Abstract
Cytokine-mediated JAK/STAT signaling controls numerous important biologic responses like immune function, cellular growth, and differentiation. Inappropriate activation of this signaling pathway is associated with a range of malignancies. Kaposi's sarcoma-associated herpesvirus (KSHV) is the infectious viral agent associated with Kaposi's sarcoma and may also contribute to B-cell disorders, which include primary effusion lymphoma (PEL) and multicentric Castleman's disease. However, regulation of cytokine-mediated lymphocytic immune response by KSHV is not fully understood. In this report, we demonstrate that KSHV suppresses the interleukin-4 (IL-4)-stimulated immune response of B-lymphocyte activation and cell proliferation. Moreover, we show that the latency-associated nuclear antigen (LANA) encoded by KSHV is essential for viral blocking of IL-4-induced signaling. LANA reduces phosphorylation of the signal transducers and activators of transcription 6 (STAT6) on Y-641 and concomitantly its DNA binding ability. Importantly, knockdown of endogenous STAT6 dramatically increases the sensitivity of PEL cells to low-serum stress or chemical-mediated cellular apoptosis and reactivation of KSHV from latent replication. Thus, these findings suggest that the IL-4/STAT6 signaling network is precisely controlled by KSHV for survival, maintenance of latency, and suppression of the host cytokine immune response of the virus-infected cells.
Kaposi's sarcoma-associated herpesvirus (KSHV; also known as human herpesvirus 8) is a lymphotropic γ-2 herpesvirus which was originally discovered in 1994 in lesions of AIDS-related Kaposi's sarcoma (KS) patients (15, 63). In addition to KS tumors, KSHV has also been causally associated with two types of lymphoproliferative diseases, primary effusion lymphoma (PEL; also referred to as body cavity-based lymphoma [BCBL]) (14) and multicentric Castleman's disease (MCD) (57), implicating this virus in multiple B-cell lineage malignancies. Like other herpesviruses, KSHV has two distinct life stages: latent infection and lytic infection. To date, it is widely accepted that latent infection by the virus plays a central role in viral pathogenesis with the expression of select genes which are responsible for targeting and controlling selective cellular pathways (25, 32). Of the at least 90 genes encoded by KSHV, only a few are expressed during latency (52). The latency-associated nuclear antigen (LANA) encoded by open reading frame 73 (ORF73) has been documented as one of the major latent proteins consistently present in all KSHV-associated tumors (23). LANA was found to play a crucial role in persistence and maintenance of the latent viral episome (5, 18, 28). Recently, LANA has also been shown to participate in regulation of a number of cellular signaling activities by functioning as a transcriptional coactivator or corepressor (1, 11, 25, 26, 33, 35, 40, 43, 49, 50, 62). The evidence showing that LANA can induce B-cell hyperplasia and lymphoma in transgenic mice (24), as well as inhibit transforming growth factor β (TGF-β) signaling through epigenetic silencing (22), suggests a potential role for LANA in dysregulation of B-lymphocyte immune response beyond its functions related to viral genome maintenance.
Interleukin-4 (IL-4) is a multifunctional cytokine that controls cell growth and is involved in regulation of the immune system (34, 46). IL-4, predominantly produced by Th2 lymphocytes and mast cells, has been shown to have diverse biological activities in many cell types, including B cells, T cells, macrophages, mast cells, and endothelial cells (38, 47, 61, 66). IL-4 exerts its effect by binding to and stimulating its transmembrane receptor, IL-4R, which is comprised of two chains, a cytokine-specific α chain (IL-4Rα) and a common γ chain (44) shared by several cytokines, including IL-2 (60) and IL-7 (45). The IL-4Rα chain can dimerize with γ chain to form the type I IL-4R, which binds IL-4 specifically or it can dimerize with the IL-13 receptor α1 chain to form the type II IL-4R, which binds either IL-4 or IL-13. Interaction of IL-4 with its receptor induces IL-4Rα receptor chain dimerization, resulting in Jak (JAK1 and JAK2) activation followed by phosphorylation of tyrosine residues within the IL-4Rα receptor chain (56). This receptor-kinase complex recruits and activates signal transducer and activator of transcription 6 (STAT6) on Y-641 (56). Subsequently, the phosphorylated STAT6 homodimerizes and translocates to the nucleus, where it binds distinct promoter regions (consensus TTCN4GAA) on DNA and activates specific gene transcription, which includes IL-4 and IL-4Rα (31, 59).
IL-4 induces Ig class switching to IgE and IgG4 (IgG1 mouse), and germ line transcription of their respective heavy chain genes (ɛ and γ) in B lymphocytes (21). In a STAT6-dependent manner, IL-4 can regulate expression of IL-4Rα, CD40, CD23, and major histocompatibility complex (MHC) class II (31, 37) and can increase B-cell activation by cytoskeleton rearrangements that influence cell morphology (17, 19, 64). In conjunction with other B-cell stimulants, IL-4 increases proliferation of B lymphocytes, cell aggregation, and cell spreading (19, 53). In addition to the effect on B lymphocyte, IL-4 also directly regulates the cytoskeleton in endothelial cells (36) and neutrophils (29). It exerts anti-inflammatory effects by downregulating T-helper Th1 cell activity and promotes T-cell differentiation toward the Th2 phenotype (58). Furthermore, despite the fact that IL-4-mediated STAT6 signaling is known to protect primary B cells susceptible to growth factor withdrawal or Fas ligation (65), IL-4 can also induce apoptosis of human endothelial cells (39), hepatocytes (4), and stimulated monocytes (41). Therefore, IL-4 plays a critical role in regulation of immune responses in different cell types, and the magnitude and duration of STAT6-induced responses will be stringently controlled.
To date, in KSHV-associated B-lymphoma diseases, the role of IL-4/STAT6 signaling in viral modulation of B-lymphoma immune response in terms of viral latent infection is still unclear. In this report, based on the data obtained from screening of immunology gene microarrays with RNA from B-lymphoid cells stably expressing LANA, we demonstrated that IL-4/STAT6 signaling is dysregulated in most PEL cells and that KSHV targets STAT6 not only for LANA-dependent repression of IL-4-mediated immune response like B-lymphocyte activation and cell proliferation, but also contributes to virus-infected cell survival and maintenance of latency.
MATERIALS AND METHODS
Reagents and antibodies.
Plasmids expressing full-length LANA with myc tag (LANA-myc) were described previously (13), and FLAG-STAT6 was a gift from Jaharul S. Haque (Lerner Research Institute). Cytokine IL-4 was from Invitrogen, and lipopolysaccharide (LPS) was from Sigma. Antibodies to STAT6 and phosphorylated STAT6 (p-STAT6) (Tyr-641) from Cell Signaling Technology were used as specified by the manufacturer. Antibodies to β-actin (#8226; Abcam), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (G8140-01; United States Biological), hemagglutinin (12CA5), and myc (9E10) were used.
Cell culture and transfection.
