for in vitro differentiation towards chondroblastic, osteoblastic, and adipocytic lineages [63]. Several groups have demonstrated that expression of EWS-FLI1 in murine MSCs resulted in transformation of the cells, and when these cells were implanted into mice, sarcomas formed [64, 65]. The tumors shared some characteristics with Ewing's sarcoma, including cell surface markers and cell morphology. In a related set of experiments, expression of EWS-FLI1 in the pluripotential murine cell line C3H10T1/2 inhibited the cells ability to differentiate into osteoblasts and adipocytes while upregulating neural genes [66]. When injected into mice, these cells formed metastatic sarcomas.
In contrast to the mouse studies, human MSCs that were infected with a retrovirus containing EWS-FLI1 failed to form tumors when injected into immunodeficient mice [67]. Although somewhat disappointing, the results are not at all surprising since it has been reported that transformation of normal human mesenchymal cells requires multiple mutations [68]. Murine mesenchymal cells are more apt to transform spontaneously in culture and form sarcomas [69]. It is worthwhile to note that human cells expressing EWS-FLI1 take on a rounded morphology and express some of the neuroectodermal markers seen in Ewing's sarcoma [67, 70]. Furthermore, the expression of CD99, a relatively specific marker for Ewing's sarcoma, was found to be present at a low level in MSCs and upregulated by EWS-FLI1 [67]. This addressed a previously raised concern against the theory of an MSC origin of Ewing's sarcoma regarding expression of CD99 in MSCs [71].
The relationship between Ewing's sarcoma and human mesodermal cells has been strengthened further in other experiments. Ewing's sarcoma cell lines with knockdown of EWS-FLI1 have a transcription profile similar to that of the human fetal fibroblast cell line IMR-90 [75]. Similarly, Tirode et al. showed that in an EWS-FLI1 knock-down experiment, the gene expression profile of the cells began to converge upon that of mesenchymal stem cells [63]. This was statistically significant for the subset of genes that were upregulated or downregulated by EWS-FLI1, but it is noteworthy that the overall pattern of gene expression appeared to have distinct differences between MSCs and Ewing's sarcoma cells with EWS-FLI1 knockdown. Comparison of cell surface markers has also shown some similarities and differences between MSCs and Ewing's sarcoma (Table 1). A complementary set of observations was made by Burns et al., who reported that late passage human mesenchymal cells that spontaneously transformed after introduction of telomerase (hMSC-TERT20) took on an immunohistochemical profile that was reminiscent of Ewing's sarcoma, namely, CD99+, vimentin+, CD45−, cytokeratin−, and desmin− [72]. Finally, in a meta-analysis of studies on genes affected by EWS-FLI1, a “core EWS-FLI1 transcriptional signature” was identified which shared similarities with published mesenchymal stem cell data [76]. While these collective data are supportive of a mesenchymal origin of Ewing's sarcoma, it is clear that simple knock-down of EWS-FLI1 in tumor cells does not cause them to revert to a normal mesenchymal cell. Other changes have accumulated in the cells that make them distinctly different from recognizable normal cells.
Table 1.
Cell surface marker analysis of mesenchymal cells and Ewing's sarcoma.
| Marker | Mes. Cells | Mes. Cells + EWS-FLI1 | ESFT | ESFT + EWS-FLI1 knockdown | Refs. |
|---|---|---|---|---|---|
| CD10 | + | − | − | [70] | |
| CD13 | + | − | − | [70] | |
| CD29 | + | − | + | [63] | |
| CD44 | + | + | − | + | [57, 63, 70] |
| CD45 (leukocyte common antigen) | − | − | [57, 72] | ||
| CD54 | ∓ | + | ∓ | + | [63, 70] |
| CD59 | + | − | + | [63] | |
| CD73 | + | − | + | [63] | |
| CD99 (MIC2) | ∓ | + | ++ | + | [67, 70, 71, 73] |
| CD105 | + | + | ∓ | + | [57, 63, 70] |
| CD 117 (c-kit) | − | + | + | [70] | |
| CD166 | + | + | + | + | [63, 70] |
| CD271 | − | + | + | [70] | |
| Vimentin | + | + | [72] | ||
| Caveolin-1 | + | + | [73, 74] | ||
| Desmin | − | − | [72] | ||
| Cytokeratin | − | − | [72] |
Mes. cells: human bone marrow-derived mesenchymal cells.
