Abstract
Cellular adaptation to environmental stress conditions requires rapid and specific changes in gene expression. During heat shock, most polyadenylated mRNAs are retained in the nucleus, whereas the export of heat shock-induced mRNAs is allowed. Although essential mRNA export factors are known, the precise mechanism for regulating transport is not fully understood. Here we find that during heat shock in Saccharomyces cerevisiae, the mRNA-binding protein Nab2 is phosphorylated on threonine 178 and serine 180 by the mitogen-activated protein (MAP) kinase Slt2/Mpk1. Slt2 is required for nuclear poly(A+) mRNA accumulation upon heat shock, and thermotolerance is decreased in a nup42 nab2-T178A/S180A mutant. Coincident with phosphorylation, Nab2 and Yra1 colocalize in nuclear foci with Mlp1, a protein involved in mRNA retention. Nab2 nuclear focus formation and Nab2 phosphorylation are independent, suggesting that heat shock induces multiple cellular alterations that impinge upon transport efficiency. Under normal conditions, we find that the mRNA export receptor Mex67 and Nab2 directly interact. However, upon heat shock stress, Mex67 does not localize to the Mlp1 nuclear foci, and its association with Nab2 complexes is reduced. These results reveal a novel mechanism by which the MAP kinase Slt2 and Mlp1 control mRNA export factors during heat shock stress.
In eukaryotes, intracellular signaling events are essential for transmitting information from the external environment to gene expression networks (reviewed in reference 7). This communication is particularly important for allowing rapid cellular adaption and survival during stress conditions. It is well established that environmental stress results in an evolutionarily conserved global alteration of the nuclear mRNA export pathway (6, 15, 30, 35). In response to heat shock, transcripts from genes encoding heat shock proteins (hsp) are exported, whereas most non-hsp poly(A+) mRNAs are retained and accumulate in the nucleus (53, 54). Together with the coincident increased transcription of heat shock genes, the retention of non-hsp mRNAs results in a coordinated mechanism for the rapid production of the heat shock proteins essential for survival and stress recovery (6). How signal transduction pathways modulate the mRNA export mechanism is not fully defined.
For mRNA export under normal growth conditions, the transcription and mRNA export (TREX) complex is critical for coupling mRNA biogenesis with messenger ribonucleoprotein particle (mRNP) packaging to allow formation of an export-competent mRNP (1, 61, 62). The TREX complex includes the essential mRNA-binding protein Yra1 and the export receptor Mex67-Mtr2 in Saccharomyces cerevisiae (TAP/NXF1-p15/NXT1 in vertebrates) (22, 56, 59). Recruitment of Mex67-Mtr2 appears to be the penultimate step, which stimulates release of the mature mRNP from chromatin-associated biogenesis factors and the transition to early mRNA export steps (31, 48). Association of Mex67-Mtr2 with the mRNP is key for export and directly mediates subsequent targeting to nuclear pore complexes (NPCs) in the nuclear envelope and NPC translocation via interactions between Mex67-Mtr2 and NPC proteins (nucleoporins [Nups]) (reviewed in reference 34) (68). Thus, defining the mechanisms that control interactions between mRNA-binding proteins and Mex67-Mtr2 will likely reveal important regulatory steps.
Several essential mRNA-binding proteins have been implicated in the mRNA export mechanism (32). In the budding yeast S. cerevisiae, this includes Npl3, Yra1, and Nab2 (21, 37, 60). These factors couple mRNA biogenesis steps, such as transcriptional elongation, pre-mRNA splicing, and 3′-end formation with assembly of an export-competent mRNP (4, 8, 14, 36, 84). Other key S. cerevisiae components involved in early mRNA export are Mlp1 and Mlp2 (18, 74). The Mlp proteins associate with the NPC and promote docking of mRNPs to the nuclear envelope. Studies have shown that the poly(A+)-binding protein Nab2 interacts directly with Mlp1, with loss of the Nab2-Mlp1 interaction enhancing the growth and mRNA export defects of mex67 mutants (13, 18). This suggests that a Nab2-Mlp1 step is central to efficient mRNA export. Importantly, the Mlp proteins also function in a nuclear quality control mechanism that acts to retain unspliced or aberrantly processed mRNAs in the nucleus (16, 47, 74). Although the precise role for the Mlp proteins in quality control is unknown, genetic and biochemical evidence suggests that Nab2 and Yra1 are linked to this process.
Many of the factors required for normal, non-hsp mRNA export are also essential for hsp export; specifically, Mex67, the DEAD-box protein Dbp5, and the Dbp5 activator Gle1 (3, 25, 44, 50, 54, 56, 58, 72, 73, 78). Altering the functions of these proteins results in the impaired export of all mRNAs, consistent with their general roles in the mRNA export process. In contrast, hsp mRNA export is independent of some factors required for normal mRNA export, including the mRNA-binding proteins Yra1 and Npl3 (35, 51). Furthermore, hsp mRNAs show a requirement for the NPC protein Nup42, whereas this factor is dispensable for efficient mRNA export under normal growth conditions (50, 53, 54, 64, 73). Given that several studies have found that specific mRNA-binding proteins associated with transcripts whose protein products are functionally linked, regulating the mRNP composition for transcripts might be a mechanism for controlling mRNA export (12, 20, 23, 24). However, it is not clear how mRNP composition is affected during heat shock or whether other molecular signals differentiate between hsp and non-hsp mRNAs during mRNA export.
The stress induced by heat shock is known to trigger a cascade of intracellular signaling events, including stimulation of the mitogen-activated protein kinase (MAPK) pathways (7). S. cerevisiae has five distinct MAP kinase pathways, each of which can be activated by specific extracellular stimuli. The Bck1-Slt2/Mpk1 pathway is specifically initiated by certain forms of cellular stress, including cell wall stress and heat shock (42, 69, 71). When cells undergo heat shock, Pkc1 initiates the kinase cascade by phosphorylating the MAPK kinase kinase (MAPKKK) (Bck1), which then phosphorylates two redundant MAPKKs (Mkk1 and Mkk2) for phosphorylation of the MAPK (Slt2/Mpk1) (38). Activated Slt2 phosphorylates both transcriptional activators and repressors, altering the gene expression pattern to allow cell survival (10, 27, 49). Interestingly, the vertebrate ortholog of the Mlp proteins, designated Tpr1, is a target of a MAP kinase pathway in vertebrate cells (76). Thus, there is a potential connection between these signaling pathways and mRNA export during heat shock.
In this study, we demonstrate that the essential poly(A+) mRNA-binding protein Nab2 is a target for heat shock-dependent phosphorylation by the MAP kinase Slt2. Unlike wild-type cells, slt2 null (Δ) mutant cells fail to accumulate poly(A+) RNA in the nucleus during heat shock. Further, Nab2 and Mlp1 form intranuclear foci upon heat shock, whereas Mex67 retains a normal cellular distribution. On the basis of a direct physical interaction between Nab2 and Mex67-Mtr2 and their differential association in complexes from heat-shocked cells, we propose a model whereby Slt2 and Mlp1 promote nuclear retention of non-hsp mRNPs by uncoupling the mRNA export receptor, Mex67, from specific mRNA-binding proteins during heat shock. This mechanism allows selectivity for hsp mRNP export during stress and facilitates thermotolerance and rapid recovery.
MATERIALS AND METHODS
S. cerevisiae strains, plasmids, and growth.
S. cerevisiae strains and plasmids used in this study are listed in Table S2 and Table S3 in the supplemental material, respectively. Unless indicated otherwise, yeast cells were grown at 23°C in rich growth medium (YPD) containing 1% yeast extract, 2% Bacto peptone, and 2% glucose. For assaying thermotolerance and recovery from heat shock, the cells were grown at 23°C to log phase and then shifted to 52°C in a shaking water bath. At each time point, aliquots of 5 × 106 cells were taken and resuspended for serial dilution and spotting for plate growth at 23°C for 2 or 3 days. Site-directed mutagenesis of NAB2 was performed using pAC636 (NAB2) (19) and CP436 (glutathione S-transferase [GST]-tagged Nab2 [GST-Nab2]) (2) as templates. To construct T178A/S180A mutants, PCR was performed with oligonucleotides 5′-ATGCAGACAGATGCTCCTGCAGCTCCAGCCCCCATATCAGCCTTTTCCGG-3′ and 5′-CCGGAAAAGGCTGATATGGGGGCTGGAGCTGCAGGAGCATCTGTCTGCAT-3′, and to construct T178E/S180E mutants, oligonucleotides 5′-ATGCAGACAGATGCTCCTGCAGAGCCAGAGCCCATATCAGCCTTTTCCGG-3′ and 5′-CCGGAAAAGGCTGATATGGGCTCTGGCTCTGCAGGAGCATCTGTCTGCAT-3′ were used. The resulting plasmids were sequenced to confirm mutations.
Immunoblot analysis of Nab2 phosphorylation.
Cultures were grown to log phase in YPD at 30°C and shifted to 42°C for 1 h or to YPD containing 10% ethanol or 0.4 M sodium chloride. Crude cell lysates were prepared by a protocol adapted from reference (80a). The cells (30 mg) were harvested, washed once with water, and lysed in 160 μl of 1.85 M sodium hydroxide and 7.4% β-mercaptoethanol on ice for 10 min. Protein was precipitated on ice for 10 min by the addition of an equal volume of 50% trichloroacetic acid. The samples were centrifuged for 2 min at 15,000 relative centrifugal force, and the pellets were washed with 500 μl Tris base and resuspended in SDS sample buffer. The samples were separated by SDS-PAGE and transferred to nitrocellulose. The blots were probed with affinity-purified rabbit anti-Nab2 antibody (1:100,000) (63), followed by horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody. For phosphatase sensitivity assays, TAP-tagged Nab2 (Nab2-TAP) was immunoprecipitated from cells grown to early log phase in YPD, using IgG-coated Sepharose beads. The beads were washed, resuspended in 1× λ phosphatase buffer, and treated with either 100 U of λ phosphatase, or 100 U of λ phosphatase and phosphatase inhibitor cocktail (Calbiochem) for 30 min at 30°C.
