Abstract
An array of nano-channels was fabricated from silicon based semiconductor materials to stretch long, native dsDNA. Here we present a labeling scheme in which it is possible to identify the location of specific sequences along the stretched DNA molecules. The scheme proceeds by first using the strand displacement activity of the Vent (exo-) polymerase to generate single strand flaps on nicked dsDNA. These single strand flaps are hybridized with sequence specific fluorophore-labeled probes. Subsequent imaging of the DNA molecules inside a nano-channel array device allows for quantitative identification of the location of probes. The highly efficient DNA hybridization on the ss-DNA flaps is an excellent method to identify the sequence motifs of dsDNA as it gives us unique ability to control the length of the probe sequence and thus the frequency of hybridization sites on the DNA. We have also shown that this technique can be extended to a multi color labeling scheme by using different dye labeled probes or by combining with a DNA- polymerase-mediated incorporation of fluorophore-labeled nucleotides on nicking sites. Thus this labeling chemistry in conjunction with the nano-channel platform can be a powerful tool to solve complex structural variations in DNA which is of importance for both research and clinical diagnostics of genetic diseases.
INTRODUCTION
The human genome is enriched in many forms of variants, including single nucleotide polymorphisms and structural variations. There has been an explosion of data describing newly recognized structural variants in the human genome and their associations with a variety of diseases (1,2). Despite recent advances in technologies in detection and confirmation of structural variants, there is still an urgent need for technologies to assess structural variants more accurately and rapidly. SNP array technology is frequently biased against certain genomic regions depending on the probe selection (3). Array CGH (comparative genome hybridization) has limited success in discerning copy number differences (4). Balanced translocations are particularly difficult to detect with array technology. Even with pair-end sequencing technologies on second generation sequencing platforms, it is difficult to assign the end sequencing to an unambiguous region, not to mention the laborious cloning steps that are required (5,6). Once discovered, novel structural variants still need to be confirmed and validated, generally relying on laborious and low throughput PCR or fluorescence in situ hybridization methods. DNA mapping has been an important strategy to study structures and organizations of genomes. Recent advances in linear mapping of single DNA molecules hold great promise in direct visualization of structural variants across the genome with high throughput at lower cost. Such single molecule linear DNA analyses are generally based on interrogating specific sequence motifs along long linear stretched DNA molecules. Schwartz’s group pioneered one such technique, optical mapping, which can provide linear ordered restriction maps from long individual DNA molecules (7). This approach has been successfully applied in numerous DNA mapping projects (8). Bensimon co-workers et al. (9), have developed molecular combing for high-resolution fluorescence in situ hybridization with hybridization probes. Chan et al. (10), reported a DNA linear analysis method, in which the dsDNA molecules were tagged at sequence-specific motif sites with fluorescent bisPNA (peptide nucleic acid) tags, and the labeled DNA molecules were then stretched in a microfluidic device and labeled sequence motifs are analyzed with fluorescence detectors. More recently, Jo et al. (11) presented a DNA mapping strategy based on DNA linearization in a nano-slit. However, two key issues still prevent the rapid adoption of DNA linear mapping technology, uniform DNA linearization and flexible sequence specific labeling. Here we report an integrated approach for linear DNA analysis, which makes significant advances on these two critical components of DNA linear analysis. Our method starts with sequence specific labeling of long genomic DNA molecules with fluorophores. The labeled DNA molecules are then linearized inside the nano-channel array and imaged with high resolution fluorescence microscopy. By determining the order of the fluorescent labels on the backbone, the distribution of specific sequence motifs of an individual DNA molecule can be inferred with great accuracy, in a manner similar to reading a bar code. This highly miniaturized nano-array device together with the flexible and efficient labeling chemistry enables analysis of single DNA molecules preserved in long linear state during the investigation. The preservation of long linear DNA molecule for single molecule analysis is imperative for obtaining some critical genetic information such as haplotype and copy number variation (CNV), which are still difficult to obtain with current short read next-generation sequencing technologies. We demonstrate its capabilities in mapping various DNA molecules and structural variants in a 115 kb human BAC clone.
