Abstract
The transcriptional coactivator Sub1 has been implicated in several aspects of mRNA metabolism in yeast, such as activation of transcription, termination, and 3′-end formation. Here, we present evidence that Sub1 plays a significant role in controlling phosphorylation of the RNA polymerase II large subunit C-terminal domain (CTD). We show that SUB1 genetically interacts with the genes encoding all four known CTD kinases, SRB10, KIN28, BUR1, and CTK1, suggesting that Sub1 acts to influence CTD phosphorylation at more than one step of the transcription cycle. To address this directly, we first used in vitro kinase assays, and we show that, on the one hand, SUB1 deletion increased CTD phosphorylation by Kin28, Bur1, and Ctk1 but, on the other, it decreased CTD phosphorylation by Srb10. Second, chromatin immunoprecipitation assays revealed that SUB1 deletion decreased Srb10 chromatin association on the inducible GAL1 gene but increased Kin28 and Ctk1 chromatin association on actively transcribed genes. Taken together, our data point to multiple roles for Sub1 in the regulation of CTD phosphorylation throughout the transcription cycle.
A prominent feature of the largest subunit of RNA polymerase II (RNAP II), Rpb1, is the presence of a highly conserved carboxy-terminal domain (CTD) that has an essential role in transcription regulation in vivo (12, 17, 54). Although the RNAP II CTD is not required for transcription in promoter-independent assays in vitro, it is essential in vivo (50), and it is required for efficient capping, splicing, and cleavage/polyadenylation of pre-mRNAs (15, 29, 47). In fact, the CTD has been described as a platform that recruits RNA processing/export and histone-modifying factors to the transcription complex, coupling mRNA metabolism to chromatin function (8, 54).
The CTD is characterized by repetition of the consensus heptapeptide sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser, ranging from 26 repeats in yeast to 52 in mammals, which is subjected to highly regulated phosphorylation (14, 15, 47). Unphosphorylated RNAP II is mostly recruited to the preinitiation complex (PIC) (45), and hyperphosphorylated RNAP II is associated with initiation and elongation complexes (42). The CTD is phosphorylated on serine 5 of the heptapeptide repeat predominantly during promoter escape and early elongation, while serine 2 becomes phosphorylated principally during elongation (16, 38). In addition, it has recently been demonstrated that the CTD can be also phosphorylated on serine 7 (3, 13, 25).
Phosphorylation of the CTD is achieved primarily by members of the cyclin-dependent kinase (CDK) family, which typically consist of a catalytic subunit and a regulatory cyclin subunit (47). In Saccharomyces cerevisiae, at least four Cdk complexes, composed of Kin28-Ccl1-Tfb3, Srb10-Srb11, Ctk1-Ctk2-Ctk3, and Bur1-Bur2, are able to phosphorylate the CTD, and all of them have a role in transcription regulation (47, 54). Srb10 (Cdk8 in higher eukaryotes) provides the kinase activity of the Mediator CDK8 module. Genetically, Srb10 has been found to act both positively and negatively in gene expression (12, 30). It has also been shown through in vitro studies to phosphorylate the CTD, on the one hand inactivating RNAP II prior to PIC formation (27) but, on the other, promoting transcription and formation of the scaffold complex (43). Kin28 (mammalian Cdk7), with Ccl1 and Tfb3, forms the transcription factor TFIIK subcomplex of the TFIIH initiation complex (reference 34 and references therein). Phosphorylation on Ser5 of the CTD by Kin28 is required for efficient cotranscriptional recruitment of 5′ capping enzymes and the placement of the 7-methyl guanosine cap on pre-mRNAs (38, 59, 63), although it is not essential for transcription (31). Kin28, as well as Cdk7, can also phosphorylate the Ser7 residue of the CTD repeats (3, 25), and Cdk7 functions in promoter-proximal pausing and, perhaps, termination by RNA polymerase II (25).
The phosphorylation of Ser2 is more complex. In mammalian and Drosophila cells, Cdk9/cyclinT, or P-TEFb, phosphorylates Ser2 and functions to promote transcription elongation (56). In S. cerevisiae, P-TEFb activity is split between two separate complexes: the CTD kinase 1 complex (CTDK1), consisting of Ctk1, Ctk2, and Ctk3, and the Bur1/Bur2 complex (16, 71). Both complexes have been implicated in CTD phosphorylation during elongation. While Ctk1 is the main kinase for Ser2 phosphorylation (16), the specificity of the Bur1/Bur2 complex kinase is not yet clear. Murray et al. (49) examined the phosphorylation site specificity of Bur1 and showed that Bur1 associates primarily with Rpb1 and phosphorylates Ser5. However, it has been shown recently that Bur1/Bur2, in addition to phosphorylating Ser2 near promoters, also stimulates Ser2 phosphorylation by Ctk1 during elongation (57). In addition, Bur1 phosphorylates the histone modifier Rad6/Bre1 (69, 70) and the carboxyl-terminal domain of the elongation factor Spt5, stimulating recruitment of the PAF1 elongation complex (44, 73).
Another factor involved in modulation of CTD phosphorylation in S. cerevisiae is Sub1. Sub1 was originally identified as a suppressor of TFIIB mutations and as a transcriptional stimulatory protein, homologous to human positive coactivator PC4 (24, 33, 40, 46, 68), that physically interacts with TFIIB, arguing for a role as coactivator in transcription initiation by RNAP II (28, 37). In that sense, Rosonina et al. (60) showed that Sub1 contributes to the activation of osmoresponse genes during osmotic shock through the assembly or stabilization of promoter-associated complexes. On the other hand, Koyama et al. (39) proposed a role for Sub1 as a repressor of the inducible IMD2 gene. Sub1 has also been implicated in other aspects of mRNA metabolism, such as transcription termination and 3′-end formation (10, 26). Moreover, several years ago, we described allele-specific interactions between SUB1 and both KIN28 and FCP1, which encodes the CTD phosphatase Fcp1. We showed that cells lacking Sub1 displayed decreased accumulation of Fcp1, altered RNAP II phosphorylation, and decreased cross-linking of RNAP II to transcribed genes (11). These results indicated that Sub1 has a role in RNAP II CTD phosphorylation and in transcription elongation.
Here, we present evidence that Sub1 indeed plays a universal role in CTD phosphorylation. We show that SUB1 genetically interacts with the genes encoding all four of the CTD kinases, SRB10, KIN28, BUR1, and CTK1, suggesting that Sub1 acts to influence CTD phosphorylation at more than one step of the transcription cycle. Supporting this view, we show first, by the results of in vitro kinase assays, that SUB1 deletion increases CTD phosphorylation by Kin28, Bur1, and Ctk1 but decreases CTD phosphorylation by Srb10, arguing for distinct roles of Sub1 in transcription preinitiation and elongation. Second, by the results of chromatin immunoprecipitation, we find that SUB1 deletion increases Kin28 and Ctk1 chromatin association while decreasing Srb10 chromatin association, indicating that Sub1 is involved in regulating the association of these kinases with the transcriptional machinery. Taken together, our data point to multiple roles for Sub1 in the regulation of RNAP II CTD phosphorylation all along the transcription cycle.
MATERIALS AND METHODS
Yeast strains and media.
The strains used in this study are listed in Table 1. Yeast strain construction and other genetic manipulations were performed by standard procedures (9). The 2% galactose and 2% raffinose media were prepared as described previously (41).
TABLE 1.