Nab2-TAP purifications and mass spectrometry.
S. cerevisiae cells expressing Nab2-TAP were grown in YPD at 23°C to an optical density at 600 nm (OD600) of 0.75. Samples for analysis of 23°C growth were directly harvested by centrifugation, washed twice in 4°C H2O, and once in resuspension buffer (20 mM HEPES [pH 7.4], 1.2% polyvinylpyrrolidone, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 4 μg/ml pepstatin A). Samples for analysis of 42°C growth were obtained by shifting a 23°C culture rapidly to heat shock conditions by the addition of YPD at 65°C to the 23°C culture at a ratio of 1:1 and allowing the culture to grow at 42°C for 1 h before harvesting. Isolated cell pellets were injected into liquid nitrogen, and frozen cells were lysed in the solid phase by milling, using a planetary ball mill (by the method of Oeffinger et al. [46]). Frozen cell lysate was thawed and resuspended in extraction buffer (20 mM HEPES [pH 7.4], 110 mM potassium acetate, 100 mM NaCl, 2 mM MgCl-2, 0.5% Triton X-100, 0.1% Tween 20, 1 mM PMSF, 4 μg/ml pepstatin A, 0.06 μl/ml antifoam B emulsion [Sigma]) and centrifuged at 900 × g for 2 min. The supernatant was incubated with rabbit IgG-coated Dynabeads (Invitrogen) for 30 min at 4°C. The beads were collected magnetically, washed 3 times in extraction buffer and 1 time in final wash buffer (100 mM ammonium acetate [pH 7.5], 0.2 mM MgCl2, 0.05% Tween 20). The beads were then washed 3 times rapidly and 1 time for 5 min in final wash buffer lacking Tween 20. Protein was eluted by two washes with 500 μl of 0.5 M NH4-OH and 5 mM EDTA, with shaking at room temperature. Elutions were pooled and lyophilized.
Multidimensional protein identification technology (MudPIT) mass spectrometry and phosphorylation site analysis were performed essentially by the method of MacCoss et al. (40). Briefly, purified protein complexes were digested in parallel with trypsin, elastase, and subtilysin. Peptides from these digestions were acidified and loaded via pressure injection platform (New Objective, Woburn, MA) onto a split-phase MudPIT column (internal diameter of 100 μm) (42a, 77, 80) consisting of a ∼4-cm Aqua C18 reverse-phase column and ∼4-cm Luna SCX (Phenomonex) fritted into an M520 filter union (IDEX, Oak Harbor, WA). After loading, this column was placed in-line with an 18-cm Jupiter (3-μm, 300-A) self-packed analytical column (internal diameter of 100 μm). Peptides were resolved using an Eksigent 1D+ high-performance liquid chromatographic (HPLC) system through a 16-h MudPIT separation with eight 5-μl salt pulses (0 mM, 50 mM, 75 mM, 100 mM, 150 mM, 300 mM, 500 mM, and 1 M ammonium acetate) followed by an organic gradient to resolve each eluted set of peptides. Tandem mass spectra were collected in a data-dependent manner using an LTQ mass spectrometer equipped with a nanoelectrospray source. Spectra were searched with SEQUEST (82) considering possible phosphate modifications to serine, threonine, and tyrosine against an S. cerevisiae protein database containing a reversed-sequence decoy database. The results from these searches were filtered to a 5% peptide false discovery rate, collated, and accounted to a given analysis (23°C versus 42°C) using IDPicker (39). Spectra corresponding to potential sites of phosphorylation were confirmed by manual evaluation.
In vitro kinase assays.
Yeast strains expressing TAP-tagged kinases were grown at 23°C to early log phase in YPD. The cells were harvested by centrifugation, washed once, and resuspended in 300 μl in 1× TBT buffer (20 mM HEPES [pH 7.4], 110 mM potassium acetate [KOAc], 100 mM NaCl, 0.5% Triton X-100, 0.1% Tween 20, 0.18 mg/ml PMSF, 4 μg/ml pepstatin A, 0.06% antifoam B [Sigma]). The cells were lysed with 0.5-mm glass beads. The supernatant was isolated by centrifugation and incubated with IgG (Sigma)-coated magnetic beads (Invitrogen) for 30 min at 4°C. The beads were washed three times in TBT buffer and once in kinase buffer (20 mM HEPES [pH 7.5], 20 mM β-glycerol phosphate, 10 mM MgCl2, 10 mM p-nitrophenylphosphate [PNPP], 100 μM Na3VO4, 2 mM DTT, 20 μM ATP). The beads were resuspended in 40 μl kinase buffer containing 1 μg of recombinant Nab2 and 0.5 μl of [γ-32P]ATP. The samples were incubated for 30 min at 30°C. The recombinant wild-type Nab2, nab2-T178A/S180A, and nab2-T178E/S180E proteins were purified as described below. Supernatants were removed, and the beads were resuspended in 1× SDS loading buffer. After the samples were heated at 100°C for 5 min, they were separated by SDS-PAGE, and the gel was exposed to autoradiography film.
Microscopy.
All images were acquired using a microscope (BX50; Olympus) with a UPlanF1 100× oil immersion objective (Olympus) with a numerical aperture (NA) of 1.30 and a camera (CoolSNAP HQ; Photometrics). Within each experiment, all images were collected and scaled identically. The images were collected using Image-Pro Express 6.0 (Media Cybernetics) and processed with Adobe Photoshop 9.0. For in situ hybridization, yeast cells were grown in YPD to early log phase at 23°C, and aliquots were shifted to 42°C for 45 min. The cells were fixed for 10 min and processed as previously described (28, 79). The digoxigenin-dUTP-labeled oligo[d(T)] probe was detected with fluorescein-labeled antidigoxigenin Fab fragments (1:50; Roche). DNA was stained with 0.1 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI), and the samples were mounted for imaging in 90% glycerol and 1 mg/ml p-phenylenediamine (Sigma-Aldrich), pH 8.0.
Assay for heat shock protein production.
Yeast cells were grown to early log phase in synthetic complete medium lacking methionine (SC−Met) at 23°C, isolated by centrifugation, and resuspended in 1 ml SC−Met. For heat shock, 250 μl of 59°C SC−Met was added to 250 μl cell suspension and placed at 42°C for 15 min. For controls, 250 μl of 23°C SC−Met was added to 250 μl cell suspension and maintained at 23°C for 15 min. The cells were radiolabeled by the addition of 50 μCi of [35S]methionine to each sample, and the cells were incubated an additional 15 min before harvesting by centrifugation at 4°C. The cells were washed two times with 4°C SC−Met and lysed in 50 μl SDS loading buffer at 100°C for 5 min. Samples were separated by SDS-PAGE, and the resulting gel was dried and exposed to autoradiography film.
Poly(A+) tail length determination.
The cells were grown in YPD at 23°C until early log phase and shifted to 42°C for 1 h. Total RNA was end labeled with 32P-labeled pCp and T4 RNA ligase. To digest non-poly(A+) RNA, the labeled RNA was treated simultaneously with RNases A and T1 and then ethanol precipitated. Resuspended RNA was resolved by denaturing urea-acrylamide gel electrophoresis and imaged using a phosphorimager.
In vitro binding assays.
Soluble binding assays were performed using recombinant purified Mex67-Mtr2 heterodimer, recombinant purified Dbp5, recombinant purified untagged Nab2 versions, and GST-Nab2 from bacterial lysate. The Dbp5 protein was purified as previously described (3, 70). For the Mex67-Mtr2 purification, the coding regions for MEX67 and MTR2 were subcloned into a single pET-Duet1 (Novagen) plasmid such that Mtr2 would be N terminally tagged with six-histidine (6×His-Mtr2) and Mex67 was untagged. This allowed coexpression of both proteins from a single vector. The heterodimer was expressed in Escherichia coli Rosetta/DE3 by induction with isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested and lysed by sonication under native conditions (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0], including RNase, DNase, 1 mM PMSF, 1× protease inhibitor tablet [ETDA-free; Roche]). Protein was purified using nickel-nitrilotriacetic acid (Ni-NTA) resin per the manufacturer's recommendations (Qiagen) and eluted with 250 mM imidazole. Purified Mex67-Mtr2 was dialyzed into buffer B (20 mM HEPES [pH 7.5], 150 mM NaCl, 20% [wt/vol] glycerol).
For the soluble binding assays with wild-type GST-Nab2 and glutathione resin, bacterial cells harboring Nab2-pGEX2TK were induced for expression of GST-Nab2 with IPTG. The cells were harvested, and total cell lysate was incubated with glutathione resin, washed, and subsequently incubated with the indicated Mex67-Mtr2 or Dbp5 proteins for 10 min at room temperature. The resin was then washed, and bound proteins were eluted with 20 mM glutathione. Input protein (10%) and 50% of bound protein were resolved by gel electrophoresis, and proteins were visualized by Coomassie blue staining.
For soluble binding assays with untagged recombinant Nab2 proteins, previously published methods were used to purify the proteins (70). Briefly, GST-Nab2, GST-nab2-T178A/S180A, and GST-nab2-T178E/S180E were purified from bacterial lysates with glutathione Sepharose fast flow resin by following the manufacturer's protocol (GE Healthcare, Piscataway, NJ). GST tag removal was performed by thrombin protease cleavage and separation by affinity chromatography. Purified 6×His-Mtr2/Mex67 was incubated with Ni2+ agarose beads at 4°C for 1 h in MHL buffer (50 mM sodium phosphate [pH 7.4], 150 mM NaCl2, 10 mM imidazole, 10% glycerol) in batches. Resin was washed in MHL buffer, resuspended in MHL buffer, and split evenly in nine tubes. To each tube, 0.5 μg, 0.25 μg, and 0.1 μg of purified wild-type Nab2, nab2-T178A/S180A, or nab2-T178E/S180E protein was added, and incubated for 1 h at 4°C. Resin was washed three times in MHL buffer, and samples were collected in SDS sample buffer. Mex67 levels were detected by Coomassie blue-stained SDS-PAGE, and Nab2 levels were detected by immunoblotting.