ESFT: Ewing's sarcoma family of tumors.
∓ indicates weak or variable staining.
6. Knock-in and Transgenic Mouse Models
The importance of cellular context has been highlighted in recent mouse models. Several knock-in and transgenic mice expressing EWS-FLI1 have now been created. In all of these models, a strategy of conditional expression has been employed to avoid lethal effects of EWS-FLI1. Constitutive expression of EWS-FLI1 protein in embryonic stem cells causes cell death [77], and mice with expression of EWS-FLI1 in the whole body have an embryonic lethal phenotype [78, 79]. To circumvent this problem, investigators have used cre-loxP recombination to achieve conditional expression of the protein in vivo. Tissue-specific cre transgenic mice are crossed to the EWS-FLI1 mice to restrict expression of the protein to certain tissues or cells.
Torchia et al. developed a knock-in EWS-FLI1 mouse at the Rosa26 locus [78]. When EWS-FLI1 was expressed by use of the Mx1-cre mouse, myeloid/erythroid leukemias developed. These malignancies were similar to but distinct from the erythroleukemias induced by F-MuLV. The Mx1-cre mouse expresses cre recombinase in the liver, spleen, bone marrow, and lymphoid tissues after induction with alpha/beta interferon or polyinosinic*poly(C) (pIpC) [80]. While the original strategy was to target expression to bone marrow progenitor cells, it is likely that the gene was preferentially expressed in hematopoietic precursors, thereby producing leukemias.
Codrington et al. developed an interesting Ews-ERG mouse, which possesses an inverted ERG cassette flanked by loxP sites in intron 8 of the Ews gene, to simulate the t(21;22) translocation that produces the EWS-ERG fusion [81]. They obtained T cell lymphomas in all mice but this may have been due to their choice of the Rag1-cre mouse, which expresses cre recombinase in lymphocytes only [82].
Although it may be somewhat surprising that leukemias and lymphomas were found in the previous two mouse models, there is reason to believe that this has clinical relevance to human disease. While it was once believed that EWS-FLI1 was specific to Ewing's sarcoma, it is now known that EWS-FLI1 and related EWS-ETS fusions occur sporadically in other malignancies, including leukemias [83–87] and biphenotypic tumors, which have features of both myogenic and neuroectodermal differentiation [88]. These cases indicate that EWS-FLI1 can potentially contribute to neoplastic growth outside of the typical cellular context in which Ewing's sarcoma arises. Quite interestingly, the CD99 marker, which is commonly used for immunohistochemical testing for Ewing's sarcoma, is actually considered a normal leukocyte marker and is uniformly expressed on thymocytes [89].
In a different transgenic mouse model of EWS-FLI1, conditional expression of the fusion protein was achieved in the mesoderm-derived tissues of the limbs by crossing to the Prx1-cre mouse, which expresses cre in the primitive mesenchyme of the early limb bud [79]. Part of the rationale behind this cross was to spare the hematopoietic cells and thereby increase the likelihood of inducing sarcomas. Limb shortening, muscle atrophy, osseous dysplasia, and other developmental abnormalities were observed, reinforcing the idea that EWS-FLI1 impairs growth and differentiation of cells. However, in this model, sarcomas did not spontaneously form unless the p53 gene was simultaneously mutated [79]. In contrast to the more differentiated osteosarcomas that formed without EWS-FLI1 in mice with conditional mutation of p53 induced by Prx1-cre, the tumors that arose with EWS-FLI1 were undifferentiated sarcomas, similar to Ewing's sarcoma.