MATERIALS AND METHODS
DNA sample preparation
λ-DNA was purchased from NEB (New England Biolabs Inc., Ipswich, MA, USA). Fosmid G248P8446G6 was a gift from Dr Eichler of University of Washington. BAC clone 3F5 was a gift from Dr Milosavljevic of Baylor College of Medicine, Houston. All the oligo probes listed in Table 1 were synthesized by IDT (Integrated DNA Technology, San Jose, CA, USA). Fosmid and BAC Clone culture and purification follow the protocol for the QIAGEN Large-Construct Kit. Cells were isolated from a streaked plate and incubated in 5 ml of LB media for 8 h at 37°C with shaking at 300 rpm. The starter culture was added to 500 ml LB in a 1 l Erlenmeyer flask, then 1 ml of Fosmid autoinduction solution was added and the flask was shaken at 37°C for 16 h with shaking at 300 rpm. In case of MCF7 BAC 3F5, no autoinduction fluid solution was added. The Escherichia coli cells were harvested by centrifugation at 6000g in a Beckman JA-10 rotor. The supernatant was removed and the pellet re-suspended in 20 ml Buffer P1, mixed with 20 ml Buffer P2 and after 5 min mixed with chilled 20 ml Buffer P3 and 10 min afterwards centrifuged at 20 000g for 30 min at 4°C. The supernatant containing the lysate was filtered through filter paper. The DNA was precipitated by adding 0.6 volume of isopropanol, spinning at 15 000g for 30 min at 4°C. The DNA pellet was washed with 5 ml 70% ethanol and then centrifuged at 15 000g for 30 min at 4°C. The pellet was air-dried and then re-dissolved in 9.5 ml Buffer EX. The DNA was then digested by adding 200 μl of ATP-dependent Exonuclease and 300 μl of ATP solution and incubating at 37°C for 1 h. The DNA was then filtered in a QIAGEN-tip 500 prewetted with 10 ml Buffer QBT, the column was washed with 60 ml Buffer QC and eluted with 15 ml Buffer QF warmed to 65°C. The DNA was then precipitated by adding 0.7 vol of isopropanol, spinning at 15 000g for 30 min at 4°C. The DNA pellet was washed with 5 ml 70% ethanol and then centrifuged at 15 000g for 30 min at 4°C. The pellet was air-dried and then re-dissolved in 50 μl TE Buffer and concentration was checked on a Nanodrop.
Table 1.
Fluorescent probe sequences
| DNA templates | Target locations | Probe sequencesa |
|---|---|---|
| λ-DNA | 8.0 kb | TCCAACTATATAATTTGACCAGAGAACAAG |
| 35.8 kb | AAGGTCTTGAGCAGGCCGTT | |
| Fosmid G248P8446G6 | Multiple repetitive sequences | TGCCTGTGAGAGGAAATCTCAACTCTCTT |
| MCF7 BAC clone 3F5 | ||
| Universal probe | Mutiple conserved sequences | ATT+CTCCTGCC+TCAb |
| Sequence specific probes | 1.3 kb (segment from 3p14.1) | TCCTTGGTTGACCTAACAACACA |
| 30.5 kb (segment from 20q12) | TGCCACCTACCCCT | |
| 83.0 kb (segment from 20q13.2) | TCCAAGTCTCAGTGACCCT | |
| 95.2 kb (segment from 20q13.2) | ACTGTAGTCTTGAATTCCTGA |
aSequence starts from 5′-end, both ends are labeled with cy3 dyes.
b+C and +T are LNA bases to increase TM.
Sequence specific labeling
(i) Nicking: Duplex DNA samples 50 ng/µl (λ-DNA, Fosmid G248P8446G6, MCF7 BAC clone 3F5) were incubated with Nb. BbvCI (0.5 U/µl) (NEB Cat #R063) 1 µl in 1× NEB buffer 2 (Cat #B7002S) in 20 µl volume for 1 h at 37°C and 20 min at 65°C. (ii) Digestion: After nicking reaction, the circular nicked DNA samples (25 ng/µl) (Fosmid G248P8446G6, MCF7 BAC clone 3F5) were digested with NotI (1 U/µl) (NEB Not1.HF Cat #R3189S) in 1× NEB2 buffer in presence of 1× BSA (NEB BSA, Cat #B90015) to make them linear. Typically the incubation was performed for 2 h at 37°C followed by 20 min at 65°C. (iii) DNA strand displacement and Flap generation: In this procedure the nicked and cut DNA samples (12.5 ng/µl) were incubated for 30 min at 50°C in 1× NEB thermopol buffer with Vent (exo-) at 0.5 U/µl in presence of 75 nM dNTP mixture. For DNA-polymerase-mediated incorporation of fluorophore-labeled nucleotide, dNTP mixture is replaced by a mixture of 75 nM dAGC and 75 nM Alexa 647 labeled dUTP. (iv) Flap-labeling: In a typical procedure of probe hybridization of flaps, 8 ng/µl DNA molecules was incubated with the dye labeled probe oligonucleotides (200 nM) (labeled with two Cy3 dyes at two ends) at 75°C for 2 min followed by 30 min at 48°C. (v) Nick–flap labeling: DNA molecules were nick-labeled according to the procedures mentioned above using 75 nm Alexa 647 dUTP and 75 nM unlabeled dAGC. After nick-labeling, the flaps were further extended by incubation with additional 200 nM unlabeled dNTP. In this procedure the DNA sample (after nicked and cut) concentration was at 12.5 ng/µl. The flap labeling was done in the same way as mentioned above.