RESULTS
Nab2 is phosphorylated under heat shock stress.
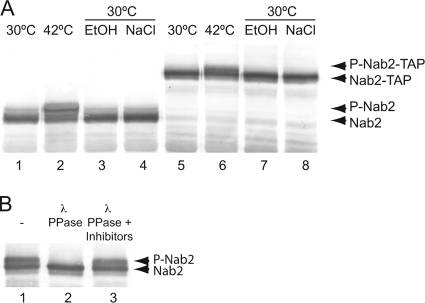
The role of the essential mRNA-binding protein Nab2 in hsp mRNA export has not been examined; yet, Nab2 is emerging as a key factor involved in coupling mRNA processing and export events (33, 52, 75). Our previous work defined Nab2 as essential for bulk mRNA export and a physiological target for release from mRNPs during NPC translocation (70). Therefore, we examined whether Nab2 was altered during cell stress in S. cerevisiae cells. To this end, we focused on identifying Nab2 posttranslational modifications that were dependent upon specific cellular stresses. Briefly, wild-type NAB2 and chromosomally tagged NAB2-TAP strains were grown to early logarithmic phase at 30°C in rich media and then exposed to multiple distinct stress conditions for 1 h. These conditions included an osmolarity shift (to 0.4 M NaCl), addition of ethanol (to 10%), and heat shock (at 42°C). Upon completion of the stress condition shifts, whole-cell lysates were prepared, resolved by gel electrophoresis, and analyzed by immunoblotting with anti-Nab2 antibodies. Neither the osmolarity nor ethanol shifts resulted in detectable changes in Nab2 electrophoretic mobility (Fig. 1 A, lanes 3, 4, 7, and 8). In contrast, when the cells were shifted to 42°C, a second, slower-migrating Nab2 band was observed for both untagged Nab2 (Fig. 1A, lane 2) and TAP-tagged Nab2 (Fig. 1A, lane 6).
FIG. 1.
Nab2 is phosphorylated upon heat shock. (A) Immunoblotting for Nab2 (lanes 1 to 4) or Nab2-TAP (lanes 5 to 8) was conducted with cell lysates isolated after shifting cells from growth in YPD at 30°C to 42°C (lanes 1 and 2 and lanes 5 and 6) or shifting to growth in YPD with 10% ethanol (EtOH) (lanes 3 and 7) or YPD with 0.4 M NaCl (lanes 4 and 8) for 1 h. P-Nab2-TAP, phosphorylated Nab2-TAP; P-Nab2, phosphorylated Nab2. (B) Nab2-TAP was immunoprecipitated from cells in early log phase grown in YPD at 30°C and shifted to 42°C for 1 h. Immunoprecipitates were treated with λ phosphatase (λ PPase) (lane 2), λ phosphatase plus phosphatase inhibitors (lane 3), or mock treatment (−) (lane 1) for 30 min at 30°C.
We speculated that the slower-migrating Nab2 isoform resulted from phosphorylation. To test this hypothesis, endogenous Nab2-TAP was isolated by affinity purification from cell lysates following heat shock, and phosphatase sensitivity was analyzed. Nab2-TAP was incubated with buffer alone (Fig. 1B, lane 1), λ phosphatase (Fig. 1B, lane 2), or λ phosphatase and a phosphatase inhibitor cocktail (Fig. 1B, lane 3). The samples were then resolved by gel electrophoresis, and the presence of the slower-migrating Nab2 band was visualized by immunoblotting as described above. Strikingly, after incubation with λ phosphatase, Nab2 in the heat-shocked isolate was present only as a single, faster-migrating band. Importantly, collapse of the Nab2 doublet was blocked by phosphatase inhibitors. These results demonstrate that Nab2 is phosphorylated specifically in response to heat shock stress.
The Bck1/Slt2 pathway is required for heat shock-induced Nab2 phosphorylation.
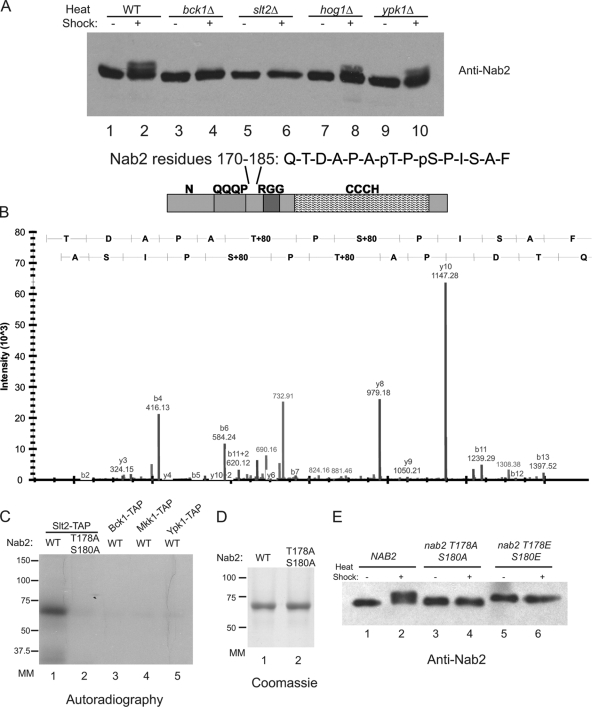
To identify the kinase responsible for Nab2 phosphorylation, we performed a screen using null (Δ) strains from the S. cerevisiae null collection. Based on the Protein Kinase Resource (http://www0.nih.go.jp/mirror/Kinases/pkr/pk_class/kinases_sc.html) and Saccharomyces Genome Database (http://www.yenstgenome.org/) annotations, 87 null mutants for the genes encoding nonessential protein kinases were selected and tested (see Table S1 in the supplemental material). Each strain was grown in rich medium at 23°C and then shifted to 42°C for 1 h. Whole-cell lysates were prepared, and immunoblotting for endogenous Nab2 was conducted. Strikingly, only two of the tested mutants showed no Nab2 doublet formation after growth at 42°C, bck1Δ and slt2Δ mutants (Fig. 2 A, lanes 4 and 6). Both Bck1 (the MAPKKK) and Slt2 (the MAPK) are components of the MAP kinase cell wall integrity pathway that is activated in response to heat shock and other cellular stresses (7). Consistent with the redundant functions of Mkk1 and Mkk2 (MAPKKs), the Nab2 doublet was still observed in the respective mkk1Δ and mkk2Δ single mutants (data not shown). Examples of other kinase null mutants that showed maintained Nab2 doublet formation are shown in Fig. 2A (lanes 7 to 10). Taken together, these data implicate Slt2, the downstream kinase in the Bck1 pathway, as the in vivo kinase targeting Nab2 during heat shock stress.
FIG. 2.
Nab2 phosphorylation occurs via the MAP kinase, Slt2, on residues 178 and 180. (A) S. cerevisiae kinase null mutants were screened by immunoblotting with anti-Nab2 antibodies for the Nab2 mobility shift. Cells were grown at 23°C (−) (lanes 1, 3, 5, 7, and 9), and shifted to 42°C for 1 h (+) (lanes 2, 4, 6, 8, and 10). WT, wild type; pS, phosphorylated serine; pT, phosphorylated threonine. (B) Diagram of Nab2 domain structure (41), with Nab2 LC-MS data for a peptide from residues 170 to 185 shown. Mass spectrometry was used to determine the phosphorylation sites on Nab2-TAP isolated from heat-shocked cells. In the graph, mass spectrum intensity for the product b and y ions from the peptide are all shown. Overall, the mass spectrum indicates that two sites are phosphorylated. T178 and S180 were identified because ions b4 (Q-T-D-A) and b6 (Q-T-D-A-P-A) have masses indicative of the absence of phosphorylation on T171, and y3 (F-A-S) is indicative of the absence of phosphorylation on S183, leaving T178 and S180 as the only possible phosphorylation targets. (C) In vitro kinase reactions were conducted with Slt2-TAP (lanes 1 and 2), Bck1-TAP (lane 3), Mkk1-TAP (lane 4), and Ypk1-TAP (lane 5) purified from S. cerevisiae on IgG-coated magnetic beads and with wild-type recombinant Nab2 (lane 1 and lanes 3 to 5) or nab2-T178A/S180A (lane 2), with [γ-32P]ATP for 30 min at 30°C. Reaction products were separated by SDS-PAGE and exposed to autoradiography film. The positions (in kilodaltons) of molecular mass markers (MM) are indicated to the left of the gel. (D) Coomassie blue-stained SDS-polyacrylamide gel of Nab2 protein used in vitro kinase reactions in panel C. (E) Immunoblotting for Nab2 was performed on lysates made from cells grown at either 23°C (−) (lanes 1, 3, and 5) or shifted to 42°C for 1 h (+) (lanes 2, 4, and 6) in nab2Δ strains containing plasmids expressing either wild-type NAB2 (lanes 1 and 2), nab2-T178A/S180A (lanes 3 and 4), or nab2-T178E/S180E (lanes 5 and 6).
Slt2 directly phosphorylates Nab2 in vitro.
To determine whether Nab2 is a direct target of Slt2, in vitro kinase assays were conducted using purified, bacterially expressed recombinant Nab2 and selected kinases from S. cerevisiae. Briefly, Slt2, Mkk1, Bck1, and Ypk1 were isolated from yeast cell lysates by TAP tag affinity isolation and subsequently incubated with purified recombinant Nab2 and [γ-32P]ATP for 30 min at 30°C. Following separation by gel electrophoresis, Nab2 phosphorylation was visualized by autoradiography. As shown in Fig. 2C (lane 1), the Slt2-TAP incubation resulted in Nab2 phosphorylation. However, the Mkk1-TAP, Bck1-TAP, and Ypk1-TAP did not promote 32P incorporation (Fig. 2C, lanes 3, 4, and 5). This result confirms that Slt2 directly phosphorylates Nab2.