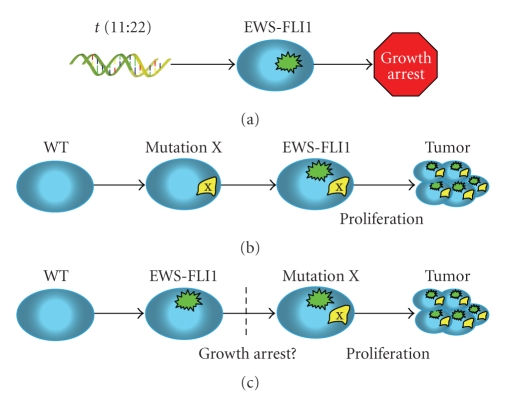
Although concern has been raised regarding the validity of mouse models for Ewing's sarcoma [76], the transgenic mouse model employing Prx1-cre does underscore one important feature of Ewing's sarcoma, namely, that EWS-FLI1 alone does not appear to be sufficient to confer sarcomatous change in an in vivo setting. Similar to the results obtained with human MSCs expressing EWS-FLI1, additional cooperating mutations seem to be required for transformation to occur. The nature and timing of these mutations may be an interesting subject for future research (Figure 1). The limb shortening of transgenic mice [79] and experimental cell culture work of various groups [39, 50] suggest that EWS-FLI1 impairs growth of normal cells, and this may be problematic in envisioning a pathway for neoplastic transformation to occur. While many have postulated that mutations occur after the t(11;22) translocation event, it is also possible that EWS-FLI1 transforms a cell that carries a silent but critical mutation in another gene.
Figure 1.
Cooperative mutations in the development of Ewing's sarcoma. (a) A t(11;22) reciprocal translocation produces the EWS-FLI1 gene, but this tends to cause growth arrest in normal cells. (b) A mutation randomly occurring prior to the t(11;22) translocation might cooperate with EWS-FLI1 to permit escape from growth arrest (or even promote cell proliferation) and subsequent transformation to Ewing's sarcoma. (c) The cooperative mutation may occur after the t(11;22) translocation; this would necessarily imply a mechanism for continued cell growth after EWS-FLI1 is expressed.
The requirement for additional mutational events has precedent in other mouse models. For example, in a knock-in model of alveolar rhabdomyosarcoma, mutations in Ink4A/Arf or Trp53 were needed for the Pax3;Fkhr fusion to transform cells effectively [90]. Interestingly, conditional expression in the correct cell of origin—postnatal differentiating myofibers—was also found to be important for rhabdomyosarcomas to develop in this particular model. TRP53, however, is unlikely to be the critical cooperating mutated gene in Ewing's sarcoma since TRP53 mutations are found only in about 10% of Ewing's sarcoma cases [91].
7. Conclusions
Identification of the correct cell of origin is crucial to the understanding of Ewing's sarcoma. While there has been steadily accumulating evidence that the cell of origin may be a primitive pluripotential cell that resides in bone marrow, one cannot state unequivocally that it is either a neuroectodermal cell or a “mesenchymal stem cell.” One important simple reason for this is that the concept of the MSC has yet to be rigorously defined, and the term is used somewhat loosely in the scientific literature. MSCs, as they are typically prepared, are composed of a mixture of different cells, and some of these do exhibit neuroectodermal properties. It is possible that a subpopulation of the pluripotential cells in bone marrow is especially vulnerable to the transforming effects of EWS-FLI1.
Even if the correct cell of origin can be identified with precision, much has yet to be learned. Expression of EWS-FLI1 alone is not sufficient for transformation of normal human cells. Instead, EWS-FLI1 tends to cause growth arrest. In the early excitement over the discovery of EWS-FLI1, researchers focused largely upon downstream transcriptional targets. Current research suggests that there may be critically important cooperating mutations that work in concert with EWS-FLI1. The “cell of origin” concept may be a simplistic one in that it might not be a normal cell from which Ewing's sarcoma arises. EWS-FLI1 may be fully transforming only in a mutated cell that possesses additional genetic alterations. The tumor that eventually arises, whether it is a conventional Ewing's sarcoma, an unusual variant, a biphenotypic tumor, lymphoblastic leukemia, or some other disease, will probably depend upon an interplay between the cellular background in which it arises and the set of cooperating mutations that occur.