Probe designs
The probes are designed based on uniform TM, which is set to be 55° in this study. The universal probes are designed by aligning all 50 bp flap sequences and determine the optimal probes sequences based on the TM and the distribution of the probes on the DNA backbone. To increase the probe TM, modified LNA bases are incorporated in the probes, for example +C and +T are modified LNA bases in the probe ATT + CTCCTGCC + TCA.
DNA loading into nano-channel
All the DNA (4 ng/µl) samples were stained with intercalating dye YOYO-1 iodide (Invitrogen, Carlsbad, CA; Cat #Y3601) (1 molecule/10 bp) in presence of 0.4 M DTT (Promega Inc, Madison, WI, USA; Cat #V3151). The sample was diluted by two times using the flow buffer consisting of 1× TBE, 3.6% Tween and 10% polyvinyl pyrrolidone (PVP). For all the solution preparation ultrapure distilled water was used. (Invitrogen, Carlsbad, CA, USA; Cat #10977-015). DNA molecules are electrokinetically driven at 3–5 V at the port of entrance of the chip (Figure 1A in text) and allowed to populate (Figure 1B in text) there for 2–3 min. Applying higher voltage (∼10 V) the populated molecules are moved through the micro pillar structure of the chip to transform the compact golubular DNA structure to an open structure (Figure 1B in text). At the 300 nm channel area the molecules adopt relaxed linear form with some heterogenity on the backbone. There, as one end of a molecule enters the nano-channel, it transiently elongates, adopting a linear conformation with almost homogeneous backbone. The remaining structural heterogeneity progressively disappears as it interacts with the nano-channels, adopting fully confined equilibrium conformation after the field is switched off (relaxation time 10–15 s). Figure 1C shows an image for DNA molecules at equilibrium. A buffer consisting 0.5× TBE, 1.8% Tween, 5% PVP have been used to flow the DNA molecules resulting in a stretch of 65% of 0.34 nm/bp (25.3 ± 1.7 μm; Figure 1D).
Figure 1.
Image of the nano-channel array in a chip that has been used for the linearization of DNA. (A) Different regions of a nano-channel device. (B) Image showing the translocation of DNA through different areas (microstructure and nano-channel) of the chip. (C) Image of the relaxed and linearized DNA molecules inside nano-channel array. (D) Size distribution of BAC 3F5 DNA molecules inside nano-channel array.
Microscopy and image processing
The imaging was done in epi-fluorescence mode using Olympus microscope model IX-71 (Olympus America Inc, Melville, NY, USA) with a 100X SAPO objective (Olympus SApo 100×/1.4 oil). YOYO-1 iodide (491 nm, absorption; 509 nm, emission), the DNA backbone staining dye was excited using 488 nm laser (BCD1, Blue DDD Laser Systems, CVI Melles Griot, Rochester, NY, USA) whereas Cy3 or Alexa 546 (∼550 nm absorption, ∼570 nm emission) was excited using 543 nm green laser (Voltex Inc, Colorado Springs, CO, USA). For Alexa 647 (∼650 nm, absorption; ∼665 nm emission) 633 nm helium–neon laser was used for excitation (JDS Uniphase, San Jose, CA, USA). We used a filter cube consisting of a custom made triple band dichroic and dual band pass emission filters (z488/532/633rpc, z488/543m, respectively) (Chroma Technology Corp., Rockingham, VT, USA) for detection of YOYO-1 and Cy3/Alexa 546 emission by alternative laser excitation with external laser shutters (Thorlabs, Newton, NJ, USA). For detection of Alexa 647, the single band filter cube Cy5-4040A from Semrock Inc., (Rochester, NY, USA) has been used. The emission signal was magnified to 1.6× and detected by a back-illuminated, thermoelectric cooled charge coupled device (EMCCD) detector (iXon) (Andor, Ireland). The data were recorded at 60–80 ms exposure time. All the image analyses were done using Image J (National Institutes of Health, USA) or custom in-house software.