KSHV-negative (BJAB and DG75) and -positive (BC3, BCBL1, and JSC) B-lymphoma cells and KSHV-infected BJAB cells (BJAB-K; a gift from Michael Lagunoff, University of Washington) were maintained in RPMI 1640 medium (Invitrogen) with 7% fetal bovine serum (FBS), 4 μM l-glutamine, penicillin, and streptomycin. All cells were incubated at 37°C in a humidified environmental incubator supplemented with 5% CO2. B cells were transfected by electroporation with a Bio-Rad Gene Pulser in 0.4-cm-gap cuvettes at 220 V and 975 μF.
RNA isolation and cDNA synthesis.
Ten million cells were rinsed twice with ice-cold phosphate-buffered saline (PBS) before RNA extraction. Total RNA was extracted in 1 ml Trizol (Invitrogen, Frederick, MD) and then treated with DNase I at 37°C for 1 h after phenol-chloroform extraction. The quality of total RNA was evaluated by measuring the A260/A280 ratio and by gel-electrophoresis pattern, which revealed two major bands of 28S and 18S RNA. Approximately 5 μg of RNA was reverse transcribed for first-strand cDNA synthesis, using the SuperScript III reverse transcription system (Invitrogen, Frederick, MD).
Quantitative PCR.
Quantitative real-time PCR was performed on the DNA Engine Opticon 2 real-time PCR detection system (Bio-Rad), using multiple-stage program parameters provided by the manufacturer. The 20-μl reaction mixture contained 10 μl of Power SYBR green PCR master mix (New England Biolabs, Beverly, MA), 1 μl (20 μM) forward primer, 1 μl (20 μM) reverse primer, 1 μl cDNA, and 7 μl sterile water. The thermocycling program consisted of 95°C for 3 min, followed by 40 cycles of 94°C for 25 s and 50°C for 25 s. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts in each sample were first amplified as internal controls to normalize mRNA input for target gene amplification. The specificity and purity of the amplification reaction were determined by performing a melting-curve analysis. Primers for STAT6 (5′-TCTCAGCCCTAGGGGAATG-3′ and 5′-ACAGGAATTTGGGGCTTTG-3′) and for GAPDH (5′-ACGACCACTTTGTCAAGCTC-3′ and 5′-GGTCTACATGGCAACTGTGA-3′) (Integrated DNA Technologies, Inc., San Diego, CA) were used in the present study. Each sample was tested in triplicate, and data obtained from three independent experiments were expressed as a subtraction of the quantity of specific transcripts from the quantity of the control gene (GAPDH) in mean arbitrary units. The fold change in expression of each target mRNA relative to GAPDH was calculated based on the threshold cycle (CT) as 2−Δ(ΔCT), where ΔCT = CT target − CT GAPDH and Δ(ΔCT) = ΔCT LANA − ΔCT vector.
Immunoprecipitation, immunoblotting, and immunofluorescence.
Cells were harvested and washed once with ice-cold phosphate-buffered saline (PBS) and lysed in 1 ml cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM Na3VO3, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) on ice and homogenized. The supernatants of lysates were precleared with protein A/G Sepharose beads (Amersham Biosciences) and then incubated with primary antibody for 2 h at 4°C with constant rotation and then with protein A/G Sepharose beads for 1 h. Beads were washed 5 times with Tris-buffered saline (TBS) buffer, resuspended in 50 μl of 1× SDS Laemmli buffer, and heated at 95°C for 5 min. The sample was subjected to SDS-PAGE and transferred to a membrane that was probed with specific antibody. For immunofluorescence, PBS-washed B cells were incubated on polylysine-treated coverslips for 20 min and then fixed in 3% paraformaldehyde for 20 min at room temperature. After fixation, cells were washed three times in PBS and permeabilized in PBS containing 0.2% fish skin gelatin (G-7765; Sigma) and 0.2% Triton X-100 for 5 min, followed by staining with the the primary and secondary antibodies as previously described (11).
Luciferase reporter assay.
Luciferase reporter plasmids 3×stat6-Luc (pTransLucent, containing three tandem repeats of pSTAT6-binding sites) was a gift from Karen Leroy (Université Paris, France) (51). The luciferase reporter assays were performed as described previously (13). After transfection for 48 h, cells were lysed in 200 μl of reporter lysis buffer (Promega, Inc.). Luciferase activities and β-galactosidase were individually measured using luciferase assay reagent (Promega, Inc., Madison, WI) and the OpticompI Luminometer (MGM Instruments, Inc.) according to the suppliers’ instructions. Luciferase activities were normalized with β-galactosidase activities. Relative luciferase activity (in relative luciferase units [RLU]) was expressed as fold activation relative to the reporter construct alone. Assays were performed in triplicate.
Lentivirus-mediated protein expression and RNA interference.
For BJAB with LANA stable expression, the LANA-expressing plasmid LVX-YLF or vector alone, LVX-AcGFP-C1, was individually cotransfected with lentivrus package-expressing plasmids (Rev, vesicular stomatitis virus G [VSVG], and glycoprotein) into CoreT cells to generate virus. The packaged viruses were used to individually transduce BJAB cells, with selection by 1 μg/ml puromycin. For RNA interference, the STAT6 short hairpin RNA (shRNA) sequence (5′-GGGAGAAGATGTGTGAAACTCTGAA-3′) (9), LANA shRNA sequence (5′-GCTAGGCCACAACACATCT-3′) (13), and the control, nontargeting sequence (5′-TGCGTTGCTAGTACCAAC-3′) were individually inserted into pGIPz vector according to the manufacturer's instructions (Clonetech). Similarly, BC3 and BJAB cells were individually transducted by lentivirus containing STAT6 or control shRNA and then selected by puromycin. The RNA interfering efficiency was assessed by Western blot analysis with specific antibodies.
Infection of primary B cells with GFP-KSHV.
For virus induction, five hundred million exponentially growing HEK293-Bac36 (KSHV-green fluorescent protein [GFP]) cells were induced with 20 ng/ml of tetradecanoyl phorbol acetate (TPA) and 1.5 mM sodium butyrate (Sigma-Aldrich, St. Louis, MO) for 2 days at 37°C with 5% CO2. The supernatant was collected and filtered through a 0.45-μm filter, and viral particles were spun down at 25,000 rpm for 2 h at 4°C. The concentrated virus was collected and used for infection experiments. For infection, primary B cells (1 × 106) were incubated with virus suspension in 1 ml of RPMI 1640 medium (10% fetal bovine serum) in the presence of cyclosporine (Sigma, Marlborough, MA) for 4 h in 37°C. Cells were centrifuged for 5 min at 1,500 rpm, the supernatant was discarded, and pelleted cells were resuspended in fresh RPMI 1640 medium (10% FBS) in 48-well plates. The infection was checked by the visualization of GFP expression using fluourescence microscopy as described previously (12).
In vitro DNA binding affinity assay of phosphorylated STAT6.