References
- 1.Ordóñez JL, Osuna D, Herrero D, de Álava E, Madoz-Gúrpide J. Advances in Ewing’s sarcoma research: where are we now and what lies ahead? Cancer Research. 2009;69(18):7140–7150. doi: 10.1158/0008-5472.CAN-08-4041. [DOI] [PubMed] [Google Scholar]
- 2.Nesbit ME, Jr., Gehan EA, Burgert EO, Jr., et al. Multimodal therapy for the management of primary, nonmetastatic Ewing’s Sarcoma of bone: a long-term follow-up of the first intergroup study. Journal of Clinical Oncology. 1990;8(10):1664–1674. doi: 10.1200/JCO.1990.8.10.1664. [DOI] [PubMed] [Google Scholar]
- 3.Grier HE, Krailo MD, Tarbell NJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. New England Journal of Medicine. 2003;348(8):694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Galindo C, Billups CA, Kun LE, et al. Survival after recurrence of ewing tumors: the St. Jude children’s research hospital experience, 1979–1999. Cancer. 2002;94(2):561–569. doi: 10.1002/cncr.10192. [DOI] [PubMed] [Google Scholar]
- 5.Ban J, Siligan C, Kreppel M, Aryee D, Kovar H. EWS-FLI1 in Ewing’s sarcoma: real targets and collateral damage. Advances in Experimental Medicine and Biology. 2006;587:41–52. doi: 10.1007/978-1-4020-5133-3_4. [DOI] [PubMed] [Google Scholar]
- 6.Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Letters. 2007;254(1):1–10. doi: 10.1016/j.canlet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Seminars in Cancer Biology. 2005;15(3):189–196. doi: 10.1016/j.semcancer.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Toretsky JA, Connell Y, Neckers L, Bhat NK. Inhibition of EWS-FLI-1 fusion protein with antisense oligodeoxynucleotides. Journal of Neuro-Oncology. 1997;31(1-2):9–16. doi: 10.1023/a:1005716926800. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing’s sarcoma and primitive neuroectodermal tumor cells. Journal of Clinical Investigation. 1997;99(2):239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Üren A, Tcherkasskaya O, Toretsky JA. Recombinant EWS-FLI1 oncoprotein activates transcription. Biochemistry. 2004;43(42):13579–13589. doi: 10.1021/bi048776q. [DOI] [PubMed] [Google Scholar]
- 11.May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(12):5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessnick SL, Braun BS, Denny CT, May WA. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10(3):423–431. [PubMed] [Google Scholar]
- 13.Ben-David Y, Giddens EB, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(4):1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truong AHL, Ben-David Y. The role of Fli-1 in normal cell function and malignant transformation. Oncogene. 2000;19(55):6482–6489. doi: 10.1038/sj.onc.1204042. [DOI] [PubMed] [Google Scholar]
- 15.Cironi L, Riggi N, Provero P, et al. IGF1 is a common target gene of Ewing’s sarcoma fusion proteins in mesenchymal progenitor cells. PLoS One. 2008;3(7) doi: 10.1371/journal.pone.0002634. Article ID e2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sollazzo MR, Benassi MS, Magagnoli G, et al. Increased c-myc oncogene expression in Ewing’s sarcoma: correlation with Ki67 proliferation index. Tumori. 1999;85(3):167–173. doi: 10.1177/030089169908500304. [DOI] [PubMed] [Google Scholar]
- 17.Herrero-Martín D, Osuna D, Ordóñez JL, et al. Stable interference of EWS-FLI1 in an Ewing sarcoma cell line impairs IGF-1/IGF-1R signalling and reveals TOPK as a new target. British Journal of Cancer. 2009;101(1):80–90. doi: 10.1038/sj.bjc.6605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9(5):405–416. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Fukuma M, Okita H, Hata J-I, Umezawa A. Upregulation of Id2, an oncogenic helix-loop-helix protein, is mediated by the chimeric EWS/ets protein in Ewing sarcoma. Oncogene. 2003;22(1):1–9. doi: 10.1038/sj.onc.1206055. [DOI] [PubMed] [Google Scholar]