Nab2 is dually phosphorylated in vivo on threonine 178 and serine 180.
As diagrammed in Fig. 2B, Nab2 is composed of distinct domains that promote protein-protein interactions or RNA-binding activity in vivo (summarized in reference 41): the C-terminal region contains seven zinc finger motifs that facilitate RNA binding, the N-terminal region associates with Mlp1 to promote NPC docking, the central RGG motif is recognized by the karyopherin Kap104 for nuclear import, and the function of a central QQQP span is not known. To understand the functional significance of Slt2-dependent modification, we determined the heat shock-induced in vivo phosphorylation sites on Nab2 using mass spectrometry (liquid chromatography coupled to mass spectrometry [LC-MS]). Briefly, Nab2-TAP protein was isolated from heat-shocked yeast cells. The purified protein was digested with proteases, and the resulting peptides were analyzed by LC-MS. Two sites of phosphorylation were detected by analyzing the resulting peptide fragments. These sites were threonine (T) 178 and serine (S) 180, both of which were identified on a single peptide spanning residues 170 to 185 (Fig. 2B). While this peptide has four possible sites of phosphorylation (T171, T178, S180, and S183), T178 and S180 were identified as the correct sites due to the presence of ions harboring the other potential sites (b4 [Q-T-D-A] and b6 [Q-T-D-A-P-A] for T171 and y3 [F-A-S] for S183) but that did not have the mass shift indicative of phosphorylation. Strikingly, the T178 and S180 residues lie in a region outside the functionally mapped domains of Nab2, suggesting that this region might be important for previously unrecognized molecular interactions in the mRNA export pathway.
Both of the phosphorylated residues were in sequence regions that matched published predictions for MAP kinase consensus sites (as determined by the search algorithms available at the Scansite website [http://scansite.mit.edu]). Thus, we speculated that Slt2 catalyzes the modification of the T178 and S180 sites in vivo. We used site-directed mutagenesis to construct a nonphosphorylatable Nab2 isoform (nab2-T178A/S180A). Recombinant nab2-T178A/S180A was expressed and purified from bacteria and subjected to in vitro kinase assays using isolated Slt2-TAP from yeast and [γ-32P]ATP (Fig. 2C). In contrast to wild-type Nab2 (Fig. 2C, lane 2), no 32P was incorporated into the nab2-T178A/S180A protein. This change in phosphorylation status was not due to differences in protein levels (Fig. 2D). We conclude that Nab2 is phosphorylated at T178 and S180 by Slt2.
To verify that Nab2 T178 and S180 sites are phosphorylated in vivo during heat shock, yeast strains expressing nonphosphorylatable (T178A/S180A) or phosphomimetic (T178E/S180E) Nab2 isoforms were constructed. Briefly, yeast LEU2/CEN plasmids harboring nab2-T187A/S180A or nab2-T178E/S180E mutant alleles were transformed into a nab2Δ yeast strain maintained by a wild-type NAB2/URA3/CEN plasmid. The resulting strains were streaked on media containing 5-fluoroorotic acid, yielding viable strains with only the nab2-T178A/S180A or nab2-T178E/S180E gene. Testing for cell growth across a range of temperatures revealed that T178 and S180 were not required for general cell viability (data not shown) (see Fig. 5). Wild-type and mutant strains were grown at 23°C and shifted to 42°C, and whole-cell lysates were generated and tested by immunoblotting with anti-Nab2 antibodies. Unlike wild-type Nab2, the nab2-T178A/S180A protein did not show a shift in migration following growth at 42°C. This finding further supports the conclusion that T178 and S180 are in vivo phosphorylation sites targeted during heat shock (Fig. 2E, lanes 3 and 4). In addition, the phosphomimetic nab2-T178E/S180E protein exhibited reduced gel migration at both 23°C and 42°C (Fig. 2E, lanes 5 and 6), most likely reflecting the overall change in the Nab2 protein isoelectric charge. Taken together, these studies demonstrate that Nab2 is phosphorylated during heat shock at T178 and S180 by the MAP kinase Slt2.
Poly(A+) mRNA does not accumulate in the nucleus upon heat shock in slt2Δ cells.
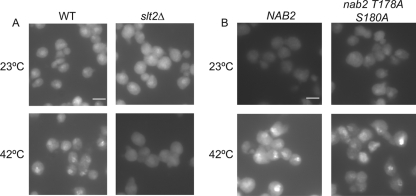
When S. cerevisiae cells undergo heat shock, most poly(A+) mRNA is retained in the nucleus, while hsp mRNAs are exported to the cytoplasm. Since heat shock-dependent posttranslational modification of an essential mRNA export factor might provide a means for differential mRNA transport, we hypothesized that Slt2 plays a critical role in this process. To test this idea, in situ hybridization experiments were conducted to localize poly(A+) mRNA in wild-type and slt2Δ cells at both the permissive temperature and after heat shock at 42°C. Strikingly, whereas wild-type cells showed strong nuclear accumulation of poly(A+) RNA indicative of normal heat shock-induced retention of non-hsp mRNA, slt2Δ cells did not show nuclear accumulation of poly(A+) mRNA (Fig. 3 A). In fact, the poly(A+) mRNA localization in the slt2Δ cells was diffuse and found throughout the cell, which is consistent with a lack of nuclear poly(A+) mRNA retention. As nab2 mutants have polyadenylation defects (33), we conducted control experiments to analyze total poly(A+) tail length in slt2Δ cells. No defects were detected (see Fig. S1 in the supplemental material). This indicates that the loss of Slt2 does not lead to altered polyadenylation, which could have indirectly impacted poly(A+) localization. Thus, the lack of nuclear poly(A+) mRNA accumulation during heat shock in slt2Δ cells reflects a change in non-hsp mRNA retention.
FIG. 3.
Poly(A+) mRNA does not accumulate in the nucleus of heat-shocked slt2Δ cells. (A and B) In situ hybridization using an oligo[d(T)] probe was performed on WT and slt2Δ cells (A) and wild-type and nab2-T178A/S180A cells (B) grown to early log phase in YPD at 23°C or after shift to 42°C for 45 min. Bars = 3 μm.
To test whether Nab2 phosphorylation is the sole Slt2 target responsible for poly(A+) nuclear mRNA accumulation during heat shock, poly(A+) mRNA localization in the nab2-T178A/S180A mutant cells was analyzed by in situ hybridization. The nab2-T178A/S180A mutant cells still showed nuclear accumulation of poly(A+) mRNA at 42°C (Fig. 3B), indicating that Nab2 phosphorylation is not sufficient to mediate non-hsp mRNA retention during heat shock. Consistent with this result, the nab2-T178E/S180E mutant showed no growth defect and did not result in nuclear poly(A+) accumulation under non-heat shock conditions (data not shown). Taken together, this indicates that there are additional unidentified Slt2 kinase targets that affect the mRNA export mechanism.
Heat shock proteins are produced under stress conditions in bck1 and slt2 mutants.
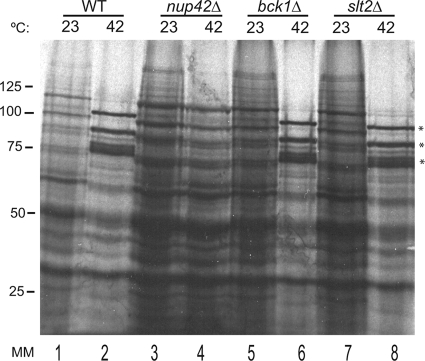
In addition to inhibiting bulk non-hsp mRNA export upon heat shock, Nab2 phosphorylation and the Slt2 MAPK pathway could also actively facilitate the export of hsp mRNA. To test this possibility, we shifted bck1Δ and slt2Δ cells to 42°C for 15 min to induce heat shock stress, and the cells were pulse-labeled with [35S]methionine to monitor heat shock protein synthesis. After an additional 15 min at 42°C, total cell lysates were prepared, the proteins were separated by gel electrophoresis, and radiolabeled proteins were visualized by autoradiography. Both wild-type and nup42Δ mutant cells were included as controls. The nup42Δ mutant has a documented defect in hsp mRNA export and subsequent heat shock protein production (53, 54, 73). Consistent with published reports, heat shock proteins were efficiently synthesized in wild-type cells but absent in nup42Δ cells (Fig. 4, lanes 2 and 4). Both slt2Δ and bck1Δ strains showed robust synthesis of heat shock proteins (Fig. 4, lanes 6 and 8), similar to wild-type cells. Thus, Slt2 is not required for synthesis of heat shock proteins and likely plays no role in facilitating export of hsp mRNA. Furthermore, these studies demonstrate that loss of Slt2 does not result in global defects in transcriptional upregulation during stress.
FIG. 4.
Production of heat shock proteins is not affected in slt2Δ or bck1Δ cells. Wild-type (WT) (lanes 1 and 2), nup42Δ (lanes 3 and 4), bck1Δ (lanes 5 and 6), and slt2Δ (lanes 7 and 8) cells were grown at 23°C, shifted to 42°C for 15 min, and labeled with [35S]methionine for an additional 15 min. Cell lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography. The positions of proteins induced upon heat shock (Hsp104, Hsp82, and Hsp70 proteins, respectively) are indicated by asterisks to the right of the gel. The positions (in kilodaltons) of molecular mass markers (MM) are indicated to the left of the gel.
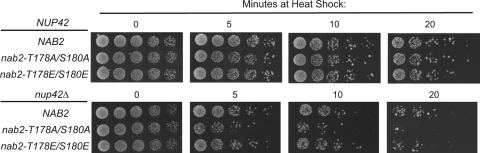
Nab2 phosphorylation is required for the recovery of a nup42Δ mutant from heat shock.