A 5′-biotinylated oligonucleotide sequence (TGTAATTCGTGTGAATTATG; the pSTAT6-binding site is underlined), isolated by binding site selection as described previously (54), was coupled to streptavidin-conjugated agarose beads. Per sample, 3 μg of biotinylated oligonucleotide was incubated with 40 μl of 50% streptavidin-conjugated agarose bead slurry (Thermo Scientific) in a total volume of 100 μl of a lysis buffer comprised of 50 mM Tris-HCl, pH 8.0, 15 mM NaCl, 0.1 mM EDTA, 10% glycerol, 10 mM NaF, 1 mM Na3VO3, 1 mM PMSF, 1 mM dithiothreitol [DTT], 1 μg/ml aprotonin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin for 2 h at 4C. Whole-cell lysates (400 μg) were incubated with 40 μl of DNA-coupled agarose beads in lysis buffer at a total volume of 500 μl for 3 h at 4C. The precipitated complexes were washed three times with lysis buffer. Purified DNA-binding proteins were boiled in SDS sample buffer and analyzed by SDS-PAGE and Western blotting. Equal input amounts of phosphorylated STAT6 were adjusted based on the immunoblotting analysis before incubation with same amount of biotinylated DNA oligonucleotide.
Flow cytometry analysis.
Human peripheral blood mononuclear cells (PBMCs) (procured from the Immuology core, University of Pennsylvania Medical School, Philadelphia, PA) were individually infected with Bac36 (KSHV-GFP) virus generated from 293/Bac36 cells. The infected cells at 7 days were treated with IL-4 for 2 h before harvest, washed once with wash buffer (1× PBS with 2% FBS), and then incubated at room temperature for 15 min with the following fluorochrome-conjugated monoclonal antibodies against cell surface markers: CD19-peridinin chlorophyll protein (PerCP)-Cy5, CD40-allophycocyanin (APC), and Ki-67-phycoerythrin (PE). The stained cells were washed with wash buffer and resuspended in 2% paraformaldehyde. Data acquisition was performed using FACScalibur flow cytometry with Cell-Quest software (Becton-Dickinson) in accordance with the manufacturer's instructions. Instrument settings were adjusted so that fluorescence of cells from uninfected controls, in the case of GFP readings, or negative controls (i.e., with antibody omitted in antibody labeling) fell within the first decade of a 4-decade logarithmic scale on which emission is displayed. Flow cytometry plots showed at least 10,000 events. The data were analyzed by FlowJo software (Becton-Dickinson).
For cell cycle profile assays, treated cells were harvested and washed twice with PBS before being fixed with 1 ml of 70% ethanol, gently vortexed, and kept at 4°C until used. Fixed cells were washed once with PBS and resuspended in a propidium iodide solution (10 μg/ml) containing RNase A (250 μg/ml). Propidium iodide-stained cells were then analyzed for their DNA contents by using a FACSCalibur cytometer (Becton Dickinson, San Jose, CA). The apoptotic cells were presented by the percentage of the sub-G1 population.
Cell proliferation assay.
Equal amounts of cells were seeded and treated with or without 5 ng/ml IL-4 in the presence of LPS. Cell numbers were counted with a hemocytometer (Beckman Coulter) at each time point. Cell counts were performed in triplicate.
RESULTS
IL-4-stimulated phosphorylation of STAT6 is significantly reduced in KSHV-infected PEL cells.
To determine whether KSHV latent infection contributes to regulation of cytokine-induced B-lymphocyte immune response, we performed immunology microarray screening by using BJAB (a Burkitt's lymphoma-derived Epstein-Barr virus [EBV]- and KSHV-negative B-cell line) stably expressing LANA. Interestingly, we found that multiple genes encoding transcription factors involved in JAK-STAT signaling, particularly cytokine IL-4/STAT6 signaling, were significantly repressed by LANA (see Fig. S1 at http://microb205.med.upenn.edu/downloads/index.htm). To verify whether KSHV latent infection does dysregulate IL-4-induced STAT6 signaling, we first determined the mRNA levels of STAT6—the key IL-4 signal transduction factor, in KSHV-positive PEL cells (BC3, BCBL1, and JSC) and the negative B-lymphoma cells (DG75 and BJAB) by real-time PCR quantitation. Intriguingly, the results showed that there was no significant difference of STAT6 mRNA levels in the KSHV-infected PEL cells when compared with KSHV-uninfected B-lymphoma cells (Fig. 1 A).
FIG. 1.
LANA contributes to the suppression of IL-4-induced tyrosine phosphorylation of STAT6 in PEL cells. (A) Levels of STAT6 mRNA transcripts in KSHV-positive PEL cells are not distinct from those in KSHV-negative B-lymphoma cells. KSHV-positive PEL cells (BC3, BCBL1, and JSC) and KSHV-negative B-lymphoma cells (BJAB and DG75) were individually subjected to quantitative PCR analysis for detection of STAT6 mRNA transcription. (B) IL-4 stimulates less phosphorylation of STAT6 in KSHV-infected B-lymphoma cells. Five million KSHV-negative cells (BJAB and DG75), KSHV-positive B-lymphoma cells (BC3, BCBL1, and JSC), or KSHV-infected BJAB cells (BJAB-K) were treated with or without IL-4 (5 ng/ml) for 2 h, respectively. The cell lysates were subjected to Western blotting with antibodies as indicated. The quantitation of tyrosine-phosphorylated STAT6 (p-STAT6) is presented at the bottom. The blank box indicates constitutive phosphorylation of STAT6 (p-STAT6c). (C) KSHV-negative and -positive B-lymphoma cells were treated with 5 ng/ml IL-4 for various times. STAT6 phosphorylation was analyzed as described in the legend to panel B. Data are quantified in a graphical format. (D) Depletion of LANA enhances the response of PEL cells to IL-4 stimulation. Assays with lentivirus-mediated LANA knockdown (shLANA) or the scramble control (shCtrl) of JSC and BC3 cells were performed similar to the assays described for panel B.
To further answer whether KSHV-mediated suppression of IL-4 signaling is due to lower response of B cells to IL-4 stimulation at the posttranslational level, we equally treated KSHV-positive PEL cells (BC3, BCBL1, and JSC) and negative B-lymphoma cells (BJAB and DG75) with IL-4 in culture media and determined the phosphorylation level of STAT6. The results showed that there was consistently lower level of phosphorylated STAT6 (p-STAT6) which was induced by IL-4 stimulation in the KSHV-positive PEL cells than the KSHV-negative cells (Fig. 1B, upper panels). Interestingly, a band showing a moderate level of constitutive phosphorylated STAT6 (p-STAT6C), which migrates at slightly higher position than the IL-4-induced phosphorylated STAT6 (p-STAT6), was seen in both BC3 and BCBL1 cells (Fig. 1B, upper panel). To exclude the possibility of the effect of other factors besides KSHV infection on inhibition of IL-4-induced phosphorylation of STAT6 in B cells, the BJAB mock-infected cells and BJAB cells infected with KSHV (BJAB-K), with or without IL-4 treatment, were analyzed. The results further support the notion that B cells infected with KSHV showed dramatically reduced levels of p-STAT6 in response to IL-4 stimulation (Fig. 1B, right panels). Furthermore, consistently lower levels of phosphorylated STAT6 were observed in the KSHV-infected cells (BC3, BCBL1, and BJAB-K) compared to KSHV-negative B-lymphoma cells (BJAB and DG75) in response to IL-4 stimulation at different time points (Fig. 1C), further confirming a potential role for KSHV in suppression of IL-4-induced STAT6 phosphorylation.