- 20.García-Aragoncillo E, Carrillo J, Lalli E, et al. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene. 2008;27(46):6034–6043. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- 21.Zwerner JP, Joo J, Warner KL, et al. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27(23):3282–3291. doi: 10.1038/sj.onc.1210991. [DOI] [PubMed] [Google Scholar]
- 22.Richter GHS, Plehm S, Fasan A, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5324–5329. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siligan C, Ban J, Bachmaier R, et al. EWS-FLI1 target genes recovered from Ewing’s sarcoma chromatin. Oncogene. 2005;24(15):2512–2524. doi: 10.1038/sj.onc.1208455. [DOI] [PubMed] [Google Scholar]
- 24.Kikuchi R, Murakami M, Sobue S, et al. Ewing’s sarcoma fusion protein, EWS/Fli-1 and Fli-1 protein induce PLD2 but not PLD1 gene expression by binding to an ETS domain of 5’ promoter. Oncogene. 2007;26(12):1802–1810. doi: 10.1038/sj.onc.1209973. [DOI] [PubMed] [Google Scholar]
- 25.Hahm K-B, Cho K, Lee C, et al. Repression of the gene encoding the TGF-β type II receptor is a major target of the EWS-FLI1 oncoprotein. Nature Genetics. 1999;23(2):222–227. doi: 10.1038/13854. [DOI] [PubMed] [Google Scholar]
- 26.Nakatani F, Tanaka K, Sakimura R, et al. Identification of p21WAF1/CIP1 as a direct target of EWS-Fli1 oncogenic fusion protein. Journal of Biological Chemistry. 2003;278(17):15105–15115. doi: 10.1074/jbc.M211470200. [DOI] [PubMed] [Google Scholar]
- 27.Dauphinot L, De Oliveira C, Melot T, et al. Analysis of the expression of cell cycle regulators in Ewing cell lines: EWS-FLI-1 modulates p57KIP2 and c-Myc expression. Oncogene. 2001;20(25):3258–3265. doi: 10.1038/sj.onc.1204437. [DOI] [PubMed] [Google Scholar]
- 28.Prieur A, Tirode F, Cohen P, Delattre O. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Molecular and Cellular Biology. 2004;24(16):7275–7283. doi: 10.1128/MCB.24.16.7275-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riggi N, Suvà M-L, De Vito C, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes and Development. 2010;24(9):916–932. doi: 10.1101/gad.1899710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Chansky HA, Hickstein DD. EWS·Fli-1 fusion protein interacts with hyperphosphorylated RNA polymerase II and interferes with serine-arginine protein-mediated RNA splicing. Journal of Biological Chemistry. 2000;275(48):37612–37618. doi: 10.1074/jbc.M005739200. [DOI] [PubMed] [Google Scholar]
- 31.Toretsky JA, Erkizan V, Levenson A, et al. Oncoprotein EWS-FLI1 activity is enhanced by RNA helicase A. Cancer Research. 2006;66(11):5574–5581. doi: 10.1158/0008-5472.CAN-05-3293. [DOI] [PubMed] [Google Scholar]
- 32.Chansky HA, Hu M, Hickstein DD, Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Research. 2001;61(9):3586–3590. [PubMed] [Google Scholar]
- 33.Gascoyne DM, Thomas GR, Latchman DS. The effects of Brn-3a on neuronal differentiation and apoptosis are differentially modulated by EWS and its oncogenic derivative EWS/Fli-1. Oncogene. 2004;23(21):3830–3840. doi: 10.1038/sj.onc.1207497. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Denny CT, Wisdom R. Cooperative DNA binding with AP-1 proteins is required for transformation by EWS-Ets fusion proteins. Molecular and Cellular Biology. 2006;26(7):2467–2478. doi: 10.1128/MCB.26.7.2467-2478.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petermann R, Mossier BM, Aryee DNT, Khazak V, Golemis EA, Kovar H. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene. 1998;17(5):603–610. doi: 10.1038/sj.onc.1201964. [DOI] [PubMed] [Google Scholar]
- 36.Spahn L, Petermann R, Siligan C, Schmid JA, Aryee DNT, Kovar H. Interaction of the EWS NH2 terminus with BARD1 links the Ewing’s sarcoma gene to a common tumor suppressor pathway. Cancer Research. 2002;62(16):4583–4587. [PubMed] [Google Scholar]
- 37.Watson DK, Robinson L, Hodge DR, Kola I, Papas TS, Seth A. FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene. 1997;14(2):213–221. doi: 10.1038/sj.onc.1200839. [DOI] [PubMed] [Google Scholar]