Our results are consistent with a role for Slt2 in retention of non-hsp mRNAs during heat shock. Moreover, others have reported that Slt2 is required for cellular response to stress (42, 43, 71). To understand the function of Nab2 phosphorylation in cell stress, we speculated that incorporation of nab2 mutants in strains with deficient hsp mRNA export would result in growth and thermotolerance defects. To test this hypothesis, we asked whether nab2-T178A/S180A and nab2-T178E/S180E mutants alone or in combination with nup42Δ mutant exhibited enhanced temperature sensitivity under normal growth conditions (Fig. 5). To assay for defects in heat shock recovery, the mutant cells were exposed to extreme heat shock (52°C) (45) for 5- to 20-min time periods and then assayed for growth recovery at 23°C by serial dilution. Strikingly, the nup42Δ nab2-T178A/S180A strain showed a substantial reduction in viability after heat shock compared to the wild-type and single mutant strains (Fig. 5). Importantly, the nup42Δ nab2-T178E/S180E double mutant did not have diminished recovery from heat shock. Taken together, this indicates that Nab2 phosphorylation is needed for optimal cellular heat shock survival. Thus, we speculate that Nab2 phosphorylation is linked to non-hsp RNA retention in the nucleus, and this promotes recovery following return to normal growth conditions.
FIG. 5.
Nab2 phosphorylation is required for efficient recovery from heat shock. The cells were grown at 23°C prior to shifting the cells to 52°C for the times indicated (0, 5, 10, and 20 min). Samples with the same number of cells were then serially diluted and plated for growth at 23°C on YPD for 2.5 days.
Nab2, Mlp1, and Yra1 form heat shock-dependent intranuclear foci.
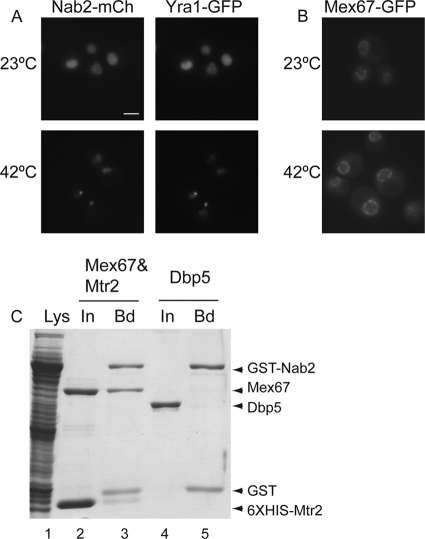
Nab2 shuttles between the nucleus and cytoplasm in a RNA polymerase II-dependent manner (11, 41). Thus, Nab2 cellular localization and/or shuttling activity might be altered by phosphorylation and upon heat shock. To test this possibility, we analyzed Nab2 localization under normal and heat shock conditions by live cell imaging using a mCherry-tagged Nab2 (Nab2-mCherry) or green fluorescent protein (GFP)-tagged Nab2 (Nab2-GFP). Strikingly, Nab2-mCherry or Nab2-GFP accumulated in nuclear foci upon heat shock (Fig. 6 A and B, respectively, left panels). These foci were not due to the epitope tag, as Nab2 foci were also observed by indirect immunofluorescence microscopy with anti-Nab2 antibodies (data not shown). Because of the documented Nab2-Mlp1 interaction and the role of Mlp1 in retention of aberrant mRNAs during normal cellular growth, we next asked whether Mlp1, Mlp2, and/or Nups localize to nuclear foci upon heat shock. Whereas the nuclear basket Nup60-GFP (GFP-tagged Nup60) and centrally localized Nup49-GFP remained associated with the nuclear envelope (see Fig. S2 in the supplemental material), both Mlp1 and Mlp2 formed nuclear foci upon heat shock (Fig. 6A, right panels; see Fig. S2 in the supplemental material). Indeed, by colocalization analysis, Nab2-mCherry and Mlp1-GFP colocalized in intranuclear foci during heat shock (Fig. 6A).
FIG. 6.
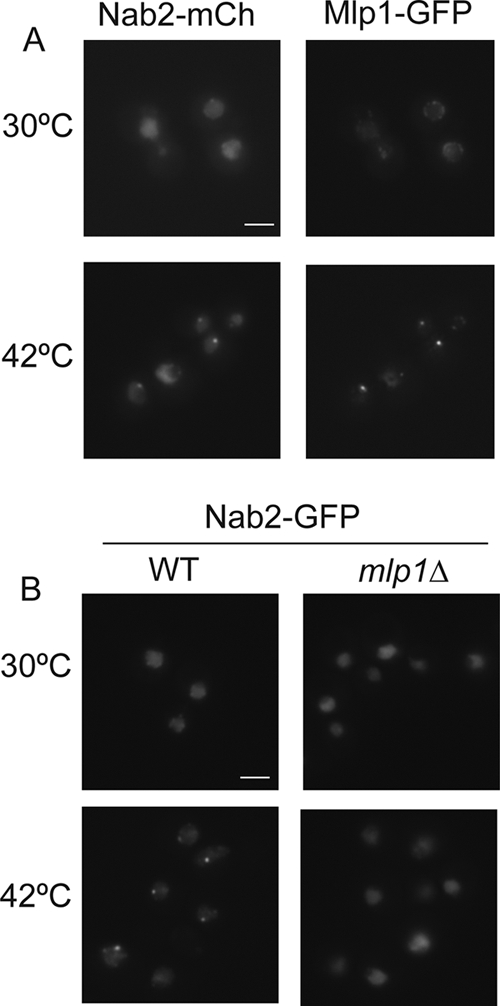
Nab2 forms MLP1-dependent foci during heat shock. Cells were grown to early log phase at 23°C and shifted to 30°C for 1 h. In wild-type cells (left panels), Nab2-GFP shifts from total nuclear localization at 30°C to nuclear foci at 42°C. (A) Nab2-mCherry (Nab2-mCh) forms intranuclear foci at 42°C that overlap with the foci formed by Yra1-GFP. (B) In mlp1Δ cells (right panels), Nab2-GFP foci do not form at 42°C. Bars = 3 μm.
To determine whether Mlp1 and/or Mlp2 are required for Nab2 focus formation, localization was analyzed in different null mutant strains. Loss of Mlp1 clearly inhibited focus formation (Fig. 6B, right panels). However, Nab2-GFP foci were not dependent on the Mlp2, Nup60, or the Mlp1-associated spindle assembly checkpoint protein Mad1 (9, 55) (see Fig. S3 in the supplemental material). Thus, Mlp1 is necessary for formation of heat shock-induced Nab2 foci.
To test whether Slt2 promotes formation of Nab2 foci, a NAB2-mCherry slt2Δ strain was tested. Nab2-mCherry foci were observed in the slt2Δ cells after heat shock (Fig. 7 A). To determine whether focus formation is linked to Nab2 phosphorylation, immunoblotting for Nab2 was conducted in different null mutants. The Nab2 doublet was present in the mlp1Δ mutant after heat shock, as well as in the nup42Δ mutant (Fig. 7B). Lack of GFD1, which encodes another Nab2-binding protein (66), also had no apparent effect on phosphorylation. This indicates that Nab2 phosphorylation and focus formation are independent. For controls, we tested whether mlp1Δ cells had defects in either hsp mRNA export or nuclear retention of bulk poly(A+) mRNA during heat shock. Following a shift to heat shock conditions, the mlp1Δ cells showed normal production of hsp proteins and accumulated nuclear poly(A+) mRNA (see Fig. S4 in the supplemental material; data not shown). Taken together, these studies demonstrate that Slt2 and Mlp1 mediate distinct molecular interactions during heat shock.
FIG. 7.
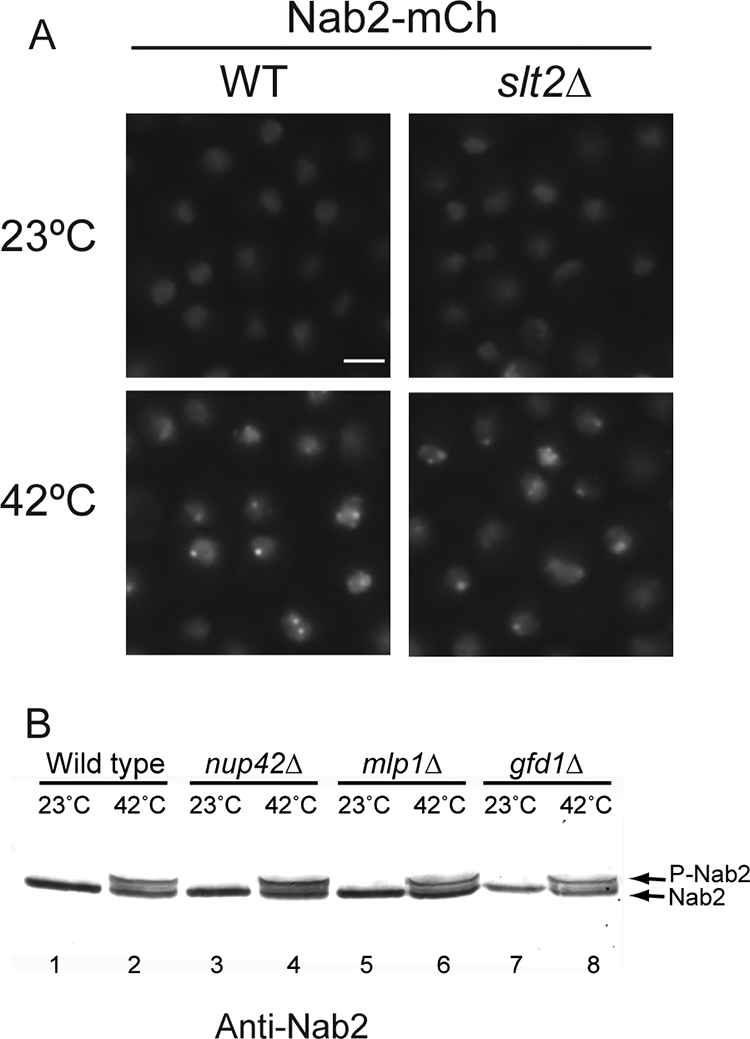
During heat shock, Nab2 phosphorylation and intranuclear focus formation are independent events. (A) Nab2-mCherry forms intranuclear foci in wild-type (WT) and slt2Δ cells shifted to growth at 42°C for 1 h. Bar = 3 μm. (B) Immunoblotting for Nab2 in lysates from yeast strains shows the mobility shift induced by heat shock (42°C for 1 h) in wild-type (lanes 1 and 2), nup42Δ (lanes 3 and 4), mlp1Δ (lanes 5 and 6), and gfd1Δ (lanes 7 and 8) cells.