LANA is important for KSHV to repress IL-4-stimulated STAT6 phosphorylation of B-cell activation and proliferation.
To determine whether the inhibitory effects of KSHV on IL-4-induced STAT6 phosphorylation are due to the expression of the key latent antigen LANA, we performed lentivirus-mediated knockdown of LANA in the PEL cells (BC3 and JSC), followed by stimulation with or without IL-4. The results, as shown by immunoblotting analysis, indicated that although there was no effect on the expression level of the native STAT6 protein, the inhibition of LANA expression dramatically enhanced the level of phosphorylated STAT6 in PEL cells with IL-4 stimulation (Fig. 1D). This indicated that LANA is important for KSHV-mediated repression of IL-4-stimulated STAT6 phosphorylation and that the STAT6 phosphorylation process might be a direct target for LANA in blocking IL-4-induced signaling.
IL-4 has been shown to elicit an immune response which includes B-lymphocyte activation and proliferation (20). To gain insights into the phenomenon of IL-4-mediated immune suppression regulated by KSHV infection, we analyzed the expression levels of B-cell surface activation markers (CD40 and Ki-67) as a result of KSHV infection by flow cytometry analysis. The results showed that CD40 expression was significantly inhibited due to KSHV infection and more dramatically so in response to IL-4 stimulation when compared with mock-infected cells without KSHV infection (Fig. 2 A). However, in the KSHV-infected cells, Ki-67 was repressed by IL-4 stimulation in KSHV-positive cells, even though it was clearly increased by KSHV infection (Fig. 2A). Importantly, in primary B cells, KSHV infection alone exerted a similar inhibitory effect on the activation of CD40 by IL-4 induction, although CD40 expression was dramatically blocked in KSHV-infected cells (Fig. 2B). Moreover, LANA knockdown greatly increased the activation of CD40 in the PEL cells (BC3 and JSC) after IL-4 stimulation (Fig. 2C).
FIG. 2.
LANA is critical for KSHV to reduce IL-4-induced expression of the B-cell activation marker. (A) BJAB cells infected with GFP-KSHV (BJAB-K) and uninfected BJAB cells, (B) primary B cells (CD19+) gated from human PBMCs with or without KSHV-GFP (Bac36) at 7 days postinfection, and (C) lentivirus-mediated LANA knockdown (shLANA) or JSC and BC3 scramble control (shCtrl) cells were individually cultured in the presence or absence of IL-4 for 2 h. The cells were fixed with and stained with anti-CD40-APC or anti-Ki-67-PE. The average ratios of each of the stained positive cells are shown on the histograms. The efficiency of KSHV infection and LANA knockdown is presented in the right panels as the percentage of GFP fluorescence detected by flow cytometry.
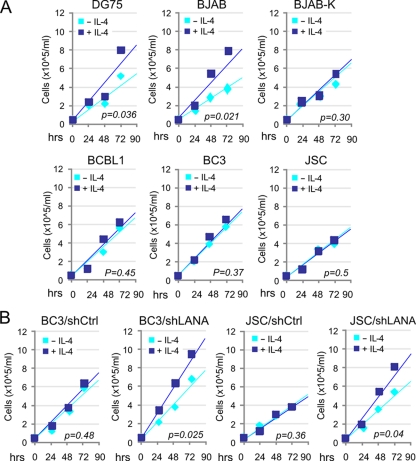
To further determine whether these changes in cell surface markers can affect the biological response of B cells to IL-4 stimulation, we evaluated the effect of IL-4 stimulation on the proliferation of KSHV-positive PEL cells (BC3, BCBL1, and JSC) and KSHV-negative B-lymphoma cells (BJAB and DG75), as well as KSHV-infected BJAB (BJAB-K) cells by monitoring different time points. In a representative experiment shown in Fig. 3 A, the growth rate of KSHV-positive cells in response to IL-4 stimulation was significantly lower than that of KSHV-negative cells. As expected, infection with KSHV (BJAB-K) efficiently suppressed B-cell proliferation in response to IL-4 stimulation (Fig. 3A, upper panel). Moreover, the inhibition of LANA expression in both BC3 and JSC cells significantly restored the growth rate induced by IL-4 (Fig. 3B), supporting an important role of LANA in suppression of IL-4-induced signaling.
FIG. 3.
KSHV infection abolishes IL-4-stimulated B-cell proliferation in vitro. (A) KSHV-negative (BJAB and DG75) and -positive (BC3, BCBL1, JSC, and BJAB-K) cell lines; (B) BC3 and JSC cells with LANA constitutively knocked down. Cells were seeded in equal amounts and treated with or without 5 ng/ml IL-4 in the presence of LPS. Proliferation was measured at 24, 48, and 72 h after treatment by triplicate cell counting. These experiments were repeated twice, and average results are shown. The P values for comparisons between the untreated and treated groups were calculated by t test.
STAT6 phosphorylation and DNA binding activity are targeted by LANA to block IL-4-signaling.
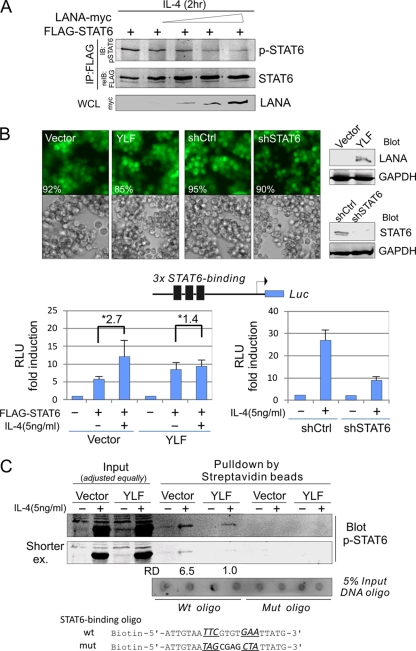
To address how LANA can directly target STAT6 posttranslational regulation to contribute to viral suppression of IL-4-induced signaling in B cells, we determined the phosphorylation level of exogenous FLAG-tagged STAT6 induced by IL-4 in the presence of increased dose of LANA. The results of immunoprecipitation with FLAG-STAT6 followed by immunoblotting against p-STAT6 showed that LANA did directly suppress IL-4-induced STAT6 phosphorylation in a dose-dependent manner (Fig. 4 A). To further demonstrate that LANA-mediated suppression of exogenous STAT6 phosphorylation does lead to less transcriptional activity of STAT6 when induced by IL-4, we generated two BJAB stable cell lines expressing LANA tagged with yellow fluorescent protein (YFP) (YLF) or the GFP vector control (vector) by using a lentivirus expression system followed by a STAT6-responsive luciferase reporter assay. As shown in the upper panels of Fig. 4B, both cell lines contained more than 85% fluorescent cells, suggestive of a high efficiency of transduction, and the results of immunoblotting to detect LANA showed strong expression of LANA in the YLF cell line. Similarly, to ascertain that the reporter was specifically recognized and bound by active STAT6, we established BJAB stable cells expressing GFP with STAT6 knockdown (shSTAT6) or control shRNA (shCtrl) used as a control in parallel (Fig. 4B, right panels). We individually transfected 3×stat6-luc plasmid DNA in the presence or absence of FLAG-STAT6 into BJAB/YLF and BJAB/vector cells, as well as BJAB/shSTAT6 and BJAB/shCtrl cells followed by IL-4 induction. The results further showed that LANA can dramatically shut down the transcriptional activity of IL-4-induced active STAT6 (Fig. 4B, lower panels).