- 38.Deneen B, Denny CT. Loss of p16 pathways stabilizes EWS/FLI1 expression and complements EWS/FLI1 mediated transformation. Oncogene. 2001;20(46):6731–6741. doi: 10.1038/sj.onc.1204875. [DOI] [PubMed] [Google Scholar]
- 39.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 40.Cavazzana AO, Miser JS, Jefferson J, Triche TJ. Experimental evidence for a neural origin of Ewing’s sarcoma of bone. American Journal of Pathology. 1987;127(3):507–518. [PMC free article] [PubMed] [Google Scholar]
- 41.Suh C-H, Ordóñez NG, Hicks J, Mackay B. Ultrastructure of the Ewing’s sarcoma family of tumors. Ultrastructural Pathology. 2002;26(2):67–76. doi: 10.1080/01913120252959236. [DOI] [PubMed] [Google Scholar]
- 42.Rettig WJ, Garin-Chesa P, Huvos AG. Ewing’s sarcoma: new approaches to histogenesis and molecular plasticity. Laboratory Investigation. 1992;66(2):133–137. [PubMed] [Google Scholar]
- 43.Franchi A, Pasquinelli G, Cenacchi G, et al. Immunohistochemical and ultrastructural investigation of neural differentiation in Ewing sarcoma/PNET of bone and soft tissues. Ultrastructural Pathology. 2001;25(3):219–225. [PubMed] [Google Scholar]
- 44.Navarro S, Gonzalez-Devesa M, Ferrandez-Izquierdo A, Triche TJ, Llombart-Bosch A. Scanning electron microscopic evidence for neural differentiation in Ewing’s sarcoma cell lines. Virchows Archiv A. 1990;416(5):383–391. doi: 10.1007/BF01605142. [DOI] [PubMed] [Google Scholar]
- 45.Rorie CJ, Thomas VD, Chen P, Pierce HH, O’Bryan JP, Weissman BE. The Ews/Fli-1 fusion gene switches the differentiation program of neuroblastomas to Ewing sarcoma/peripheral primitive neuroectodermal tumors. Cancer Research. 2004;64(4):1266–1277. doi: 10.1158/0008-5472.can-03-3274. [DOI] [PubMed] [Google Scholar]
- 46.Teitell MA, Thompson AD, Sorensen PHB, Shimada H, Triche TJ, Denny CT. EWS/ETS fusion genes induce epithelial and neuroectodermal differentiation in NIH 3T3 fibroblasts. Laboratory Investigation. 1999;79(12):1535–1543. [PubMed] [Google Scholar]
- 47.Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing’s family of tumors. Cancer Research. 2005;65(11):4633–4644. doi: 10.1158/0008-5472.CAN-04-2857. [DOI] [PubMed] [Google Scholar]
- 48.Takashima Y, Era T, Nakao K, et al. Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell. 2007;129(7):1377–1388. doi: 10.1016/j.cell.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 49.Olsen BR, Reginato AM, Wang W. Bone development. Annual Review of Cell and Developmental Biology. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 50.Torchia EC, Jaishankar S, Baker SJ. Ewing tumor fusion proteins block the differentiation of pluripotent marrow stromal cells. Cancer Research. 2003;63(13):3464–3468. [PubMed] [Google Scholar]
- 51.Kopen GC, Prockop DJ, Phinney DG. Marrow stromal cells migrate throughout forebrain and cerebellum, and they differentiate into astrocytes after injection into neonatal mouse brains. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10711–10716. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodbury D, Schwarz EJ, Prockop DJ, Black IB. Adult rat and human bone marrow stromal cells differentiate into neurons. Journal of Neuroscience Research. 2000;61(4):364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 53.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beyer Nardi N, da Silva Meirelles L. Mesenchymal stem cells: isolation, in vitro expansion and characterization. Handbook of Experimental Pharmacology. 2006;(174):249–282. [PubMed] [Google Scholar]