RESULTS AND DISCUSSION
For linear DNA mapping techniques, uniform linearization of DNA is critical. In free solution, DNA polymer chains adopt a coiled configuration and are generally many times smaller than the length of the fully stretched molecule. However, when that same polymer is confined in a channel whose diameter is on the order of its persistence length, self-avoidance forces the polymer to extend, distributing its mass along the channel. We have shown previously that long DNA molecules can be confined and linearized in nano-channels less than 100 nm in diameter (12–14). Figure 1A shows an optical image of the nano-arrary structure, which consists of different sections with different functionalities. The device was fabricated in silicon using conventional 192 nm photolithography and then capped using anodic bonding with a glass substrate. DNA molecules are loaded into the port, and flow past the pillar structures under the electric field. The pillar structures are designed to untangle the randomly coiled DNA molecules before entering the nano-channel (Figure 1B). Figure 1C shows linearized 115 kb circular DNA molecules from BAC clone MCF7-3F5 in the 60 × 100 nm size nano-channel. The length distribution was shown in Figure 1D with a single peak at ∼25.3 micron, corresponding to ∼65% DNA stretching (complete elongation of 115 kb DNA molecules is 39.1 micron). The full length DNA molecules (580 such molecules out of total 1519 molecules) were selected for mapping analysis within the SD of ±1.7 micron. At the current imaging conditions, it takes ∼30 s to collect such amount of molecules. Though the throughput could be dramatically improved with optimized DNA loading and imaging conditions. As reported earlier, DNA stretching >60% is sufficient to provide valuable DNA mapping data (15). This consistent and uniform DNA linearization forms the foundation for single molecule DNA linear analysis.
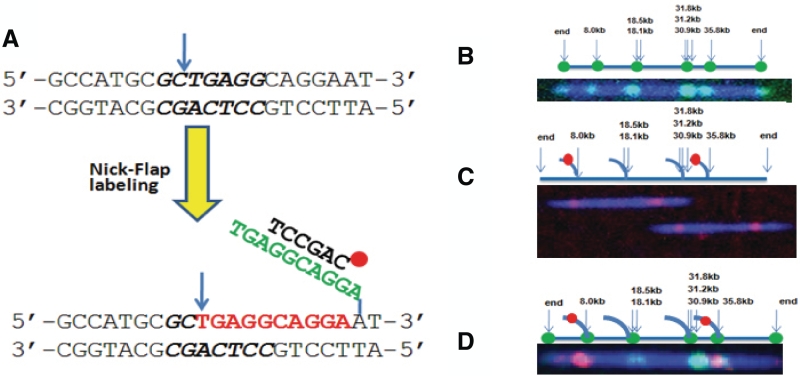
As the linear single DNA molecule mapping scheme requires intact, dsDNA molecules, traditional DNA oligo hybridization techniques are not suitable for marking sequence specific motifs. We adopted a nick–flap labeling scheme for tagging specific sequences along the dsDNA molecules as shown in Figure 2A. This process utilizes hybridization probes capable of recognizing any sequences across the whole genome on dsDNA molecules under non-denaturing conditions (16). In brief, the nicks are introduced in dsDNA at specific sequence motifs recognized by nicking endonucleases, which cleave only one strand of a dsDNA substrate (17). In the direct nick-labeling scheme, fluorescent dye nucleotides can be directly incorporated by DNA polymerase extension, which indicate the presence of nicking endonuclease recognition sequences. In the flap-labeling scheme, a polymerase with 5′-3′ displacement activity but lacking 5′-3′ exonuclease activity such as Vent (exo-) is used for strand extension and displacement of the downstream strand from the nicking sites. The displaced ssDNA sequence segments form flap structures attached to intact dsDNA molecules, which open up more sequences for further information beyond the nicking endonuclease recognition sequences. The nicking sites and flap sequences can be labeled at the same time, a process we term nick–flap labeling. Figure 2A–D demonstrates various labeling schemes, using nicking endonuclease Nb.BbvCI on λ-DNA as a model system. The distributions of the seven nick endonuclease Nb.BbvCI recognition sequences (GCTGAGG) of λ-DNA are shown in the top graph of Figure 2B. There are two nicking sites at ∼18.3 kb and three nicking sites at ∼31.3 kb, which are separated by no more than 1000 bp and thus clustered as one optically resolvable spot at each of these locations. Accordingly, the 7 Nb.BbvCI sites of λ-DNA are collapsed to four resolvable sites