Nab2-Mex67 interactions are uncoupled during heat shock.
Because Slt2 is needed for nuclear accumulation of non-hsp mRNA, we speculated that stress signaling might alter interactions between Nab2 and other essential mRNA export factors during heat shock. A recent proteomic study reported Nab2 association with members of the TREX complex, including Yra1 and Mex67 (5). To determine whether heat shock uncouples Nab2 from the TREX complex, first, the subcellular localizations of Yra1-GFP and Mex67-GFP were determined. Whereas Yra1-GFP localized to nuclear foci during heat shock (Fig. 8 A), Mex67-GFP remained associated with the nuclear rim (Fig. 8B). Importantly, Yra1-GFP and Nab2-mCherry foci colocalized (Fig. 8A). Second, we employed biochemical approaches to analyze changes in Nab2 complexes from normal cells (not heat shocked) and heat shock-induced cells. Nab2-TAP protein complexes were isolated from yeast cells grown at 23°C or following a shift to growth at 42°C for 1 h. Cells were lysed using a liquid nitrogen grinding method that preserves native mRNP interactions (5, 46). Purified complexes were then analyzed by multidimensional protein identification technology (MudPIT) mass spectrometry to determine protein composition. Included in Table 1 are all copurifying proteins with documented mRNP or NPC links. The copurifying proteins at 23°C largely paralleled those reported by Batisse et al. (5), with the exception of Sub2. We did not detect Sub2 in our Nab2-TAP isolation, potentially due to strain, isolation, or detection differences. Interestingly, we found that the Nab2-TAP complexes varied greatly in composition when isolated from heat-shocked versus non-heat-shocked cells. To quantify each copurifying protein, total spectral counts of each were taken and normalized to the total spectral counts for Nab2 in the same sample (Table 1). The resulting ratio reflected the relative level of each protein to Nab2-TAP (Table 1, Relative 42°C/23°C level column). At 23°C, NPC-associated proteins were the largest class found to be copurifying with Nab2-TAP. Strikingly, the relative amounts of all the detected NPC proteins decreased when Nab2-TAP was isolated from heat-shocked cells (with a value of <0 in the Relative 42°C/23°C level column). Moreover, the Mex67 levels were also decreased after heat shock. In parallel, after heat shock, increased amounts of Mlp1, Mlp2, and Yra1 were isolated in Nab2-TAP complexes (those with a value of >0 in the Relative 42°C/23°C level column). Overall, the biochemical analysis was consistent with the distinct localization patterns of these factors upon heat shock. We conclude that Mex67 association with Nab2-complexes is inhibited during heat shock stress.
FIG. 8.
Nab2-Mex67 directly interact and are not colocalized during heat shock. (A) Cells coexpressing Nab2-mCherry and Yra1-GFP were tested for formation of heat shock-induced intranuclear foci. Nab2-mCherry and Yra1-GFP colocalize in foci at 42°C. Bar = 3 μm. (B) Mex67-GFP localization does not change during heat shock and remains localized in the nuclear envelope. (C) Nab2 and Mex67-Mtr2 directly interact in vitro. Soluble binding assays with glutathione resin were conducted with purified recombinant Mex67-Mtr2 (lanes 2 and 3) or with Dbp5 (lanes 4 and 5) with lysate (Lys) from bacterial cells expressing GST-Nab2 (lane 1). Samples were separated by SDS-PAGE, and proteins were visualized by Coomassie blue staining. Input (In) and bound (Bd) fractions are shown. GST alone as a proteolytic product from GST-Nab2 was observed in lanes 3 and 5.
TABLE 1.
Mass spectrometry analysis of Nab2-TAP complexes isolated from normal (non-heat-shocked) cells and heat-shocked cells
| Protein | Raw spectral count |
Normalized spectral count |
Relative 42°C/23°C levela | ||
|---|---|---|---|---|---|
| 23°C | 42°C | 23°C | 42°C | ||
| Nab2 | 561 | 257 | 100 | 100 | 1 |
| Mlp1 | 113 | 349 | 20.1 | 135.8 | 6.8 |
| Mlp2 | 77 | 158 | 13.7 | 61.5 | 4.5 |
| Yra1 | 56 | 60 | 10 | 23.4 | 2.3 |
| Pab1 | 218 | 209 | 38.9 | 81.3 | 2.0 |
| Mex67 | 45 | 10 | 8.0 | 3.9 | 0.5 |
| Gfd1 | 40 | 51 | 7.1 | 19.8 | 2.8 |
| Stm1 | 16 | 15 | 2.9 | 5.8 | 2 |
| Tho2 | 9 | 6 | 1.6 | 2.3 | 1.4 |
| Nsp1 | 185 | 17 | 33 | 6.6 | 0.2 |
| Pom152 | 116 | 13 | 20.7 | 5.0 | 0.24 |
| Nup170 | 94 | 3 | 16.7 | 1.2 | 0.07 |
| Nup159 | 76 | 9 | 13.5 | 3.5 | 0.26 |
| Nup188 | 72 | 6 | 12.8 | 2.3 | 0.18 |
| Nup145 | 83 | 3 | 14.8 | 1.2 | 0.08 |
| Nup60 | 76 | 7 | 13.5 | 2.7 | 0.2 |
| Nup133 | 76 | 0 | 13.5 | 0 | 0 |
| Nup2 | 66 | 3 | 11.7 | 1.2 | 0.10 |
| Nic96 | 67 | 4 | 11.9 | 1.6 | 0.13 |
| Nup157 | 63 | 5 | 11.2 | 2.0 | 0.18 |
| Nup192 | 63 | 3 | 11.2 | 1.2 | 0.10 |
| Nup85 | 53 | 1 | 9.5 | 0.9 | 0.09 |
| Nup82 | 50 | 7 | 8.9 | 2.7 | 0.30 |
| Sac3 | 53 | 1 | 9.5 | 0.4 | 0.04 |
| Nup120 | 47 | 0 | 7.7 | 0 | 0 |
| Nup1 | 29 | 3 | 5.2 | 1.2 | 0.23 |
| Gle2 | 30 | 0 | 5.4 | 0 | 0 |
| Nup59 | 59 | 0 | 2.6 | 0 | 0 |
| Pom34 | 16 | 2 | 2.9 | 0.8 | 0.28 |
| Nup53 | 14 | 1 | 2.5 | 0.4 | 0.16 |
The relative 42°C/23°C level is the normalized spectral count at 42°C divided by the normalized spectral count at 23°C.
Current models predict that Yra1 acts as an adaptor protein for Mex67 recruitment to mature mRNPs (60). However, Yra1 is constitutively nuclear and thus is not exported to the cytoplasm with a Mex67-bound mRNP. In contrast, Nab2 shuttles to the cytoplasm in an export-dependent manner (41, 66). On the basis of the differential association of Mex67 with Nab2 complexes, we speculated that Nab2 might be a previously unrecognized adaptor for Mex67-Mtr2. To test for direct physical interactions, in vitro binding assays were conducted with purified recombinant Mex67-Mtr2 heterodimer as previously described (81). Briefly, glutathione S-transferase (GST)-tagged Nab2 (GST-Nab2) was expressed in bacteria (Fig. 8C, lane 1), and lysate was bound to glutathione resin. Purified Mex67-Mtr2, or as a control the DEAD box protein Dbp5, was applied to the GST-Nab2 resin. After the column was washed, bound proteins were eluted with glutathione and separated by gel electrophoresis. Coomassie blue staining showed isolation of Mex67-Mtr2 with the GST-Nab2 (Fig. 8C, lane 3), whereas Dbp5 was not isolated (Fig. 8C, lane 5). Previous studies have documented that Mex67 does not bind to GST or glutathione resin nonspecifically (67), and the Mex67-Mtr2 interaction with Nab2 was also verified by an alternative binding assay described below. The direct in vitro binding of recombinant Mex67-Mtr2 to Nab2 suggests that Nab2 could serve as a direct adaptor for Mex67-Mtr2 recruitment to mRNPs.
Taking into account both the in vitro Nab2 interaction with Mex67-Mtr2 and the decreased in vivo Mex67-Mtr2 association with Nab2 complexes isolated from heat-shocked cells, we speculated that Nab2 phosphorylation during heat shock stress might impact Nab2 interaction with Mex67-Mtr2. To test this, in vitro soluble binding assays were conducted with purified recombinant nab2-T178E/S180E protein and Mex67-Mtr2. For these assays, Mex67-Mtr2 was immobilized on Ni2+ agarose beads via the 6×His epitope tag on Mtr2. Decreasing amounts of untagged wild-type Nab2, nab2-T178A/S180A, or nab2-T178E/S180E protein were incubated with the Mex67-Mtr2 beads, and after the column was washed, the bound proteins were eluted, and the levels of bound proteins were analyzed by immunoblotting. No difference in Mex67-Mtr2 interaction was detected between the Nab2 proteins (see Fig. S5 in the supplemental material). Thus, Nab2 phosphorylation is potentially not sufficient to inhibit Mex67 recruitment to Nab2 complexes during heat shock, and other potential Slt2 targets and Mex67 interaction partners are playing roles.