- 55.Otto WR, Rao J. Tomorrow’s skeleton staff: mesenchymal stem cells and the repair of bone and cartilage. Cell Proliferation. 2004;37(1):97–110. doi: 10.1111/j.1365-2184.2004.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deans RJ, Moseley AB. Mesenchymal stem cells: biology and potential clinical uses. Experimental Hematology. 2000;28(8):875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 57.Lodie TA, Blickarz CE, Devarakonda TJ, et al. Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue Engineering. 2002;8(5):739–751. doi: 10.1089/10763270260424105. [DOI] [PubMed] [Google Scholar]
- 58.Anjos-Afonso F, Siapati EK, Bonnet D. In vivo contribution of murine mesenchymal stem cells into multiple cell-types under minimal damage conditions. Journal of Cell Science. 2004;117(23):5655–5664. doi: 10.1242/jcs.01488. [DOI] [PubMed] [Google Scholar]
- 59.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276(5309):71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 60.Colter DC, Sekiya I, Prockop DJ. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(14):7841–7845. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418(6893):41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 62.Beltrami AP, Cesselli D, Bergamin N, et al. Multipotent cells can be generated in vitro from several adult human organs (heart, liver, and bone marrow) Blood. 2007;110(9):3438–3446. doi: 10.1182/blood-2006-11-055566. [DOI] [PubMed] [Google Scholar]
- 63.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11(5):421–429. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 64.Castillero-Trejo Y, Eliazer S, Xiang L, Richardson JA, Ilaria RL., Jr. Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells results in EWS/FLI-1-dependent, Ewing sarcoma-like tumors. Cancer Research. 2005;65(19):8698–8705. doi: 10.1158/0008-5472.CAN-05-1704. [DOI] [PubMed] [Google Scholar]
- 65.Riggi N, Cironi L, Provero P, et al. Development of Ewing’s sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Research. 2005;65(24):11459–11468. doi: 10.1158/0008-5472.CAN-05-1696. [DOI] [PubMed] [Google Scholar]
- 66.González I, Vicent S, de Alava E, Lecanda F. EWS/FLI-1 oncoprotein subtypes impose different requirements for transformation and metastatic activity in a murine model. Journal of Molecular Medicine. 2007;85(9):1015–1029. doi: 10.1007/s00109-007-0202-5. [DOI] [PubMed] [Google Scholar]
- 67.Riggi N, Suvà M-L, Suvà D, et al. EWS-FLI-1 expression triggers a ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Research. 2008;68(7):2176–2185. doi: 10.1158/0008-5472.CAN-07-1761. [DOI] [PubMed] [Google Scholar]
- 68.Funes JM, Quintero M, Henderson S, et al. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(15):6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou YF, Bosch-Marce M, Okuyama H, et al. Spontaneous transformation of cultured mouse bone marrow-derived stromal cells. Cancer Research. 2006;66(22):10849–10854. doi: 10.1158/0008-5472.CAN-06-2146. [DOI] [PubMed] [Google Scholar]
- 70.Miyagawa Y, Okita H, Nakaijima H, et al. Inducible expression of chimeric EWS/ETS proteins confers Ewing’s family tumor-like phenotypes to human mesenchymal progenitor cells. Molecular and Cellular Biology. 2008;28(7):2125–2137. doi: 10.1128/MCB.00740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kovar H, Bernard A. CD99-positive “Ewing’s sarcoma” from mouse bone marrow-derived mesenchymal progenitor cells? Cancer Research. 2006;66(19):p. 9786. doi: 10.1158/0008-5472.CAN-06-0205. [DOI] [PubMed] [Google Scholar]
- 72.Burns JS, Abdallah BM, Shrøder HD, Kassem M. The histopathology of a human mesenchymal stem cell experimental tumor model: support for an hMSC origin for Ewing’s sarcoma? Histology and Histopathology. 2008;23(10):1229–1240. doi: 10.14670/HH-23.1229. [DOI] [PubMed] [Google Scholar]
- 73.Llombart-Bosch A, Machado I, Navarro S, et al. Histological heterogeneity of Ewing's sarcoma/PNET: an immunohistochemical analysis of 415 genetically confirmed cases with clinical support. Virchows Archiv. 2009;455(5):397–411. doi: 10.1007/s00428-009-0842-7. [DOI] [PubMed] [Google Scholar]