Yeast strains
| Strain | Description | Source |
|---|---|---|
| YSB756 | matα ade2-1 ade3-22 can1-100 his3-11,15 ura3-1 kin28::LEU2 [KIN28-3×HA TRP CEN] | S. Buratowski |
| OCSC154 | matα ade2-1 ade3-22 can1-100 his3-11,15 ura3-1 kin28::LEU2 [KIN28-3×HA TRP CEN] sub1::URA3 | This study |
| YSB609 | matα ade2-1 ade3-22 can1-100 his3-11,15 ura3-1 kin28::LEU2 [kin28-K36A-3×HA TRP CEN] | S. Buratowski |
| YSB776 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 SRB10-3×HA::TRP | S. Buratowski |
| OCSC159 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 SRB10-3×HA::TRP sub1::URA3 | This study |
| OCSC166 | matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 sub1::URA3 | This study |
| OCSC261 | matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 srb10::KAN | This study |
| OCSC169 | matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 srb10::KAN sub1::URA3 | This study |
| OCSC267 | matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 ADH1-SUB1 | This study |
| OCSC268 | matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 srb10::KAN [ADH1-SUB1 LEU CEN] | This study |
| YSB786 | matahis3-Δ200 leu2-3,112 ura3-52 ceg1-Δ1::his3 [CEG1-3×HA LEU CEN] | S. Buratowski |
| OCS570 | matahis3-Δ200 leu2-3,112 ura3-52 ceg1-Δ1::his3 [CEG1-3×HA LEU CEN] sub1::URA3 | This study |
| YSB770 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 BUR1-3×HA::TRP | S. Buratowski |
| OCSC157 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 BUR1-3×HA::TRP sub1::URA3 | This study |
| OCSC560 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 BUR1-3×HA::TRP SUB1-GST:: KanMX | This study |
| YSB1021 | matahis3-Δ200 leu2-Δ1 trp1-Δ63 ura3-52 lys2-Δ202 bur1Δ::HIS3 [bur1-23-3×HA LEU CEN] | S. Buratowski |
| GY458 | matahis4-912δ lys2-128δ suc2-Δuas (−1900/−390) ura3-52 trp1-Δ63 | G. Prelich |
| OCSC301 | matahis4-912δ lys2-128δ suc2-Δuas (−1900/−390) ura3-52 trp1-Δ63 sub1::URA3 | This study |
| GY170 | matahis4-912δ lys2-128δ suc2-Δuas (−1900/−390) ura3-52 trp1-Δ63 bur1-2 | G. Prelich |
| OCSC303 | matahis4-912δ lys2-128δ suc2-Δuas (−1900/−390) ura3-52 trp1-Δ63 bur1-2 sub1::URA3 | This study |
| ERYM356 | mataura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 CTK-6×HA::TRP1 | This study |
| ERYM357 | mataura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 CTK1-6×HA::TRP1 sub1::KanMX | This study |
| OCSC558 | mataura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 CTK-6×HA::TRP1 SUB1-GST::KanMX | This study |
| OCSC1077 | mataura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 CTK-6×HA::TRP1 SRB10-MYC::HIS3 | This study |
| OCSC1078 | mataura3-52 lys2-801amber ade2-101ochre trp1-Δ63 his3-Δ200 leu2-Δ1 CTK1-6×HA::TRP1 SRB10-3×MYC::HIS3 sub1::KanMX | This study |
| OCSC1160 | mataura3-52 lys801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 KIN28-6×HA::TRP1 | This study |
| OCSC1168 | mataura3-52 lys801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 KIN28-6×HA::TRP1 SUB1-3×FLAG::KanMX | This study |
| YP655 | matα ade2-1 his4-260 leu2-3,112 trp1-289 ura3-1 CDC5-3×HA::KanMX | P. San Segundo |
| OCSC1323 | matα ade2-1 his4-260 leu2-3,112 trp1-289 ura3-1 CDC5-3×HA::KanMX sub1::URA3 | This study |
In vitro kinase assays.
Cells were grown to an optical density at 600 nm (OD600) of 0.8, collected, washed, and suspended in lysis buffer (20 mM HEPES [pH 7.6], 200 mM potassium acetate [KOAc], 10% glycerol, and 1 mM EDTA) (35) with protease and phosphatase inhibitors. Yeast whole-cell extracts were prepared by glass bead disruption of cells using a FastPrep system. Protein concentrations were determined, and 150 μg was incubated with 15 μl of 12CA5 (antihemagglutinin [anti-HA]) or anti-MYC antibody coupled to protein G-Sepharose for 2 h at 4°C to immunoprecipitate HA- and MYC-tagged kinases (Kin28, Srb10, Ctk1, and Bur1). The immunoprecipitates were washed three times with lysis buffer and twice with kinase buffer (20 mM HEPES [pH 7.6], 7.5 mM MgOAc, 100 mM KOAc, 2% glycerol). The beads were resuspended in 25 μl of kinase buffer with 2.5 mM ATP and incubated with 30 ng of glutathione S-transferase (GST)-CTD for 30 min at 30°C. The reaction mixtures were run and then electrophoresed on 8% SDS-polyacrylamide gels, transferred, and immunoblotted with the following antibodies: 8WG16 (nonphosphorylated CTD; Covance), CTD4H8 (phosphorylated CTD Ser5 [CTD Ser5P]; Millipore), or ab5095 (CTD Ser2P; Abcam).
ChIPs.
The preparation of chromatin was performed as previously described (11, 35). For the Ser5P immunoprecipitations (IPs), CTD4H8 antibody was used and immunoprecipitation was performed as described in reference 11. PCR of purified chromatin was performed by quantitative real-time PCR with an ABI Prism 7000 detection system (Applied Biosystems), using SYBR Premix Ex Taq (Takara Bio, Inc.) and following the manufacturer's instructions. Four serial 10-fold dilutions of genomic DNA were amplified using the same reaction mixture as for the samples to construct the standard curves. All real-time PCRs were performed in quadruplicate and with at least three independent chromatin IPs (ChIPs). Quantitative analysis was carried out using the ABI Prism 7000 SDS software (version 1.2.3). The values obtained for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of the value for each PCR product of transcribed genes to that of a nontranscribed region of chromosome VII was calculated. Numbers on the y axis of graphs are detailed in the corresponding figure legends. ChIP assays of cells grown under conditions of galactose induction were performed as described previously (41).
RNA isolation and RT-PCR.
Total RNA was extracted as described previously (62), and reverse transcription (RT)-PCR was performed using a PrimeScript RT reagent kit (Takara Bio, Inc.) following the manufacturer's instructions.
RESULTS
SUB1 deletion enhances both CTD Ser5 phosphorylation by Kin28 and its recruitment to gene promoters.
We previously described a genetic interaction between SUB1 and KIN28 (11), suggesting that Sub1 influences CTD phosphorylation by Kin28. In order to study the significance of this genetic interaction, we followed two strategies. We first performed in vitro kinase assays to determine if Sub1 influences CTD phosphorylation by Kin28 and then carried out chromatin immunoprecipitation (ChIP) experiments to analyze whether Sub1 influences Kin28's association with gene promoters.
We first tested whether Sub1 affects Kin28 activity toward the CTD. HA-Kin28-containing complexes from SUB1 (wild type [wt]) and sub1Δ whole-cell extracts prepared from cells expressing HA-Kin28 in place of endogenous Kin28 were immunoprecipitated via the HA epitope tag and assayed in an in vitro IP kinase assay using as the substrate a GST-CTD fusion protein, as previously described (35). GST-CTD was incubated with no kinase or with HA-Kin28 immunoprecipitated from wt and sub1Δ cell extracts (Fig. 1 A). Reaction mixtures were loaded onto an 8% SDS-PAGE gel and immunoblotted with CTD4H8 antibody, which recognizes CTD phosphorylation on Ser5 (53, 64), and with 8W16G antibody, which recognizes unphosphorylated CTD (7, 65). Equivalent IP of Kin28-HA was confirmed by Western blot assay using an anti-HA antibody. As shown by the results in Fig. 1A, Kin28-HA was active on GST-CTD in both wt and sub1Δ cell IPs, since it efficiently phosphorylated the CTD on Ser5 as determined by immunoblotting. Strikingly, Kin28 activity in the IPs from sub1Δ cells was greater than its activity in those from wt cells. Since SUB1 deletion did not affect total Kin28 levels (Fig. 1A, HA blot), this result suggests that Sub1 negatively influences Kin28 kinase activity.
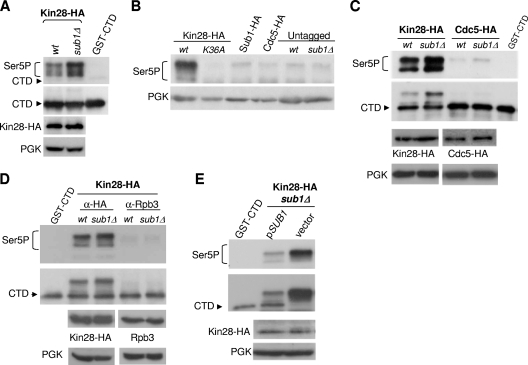
FIG. 1.