DISCUSSION
Defining the mechanism for regulated mRNA export during stress is critical to understanding cellular adaptation and survival. Here we have uncovered a functional correlation during heat shock stress between changes in Nab2 phosphorylation, localization, and association with Mex67 and Mlp1. We identify the cell wall integrity MAP kinase Slt2 signaling pathway as being key to the mRNA export mechanism during heat shock. Two specific heat shock-induced Slt2 phosphorylation sites in Nab2 are needed for thermotolerance and stress recovery in a nup42Δ mutant. As slt2Δ cells do not accumulate nuclear poly(A+) RNA upon heat shock but remain competent for hsp mRNA export and poly(A+) tail length maintenance, we conclude that Slt2 is required for nuclear non-hsp mRNA retention during stress. Further, on the basis of decreased levels of Mex67 and Nups in Nab2 complexes and independent relocalization and association of Nab2 and Yra1 with Mlp1-dependent nuclear foci, we conclude that at least two independent mechanisms control the differential export of non-hsp and hsp mRNA during heat shock stress: regulated recruitment of Mex67-Mtr2 to mRNA-binding proteins and sequestering of mRNA-binding proteins in Mlp1 intranuclear foci.
This work directly addresses the long-standing question of whether an active mechanism exists for promoting hsp mRNA transport or whether non-hsp mRNAs are selectively retained. Prior work has shown that both hsp and non-hsp mRNA export require the Mex67 mRNA export receptor as well as the direct mediators of mRNA transport, Dbp5, Gle1, and Nup159 (summarized in references 50 and 51; reviewed in reference 26). Moreover, hsp mRNA export is not dependent on Npl3 or Yra1, and others have shown that Npl3 relocalizes to the cytoplasm during heat shock to uncouple from hsp mRNA export (35). We now document a critical role for Nab2, Slt2, and Mlp1 in heat shock-induced retention of bulk poly(A+) mRNA, which reinforces the hypothesis that the hsp mRNA export mechanism is distinct from the non-hsp global poly(A+) mRNA export mechanism.
We propose a model wherein activated Slt2 promotes the retention of non-heat shock mRNAs through modulation of mRNA-binding proteins during heat shock stress. Under normal growth conditions, Mex67-Mtr2 is recruited to nascent mRNAs via the concerted action of Yra1 and Nab2. The Mex67-Mtr2/Nab2-bound mRNP is released from the site of synthesis and then docks at the nuclear face of the NPC. Interactions between Nab2 and Mlp1 at the NPC provide a quality control step to monitor the proper mRNP composition of a given mRNA prior to pore translocation. During heat shock, the MAP kinase Slt2 is activated by the cell wall integrity phosphorylation cascade. This activation results in phosphorylation of Nab2 and possibly other components of the mRNA export machinery. Other signaling pathways are also likely triggered to impact mRNA export factors independently.
We further speculate that sequestering of Nab2 and Yra1 in Mlp1-dependent intranuclear foci, in combination with Slt2 signaling, inhibits recruitment of Mex67-Mtr2 to non-hsp mRNPs and diminishes export of such mRNPs. As such, under heat shock conditions, Nab2 complexes are not found associated with NPC proteins, as they are not undergoing NPC targeting or nuclear export. Overall, this mechanism might allow Mex67-Mtr2 to associate predominantly with hsp mRNAs through an uncharacterized, Nab2-independent mechanism. Such selective mRNA transport and retention could provide a means to tightly control gene expression: by coordinating transcription and export of hsp mRNAs necessary for cellular stress responses at the same time as retaining housekeeping (non-hsp) mRNAs needed for cell growth and recovery after the stress.
Our observations also highlight future questions to address regarding the mechanism for regulated mRNA transport during cellular stress. First, how is Mex67-Mtr2 recruited to hsp mRNAs? To date, none of the characterized mRNA adaptor proteins for Mex67 (Yra1, Npl3, and Nab2) are required for hsp mRNA export. There might be an uncharacterized mRNA-binding protein that serves as a Mex67-Mtr2 adaptor protein on hsp mRNAs. Alternatively, Mex67-Mtr2 might directly bind and recognize hsp mRNAs. Indeed, Mex67-Mtr2 can associate directly with a stem-loop structure in the 5S rRNA component of the large ribosomal subunit (81). The hypothesis that a specific RNA sequence mediates hsp mRNA export was first suggested by Saavedra and coworkers, based on the discovery of 5′ and 3′ untranslated region (UTR) elements in the SSA4 hsp mRNA that are both necessary and sufficient for directing export during heat shock (53).
Another intriguing question encompasses whether and how differential mRNA transport is employed during other non-heat shock growth conditions. Although our studies demonstrate key steps required for hsp versus non-hsp mRNAs transport during stress, it is possible that similar mechanisms exist for controlling transport of specific gene transcripts under other conditions. This could include other environmental stresses, such as salt and ethanol shock (29), as well as during changes in cellular programs, such as mating, cellular differentiation, and the cell cycle. On the basis of work showing Npl3 dynamics are influenced by the Sky1 kinase and Glc7 phosphatase (17), there is tremendous potential for the role of posttranslational modifications of mRNA-binding proteins in cell regulation. There are also interesting parallels to consider between the mechanisms for controlling mRNA export during heat shock and how viruses pirate the cellular mRNA export machinery. Indeed, Nup42 (Rip1) was first identified based on its role in facilitating Rev export in yeast cells (65).
Alternatively, Nab2-Mlp1 foci might play an as-of-yet uncharacterized role that is distinct from mRNA export. Because Mlp1 and Mlp2 have been linked to downregulation of transcription (74), these foci could influence gene expression during stress. Future characterization of the Mlp1-Nab2-Yra1 foci is required to decipher these possibilities. Work will also be needed to reveal the mechanism for intranuclear focus formation and Mlp1 relocalization. As the vertebrate Tpr is targeted by extracellular signal-regulated kinase 2 (ERK2) (76), Mlp1 itself is also potentially targeted by posttranslation modification during cellular stress. We predict that multiple signaling events impinge on both mRNP and NPC components to coordinate selective transport events.
Recent studies demonstrate that the MAP kinase Ste20 plays a key role in formation of cytoplasmic RNP foci called stress granules (83). It is intriguing to consider that the Nab2-Yra1-Mlp1 intranuclear foci that form during heat shock stress parallel the role of cytoplasmic stress granules. As such, they could store non-hsp mRNAs or mRNA-binding proteins during times of stress to allow rapid recovery after stress. Overall, this places the posttranslational modifications and subcellular localization of mRNA-binding proteins as key elements for controlling selective nuclear mRNA export and retention.
Supplementary Material
Acknowledgments
We thank Michael Rout and Marlene Oeffinger for sharing cell lysis and immunopurification protocols, John Aitchison for yeast plasmids, W. Hayes McDonald for phospho-proteomic and mass spectrometry analysis, Yingna Zhou, Laura Terry, and Ryan Baldridge for assistance with experiments, and members of the Wente laboratory for discussions.
This work was supported by grants from the National Institutes of Health: R01-GM51219 (S.R.W.), R01-GM058728-11 (A.H.C.), National Research Service Awards (NRSA) 5T32-GM008554 (S.R.C.), 5T32-CA009383 (E.J.T.), and 1F32-GM075459 (E.J.T.). Mass spectrometry data were acquired by the Vanderbilt Mass Spectrometry Research Center (supported by NIH/NCI 3P30 CA068485-13 Cancer Center support grant to Vanderbilt University).