FIG. 4.
LANA suppresses IL-4-stimulated activation of STAT6 by affecting its DNA-binding ability. (A) LANA reduces the IL-4-induced phosphorylation of exogenous STAT6. DG75 cells were transiently transfected with FLAG-STAT6 (10 μg) or with LANA-myc (0, 5, 10, 15, or 20 μg). The equal amounts of DNA were made up with empty vector. Forty-eight hours posttransfection, cells were treated with 5 ng/ml IL-4 for 2 h before harvest. Whole-cell lysates (WCL) were subjected to immunoprecipitation (IP) with anti-FLAG followed by immunoblotting (IB) against p-STAT6. The membrane was stripped and reimmunoblotted (reIB) for anti-FLAG, or immunoblotting was directly performed with anti-myc as indicated on the figure. (B) Luciferase (Luc) reporter assays of IL-4-induced transcriptional activity of phosphorylated STAT6. BJAB cells with LANA (YLF) or GFP (Vector) stable expression, or with STAT6 knockdown (shSTAT6) or scramble control (shCtrl) generated by lentivirus-mediated transduction followed by 2 μg/ml puromycin selection (Western analysis was performed to detect LANA expression and STAT6 knockdown efficiency), were individually transfected with FLAG-STAT6 (0, 10 μg) in the presence of the reporter 3×stat6-luc (10 μg). At 48 h posttransfection, cells were treated or not treated with IL-4 for 2 h before harvest. Cell lysates were performed reporter assays. The results are presented as the fold change compared with vector alone. RLU, relative luciferase units. Asterisks indicate the fold change after IL-4 induction. (C) In vitro DNA-binding assays of phosphorylated STAT6 in the presence of LANA. Whole-cell extracts with preadjusted equally IL-4-induced phosphorylated STAT6 from BJAB/vector or BJAB/YLF were individually incubated with wild-type or mutant STAT6-binding DNA oligonucleotide followed by three washings, and the precipitates were Western blotted with p-STAT6 antibody. Five percent input of DNA oligonucleotide is shown at the bottom. RD, relative density.
To elucidate whether LANA also impaired the DNA binding activity of phosphorylated STAT6, which may act as a second potential mechanism to block IL-4-induced signaling, we performed an in vitro DNA binding assay by individually incubating the wild-type or mutated STAT6-binding DNA oligonucleotide, with biotinylated labeling and loading equal amounts of phosphorylated STAT6 cell extracts from BJAB/YLF or control BJAB/vector after IL-4 treatment. The results showed that the DNA binding activity of phosphorylated STAT6 in the LANA-expressing cells (YLF) was significantly (about 6.5-fold) lower than that seen in the vector control. Importantly, little or no signal was seen with the mutant oligonucleotide (Fig. 4C). This supports our hypothesis that LANA is a negative regulator of STAT6 by suppressing phosphorylation of STAT6 and so affects its binding activity to its cognate DNA sequence.
Inhibition of endogenous STAT6 expression leads to increased sensitivity of PEL cells to extracellular stress and reactivation of viral lytic replication.
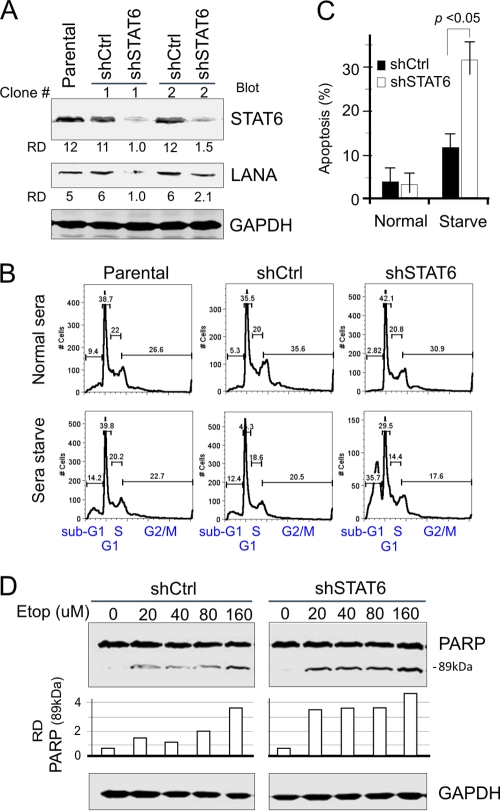
IL-4-induced phosphorylation of STAT6 is repressed in KSHV-infected cells but not the expression of native STAT6. To clarify whether KSHV selectively inhibits IL-4-induced activation of STAT6 and to take advantage of native STAT6 function for viral infection, we inhibited STAT6 expression in the KSHV-positive BC3 cells by introducing small interfering RNA and then performed apoptosis analysis by treating cells with the chemical (etoposide)-induced apoptosis or by serum starvation. As shown in Fig. 5 A to C, in the normal culture medium, lower expression of STAT6 did not affect the cell cycle profile of BC3 cells. However, in low serum, the inhibition of STAT6 dramatically arrested BC3 cells on sub-G1 and G1 populations of the cell cycle profile and induced apoptosis in 30% of the cells (Fig. 5B and C). To further confirm whether native STAT6 contributed to the survival of KSHV-infected cells under extracellular stress, we monitored the active form (89 kDa) of the apoptosis maker poly(ADP-ribose) polymerase 1 (PARP1) in the BC3/shSTAT6 and BC3/shCtrl cells with different concentrations of etoposide (a drug for inducing cell apoptosis). The results of immunoblotting against PARP1 further showed that STAT6 knockdown does increase the sensitivity of KSHV-positive BC3 cells to chemical-induced cellular apoptosis, where increased levels of PARP1 cleavage were seen in a dose-dependent manner (Fig. 5D).
FIG. 5.
STAT6 knockdown increases the sensitivity of KSHV-infected cells to extracellular stress. (A) PEL cell lines with STAT6 knockdown. Two BC3 cell lines with STAT6 knockdown (shSTAT6) or the scramble control (shCtrl) were generated from parental BC3 cells by lentivirus-mediated transduction followed by selection with 2 μg/ml puromycin, and Western analysis was performed to detect STAT6 knockdown efficiency. (B and C) STAT6 knockdown increases the response of PEL cells to low-serum stress. BC3 cells (parent, shSTAT6, or shCtrl) cultured in normal (7%) or low-serum (0.1%) medium overnight were subjected to cell cycle analysis. Apoptosis was indicated by average percentage of the sub-G1 population from three independent experiment repeats. (D) STAT6 knockdown increases the sensitivity of PEL cells to etoposide (Etop)-induced apoptosis. BC3/shSTAT6 and BC3/shCtrl cells were treated with various concentration of eptopside for 3 h. Cell lysates were subjected to immunoblotting against PARP1 and GAPDH. The relative density (RD) of the 89-kDa active form of PARP1 was quantified and is shown at the bottom.