SUB1 deletion increases Kin28 CTD kinase activity and Kin28 cross-linking to gene promoters. (A) In vitro kinase assay. Whole-cell extracts were prepared from wild-type (wt) and sub1Δ strains expressing HA-tagged Kin28. The epitope-tagged kinase complexes were immunoprecipitated with 12CA5-protein A beads, and kinase activity was assayed with 2.5 mM ATP and recombinant GST-CTD as substrate. SDS-PAGE and immunoblot analysis were performed to analyze CTD phosphorylation, using the following antibodies: CTD4H8 (anti-CTD Ser5P), 8WG16 (anti-CTD), 12CA5 (anti-HA, for Kin28-HA), and PGK (anti-PGK, as total protein level control). (B) Whole-cell extracts were prepared from the following strains: tagged Kin28-HA (wt and kin28-K36A mutant), Sub1-HA, and Cdc5-HA strains and two nontagged strains (wt and sub1Δ). In vitro kinase assays were conducted as described for panel A, and CTD Ser5 phosphorylation analyzed with CTD4H8 antibody. (C) Whole-cell extracts were prepared from wt and sub1Δ strains expressing HA-tagged Cdc5, and in vitro kinase assays performed to analyze CTD Ser5P as described above. (D) In vitro kinase assay to analyze CTD Ser5 phosphorylation was performed using immunoprecipitated Kin28-HA and Rpb3 from wt and sub1Δ Kin28-HA tagged strains. (E) Kin28-HA sub1Δ cells were transformed with an empty plasmid (vector) or with a plasmid bearing SUB1 (pSUB1) under the control of its own promoter. Kin28-HA was immunoprecipitated, kinase assays performed, and CTD Ser5P analyzed as described for panel B.
In light of the above results and of additional related results described below, we wished to verify the specificity of the IP kinase assay. To this end, we performed a number of additional control experiments. First, we analyzed the extent of nonspecific background kinase activity by using extracts from strains expressing the following proteins for anti-HA IP: Kin28-HA (wt and Kin28-K36A, a mutant devoid of kinase activity [59]), Sub1-HA, Cdc5-HA (a non-CTD kinase functioning in mitosis and cytokinesis [5]), and two nontagged strains (wt and sub1Δ). As shown by the results in Fig. 1B, only wild-type Kin28-HA was able to phosphorylate GST-CTD efficiently. Thus, IPs of HA-Sub1, HA-Cdc5, or extracts not expressing a tagged protein contained no significant Ser5 kinase activity. Second, we immunoprecipitated the HA-tagged unrelated kinase Cdc5-HA from sub1Δ as well as wt extracts and detected no Ser5 kinase activity in either case (Fig. 1C). Third, we analyzed the possibility that the kinase activities observed in wt and sub1Δ cell extracts could be due to other kinases that might coimmunoprecipitate with HA-Kin28, perhaps reflecting a bridging interaction with some other associated factor, such as RNAP II. For this purpose, we immunoprecipitated Kin28-HA and RNAP II (with anti-HA and Rpb3 antibodies, respectively) from wt and sub1Δ Kin28-HA extracts. GST-CTD kinase activity was observed only in the Kin28-HA IPs (Fig. 1D). Fourth, to provide further evidence that loss of Sub1 was indeed responsible for the observed effects on GST-CTD kinase activity, Kin28-HA sub1Δ cells were transformed with an empty plasmid or with a plasmid bearing SUB1 under the control of its own promoter and IP kinase assays were performed as described above. As shown by the results in Fig. 1E, wild-type levels of kinase activity were restored when SUB1 was expressed from the low-copy-number plasmid in sub1Δ cells, confirming that increased Kin28 kinase activity was in fact due to lack of Sub1. Finally, Sub1 did not coimmunoprecipitate with Kin28-HA (or with the three other CTD kinases analyzed below) when using extracts from wt cells (data not shown), indicating that its effects on kinase activity were indirect (see Discussion).
We next wished to determine whether Sub1 also affects Kin28 recruitment to gene promoters. We therefore analyzed the recruitment of Kin28 to the promoters of several constitutively transcribed genes, ADH1, ACT1, PMA1, and PYK1, in the presence or absence of Sub1. For this purpose, we performed ChIP coupled with quantitative PCR (qPCR) with Kin28-HA-expressing wt and sub1Δ cells. Significantly, Kin28 cross-linking was increased at the promoter regions of all genes tested in the absence of Sub1 (Fig. 2 A). The enrichment of Kin28 at promoters in sub1Δ cells compared to that in wt cells (considered 100%) ranged from 125% at the ADH1 promoter to 225% at the ACT1 promoter. These data are in agreement with the observed increase of CTD phosphorylation in sub1Δ cells shown previously (11). Taking into account that Sub1 negatively affects the recruitment of RNAP II (11, 60), as we have confirmed here for sub1Δ cells (data not shown), the Kin28-HA/Rpb1 ratio was increased at promoter regions in sub1Δ cells compared to the ratio in wild-type cells (Fig. 2B). Hence, SUB1 deletion significantly enhanced the levels of Kin28 associated with the RNAP II transcriptional machinery. Taken together, our data support the idea that Sub1 associates with the transcription complex and modulates RNAP II CTD Ser5 phosphorylation by Kin28.
FIG. 2.
SUB1 deletion increases Kin28 and Rpb1 Ser5P cross-linking to gene promoters. ChIP analysis. ChIPs were performed on wt and sub1Δ Kin28 HA-tagged strains. (A) Kin28 binding to the promoters of four constitutively expressed genes, ADH1-P, ACT1-P, PYK1-P, and PMA1-P, was examined by qRT-PCR, and quantifications (see Materials and Methods) were graphed. Numbers on the y axis represent the percentages of Kin28 cross-linked to gene promoters in sub1Δ cells relative to the levels in wt cells, where the level of cross-linking is considered to be 100%. (B) Kin28-HA/Rpb1 ratio. Percentages of Kin28-HA and Rpb1 cross-linking for sub1Δ cells relative to the levels in wt cells were independently quantified, and the ratio was calculated and then graphed. (C) ChIP analysis of Rpb1 and CTD Ser5P was performed in wild-type (wt) and sub1Δ cells using 8WG16 (anti-Rpb1) and CTD4H8 (anti-CTD Ser5P) antibodies. Rpb1 and Rpb1 CTD Ser5P binding to promoters of ACT1, PYK1, and PMA1 genes was analyzed by qRT-PCR, and the results graphed. Numbers on the y axis represent the percentages of Rpb1 and Rpb1 CTD Ser5P cross-linked to gene promoters in sub1Δ cells relative to the levels in wt cells, where the level of cross-linking is considered to be 100%. (D) Occupancy of Rpb1 and Ceg1-HA at promoters of ACT1, PYK1, and PMA1 genes was determined by ChIP in wt and sub1Δ cells using 8WG16 and HA antibodies. Numbers on the y axis represent the percentages of Rpb1 and Ceg1-HA cross-linked to gene promoters in sub1Δ cells relative to the levels in wt cells, where the level of cross-linking is considered 100%. Error bars show standard deviations.
Promoter association of Ser5-phosphorylated RNAP II and the capping enzyme Ceg1 increases in sub1Δ cells.
Ser5 phosphorylation is strongest at the promoter regions of transcribed genes and diminishes downstream (38). We therefore tested whether increased Kin28 recruitment to promoters in sub1Δ cells resulted in increased Ser5 CTD phosphorylation at the promoters of several genes. For this purpose, we performed ChIP on wt and sub1Δ cells with CTD4H8 and 8WG16 antibodies. As expected and as previously described (11), decreased RNAP II was detected at the promoters of ACT1, PMA1, and PYK1 genes in sub1Δ cells compared to the amounts in wt cells. However, Ser5P cross-linking in sub1Δ cells increased significantly at ACT1-P and PYK1-P and slightly at PMA1-P (Fig. 2C). Therefore, SUB1 deletion caused an increase in Ser5 phosphorylation associated with gene promoters relative to total RNAP II, in agreement with the observed increase in Kin28 activity and association with chromatin.