Footnotes
Published ahead of print on 7 September 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abruzzi, K. C., S. Lacadie, and M. Rosbash. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. [DOI] [PubMed] [Google Scholar]
- 3.Alcazar-Roman, A. R., E. J. Tran, S. Guo, and S. R. Wente. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8:711-716. [DOI] [PubMed] [Google Scholar]
- 4.Apponi, L. H., S. M. Kelly, M. T. Harreman, A. N. Lehner, A. H. Corbett, and S. R. Valentini. 2007. An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability. Mol. Cell. Biol. 27:6569-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batisse, J., C. Batisse, A. Budd, B. Bottcher, and E. Hurt. 2009. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 284:34911-34917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond, U. 2006. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 6:160-170. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R. E., and J. Thorner. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deka, P., M. E. Bucheli, C. Moore, S. Buratowski, and G. Varani. 2008. Structure of the yeast SR protein Npl3 and interaction with mRNA 3′-end processing signals. J. Mol. Biol. 375:136-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Souza, C. P., S. B. Hashmi, T. Nayak, B. Oakley, and S. A. Osmani. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 20:2146-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodou, E., and R. Treisman. 1997. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, K., J. G. Umen, and C. Guthrie. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 12.Farny, N. G., J. A. Hurt, and P. A. Silver. 2008. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 22:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasken, M. B., M. Stewart, and A. H. Corbett. 2008. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated protein, Mlp1, in mRNA export. J. Biol. Chem. 283:27130-27143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallardo, M., R. Luna, H. Erdjument-Bromage, P. Tempst, and A. Aguilera. 2003. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278:24225-24232. [DOI] [PubMed] [Google Scholar]
- 15.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. U. S. A. 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galy, V., O. Gadal, M. Fromont-Racine, A. Romano, A. Jacquier, and U. Nehrbass. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116:63-73. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert, W., and C. Guthrie. 2004. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell 13:201-212. [DOI] [PubMed] [Google Scholar]
- 18.Green, D. M., C. P. Johnson, H. Hagan, and A. H. Corbett. 2003. The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl. Acad. Sci. U. S. A. 100:1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, X. Cheng, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277:7752-7760. [DOI] [PubMed] [Google Scholar]
- 20.Guisbert, K. K., K. Duncan, H. Li, and C. Guthrie. 2005. Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles. RNA 11:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hector, R. E., K. R. Nykamp, S. Dheur, J. T. Anderson, P. J. Non, C. R. Urbinati, S. M. Wilson, L. Minvielle-Sebastia, and M. S. Swanson. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21:1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold, A., T. Klymenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7:1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 23.Hieronymus, H., and P. A. Silver. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33:155-161. [DOI] [PubMed] [Google Scholar]
- 24.Hogan, D. J., D. P. Riordan, A. P. Gerber, D. Herschlag, and P. O. Brown. 2008. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurt, E., K. Strasser, A. Segref, S. Bailer, N. Schlaich, C. Presutti, D. Tollervey, and R. Jansen. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275:8361-8368. [DOI] [PubMed] [Google Scholar]
- 26.Iglesias, N., and F. Stutz. 2008. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582:1987-1996. [DOI] [PubMed] [Google Scholar]
- 27.Igual, J. C., A. L. Johnson, and L. H. Johnston. 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15:5001-5013. [PMC free article] [PubMed] [Google Scholar]
- 28.Iovine, M. K., J. L. Watkins, and S. R. Wente. 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izawa, S., T. Kita, K. Ikeda, and Y. Inoue. 2008. Heat shock and ethanol stress provoke distinctly different responses in 3′-processing and nuclear export of HSP mRNA in Saccharomyces cerevisiae. Biochem. J. 414:111-119. [DOI] [PubMed] [Google Scholar]
- 30.Izawa, S., R. Takemura, T. Miki, and Y. Inoue. 2005. Characterization of the export of bulk poly(A)+ mRNA in Saccharomyces cerevisiae during the wine-making process. Appl. Environ. Microbiol. 71:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, S. A., G. Cubberley, and D. L. Bentley. 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33:215-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly, S. M., and A. H. Corbett. 2009. Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10:1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly, S. M., S. W. Leung, L. H. Apponi, A. M. Bramley, E. J. Tran, J. A. Chekanova, S. R. Wente, and A. H. Corbett. 2010. Recognition of polyadenosine RNA by the zinc finger domain of nuclear poly(A) RNA binding protein 2 (Nab2) is required for correct mRNA 3′-end formation. J. Biol. Chem. 285:26022-26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761-773. [DOI] [PubMed] [Google Scholar]
- 35.Krebber, H., T. Taura, M. S. Lee, and P. A. Silver. 1999. Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev. 13:1994-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kress, T. L., N. J. Krogan, and C. Guthrie. 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 32:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, M. S., M. Henry, and P. A. Silver. 1996. A protein that shuttles between the nucleus and cytoplasm is an important mediator of RNA export. Genes Dev. 10:1233-1246. [DOI] [PubMed] [Google Scholar]
- 38.Levin, D. E., B. Bowers, C. Y. Chen, Y. Kamada, and M. Watanabe. 1994. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell. Mol. Biol. Res. 40:229-239. [PubMed] [Google Scholar]
- 39.Ma, Z. Q., S. Dasari, M. C. Chambers, M. D. Litton, S. M. Sobecki, L. J. Zimmerman, P. J. Halvey, B. Schilling, P. M. Drake, B. W. Gibson, et al. 2009. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 8:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacCoss, M. J., W. H. McDonald, A. Saraf, R. Sadygov, J. M. Clark, J. J. Tasto, K. L. Gould, D. Wolters, M. Washburn, A. Weiss, et al. 2002. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U. S. A. 99:7900-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marfatia, K. A., E. B. Crafton, D. M. Green, and A. H. Corbett. 2003. Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 278:6731-6740. [DOI] [PubMed] [Google Scholar]
- 42.Martin, H., J. Arroyo, M. Sanchez, M. Molina, and C. Nombela. 1993. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 degrees C. Mol. Gen. Genet. 241:177-184. [DOI] [PubMed] [Google Scholar]
- 42a.McDonald, W. H., R. Ohi, D. T. Miyamoto, T. J. Mitchison, and J. R. Yates III. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrometry 219:245-251. [Google Scholar]
- 43.Millson, S. H., A. W. Truman, V. King, C. Prodromou, L. H. Pearl, and P. W. Piper. 2005. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4:849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, R., and S. R. Wente. 1996. An RNA-export mediator with an essential nuclear export signal. Nature 383:357-360. [DOI] [PubMed] [Google Scholar]
- 45.Nwaka, S., B. Mechler, O. von Ahsen, and H. Holzer. 1996. The heat shock factor and mitochondrial Hsp70 are necessary for survival of heat shock in Saccharomyces cerevisiae. FEBS Lett. 399:259-263. [DOI] [PubMed] [Google Scholar]
- 46.Oeffinger, M., K. E. Wei, R. Rogers, J. A. DeGrasse, B. T. Chait, J. D. Aitchison, and M. P. Rout. 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4:951-956. [DOI] [PubMed] [Google Scholar]
- 47.Palancade, B., M. Zuccolo, S. Loeillet, A. Nicolas, and V. Doye. 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell 16:5258-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu, X., S. Lykke-Andersen, T. Nasser, C. Saguez, E. Bertrand, T. H. Jensen, and C. Moore. 2009. Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol. Cell. Biol. 29:5327-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray, A., R. E. Hector, N. Roy, J. H. Song, K. L. Berkner, and K. W. Runge. 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33:522-526. [DOI] [PubMed] [Google Scholar]
- 50.Rollenhagen, C., C. A. Hodge, and C. N. Cole. 2004. The nuclear pore complex and the DEAD box protein Rat8p/Dbp5p have nonessential features which appear to facilitate mRNA export following heat shock. Mol. Cell. Biol. 24:4869-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rollenhagen, C., C. A. Hodge, and C. N. Cole. 2007. Following temperature stress, export of heat shock mRNA occurs efficiently in cells with mutations in genes normally important for mRNA export. Eukaryot. Cell 6:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth, K. M., J. Byam, F. Fang, and J. S. Butler. 2009. Regulation of NAB2 mRNA 3′-end formation requires the core exosome and the Trf4p component of the TRAMP complex. RNA 15:1045-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saavedra, C., K. S. Tung, D. C. Amberg, A. K. Hopper, and C. N. Cole. 1996. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 10:1608-1620. [DOI] [PubMed] [Google Scholar]
- 54.Saavedra, C. A., C. M. Hammell, C. V. Heath, and C. N. Cole. 1997. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 11:2845-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott, R. J., C. P. Lusk, D. J. Dilworth, J. D. Aitchison, and R. W. Wozniak. 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 16:4362-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skaggs, H. S., H. Xing, D. C. Wilkerson, L. A. Murphy, Y. Hong, C. N. Mayhew, and K. D. Sarge. 2007. HSF1-TPR interaction facilitates export of stress-induced HSP70 mRNA. J. Biol. Chem. 282:33902-33907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snay-Hodge, C. A., H. V. Colot, A. L. Goldstein, and C. N. Cole. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strasser, K., J. Bassler, and E. Hurt. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strasser, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 62.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 63.Strawn, L. A., T. Shen, N. Shulga, D. S. Goldfarb, and S. R. Wente. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197-206. [DOI] [PubMed] [Google Scholar]
- 64.Stutz, F., J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash. 1997. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 11:2857-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stutz, F., M. Neville, and M. Rosbach. 1995. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell 82:495-506. [DOI] [PubMed] [Google Scholar]
- 66.Suntharalingam, M., A. R. Alcazar-Roman, and S. R. Wente. 2004. Nuclear export of the yeast mRNA-binding protein Nab2 is linked to a direct interaction with Gfd1 and to Gle1 function. J. Biol. Chem. 279:35384-35391. [DOI] [PubMed] [Google Scholar]
- 67.Terry, L. J., and S. R. Wente. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178:1121-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terry, L. J., E. B. Shows, and S. R. Wente. 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318:1412-1416. [DOI] [PubMed] [Google Scholar]
- 69.Torres, L., H. Martin, M. I. Garcia-Saez, J. Arroyo, M. Molina, M. Sanchez, and C. Nombela. 1991. A protein kinase gene complements the lytic phenotype of Saccharomyces cerevisiae lyt2 mutants. Mol. Microbiol. 5:2845-2854. [DOI] [PubMed] [Google Scholar]
- 70.Tran, E. J., Y. Zhou, A. H. Corbett, and S. R. Wente. 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28:850-859. [DOI] [PubMed] [Google Scholar]
- 71.Truman, A. W., S. H. Millson, J. M. Nuttall, M. Mollapour, C. Prodromou, and P. W. Piper. 2007. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6:744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tseng, S. S., P. L. Weaver, Y. Liu, M. Hitomi, A. M. Tartakoff, and T. H. Chang. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17:2651-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vainberg, I. E., K. Dower, and M. Rosbash. 2000. Nuclear export of heat shock and non-heat-shock mRNA occurs via similar pathways. Mol. Cell. Biol. 20:3996-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vinciguerra, P., N. Iglesias, J. Camblong, D. Zenklusen, and F. Stutz. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24:813-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viphakone, N., F. Voisinet-Hakil, and L. Minvielle-Sebastia. 2008. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 36:2418-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vomastek, T., M. P. Iwanicki, W. R. Burack, D. Tiwari, D. Kumar, J. T. Parsons, M. J. Weber, and V. K. Nandicoori. 2008. Extracellular signal-regulated kinase 2 (ERK2) phosphorylation sites and docking domain on the nuclear pore complex protein Tpr cooperatively regulate ERK2-Tpr interaction. Mol. Cell. Biol. 28:6954-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 78.Weirich, C. S., J. P. Erzberger, J. M. Berger, and K. Weis. 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16:749-760. [DOI] [PubMed] [Google Scholar]
- 79.Wente, S. R., M. P. Rout, and G. Blobel. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119:705-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73:5683-5690. [DOI] [PubMed] [Google Scholar]
- 80a.Yaffe, M. P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. U. S. A. 81:4819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yao, W., D. Roser, A. Kohler, B. Bradatsch, J. Bassler, and E. Hurt. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 26:51-62. [DOI] [PubMed] [Google Scholar]
- 82.Yates, J. R., III, J. K. Eng, A. L. McCormack, and D. Schieltz. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426-1436. [DOI] [PubMed] [Google Scholar]
- 83.Yoon, J. H., E. J. Choi, and R. Parker. 2010. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189:813-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.