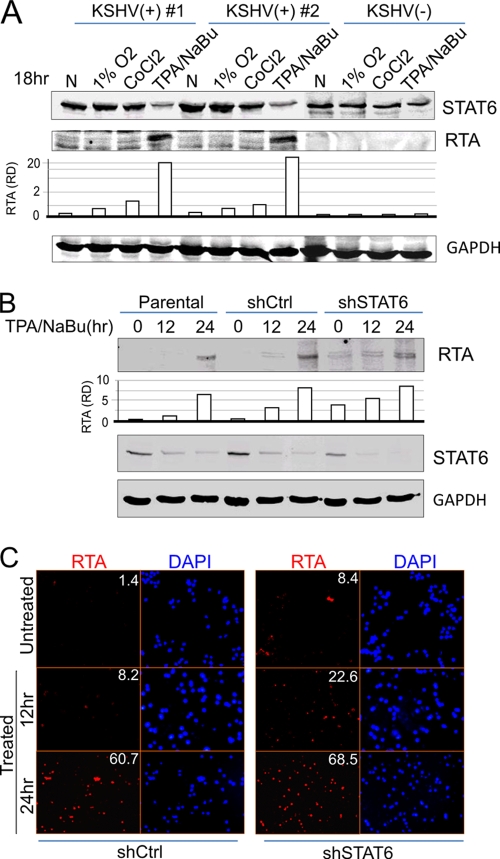
Consistently lower levels of LANA expression were observed when STAT6 is knocked down in PEL cells (Fig. 5A, lower panel); we therefore hypothesized that inhibition of STAT6 expression may contribute to reactivation of KSHV and that this may result in feedback to decrease LANA expression. We first checked whether the viral lytic reactivation agents (hypoxia, TPA, and sodium butyrate) cause less STAT6 expression with the expression of the viral gene coding for replication and transcription activator (RTA)—the key KSHV reactivation marker. As shown in Fig. 6 A, high expression of RTA induced by TPA and sodium butyrate was consistently associated with low level of native STAT6 in both KSHV-infected PEL cell lines (BC3 and BCBL1), when compared with untreated normal control. In contrast, less expression of RTA induced by hypoxia (1% O2 or CoCl2) treatment in both PEL cells maintained a higher level of STAT6. This strongly indicated that the levels of STAT6 were indirectly proportional to the level of RTA expressed. To further demonstrate that inhibition of STAT6 expression could directly trigger RTA expression, we performed immunofluorescence and immunoblotting analyses on BC3 cells with or without STAT6 knockdown after TPA/sodium butyrate treatment at various time points. The results showed that although we observed an increased level of RTA expression in the STAT6-knockdown cells without TPA/sodium butyrate treatment, we also observed an increased amount of RTA at a shorter time (12 h) with TPA/sodium butyrate treatment in the STAT6-knockdown (shSTAT6) cells, when compared to the parental or scramble control knockdown (shCtrl) cells (Fig. 6B and C). This supports our hypothesis that STAT6 contributes to the maintenance of viral latency and that inhibition of STAT6 will lead to reactivation of KSHV from latency.
FIG. 6.
Inhibition of STAT6 expression contributes to reactivation of KSHV lytic replicaton. (A) Stress-induced RTA expression in PEL cells is correlated with inhibition of STAT6 expression. PEL cells (BC3 and BCBL1) and KSHV-negative BJAB cells were individually subjected to treatment with hypoxia (1% O2 or 100 μM CoCl2) or TPA and sodium butyrate (NaBu) (20 ng/ml and 1.5 mM, respectively) for 18 h. Cell lysates were subjected to immunoblotting against STAT6, RTA, and GAPDH. The relative density (RD) of RTA was quantified and is shown at the bottom. (B and C) STAT6 knockdown enhances the expression of RTA. PEL BC3 cells (parent, shSTAT6, or shCtrl) were subjected to treatment with TPA and NaB (20 ng/ml and 1.5 mM, respectively) for various times (0, 12, or 24 h). Cell lysates were subjected to the immunoblotting assay as in panel A, or the treated cells were fixed and subjected to an immunofluorescence assay with anti-RTA and DAPI (4′,6-diamidino-2-phenylindole) staining. The numbers shown in the figure indicate the percentages of RTA-positive cells.
DISCUSSION
KSHV has been documented to infect human B cells, macrophages, endothelial cells, and epithelial cells (2, 7). The infected cells are not completely eradicated from host cells by the immune system, suggesting that KSHV has developed a series of complex strategies for evading the host's immune surveillance. In general, infection stimulates an immune response specific for the expressed antigens encoded by the virus. LANA as a major antigen is highly expressed in all KSHV latently infected cells (33, 50). To explore the potential role of LANA in regulation of global immunosuppression, we performed an immunology gene microarray analysis by using B cells stably expressing LANA. The microarray results showed that LANA dramatically and specifically downregulated cellular molecules which contribute to lymphocytes’ differentiation and activation and, more importantly, suppressed expression of cellular markers for antigen presenting and processing. This may explain why LANA as one of the dominant latent antigens encoded by KSHV may have the potential to destroy T-lymphocyte function and thereby impair induction of a significant immune response.
Cytokines regulate numerous biologic processes through their interaction with membrane-bound receptors. Despite the number and diversity of cytokines, the signaling pathways appear to be highly conserved. One of the most important pathways consists of the Janus family of tyrosine kinases (JAK) and the signal transducers and activators of transcription proteins (STAT) (56). Cytokines utilizing this pathway form a ligand-receptor complex, resulting in tyrosine phosphorylation of the receptor-associated kinase JAK, the cytoplasmic portion of the cytokine receptor, and STAT (8). The phosphorylated STATs form an activated dimer or tetramer, which translocates to the cell nucleus, where it binds to its cognate DNA sequence and/or other transcription factors to influence target gene transcription (31, 59).
KSHV latent infection can cause persistent activation of STAT3 via gp130 receptor signaling in endothelial cells (42, 48), and inhibition of STAT3 signaling was able to efficiently induce apoptosis of KSHV-positive PEL cells (3). In lytic replication, it was found that KSHV also targets STAT2 and STAT1 to block transduction of type I interferon signaling (6). Further studies have indicated that vGPCR plays a role on regulation of JAK2/STAT3 signaling to produce angiogenic factors (10), and IL-6 induced by the Tat protein of HIV-1 is dependent on activated STAT3 signaling (16, 67). In this study, we showed that LANA is essential for KSHV to repress IL-4-stimulated signal transduction during latent infection. LANA impairs tyrosine phosphorylation of STAT6 and its ability to bind to its cognate DNA sequence. This in turn blocks IL-4-induced B-lymphocyte activation and proliferation (Fig. 7). Interestingly, we also found that inhibition of STAT6 increases the sensitivity of KSHV-infected cells to apoptosis induced by extracellular low-serum stress and chemical (etoposide) treatment, as well as reactivation of KSHV latency replication (Fig. 7). Thus, to achieve the best outcome for KSHV-mediated pathogenesis, KSHV-encoded LANA selectively blocks IL-4-induced STAT6 phosphorylation and activation. However, the native STAT6 expression is maintained to support KSHV-infected cell survival and maintenance of long-term latent infection in host cells. To our knowledge, this is the first description of a virus targeting the regulation of the IL-4/STAT6 signaling pathway to regulate viral infection, although it remains unclear whether LANA posses phosphatase activity for blocking the IL-4-induced STAT6 phosphorylation.