Kin28 phosphorylation of the CTD at Ser5 mediates cotranscriptional recruitment of the capping enzyme Ceg1 (e.g., see references 38, 63, and 67). We previously reported that SUB1 genetically interacts in an allele-specific manner with several kin28 mutants (kin28-T17D, kin28-K36D, and kin28-T162D) (11). Interestingly, these interactions are comparable to interactions previously described between ceg1-250 and kin28 mutants (59). That is, the effects displayed by combining sub1Δ or ceg1-250 mutations with kin28 mutations resulted in similar phenotypes, ranging from lethality (sub1Δ kin28-T17D and ceg1-250 kin28-T17D) to slight or no effect (sub1Δ kin28-T162D and ceg1-250 kin28-T162D, respectively). Although we did not observe a genetic interaction between sub1Δ and ceg1-250 mutants (11), we hypothesized that due to the increased Kin28 activity and increased CTD Ser5P in sub1Δ cells, Ceg1 recruitment to promoters could also be increased. To test this, ChIP analysis was performed with wt and sub1Δ cells expressing Ceg1-HA. As shown by the results in Fig. 2D, recruitment of the capping enzyme was in fact decreased in sub1Δ cells compared to the level in wt cells (Fig. 2D). However, if we compare Ceg1 and Rpb1 occupancies at promoters and calculate the Ceg1/Rpb1 ratio, considered to be 1.0 in wt cells, there was a slight increase in the Ceg1/Rpb1 ratio at the PYK1 (1:1.33) and ACT1 (1:1.57) promoters in sub1Δ cells (Fig. 2D, compare the results for Rpb1 and Ceg1-HA). Notably, in the case of PMA1, where the CTD Ser5P/Rpb1 ratio was 1.0, the Ceg1/Rpb1 ratio was also 1.0; in the case of ACT1, where the CTD Ser5P/Rpb1 ratio was >1 (1.93), the Ceg1/Rpb1 ratio was also >1 (1.57), in agreement with a functional relationship between Ser5 phosphorylation and Ceg1 capping enzyme recruitment.
SUB1 genetically interacts with SRB10.
To extend the above results, we next asked whether another CTD kinase, Srb10, might also be affected by Sub1, first by testing whether SUB1 and SRB10 interact genetically. We examined the effects of both SUB1 deletion (sub1Δ) and SUB1 overexpression (ADH1SUB1) from the strong ADH1 promoter on the growth of srb10Δ cells, which display a growth defect at both 30 and 37°C. Strikingly, the growth defect of srb10Δ cells was partially suppressed by SUB1 deletion and dramatically enhanced by SUB1 overexpression, with the strongest effect at 37°C (Fig. 3 A). We observed similar effects when we overexpressed SUB1 from a GAL1-inducible promoter (data not shown). Although we have consistently observed, here (Fig. 3A) and elsewhere (11), that when Sub1 is overexpressed, cells tend to grow slightly more slowly than wt cells at 37°C and that sub1Δ cells generally grow slightly faster than wt cells at this temperature, the effects of both deletion and overexpression of SUB1 on the srb10Δ cells were much stronger than the effects observed on wt cells. This strong genetic interaction suggests that Sub1 also influences Srb10 kinase activity.
FIG. 3.
SUB1 genetically interacts with SRB10, and SUB1 deletion negatively influences Srb10 kinase activity and association with active genes. (A) Genetic interaction between SUB1 and SRB10. SUB1 deletion partially suppresses the slow-growth phenotype of srb10Δ cells and overexpression of SUB1 enhances it, as shown by spot assay. To overexpress SUB1, its open reading frame was cloned under the control of the ADH1 promoter in a plasmid that was transformed into the srb10Δ strain (ADH1SUB1). Yeast strains with the indicated genotypes were spotted onto synthetic complete medium and grown at 30 and 37°C for 2 days. (B) Results of in vitro CTD kinase assay showing that SUB1 deletion causes a decrease in CTD phosphorylation by Srb10. Whole-cell extracts were prepared from the wt and sub1Δ strains expressing MYC-tagged Srb10, and in vitro kinase assay and CTD phosphorylation analysis were performed as described in the Fig. 1 legend. CTD Ser5 and Ser2 phosphorylation was analyzed using the following antibodies: CTD4H8 (anti-CTD Ser5P), ab5095 (anti-CTD Ser2P), 8WG16 (anti-CTD), anti-MYC antibody (Srb10 levels), and anti-PGK antibody (PGK levels, as control). (C) Induction of GAL1 transcription was monitored by RT-PCR. Total RNA was isolated from wt and sub1Δ cells grown under noninducible (2% raffinose [RAF]) and inducible (2% galactose [GAL]) conditions. cDNA was synthesized, and PCR performed using specific primers for GAL1, PYK1, SUB1, and SRB10 genes. (D) ChIP analysis of Srb10 was conducted with wt and sub1Δ cells grown in raffinose- or galactose-containing medium, using anti-MYC antibody. Srb10-MYC cross-linking to the promoter of PYK1 and to the upstream activating sequence of the GAL1 (UAS-GAL1) gene was analyzed by qRT-PCR. Quantifications of the results are shown in the graph, where numbers on the y axis represent the ratio of the values obtained from specific primer products to the value for the negative control (intergenic region of chromosome VII), after normalizing to the results for the input controls. Error bars show standard deviations. (E) SRB10 deletion causes an increase in CTD Ser5 phosphorylation by Kin28, reflecting increased Kin28 levels in the srb10Δ cells. Whole-cell extracts were prepared from wt and srb10Δ strains expressing HA-tagged Kin28, and in vitro kinase assays and CTD phosphorylation analysis were performed. CTD phosphorylation was analyzed using the following antibodies: CTD4H8 (anti-CTD Ser5P) and 8WG16 (anti-CTD). Kin28-HA and PGK levels were analyzed using 12CA5 (anti-HA) and anti-PGK, respectively.
Sub1 positively influences Srb10 kinase activity and association with chromatin.
We next performed in vitro IP kinase assays as described above to determine if SUB1 deletion influences CTD phosphorylation by Srb10. For this purpose, we generated strains expressing Srb10-MYC in the wt or sub1Δ background, where we previously had HA tagged Ctk1 (see below). Srb10 has the capacity to phosphorylate the CTD on both Ser2 and Ser5 (6, 27, 58). As shown by the results in Fig. 3B, the Srb10-MYC immunoprecipitated from wt and sub1Δ cell extracts was active, since it phosphorylated the CTD on Ser5 and Ser2, as determined by immunoblotting with a monoclonal Ser5 antibody (CTD4H8) that specifically recognizes Ser5 phosphorylation (64) and a polyclonal Ser2 antibody (ab5095) that preferentially recognizes Ser2-phosphorylated, as well as unphosphorylated, CTD (57), respectively. However, in contrast to Kin28, the Srb10 kinase activity in sub1Δ cells was significantly reduced compared to that in wt cells for both Ser5 and Ser2 phosphorylation (Fig. 3B). We also performed IP kinase assays with Srb10-HA strains (data not shown). Both Srb10-MYC and Srb10-HA displayed decreased CTD phosphorylation when cells lacked Sub1. As SUB1 deletion did not affect the total Srb10 levels (Fig. 3B), our results indicate that Sub1, in contrast to its negative effect on Kin28, positively influences Srb10 kinase activity.
We next investigated whether Sub1 influences Srb10 association with chromatin. The Srb8-11 complex is known to be required for Gal4-dependent activation of GAL1 transcription (41). We therefore conducted ChIP experiments in cells grown both under conditions of GAL1 induction (2% galactose) and in noninducible conditions (2% raffinose). In addition, we extracted RNA to monitor GAL1 gene induction by RT-PCR. We also tested the expression of the constitutive gene PYK1 and of the SRB10 and SUB1 genes. As shown by the results in Fig. 3C, as expected, GAL1 is only expressed in cells grown in galactose-containing medium, while PYK1 and SRB10 are expressed similarly in both inducible and noninducible conditions. Unexpectedly, we detected an increase of SUB1 mRNA levels during galactose induction (Fig. 3C).
We next examined the effect of Sub1 on the association of Srb10 with the GAL1 and PYK1 promoters by ChIP. As expected, in wild-type cells, Srb10 was not present at the GAL1 promoter when cells were grown in raffinose medium but was rapidly and efficiently recruited when cells were transferred to galactose-containing medium (20 min of galactose induction). However, Srb10 recruitment was significantly decreased in the absence of Sub1 (Fig. 3D). In the case of PYK1, although Srb10 cross-linking to the promoter region was less efficient, it was also decreased in cells lacking Sub1 (Fig. 3D). These results indicate that Sub1 positively influences Srb10 recruitment to the GAL1 and, to a lesser extent, PYK1 promoter, which agrees with the observed decrease of Srb10 kinase activity in sub1Δ cells.
We next asked whether the effects of SUB1 deletion on Srb10 might contribute to the effects we observed on Kin28. Previous studies have shown that TFIIH is negatively regulated by CDK8-containing Mediator complexes in human cells (4) and that Srb10 inhibits KIN28 transcription in meiotic yeast cells (51). Extending these results, we observed that deletion of SRB10 resulted in an increase in Kin28-HA levels and, as a consequence, an increase in Kin28 activity, as analyzed by in vitro kinase assays (Fig. 3E). The effect of SRB10 deletion on Kin28 levels was specific, as we detected no changes in the levels of Sub1-HA, Ceg1-HA, or Ctk1-HA in srb10Δ cells compared to their levels in wt cells (data not shown). Since Srb10 promoter recruitment and activity were reduced in sub1Δ cells, this could explain, at least in part, how Kin28 recruitment and/or activity was enhanced (see Discussion).
sub1Δ and ctk1Δ mutations are synthetically lethal.