at 8, 18.3 (average of 18.1 and 18.5 kb), 31.3 (average of 30.9, 31.2 and 31.8 kb) and 35.8 kb. A typical labeled DNA molecule is shown in the bottom graph of Figure 2A, indicating the experimental data matches well with the predicted map. The resolution is better than 5 kb, as the two spots at 31.3 and 35.8 kb are clearly resolvable. The labeling is very specific due to two enzymatic reactions, DNA nicking by nicking endonuclease and fluorescent dye nucleotide incorporation by polymerase. Furthermore, the fluorescent dye molecules are covalently bound to the dsDNA. Instead of directly tagging the recognition sequences, flap structures can be generated, opening up more sequences other than the nicking enzyme recognition sequences for selective interrogation. Figure 2C shows that two λ-DNA molecules were selectively labeled at the 8 and 35.8 kb flap sites with two sequence specific hybridization probes targeting these two flap sites. Clearly, the integrity of the dsDNA molecules was maintained. This was accomplished by limiting the flap length to ∼50 bp with the combination of amount of polymerase used, reaction temperature, reaction time and especially the amount of nucleotide used in the reaction. In a control experiment, probes designed to hybridize to the sequences after the first 50 bp showed minimal hybridization events (data not shown). This clearly demonstrates that the flap length can be limited to 50 bp under above-mentioned optimized conditions. With 300 full length λ-DNA molecules analyzed, 85% of the two targeted flap sites were labeled. By combining the nick and flap labeling strategies, one can globally label all nicking sites and at the same time selectively label the individual flap sites. One such labeled lambda molecule is shown in Figure 2D. In this case, all the nicking recognition sequences of the λ-DNA molecule are tagged by incorporation of fluorescent nucleotides (green), and two flap sites at 8 and 35.8 kb were hybridized and labeled with two sequence specific probes (red).
Figure 2.
Schematic drawing of recognition sequence of nicking endonuclease Nb.BbvCI and the Nick–Flap labeling scheme. (A) After nicking (blue arrow) at the recognition sequence (GCTGAGG, Nb.BbvCI), fluorescent or non-labeled nucleotides (red) are incorporated using a polymerase with displacement activity but lacking 5′→3′ exonulcease activity. As a result, the native sequences are displaced (green) downstream. A ssDNA structure (flap) is generated and can be interrogated with various chemistry including hybridization probes. For example, an oligo probe (black) can hybridize to the flap. (B) Nick-labeling of λ-DNA molecules. The top graph shows the distribution of the seven nick endonuclease Nb.BbvCI recognition sites of λ-DNA. The solid blue line represents the backbone of the λ-DNA, the arrow indicates the positions of the predicted Nb.BbvCI sites and the green dots represent the potential tagging sites. The bottom graph shows a single labeled λ-DNA molecule. Backbone was labeled with YOYO-1 (blue) and nicking sites were labeled with Alexa-546 dUTP nucleotides and four internal labels were observed and matched well with predicted sites. (C) Flap-labeling. Two labeled flap sites are shown in red dots on a solid line peeling off the DNA backbone in top graph. The bottom graph shows two λ-DNA molecules, whose flap sites at 8 and 35.5 kb were hybridized and labeled with probes Cy3-AAGGTCTTGAGCAGGCCGTT-Cy3 and Cy3-TCCAACTATATAATTT-GACCAGAGAACAAG-Cy3, respectively. In this case the nicking sites were not labeled. (D) Nick–flap labeling. All nicking sites of λ-DNA molecules were labeled with Alexa 647 dUTP (green) and two flap sequences at nicking sites of 8 kb and 35.5 kb were selectively hybridized and labeled with green probes Cy3-AAGGTCTTGA-GCAGGCCGTT-Cy3 and Cy3-TCCAACTATATAA-TTTGACCAGAGAACAAG-Cy3, respectively.