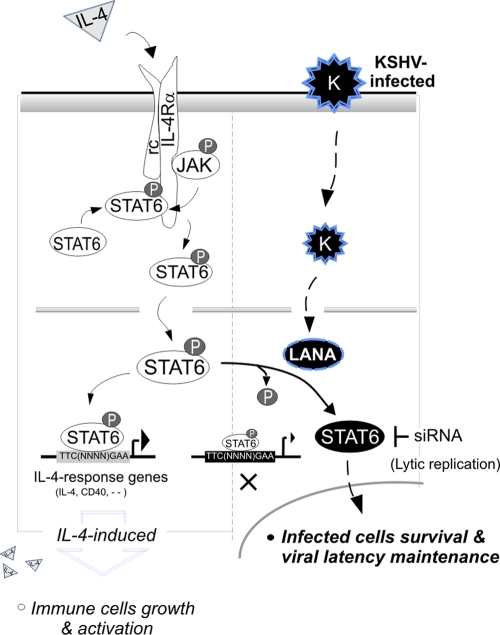
FIG. 7.
A proposed model of KSHV-mediated suppression of IL-4/STAT6 signaling in B cells. Generally, IL-4 binds to its receptor, IL-4Rα, and the activation of IL-4Rα recruits the kinase JAK1 or -2, which directly phosphorylates and activates STAT6 to translocation into the nucleus and enhances IL-4-responsive gene expression (like IL-4 and CD40) in terms of immune cell growth and activation. In KSHV-infected cells, the latent antigen LANA encoded by KSHV inhibits IL-4-induced STAT6 phosphorylation and DNA binding affinity, which blocks the immune response of the host cell to IL-4 stimulation (autocrine and paracrine). The dephosphorylated STAT6 contributes to KSHV-infected cell survival under extracellular stress and maintenance of viral latency.
Activation of STAT proteins has been observed in several tumors and frequently in hematologic pathologies (8). The constitutive activation of these transcription factors is induced by various factors, including overexpression/deregulation of kinases or inhibition of negative regulators. Cellular transformation has been linked to transcription regulation of specific genes controlling cell viability, proliferation, and angiogenesis (8). With respect to STAT6, the constitutively activation of STAT6 has been previously reported in Hodgkin's lymphoma (HL) (55), and primary mediastinal large B-cell lymphoma (PMBL) (30). In this study, we also observed a basal level of constitutive phosphorylation of STAT6 in two of three KSHV-positive cell lines (BC3 and BCBL1, but not JSC). To address whether it was the virus infection which triggered constitutive activation of STAT6 in B cells, further studies are warranted to determine the mechanism by which KSHV-encoded LANA can regulate STAT6 phosphorylation and its activity.
Additionally, the STAT6-dependent promotion of apoptosis resistance has been shown by some recent studies (27, 68). In our study, we also demonstrated that inhibition of STAT6 is able to efficiently increase apoptosis of KSHV-infected PEL cells, as well as reactivation from viral latency. Thus, KSHV infection precisely targets and regulates STAT6 phosphorylation and at the same time maintains the expression level of endogenous STAT6 protein.
In summary, IL-4/STAT6 signaling has been linked to the regulation of B-cell viability, proliferation, and activation (8). Our observations now strongly show that in KSHV-infected cells, IL-4 is incapable of inducing expression of the B-cell surface activation marker or promoting B-cell proliferation in vitro, while endogenous STAT6 expression contributes to KSHV-infected cell survival and maintenance of viral latency.
Acknowledgments
We are grateful to Jaharul S. Haque, Michael Lagunoff, and Karen Leroy for providing reagents. Special thanks goes to members of the Robertson lab for suggestions and support.
This work was supported by Public Health Service grants NCI CA072510 and CA091792, NIDCR DE014136, NIAID AI067037, and DE17338 (to E.S.R.). E.S.R. is a scholar of the Leukemia and Lymphoma Society of America. S.C.V. is supported by the NIH Pathways to Independence Award, CA126182.
Footnotes
Published ahead of print on 18 August 2010.
REFERENCES
- 1.An, J., Y. Sun, and M. B. Rettig. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222-228. [DOI] [PubMed] [Google Scholar]
- 2.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027-1038. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, Y., G. M. Feldman, and G. Tosato. 2003. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 101:1535-1542. [DOI] [PubMed] [Google Scholar]
- 4.Aoudjehane, L., P. Podevin, O. Scatton, P. Jaffray, I. Dusanter-Fourt, G. Feldmann, P. P. Massault, L. Grira, A. Bringuier, B. Dousset, S. Chouzenoux, O. Soubrane, Y. Calmus, and F. Conti. 2007. Interleukin-4 induces human hepatocyte apoptosis through a Fas-independent pathway. FASEB J. 21:1433-1444. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Bisson, S. A., A. L. Page, and D. Ganem. 2009. A Kaposi's sarcoma-associated herpesvirus protein that forms inhibitory complexes with type I interferon receptor subunits, Jak and STAT proteins, and blocks interferon-mediated signal transduction. J. Virol. 83:5056-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasig, C., C. Zietz, B. Haar, F. Neipel, S. Esser, N. H. Brockmeyer, E. Tschachler, S. Colombini, B. Ensoli, and M. Sturzl. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J. Virol. 71:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg, J. 2002. Stat proteins and oncogenesis. J. Clin. Invest. 109:1139-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buglio, D., G. V. Georgakis, S. Hanabuchi, K. Arima, N. M. Khaskhely, Y. J. Liu, and A. Younes. 2008. Vorinostat inhibits STAT6-mediated TH2 cytokine and TARC production and induces cell death in Hodgkin lymphoma cell lines. Blood 112:1424-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger, M., T. Hartmann, J. A. Burger, and I. Schraufstatter. 2005. KSHV-GPCR and CXCR2 transforming capacity and angiogenic responses are mediated through a JAK2-STAT3-dependent pathway. Oncogene 24:2067-2075. [DOI] [PubMed] [Google Scholar]
- 11.Cai, Q., K. Lan, S. C. Verma, H. Si, D. Lin, and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1α to upregulate RTA expression during hypoxia: latency control under low oxygen conditions. J. Virol. 80:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai, Q., M. Murakami, H. Si, and E. S. Robertson. 2007. A potential α-helix motif in the amino terminus of LANA encoded by Kaposi's sarcoma-associated herpesvirus is critical for nuclear accumulation of HIF-1α in normoxia. J. Virol. 81:10413-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai, Q. L., J. S. Knight, S. C. Verma, P. Zald, and E. S. Robertson. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 15.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 16.Chen, X., L. Cheng, X. Jia, Y. Zeng, S. Yao, Z. Lv, D. Qin, X. Fang, Y. Lei, and C. Lu. 2009. Human immunodeficiency virus type 1 Tat accelerates Kaposi sarcoma-associated herpesvirus Kaposin A-mediated tumorigenesis of transformed fibroblasts in vitro as well as in nude and immunocompetent mice. Neoplasia 11:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinchy, B., C. Elenstrom, E. Severinson, and G. Moller. 1991. T and B cell collaboration: induction of motility in small, resting B cells by interleukin 4. Eur. J. Immunol. 21:1445-1451. [DOI] [PubMed] [Google Scholar]