We showed previously that SUB1 deletion results in increased levels of CTD Ser2 phosphorylation on RNAP II associated with chromatin (11). To extend this result, we performed genetic experiments to investigate whether SUB1 also interacts with the gene encoding the elongating CTD Ser 2 kinase Ctk1. Although it is well established that Ser2 phosphorylation follows Ser5 phosphorylation, it is not yet clear whether Ser5 phosphorylation is required for subsequent Ser2 phosphorylation and transcriptional elongation. It has been suggested that CTD Ser5 phosphorylation by Kin28 does not affect the level of CTD Ser2 phosphorylation (16); however, CTD Ser5P is a preferential substrate for Ser2 phosphorylation by Ctk1 (32) and it has recently been suggested that Ser5 phosphorylation stimulates Ser2 phosphorylation by Bur1/Bur2 kinase (57).
We first examined whether SUB1 genetically interacts with CTK1. For this, we generated a diploid strain lacking one copy each of SUB1 and CTK1 and analyzed the meiotic progeny by tetrad dissection (Fig. 4 A). As expected, haploid progeny lacking SUB1 showed no growth defect compared to the growth of the wt, whereas the growth of cells lacking CTK1 was significantly impaired. Significantly, however, in every case, meiotic progeny lacking both CTK1 and SUB1 were nonviable, indicating a synthetic lethal genetic interaction between these genes (Fig. 4A). This interaction, together with the genetic interactions with genes encoding other CTD kinases, suggests that Sub1 regulates phosphorylation of the CTD at multiple stages of the transcription cycle, including elongation.
FIG. 4.
Sub1 influences recruitment and kinase activity of the elongation kinase Ctk1 during transcription. (A) sub1Δ and ctk1Δ are synthetically lethal. A diploid yeast strain heterozygous for both SUB1 and CTK1 (sub1Δ::URA3/SUB1 ctk1Δ::kanMX/CTK1) was sporulated, and the meiotic progeny were separated by tetrad dissection and allowed to grow for 3 days. Thirteen tetrads were dissected, with nine showing a tetratype segregation pattern (eight of which are shown), and two showing a paternal ditype segregation pattern. The genotype of the resulting colonies was inferred by growth or lack of growth on selective medium. Cells with a deletion of CTK1 alone show a slow-growth phenotype, as reported previously. (B) SUB1 deletion causes increased Ctk1 kinase activity. Whole-cell extracts were prepared from wt and sub1Δ strains with an HA-tagged Ctk1, and in vitro kinase assay was performed as described in the Fig. 1 legend to analyze CTD Ser2 phosphorylation, using anti-CTD Ser2P. (C) Schematic representation of the ACT1, PYK1, and PMA1 genes. Numbers are nucleotide positions relative to start codon (+1), and black bars represent PCR products analyzed by ChIP. (D) Increased Ctk1-HA association with chromatin in cells lacking SUB1. ChIP for Ctk1-HA was performed in wt and sub1Δ cells. Ctk1-HA association with PMA1, PYK1, and ACT1 genes was analyzed by qRT-PCR, and quantifications were graphed (see Materials and Methods). (E) Ctk1-HA/Rpb1 ratio. Ctk1-HA and Rpb1 cross-linking were independently quantified in wt and sub1Δ cells, and then Ctk1/Rpb1 ratio was calculated and graphed. Error bars show standard deviations.
Sub1 negatively regulates Ctk1 activity and recruitment.
We previously analyzed total CTD Ser2 phosphorylation by ChIP and detected an increase on chromatin-associated RNAP II in the absence of SUB1 (11). In light of the results described above, this increase could be due to an inhibitory effect of Sub1 on Ctk1 recruitment and/or activity that is alleviated in sub1Δ cells. To test these possibilities, we generated strains expressing Ctk1 with a C-terminal 6×HA tag in the wt or sub1Δ background. We then investigated whether Ctk1 kinase activity was also impaired in sub1Δ cells, again using in vitro IP kinase assays. Indeed, Ctk1 kinase activity toward GST-CTD in IPs from sub1Δ cells was increased compared to its level in the wt, as determined by Western blotting using the anti-Ser2P antibody (Fig. 4B). The Ctk1 levels, as measured by Western blotting with anti-HA antibody, were equivalent in the two IPs. We also verified the specificity of the Ser2 activity in the IPs by using extracts from two nontagged strains (wt and sub1Δ). The kinase activity in HA IPs from these strains was barely detectable compared to the Ctk1-6×HA kinase activity (data not shown). Thus, we conclude that Sub1 negatively influences Ctk1 kinase activity.
The Ctk1-6×HA strains (wt and sub1Δ) that we analyzed were designed to also express Srb10-MYC. This allowed us to determine the effect of SUB1 deletion on Srb10 and Ctk1 kinase activity in the same cells, using a single cell extract. As shown in Fig. 3B and 4B, we observed a decrease in GST-CTD phosphorylation when Srb10-MYC was immunoprecipitated from sub1Δ extracts (Fig. 3B) and an increase when Ctk1-HA was immunoprecipitated from the same extracts (Fig. 4B). These results confirm that Sub1 has a positive influence on Srb10 kinase activity, probably acting at the level of PIC formation, and a negative effect on Ctk1 activity, likely during transcription elongation.
We next examined the effect of Sub1 on the recruitment of Ctk1 to active genes. The Ctk1-6×HA association with three different genes was determined by ChIP (Fig. 4D). Compared to the levels in wt cells, we observed increases of ∼1.5- to 2.5-fold in the levels of Ctk1-6×HA at the PMA1, PYK1, and ACT1 genes in cells lacking Sub1. Considering that Sub1 negatively affects the recruitment of RNAP II (11, 60), as we have confirmed for sub1Δ cells expressing Ctk1-6×HA (data not shown), the Ctk1/Rpb1 ratio was increased in sub1Δ cells from the promoter to the 3′ regions (Fig. 4E). Thus, the deletion of SUB1 significantly increased the levels of Ctk1 associated with the RNAP II transcription machinery.
Together, our data indicate that the increased Ser2 phosphorylation observed in the absence of SUB1 is due to three factors: increased Ctk1 recruitment, increased Ctk1 kinase activity, and reduced Fcp1 phosphatase levels (11).
SUB1 deletion increases the elongation defects of bur1 mutants.
Bur1 associates with nonphosphorylated Rpb1 and phosphorylates Ser5 and Ser2 of the CTD (32, 45). More recently, it has been shown that Bur1/Bur2 phosphorylates Ser2 near promoters and stimulates Ser2 phosphorylation by Ctk1 during transcription elongation (57). To complete our genetic study aimed at understanding the connection between Sub1 and CTD kinases, we next examined possible genetic interactions between SUB1 and BUR1. For this purpose, we used two bur1 mutants, the bur1-2 and bur1-23 strains (35, 55, 72). Both bur1 mutations impair Bur1's in vitro kinase activity, while bur1-23 cells show defects in elongation efficiency as measured by ChIP, although cotranscriptional phosphorylation of CTD Ser5 and Ser2 was not strongly affected (35). The bur1-2 mutant shows an overall slow-growth phenotype, while bur1-23 presents a more serious growth defect at 28°C and strong thermosensitivity at 37°C. Both mutants are sensitive to 6-azauracil (6-AU), a drug commonly used to detect effects in elongation (23, 66).
We deleted SUB1 in wt and bur1-2 isogenic strains and analyzed the effect on growth phenotypes at 28 and 37°C and in medium containing 6-AU (Fig. 5A, bottom). In the case of bur1-23, we generated a diploid by crossing it with the sub1Δ strain. The diploid was sporulated, and the tetrads dissected and analyzed. The growth phenotypes of one tetrad type are shown in Fig. 5A (top). SUB1 deletion was found to exacerbate the 6-AU sensitivity of the bur1-2 mutant, as seen by increased sensitivity to the drug in bur1-2 sub1Δ cells compared to that of bur1-2 mutant cells. In the case of the bur1-23 mutant, SUB1 deletion strongly enhanced both the growth defect and sensitivity to 6-AU (Fig. 5A). Although SUB1 deletion alone did not result in sensitivity to 6-AU in these assays (Fig. 5A and data not shown), sensitivity to the drug was detected when cells were grown in liquid medium (Fig. 5B). In any event, these data reveal a genetic interaction between SUB1 and BUR1. Indeed, the fact that the bur1-2 sub1Δ double mutant only displayed a synthetic phenotype on 6-AU-containing medium likely indicates that SUB1 deletion mainly affects bur1 elongation defects (35, 71).