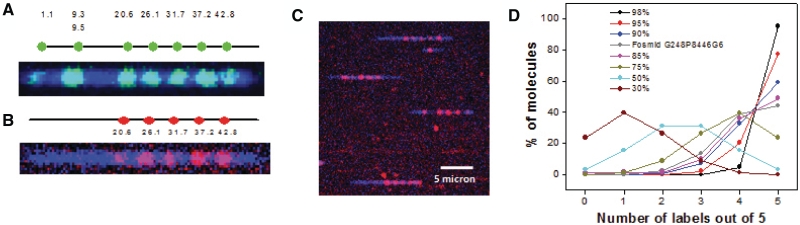
Nick-labeling depends on the nicking endonuclease recognition sequences, and all recognition sequence motifs are tagged during the labeling processes. On the other hand, the flap sequences form a unique subset of genomic sequences, and each flap can be selectively labeled with unique probes. Moreover, a subset of flaps contains repetitive sequence which makes it possible to use one probe to tag many flaps at the same time. The 50 kb circular fosmid G248P8446G6 contains an insert of a 43 kb fragment from chromosome 6, including five copies of 5 kb repetitive elements (6). The top graph in Figure 3A shows the nicking endonuclease Nb.BbvCI nicking pattern of this fosmid DNA, in which there are a total of eight sites with two sites at 9.3 and 9.5 kb merged into a single spot, and five nicking sites within the repeat sequences distributed between 20.6 and 42.8 kb. One typical nicked-labeled molecule is shown in the bottom graph of Figure 3A with seven labeling spots that matches well with predicted nicking pattern. The nick-labeling offers limited information besides the presence of the 6 bp recognition sequence GCTGAGG. To obtain more sequence information, flap structures were generated and a single probe was designed to hybridize all five flap sites within the repetitive sequences. Clearly, five spots were observed between 20.6 and 42.8 kb and indicate the presence of multiple copies of the probe sequence TGCCTGTGAGAGGAAATCTCAACTCTCTT (Figure 3B). Figure 3C shows a few typical DNA molecules in the field of view. Together with the nicking enzyme Nb.BbvCI recognition sequence, this clone can be uniquely mapped back to a segment of human reference genome chromosome 6 containing the LPA gene. A total of 468 DNA molecules were analyzed. The numbers of molecules with 5, 4, 3, 2, 1 and 0 labels were 213, 164, 52, 30, 7 and 2, respectively. Assuming that the labeling of each spot is independent with the efficiency of P, the probability of molecules with m sites labeled out of a total of n sites observes n! (1−P)(n−m)Pm/(n−m)!m!. Here n = 5, and the calculated P matches the simulated fitting of 85–90% labeling efficiency (Figure 3D, gray line).
Figure 3.
Image of Fosmid G248P8446G6 DNA in nano-channel (60 nm × 100 nm). The DNA was nicked with Nb.BbvCI and the free 3′ end is extended by Vent (exo-) in presence of a mixture of three unlabeled nucleotides (dAGC) and Alexa-546 labeled dUTP (Figure 3A) or a mixture of four unlabeled nucleotides (dNTP) (Figure 3B). The DNA backbone was stained with intercalated dye YOYO-1 iodide. (A) Eight nicked sites were thus labeled with Alexa 546 (green). Two nicking sites at ∼9.3 and ∼9.5 kb were too close to resolve optically. The DNA backbone is indicated as a blue line. The positions of the labeled dyes match well with the predicted nicking positions on the backbone. (B) The generated single strand flaps (by nick translation in presence of dNTP mixtures) were hybridized with dye labeled probe of sequences Cy3-TGCCTGTGAGAGG-AAATCTCAACTCTCTT-Cy3. Five out of the eight single strand flaps contain the complement of the probe sequence and thus get hybridized. (B) shows five labels (red) along with the blue backbone. All these positions match well with the predicted ones (6). (C) Image shows several full length flap labeled Fosmid molecules inside a nano-channel array. (D) The prediction of labeling efficiency of one site in a DNA molecule with maximum five labeling sites available in it. The lines show the changes in the number distribution of 1, 2, 3, 4 and 5 labeled molecules with change in labeling efficiencies (30, 50, 75, 85, 90, 95 and 98%). The gray line is the distribution of number of molecules that were experimentally obtained from the flap labeled (five sites) Fosmid G248P8446G6. This line shows its labeling efficiency ∼85–90%. The imaging procedure is described in the ‘Material and Methods’ section.