- 74.Tirado OM, Mateo-Lozano S, Villar J, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Research. 2006;66(20):9937–9947. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 75.Potikyan G, France KA, Carlson MRJ, Dong J, Nelson SF, Denny CT. Genetically defined EWS/FLI1 model system suggests mesenchymal origin of Ewing’s family tumors. Laboratory Investigation. 2008;88(12):1291–1302. doi: 10.1038/labinvest.2008.99. [DOI] [PubMed] [Google Scholar]
- 76.Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7(2):250–256. doi: 10.4161/cc.7.2.5229. [DOI] [PubMed] [Google Scholar]
- 77.Sohn EJ, Li H, Reidy K, Beers LF, Christensen BL, Lee SB. EWS/FLI1 oncogene activates caspase 3 transcription and triggers apoptosis in vivo. Cancer Research. 2010;70(3):1154–1163. doi: 10.1158/0008-5472.CAN-09-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Molecular and Cellular Biology. 2007;27(22):7918–7934. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin PP, Pandey MK, Jin F, et al. EWS-FLI1 induces developmental abnormalities and accelerates sarcoma formation in a transgenic mouse model. Cancer Research. 2008;68(21):8968–8975. doi: 10.1158/0008-5472.CAN-08-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 81.Codrington R, Pannell R, Forster A, et al. The Ews-ERG fusion protein can initiate neoplasia from lineage-committed haematopoietic cells. PLoS Biology. 2005;3(8, article e242) doi: 10.1371/journal.pbio.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCormack MP, Forster A, Drynan L, Pannell R, Rabbitts TH. The LMO2 T-cell oncogene is activated via chromosomal translocations or retroviral insertion during gene therapy but has no mandatory role in normal T-cell development. Molecular and Cellular Biology. 2003;23(24):9003–9013. doi: 10.1128/MCB.23.24.9003-9013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jakovljević G, Nakić M, Rogošić S, et al. Pre-B-cell acute lymphoblastic leukemia with bulk extramedullary disease and chromosome 22 (EWSR1) rearrangement masquerading as ewing sarcoma. Pediatric Blood and Cancer. 2010;54(4):606–609. doi: 10.1002/pbc.22365. [DOI] [PubMed] [Google Scholar]
- 84.Martini A, La Starza R, Janssen H, et al. Recurrent rearrangement of the Ewing’s sarcoma gene, EWSR1, or its homologue, TAF15, with the transcription factor CIZ/NMP4 in acute leukemia. Cancer Research. 2002;62(19):5408–5412. [PubMed] [Google Scholar]
- 85.Marcucci G, Baldus CD, Ruppert AS, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. Journal of Clinical Oncology. 2005;23(36):9234–9242. doi: 10.1200/JCO.2005.03.6137. [DOI] [PubMed] [Google Scholar]
- 86.Hawkins JM, Craig JM, Secker-Walker LM, Prentice HG, Mehta AB. Ewing’s sarcoma t(11;22) in a case of acute nonlymphocytic leukemia. Cancer Genetics and Cytogenetics. 1991;55(2):157–162. doi: 10.1016/0165-4608(91)90072-3. [DOI] [PubMed] [Google Scholar]
- 87.Haresh K, Joshi N, Gupta C, et al. Granulocytic sarcoma masquerading as Ewing’s sarcoma: a diagnostic dilemma. Journal of Cancer Research and Therapeutics. 2008;4(3):137–139. doi: 10.4103/0973-1482.43150. [DOI] [PubMed] [Google Scholar]
- 88.Sorensen PHB, Shimada H, Liu XF, Lim JF, Thomas G, Triche TJ. Biphenotypic sarcomas with myogenic and neural differentiation express the Ewing’s sarcoma EWS/FLI1 fusion gene. Cancer Research. 1995;55(6):1385–1392. [PubMed] [Google Scholar]
- 89.Aussel C, Bernard G, Breittmayer J-P, Pelassy C, Zoccola D, Bernard A. Monoclonal antibodies directed against the E2 protein (MIC2 gene product) induce exposure of phosphatidylserine at the thymocyte cell surface. Biochemistry. 1993;32(38):10096–10101. doi: 10.1021/bi00089a027. [DOI] [PubMed] [Google Scholar]
- 90.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes and Development. 2004;18(21):2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. Journal of Clinical Oncology. 2005;23(3):548–558. doi: 10.1200/JCO.2005.02.081. [DOI] [PubMed] [Google Scholar]