FIG. 5.
Bur1 kinase recruitment and activity is also enhanced by deletion of SUB1. (A) SUB1 deletion increases the elongation defect of bur1 mutants. Phenotypic analysis of SUB1-BUR1 genetic interaction. SUB1 was deleted in a bur1-2 strain and in the isogenic wt strain. In the case of the bur1-23 mutant, we generated a diploid strain by crossing it with a sub1Δ mutant. The diploid was sporulated, and the tetrads dissected and analyzed. The growth phenotype of one tetrad is shown. Yeast strains with the indicated genotypes were spotted on yeast extract-peptone-dextrose or synthetic complete (SC) medium containing 50 μg/ml of 6-azauracil (6-AU), and plates were incubated for 3 days. As shown, SUB1 deletion increases the growth and elongation defects of bur1-23 cells and the elongation defect of bur1-2 cells. (B) sub1Δ cells are sensitive to 6-AU in liquid medium. Growth curves of wt and sub1Δ strains in SC medium without or with 75 μg/ml of 6-AU. (C) Bur1 kinase activity is increased in sub1Δ mutant. In vitro kinase assay was performed as described in the Fig. 1 legend in wt and sub1Δ cells expressing an HA-tagged Bur1. CTD phosphorylation was analyzed using anti-CTD Ser5P (CTD4H8, top left), anti-Ser2P (ab5095, top right) and anti-nonphosphorylated CTD (8WG16) antibodies. Bur1-HA levels were tested with anti-HA antibody. For both Ser2P and Ser5P, we observe increased phosphorylation of the CTD in the absence of Sub1. (D) SUB1 deletion increases Bur1 autophosphorylation and Bur1/RNAP II-CTD interaction. Whole-cell extracts were prepared from wt and sub1Δ Bur1-HA strains. Epitope-tagged kinase complexes were immunoprecipitated with 12CA5-protein A beads, and kinase activity with or without ATP was assayed. SDS-PAGE and immunoblot analysis were performed to analyze Bur1 autophosphorylation using HA antibody and CTD Ser5P/Bur1-HA coimmunoprecipitation using anti-Ser5P (CTD4H8) antibody. (E) SUB1 deletion slightly influences Bur1 association with coding gene regions compared to its influence on Rpb1 association. Bur1 occupancy at PMA1, PYK1, and ACT1 genes was assayed by ChIP in wt and sub1Δ cells. Graph shows qRT-PCR quantifications performed as described in the Fig. 4 legend.
SUB1 deletion alters Bur1 kinase activity.
The genetic interaction of SUB1 with BUR1 suggested that, similarly to Kin28, Srb10, and Ctk1, Sub1 may influence Bur1 kinase activity. Given that phosphorylation of the RNAP II CTD by Kin28 was reported to enhance Bur1/Bur2 recruitment and Ser2 CTD phosphorylation near promoters (57), it is possible that the increased Kin28 activity in sub1Δ cells can lead to increased CTD phosphorylation by Bur1. On the other hand, Bur1 was observed to associate primarily with Rpb1 containing unphosphorylated CTD repeats and then to phosphorylate Rpb1 on Ser5 (49). Therefore, we decided to analyze both Ser5 and Ser2 CTD phosphorylation by in vitro IP kinase assays with wt and sub1Δ cells containing HA epitope-tagged Bur1, again using the GST-CTD fusion protein as the substrate. Significantly, the deletion of SUB1 resulted in increased Ser5 and Ser2 phosphorylation in the Bur1-HA IPs (Fig. 5C). Consistent with the results of Murray et al. (49), Ser5 phosphorylation by Bur1 was more efficient than the Ser2 phosphorylation (Fig. 5C, compare results for wt cells in left and right panels, respectively). It has been shown that Bur1 has the capacity for autophosphorylation and that Bur1 phosphorylation promotes CTD phosphorylation (49, 72). We therefore tested whether SUB1 deletion affects Bur1 autophosphorylation by immunoprecipitating Bur1-HA from wt and sub1Δ cell extracts and performing in vitro kinase assays with coimmunoprecipitating proteins. As shown by the results in Fig. 5D, top, Bur1 coimmunoprecipitated and phosphorylated Rpb1 and, consistent with the results obtained with GST-CTD, this phosphorylation was increased in sub1Δ extracts. Significantly, Bur1-HA autophosphorylation was also increased in the absence of Sub1 (Fig. 5D, bottom). Therefore, it is possible that increased CTD phosphorylation in sub1Δ cells is due to an increase of Bur1 kinase activation.
Like the cell cycle CDKs, with the exception of Srb10, transcriptional CDKs undergo activating phosphorylation within their T loops. Thus, previous studies demonstrated that Cak1 phosphorylates Kin28, Bur1, and Ctk1 within their T loops and stimulates their activities (22, 36, 52, 72). Therefore, we tested whether Cak1 levels were altered in the absence of SUB1. However, we did not detect any significant variation of Cak1 levels in sub1Δ cells compared to the level in wt cells, nor did we detect Cak1 coimmunoprecipitating with Sub1 (results not shown).
ChIP analysis of Bur1-HA performed in wt and sub1Δ cells showed no significant difference in total Bur1 cross-linked to genes (Fig. 5E). However, again if we consider that Rpb1 cross-linking is reduced in sub1Δ cells (see above and reference 11), the Bur1-HA/Rpb1 ratio was slightly increased (∼1.2- to 2.0-fold [data not shown]). But in this case, increased recruitment could be an indirect effect due to increased Kin28 activity, because, as mentioned above, phosphorylation by Kin28 enhances Bur1/Bur2 recruitment (57).
DISCUSSION
We have presented evidence that Sub1 influences RNAP II CTD phosphorylation via interactions with all four CTD kinases, Kin28, Srb10, Bur1, and Ctk1. These effects were observed both genetically and biochemically, including effects on kinase activity and/or recruitment to chromatin. Our results thus indicate that Sub1 can act throughout the transcription cycle as a general regulator of CTD phosphorylation. Below, we discuss the implications of Sub1's interactions with these kinases and how Sub1 can influence CTD phosphorylation and transcription by RNAP II.
Opposing effects of Sub1 on CTD phosphorylation during transcription initiation.
It has been proposed that the timing of Srb10 and Kin28 activation can regulate transcription. Srb10 phosphorylates the CTD prior to PIC formation and inhibits transcription, while Kin28 promotes transcription by phosphorylating the CTD after PIC formation (27). However, Srb10, together with Kin28, can also promote transcription and contribute to PIC dissociation and scaffold complex formation (43). Here, we have shown that the presence of Sub1 has a negative effect on the growth of cells lacking Srb10. But our in vitro kinase assays showed that Sub1 positively influences CTD phosphorylation by Srb10, implying a negative effect on transcription inhibition according to Hengartner et al. (27). In contrast, we have detected additive effects on cell growth when combining mutations of KIN28, CTK1, and BUR1 with SUB1 deletion, consistent with a positive role of Sub1 in transcription (28) and with the fact that sub1Δ decreases RNAP II recruitment to gene promoters (11). However, the results of our kinase and ChIP assays indicate a repressive role for Sub1 on CTD phosphorylation by Kin28, Ctk1, and Bur1. How can these disparate results be reconciled?
In the absence of Kin28 activity, Srb10 activity is important to promote transcription (43). It is possible, then, that when SUB1 is overexpressed in srb10Δ cells, Kin28 activity at the PIC is inhibited by Sub1, which in this case cannot be compensated by the action of Srb10. Therefore, PIC dissociation, scaffold complex formation, and consequently, transcription are impaired, giving rise to the observed growth defect. Altogether, our data are consistent with a negative role for Sub1 in transcription preinitiation, favoring Srb10 kinase activity and negatively influencing CTD phosphorylation by Kin28 (Fig. 6).
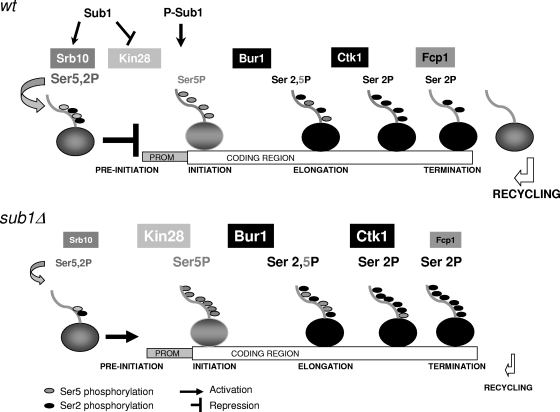
FIG. 6.