To demonstrate the capabilities of our linear DNA mapping method, we mapped a 115 kb BAC clone 3F5 with flap labeling. BAC clone 3F5 is from a breast cancer cell line, which displays highly rearranged structures. Based on pair end sequencing data, it consists of four segments: an inverted 14.1 kb block from 3p14.1; an inverted 22.3 kb block from 20q12 containing exon 6 of the PTPRT gene; a 45.5 kb block from 20q13.31 containing exon 1 of the truncated BMP7 gene along with its intact promoter and a 23.4 kb block from 20q13.2 containing the complete ZNF217 gene (18). There are a total of 52 endonuclease Nb.BbvCI nicking sites on this BAC clone, which is similar to the density of the whole genome. To selectively tag these nicking sites, a universal hybridization probe was designed to target a subset of nicking sites based on the conserved flap sequences (Table 1). Figure 4A is a false color two-channel composite image showing several DNA molecules (YOYO-1 in blue) in the nano-channel array with the universal probe (red) hybridized to selective nicking sites. The predicted sequence tagging motif map is shown in Figure 4B, the square indicates the perfect sequence match between probe and flap sequences, while the circle represents one mismatch. Due to our current 2.5 kb resolution, some labels closer than 2.5 kb will be unresolvable and cluster as large spots. Four such typical full-length DNA molecules are shown in Figure 4C. At 33 and 70 kb loci, the probe hybridized to flaps with one base mismatch, which indicates the hybridization under this condition tolerates single base mismatch. Figure 4D is the sequence motif map of the BAC clone DNA. Three individual peaks were calculated as 33, 39 and 77 kb from one end, and three clusters also exist in between them, which show good agreement with the predicted distribution of sequence motif (Figure 4B).
Figure 4.
Mapping BAC clone 3F5. (A) A two color superimposed image shows several dsDNA molecules (blue, YOYO-1 stained backbone) and selectively labeled flaps (red) with one universal probe Cy3-ATT+CTCCTGCC+TCA-Cy3 (+C and +T are LNA bases) in the nano-channel array (60 × 100 nm channel). (B) The predicted sequence tagging motif map, square indicating the perfect sequence match between probe and flap sequences, while the circle representing one mismatch. (C) Several full length dsDNA molecules (blue, YOYO-1 stained backbone) and hybridized labels (red) are lined up against the predicted map in Figure 4B (18). (D) Sequence motif map calculated based on the distance from one end and matchs well with the predicted map.
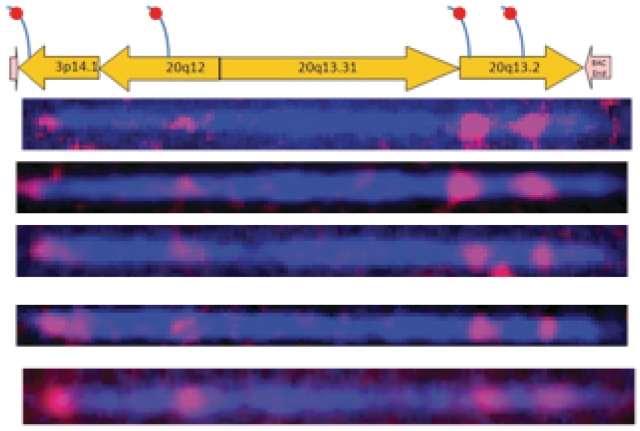
To further validate the presence of different rearrangements on this clone, we selectively labeled different segments with segment specific probes, shown in Figure 5A as a schematic drawing. One such probe was designed to hybridize the flap structure of a 14.1 kb block from chromosome 3p14.1, another probe designed for a flap on a 22.3 kb block from chromosome 20q12, and two other probes were hybridized on a 23.4 kb block from chromosome 20q13.2 containing the complete ZNF217 gene. The resulting image in Figure 5B shows four labels on 115 kb dsDNA at locations of 1.3, 30.0, 83.3 and 95.5 kb, respectively. The spatial distribution matches well with the known locations of the flap sequences. The presence of two ZNF217 region specific probes separated by 12 kb confirms the clone contains 23.4 kb ZNF217 block from 20q13.2 and also confirms the translocation of two other segments from chromosome 3p14.1 and chromosome 20q13.2. The detection of the presence of a particular gene such as ZNF217 could be useful in clinical diagnostics, as oncogen ZNF217 gene has been used as a prognostic marker in human tumors, and amplification of this gene has been associated with reduced survival duration in human breast cancer (19,20).
Figure 5.
Validation of structural variations. Top graph indicates the predicted locations of probe sequences on different segments. Bottom graph shows several labeled dsDNA molecules match well with the predicted map with four probes hybridized to different segments (i) TCCTTGGTTGACCTAACAACACA (3p14.1), (ii) TGCCACCTACCCCT (20q12), (iii) TCCAAGTCTCAGTGACCCT (20q13.2) and (iv) ACTGTAGTCTTGAATTCCTGA (20q13.2).