Model showing how Sub1 might function to regulate RNAP II CTD phosphorylation. In wild-type cells, nonphosphorylated Sub1 joins the promoter (PROM) (possibly via TFIIB [see references 28, 37, and 60]), contacting the promoter via its DNA binding domain. At that point, Sub1 interacts with the CDK8 (Srb10) Mediator complex, helping to maintain the PIC in a stable but inactive conformation. Sub1 is then phosphorylated (possibly by the action of kinases at the PIC, similarly to PC4), losing its DNA binding capacity and promoting clearance of TFIIB (26, 35). The PIC next changes conformation such that Kin28 can be activated and, with the help of Srb10, promotes PIC dissociation into the scaffold complex, as well as the recruitment of elongating kinases Ctk1 and Bur1. In contrast, in sub1Δ cells, Srb10 activity and recruitment are decreased, while Kin28 recruitment and activity increase, in agreement with TFIIH being negatively regulated by CDK8-containing Mediator complexes (4, 51). As a result, Ser5P levels are increased, and consequently, Bur1 and Ctk1 association with chromatin is also enhanced (19, 57). Furthermore, in sub1Δ cells, there is a reduction in Fcp1 phosphatase levels and its association with chromatin, which induces an additional increase in Ser2P, impairing RNAP II recycling after transcription termination. Thus, a decrease in RNAP II recruitment is observed in cells lacking Sub1 (11). Different font sizes in the figure labels indicate the increase or decrease of the corresponding CTD-modifying enzymes in sub1Δ versus wt cells.
Sub1 also negatively affects Ctk1 association with chromatin. Recently, it has been suggested that Ctk1 contributes to scaffold maintenance, as it promotes the dissociation of basal transcription factors from elongating polymerase independent of its kinase activity (1). It is thus probable that, as suggested by in vitro studies for its human homolog PC4 (46), Sub1 has the capacity to repress transcription while promoting PIC formation and, possibly, PIC dissociation into the scaffold complex through the action of CTD kinases, RNAP II, and TFIIB. In agreement with this, it has been suggested that Sub1 is a clearance factor, since it promotes the release of TFIIB from the promoter by disrupting the interaction between TFIIB and TATA-binding protein (37).
Additionally, PC4 transcription inhibition correlates with its ability to inhibit RNAP II phosphorylation by cdk-1, cdk-2, and cdk-7 in vitro (61). This inhibition is regulated by phosphorylation, as unphosphorylated PC4 displayed the kinase inhibitory activity, whereas phosphorylated PC4 was devoid of it. Sub1 is also likely regulated by phosphorylation, as it can be phosphorylated in vitro, regulating its capacity to bind DNA (28).
Sub1 influences CTD Ser5P and CTD Ser2P by different mechanisms.
Our genetic and biochemical studies have provided evidence that Sub1 has opposing effects on CTD phosphorylation at the preinitiation step versus the initiation/elongation steps. Increased Ser5 phosphorylation in cells lacking SUB1 is the result of increased Kin28 recruitment and kinase activity. Although SUB1 deletion reduces the levels of the Ser2 phosphatase Fcp1 (11), we have not observed a similar effect for the Ser5 phosphatase Ssu72 (data not shown). In addition, we have observed no genetic interaction between SUB1 and the recently described RNAP II CTD Ser5 phosphatase RTR1 (48; data not shown). These results suggest that Sub1 influences Ser5 and Ser2 phosphorylation/dephosphorylation by different mechanisms.
Taken together, our data indicate that the increase in Ser2 phosphorylation observed in the absence of SUB1 is due to as many as four distinct factors: (i) increased Ctk1 recruitment, probably due to the effects of Sub1 on PIC formation and/or dissociation of the scaffold complex; (ii) increased Ctk1 kinase activity and reduced Fcp1 phosphatase levels (11), in agreement with the fact that Fcp1 and Ctk1 play opposite roles in CTD Ser2 phosphorylation (16); (iii) the likelihood that some of the increase in Ser2 phosphorylation is due to the Ser5 phosphorylation increase, as CTD Ser5P is a preferential substrate for Ser2 phosphorylation by Ctk1 (32); and (iv) an increase in Bur1 kinase activity.
A model for Sub1 regulation of the CTD kinases.
An important question concerns the mechanism(s) by which Sub1 influences the activity and chromatin recruitment of the four CTD kinases. Based on our results and those of previous studies, we propose that the Sub1-Srb10 connection provides the key to explaining the Sub1 effect on recruitment. Srb10 provides the kinase activity of the CDK8 (Srb8-11) module of Mediator, which plays negative roles in the recruitment of RNAP II and TFIIH. CDK8 sterically blocks Mediator interactions with RNAP II (20), and Mediator appears to play a critical role in PIC assembly at the level of TFIIH and TFIIE recruitment (21). In addition, TFIIH is negatively regulated by CDK8-containing Mediator complexes in human cells (4), Srb10 inhibits KIN28 transcription in yeast (51), and we have observed that SRB10 deletion increases Kin28 levels and, as a consequence, Kin28-HA kinase activity. And finally, Sub1 is genetically and functionally linked to Mediator (18). Thus, Sub1 might influence CTD kinase recruitment via effects on the Mediator CDK8 complex. In fact, Sub1 has a positive role in promoting Srb10 recruitment to the inducible GAL1 gene, the transcription of which depends on the Srb8-11 and SAGA complexes (41).
Since Srb10 promoter recruitment and kinase activity were reduced in sub1Δ cells, this might suggest that SUB1 deletion should increase Kin28 levels. However, our results showed that Kin28 levels were in fact unaltered in cells lacking Sub1. One possibility is that the decrease in Srb10 activity and/or recruitment due to SUB1 deletion is not sufficient to significantly affect KIN28 expression, as was observed with SRB10 deletion. The levels of Srb10 in sub1Δ cells remained at wt levels, and this could be sufficient to maintain KIN28 expression at wt levels. However, the reduction in Srb10 kinase activity brought about by SUB1 deletion could in turn affect Kin28 activity. For example, if Srb10 phosphorylates Kin28 or its cyclin partner, as is the case in mammals (4), this could explain, at least in part, how SUB1 deletion enhances Kin28 activity and/or promoter recruitment without increasing its levels.
It is possible, then, that in sub1Δ cells, Srb10's inhibitory effect on Kin28 is reduced, enhancing the recruitment of Kin28 and thereby enhancing the recruitment of Ctk1 subsequent to PIC formation. This idea is in agreement not only with the above-described studies indicating an evolutionarily conserved negative effect of Srb10/Cdk8 on Kin28 but also with recent work showing that Mediator CDK8 can interact with and recruit P-TEFb to the transcriptional machinery in mammals (19). Increased Ctk1 activity, along with decreased Fcp1 levels (11), in sub1Δ cells will then reduce RNAP II recycling and, as a result, its recruitment to gene promoters (Fig. 6) (11, 60).
Perhaps most strikingly, the results of our in vitro IP kinase assays indicate that, in addition to regulating the recruitment of CTD kinases, Sub1 affects CTD phosphorylation by influencing the activity of all four CTD kinases. We currently do not understand the biochemical basis for these effects. The altered kinase activities were not due to the absence of Sub1 in the sub1Δ cell IPs because Sub1 did not immunoprecipitate with any of the CTD kinases in extracts from wt cells, indicating that Sub1 plays an indirect role in regulating the activities of the kinases. We also have found no evidence that Sub1 modifies CTD kinase activities by influencing posttranslational modifications of the kinases. We thus consider two possible explanations for the effects of Sub1 on the activities of the kinases. One is that Sub1 enhances the association (or dissociation) of an unidentified common regulator with the kinases, while the second is that Sub1 in some way influences kinase conformation and, thus, accessibility to the CTD. We are currently investigating these possibilities. In any case, the fact that Sub1 coordinates both the activities and recruitment to active genes of all four CTD kinases strongly supports an important role for Sub1 in regulating CTD phosphorylation throughout the transcription cycle.
Acknowledgments
We thank S. Buratowski, G. Prelich, and P. San Segundo for yeast strains. O.C. thanks R. Jiménez for laboratory facilities and support and María Gómez for technical support on qPCR.
This work was supported by grants number BFU 2006-09041 and BFU 2009-07179 from the Spanish Ministerio de Ciencia e Innovación and SA012A08 from the Junta de Castilla y León to O.C. and a grant from the NIH to J.L.M. A.G. was supported by a fellowship from the Junta de Castilla y León.
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Ahn, S. H., M. C. Keogh, and S. Buratowski. 2009. Ctk1 promotes dissociation of basal transcription factors from elongating RNA polymerase II. EMBO J. 28:205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 3.Akhtar, M. S., M. Heidemann, J. R. Tietjen, D. W. Zhang, R. D. Chapman, D. Eick, and A. Z. Ansari. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 5.Alexandru, G., F. Uhlmann, K. Mechtler, M. A. Poupart, and K. Nasmyth. 2001. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105:459-472. [DOI] [PubMed] [Google Scholar]
- 6.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 7.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski, S. 2005. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 17:257-261. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 11.Calvo, O., and J. L. Manley. 2005. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24:1009-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson, M. 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13:1-23. [DOI] [PubMed] [Google Scholar]
- 13.Chapman, R. D., M. Heidemann, T. K. Albert, R. Mailhammer, A. Flatley, M. Meisterernst, E. Kremmer, and D. Eick. 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780-1782. [DOI] [PubMed] [Google Scholar]
- 14.Chapman, R. D., M. Heidemann, C. Hintermair, and D. Eick. 2008. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 24:289-296. [DOI] [PubMed] [Google Scholar]
- 15.Cho, E. J. 2007. RNA polymerase II carboxy-terminal domain with multiple connections. Exp. Mol. Med. 39:247-254. [DOI] [PubMed] [Google Scholar]
- 16.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 18.Dettmann, A., Y. Jaschke, I. Triebel, J. Bogs, I. Schroder, and H. J. Schuller. 2010. Mediator subunits and histone methyltransferase Set2 contribute to Ino2-dependent transcriptional activation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 283:211-221. [DOI] [PubMed] [Google Scholar]
- 19.Donner, A. J., C. C. Ebmeier, D. J. Taatjes, and J. M. Espinosa. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elmlund, H., V. Baraznenok, M. Lindahl, C. O. Samuelsen, P. J. Koeck, S. Holmberg, H. Hebert, and C. M. Gustafsson. 2006. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 103:15788-15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esnault, C., Y. Ghavi-Helm, S. Brun, J. Soutourina, N. Van Berkum, C. Boschiero, F. Holstege, and M. Werner. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31:337-346. [DOI] [PubMed] [Google Scholar]
- 22.Espinoza, F. H., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi, and D. O. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Exinger, F., and F. Lacroute. 1992. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22:9-11. [DOI] [PubMed] [Google Scholar]
- 24.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 25.Glover-Cutter, K., S. Larochelle, B. Erickson, C. Zhang, K. Shokat, R. P. Fisher, and D. L. Bentley. 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29:5455-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, X., A. U. Khan, H. Cheng, D. L. Pappas, Jr., M. Hampsey, and C. L. Moore. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 28.Henry, N. L., D. A. Bushnell, and R. D. Kornberg. 1996. A yeast transcriptional stimulatory protein similar to human PC4. J. Biol. Chem. 271:21842-21847. [DOI] [PubMed] [Google Scholar]
- 29.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 30.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 31.Hong, S. W., S. M. Hong, J. W. Yoo, Y. C. Lee, S. Kim, J. T. Lis, and D. K. Lee. 2009. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U. S. A. 106:14276-14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279:24957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser, K., G. Stelzer, and M. Meisterernst. 1995. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 14:3520-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keogh, M. C., E. J. Cho, V. Podolny, and S. Buratowski. 2002. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keogh, M. C., V. Podolny, and S. Buratowski. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knaus, R., R. Pollock, and L. Guarente. 1996. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 15:1933-1940. [PMC free article] [PubMed] [Google Scholar]
- 38.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koyama, H., E. Sumiya, M. Nagata, T. Ito, and K. Sekimizu. 2008. Transcriptional repression of the IMD2 gene mediated by the transcriptional co-activator Sub1. Genes Cells 13:1113-1126. [DOI] [PubMed] [Google Scholar]
- 40.Kretzschmar, M., K. Kaiser, F. Lottspeich, and M. Meisterernst. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78:525-534. [DOI] [PubMed] [Google Scholar]
- 41.Larschan, E., and F. Winston. 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laybourn, P. J., and M. E. Dahmus. 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J. Biol. Chem. 265:13165-13173. [PubMed] [Google Scholar]
- 43.Liu, Y., C. Kung, J. Fishburn, A. Z. Ansari, K. M. Shokat, and S. Hahn. 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24:1721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, Y., L. Warfield, C. Zhang, J. Luo, J. Allen, W. H. Lang, J. Ranish, K. M. Shokat, and S. Hahn. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29:4852-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, H., O. Flores, R. Weinmann, and D. Reinberg. 1991. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc. Natl. Acad. Sci. U. S. A. 88:10004-10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. U. S. A. 95:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 19:1401-1415. [DOI] [PubMed] [Google Scholar]
- 48.Mosley, A. L., S. G. Pattenden, M. Carey, S. Venkatesh, J. M. Gilmore, L. Florens, J. L. Workman, and M. P. Washburn. 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nonet, M., D. Sweetser, and R. A. Young. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50:909-915. [DOI] [PubMed] [Google Scholar]
- 51.Ohkuni, K., and I. Yamashita. 2000. A transcriptional autoregulatory loop for KIN28-CCL1 and SRB10-SRB11, each encoding RNA polymerase II CTD kinase-cyclin pair, stimulates the meiotic development of S. cerevisiae. Yeast 16:829-846. [DOI] [PubMed] [Google Scholar]
- 52.Ostapenko, D., and M. J. Solomon. 2005. Phosphorylation by Cak1 regulates the C-terminal domain kinase Ctk1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:3906-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patturajan, M., R. J. Schulte, B. M. Sefton, R. Berezney, M. Vincent, O. Bensaude, S. L. Warren, and J. L. Corden. 1998. Growth-related changes in phosphorylation of yeast RNA polymerase II. J. Biol. Chem. 273:4689-4694. [DOI] [PubMed] [Google Scholar]
- 54.Phatnani, H. P., and A. L. Greenleaf. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922-2936. [DOI] [PubMed] [Google Scholar]
- 55.Prelich, G., and F. Winston. 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu, H., C. Hu, and A. G. Hinnebusch. 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramanathan, Y., S. M. Rajpara, S. M. Reza, E. Lees, S. Shuman, M. B. Mathews, and T. Pe'ery. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913-10920. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez, C. R., E. J. Cho, M. C. Keogh, C. L. Moore, A. L. Greenleaf, and S. Buratowski. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosonina, E., I. M. Willis, and J. L. Manley. 2009. Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol. Cell. Biol. 29:2308-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schang, L. M., G. J. Hwang, B. D. Dynlacht, D. W. Speicher, A. Bantly, P. A. Schaffer, A. Shilatifard, H. Ge, and R. Shiekhattar. 2000. Human PC4 is a substrate-specific inhibitor of RNA polymerase II phosphorylation. J. Biol. Chem. 275:6071-6074. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stock, J. K., S. Giadrossi, M. Casanova, E. Brookes, M. Vidal, H. Koseki, N. Brockdorff, A. G. Fisher, and A. Pombo. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9:1428-1435. [DOI] [PubMed] [Google Scholar]
- 65.Thompson, N. E., T. H. Steinberg, D. B. Aronson, and R. R. Burgess. 1989. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264:11511-11520. [PubMed] [Google Scholar]
- 66.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 67.Valay, J. G., M. Simon, M. F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249:535-544. [DOI] [PubMed] [Google Scholar]
- 68.Werten, S., G. Stelzer, A. Goppelt, F. M. Langen, P. Gros, H. T. Timmers, P. C. Van der Vliet, and M. Meisterernst. 1998. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 17:5103-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20:589-599. [DOI] [PubMed] [Google Scholar]
- 70.Wood, A., and A. Shilatifard. 2006. Bur1/Bur2 and the Ctk complex in yeast: the split personality of mammalian P-TEFb. Cell Cycle 5:1066-1068. [DOI] [PubMed] [Google Scholar]
- 71.Yao, S., A. Neiman, and G. Prelich. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao, S., and G. Prelich. 2002. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol. Cell. Biol. 22:6750-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou, K., W. H. Kuo, J. Fillingham, and J. F. Greenblatt. 2009. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U. S. A. 106:6956-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]