CONCLUSIONS
In conclusion, we have demonstrated the capabilities of our integrated approach of single molecule DNA linear analysis. By confining and stretching long DNA molecules in nano-channel arrays, sequence information can be extracted through single molecule linear analysis. The flexible and efficient fluorescent tagging of specific sequences allow us to obtain context specific sequence information along the long linear DNA molecules within the nano-channel. Our global nick-labeling scheme tags short recognition sequences, whose spatial relation can be translated into a genomic map. By creating single-stranded flap sequences at the nicking sites, the 3 Gbp human genome is reduced to ∼50 Mbp of target sequences available for probe hybridization (considering the 50 bp flap length and 1 million total nicking sites), with the rest of the double stranded genome protected from labeling events. The content and distribution of these flap sequences offer a very interesting picture of the whole genome, and can be utilized for various applications. For example, a large percentage of the flap sequences are unique in the genome and can be used to study targeted regions in the genome for CNVs, inversions or specific translocations. In addition, a significant fraction of flap sequences generated by some enzymes (such as Nb.BbvCI, with recognition site CCTCAGC) contains conserved sequences such that one or more probes can target many of these flaps. This integrated approach of single molecule DNA linear analysis will find wide range applications in mapping structural variations over long intact strands that are associated with complex genetic traits in individual human or cancer genomes. Such complex genomic structural information is often difficult to be assembled accurately and cost effectively through short read sequencing method.
FUNDING
National Institutes of Health (grant R43HG004817). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. All authors are employees of the commercial company, Bionanomatrix. They have filed patents based on these findings and own the company’s stocks.
ACKNOWLEDGEMENTS
Fosmid G248P8446G6 was a gift from Dr E. Eichler of University of Washington, Seattle, WA USA and MCF7 BAC clones were gift from Dr A. Milosavljevic of Baylor College of Medicine, Houston, USA.
REFERENCES
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 2002;11:1987–1995. doi: 10.1093/hmg/11.17.1987. [DOI] [PubMed] [Google Scholar]
- 4.Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 7.Jing JP, Reed J, Huang J, Hu XH, Clarke V, Edington J, Housman D, Anantharaman TS, Huff EJ, Mishra B, et al. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc. Natl Acad. Sci. USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin JY, Qi R, Aston C, Jing JP, Anantharaman TS, Mishra B, White O, Daly MJ, Minton KW, Venter JC, et al. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 9.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, vanSlegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 10.Chan EY, Goncalves NM, Haeusler RA, Hatch AJ, Larson JW, Maletta AM, Yantz GR, Carstea ED, Fuchs M, Wong GG, et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo K, Dhingra DM, Odijk T, de Pablo JJ, Graham MD, Runnheim R, Forrest D, Schwartz DC. A single-molecule barcoding system using nanoslits for DNA analysis. Proc. Natl Acad. Sci. USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao H, Tegenfeldt JO, Austin RH, Chou SY. Gradient nanostructures for interfacing microfluidics and nanofluidics. Appl. Phys. Lett. 2002;81:3058–3060. [Google Scholar]
- 13.Cao H, Yu ZN, Wang J, Tegenfeldt JO, Austin RH, Chen E, Wu W, Chou SY. Fabrication of 10 nm enclosed nanofluidic channels. Appl. Phys. Lett. 2002;81:174–176. [Google Scholar]
- 14.Tegenfeldt JO, Prinz C, Cao H, Chou S, Reisner WW, Riehn R, Wang YM, Cox EC, Sturm JC, Silberzan P, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc. Natl Acad. Sci. USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimalanta ET, Lim A, Runnheim R, Lamers C, Churas C, Forrest DK, de Pablo JJ, Graham MD, Coppersmith SN, Goldstein S, et al. A microfluidic system for large DNA molecule arrays. Analytical Chemistry. 2004;76:5293–5301. doi: 10.1021/ac0496401. [DOI] [PubMed] [Google Scholar]
- 16.Xiao M, Phong A, Ha C, Chan TF, Cai DM, Leung L, Wan E, Kistler AL, DeRisi JL, Selvin PR, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2007;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan RD, Calvet C, Demeter M, Agra R, Kong HM. Characterization of the specific DNA nicking activity of restriction endonuclease N.BstNBI. Biol. Chem. 2000;381:1123–1125. doi: 10.1515/BC.2000.137. [DOI] [PubMed] [Google Scholar]
- 18.Hampton OA, Den Hollander P, Miller CA, Delgado DA, Li J, Coarfa C, Harris RA, Richards S, Scherer SE, Muzny DM, et al. A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome. Genome Res. 2009;19:167–177. doi: 10.1101/gr.080259.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang S, Polikoff D, Nonet G, Cochran J, Myambo K, et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc. Natl Acad. Sci. USA. 1998;95:8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray JW, Kallioniemi OP, Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: Prognostic implications. Clin. Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]