- 18.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 19.Davey, E. J., G. Greicius, J. Thyberg, and E. Severinson. 2000. STAT6 is required for the regulation of IL-4-induced cytoskeletal events in B cells. Int. Immunol. 12:995-1003. [DOI] [PubMed] [Google Scholar]
- 20.Davey, E. J., J. Thyberg, D. H. Conrad, and E. Severinson. 1998. Regulation of cell morphology in B lymphocytes by IL-4: evidence for induced cytoskeletal changes. J. Immunol. 160:5366-5373. [PubMed] [Google Scholar]
- 21.Delphin, S., and J. Stavnezer. 1995. Characterization of an interleukin 4 (IL-4) responsive region in the immunoglobulin heavy chain germline epsilon promoter: regulation by NF-IL-4, a C/EBP family member and NF-kappa B/p50. J. Exp. Med. 181:181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Bartolo, D. L., M. Cannon, Y. F. Liu, R. Renne, A. Chadburn, C. Boshoff, and E. Cesarman. 2008. KSHV LANA inhibits TGF-beta signaling through epigenetic silencing of the TGF-beta type II receptor. Blood 111:4731-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U. S. A. 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fakhari, F. D., J. H. Jeong, Y. Kanan, and D. P. Dittmer. 2006. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Invest. 116:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 26.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 27.Galka, E., J. L. Thompson, W. J. Zhang, L. S. Poritz, and W. A. Koltun. 2004. Stat6(null phenotype) human lymphocytes exhibit increased apoptosis. J. Surg. Res. 122:14-20. [DOI] [PubMed] [Google Scholar]
- 28.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard, D., R. Paquin, and A. D. Beaulieu. 1997. Responsiveness of human neutrophils to interleukin-4: induction of cytoskeletal rearrangements, de novo protein synthesis and delay of apoptosis. Biochem. J. 325:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guiter, C., I. Dusanter-Fourt, C. Copie-Bergman, M. L. Boulland, S. Le Gouvello, P. Gaulard, K. Leroy, and F. Castellano. 2004. Constitutive STAT6 activation in primary mediastinal large B-cell lymphoma. Blood 104:543-549. [DOI] [PubMed] [Google Scholar]
- 31.Hebenstreit, D., G. Wirnsberger, J. Horejs-Hoeck, and A. Duschl. 2006. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 17:173-188. [DOI] [PubMed] [Google Scholar]
- 32.Katano, H., Y. Sato, and T. Sata. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92:3076-3084. [DOI] [PubMed] [Google Scholar]
- 33.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keegan, A. D., and J. Zamorano. 1998. Regulation of gene expression, growth, and cell survival by IL-4: contribution of multiple signaling pathways. Cell Res. 8:1-13. [DOI] [PubMed] [Google Scholar]
- 35.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 36.Klein, N. J., K. P. Rigley, and R. E. Callard. 1993. IL-4 regulates the morphology, cytoskeleton, and proliferation of human umbilical vein endothelial cells: relationship between vimentin and CD23. Int. Immunol. 5:293-301. [DOI] [PubMed] [Google Scholar]
- 37.Kretsovali, A., and J. Papamatheakis. 1995. A novel IL-4 responsive element of the E alpha MHC class II promoter that binds to an inducible factor. Nucleic Acids Res. 23:2919-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, F., T. Yokota, T. Otsuka, P. Meyerson, D. Villaret, R. Coffman, T. Mosmann, D. Rennick, N. Roehm, C. Smith, et al. 1986. Isolation and characterization of a mouse interleukin cDNA clone that expresses B-cell stimulatory factor 1 activities and T-cell- and mast-cell-stimulating activities. Proc. Natl. Acad. Sci. U. S. A. 83:2061-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, Y. W., H. Kuhn, B. Hennig, and M. Toborek. 2000. IL-4 induces apoptosis of endothelial cells through the caspase-3-dependent pathway. FEBS Lett. 485:122-126. [DOI] [PubMed] [Google Scholar]
- 40.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 41.Mangan, D. F., B. Robertson, and S. M. Wahl. 1992. IL-4 enhances programmed cell death (apoptosis) in stimulated human monocytes. J. Immunol. 148:1812-1816. [PubMed] [Google Scholar]
- 42.Morris, V. A., A. S. Punjabi, and M. Lagunoff. 2008. Activation of Akt through gp130 receptor signaling is required for Kaposi's sarcoma-associated herpesvirus-induced lymphatic reprogramming of endothelial cells. J. Virol. 82:8771-8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muromoto, R., K. Okabe, M. Fujimuro, K. Sugiyama, H. Yokosawa, T. Seya, and T. Matsuda. 2006. Physical and functional interactions between STAT3 and Kaposi's sarcoma-associated herpesvirus-encoded LANA. FEBS Lett. 580:93-98. [DOI] [PubMed] [Google Scholar]
- 44.Nelms, K., A. D. Keegan, J. Zamorano, J. J. Ryan, and W. E. Paul. 1999. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 17:701-738. [DOI] [PubMed] [Google Scholar]
- 45.Noguchi, M., Y. Nakamura, S. M. Russell, S. F. Ziegler, M. Tsang, X. Cao, and W. J. Leonard. 1993. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science 262:1877-1880. [DOI] [PubMed] [Google Scholar]
- 46.Paul, W. E. 1991. Interleukin-4: a prototypic immunoregulatory lymphokine. Blood 77:1859-1870. [PubMed] [Google Scholar]
- 47.Paul, W. E., and J. Ohara. 1987. B-cell stimulatory factor-1/interleukin 4. Annu. Rev. Immunol. 5:429-459. [DOI] [PubMed] [Google Scholar]
- 48.Punjabi, A. S., P. A. Carroll, L. Chen, and M. Lagunoff. 2007. Persistent activation of STAT3 by latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells. J. Virol. 81:2449-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 50.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritz, O., C. Guiter, K. Dorsch, I. Dusanter-Fourt, S. Wegener, H. Jouault, P. Gaulard, F. Castellano, P. Moller, and K. Leroy. 2008. STAT6 activity is regulated by SOCS-1 and modulates BCL-XL expression in primary mediastinal B-cell lymphoma. Leukemia 22:2106-2110. [DOI] [PubMed] [Google Scholar]
- 52.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santos-Argumedo, L., P. W. Kincade, S. Partida-Sanchez, and R. M. Parkhouse. 1997. CD44-stimulated dendrite formation (′spreading') in activated B cells. Immunology 90:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindler, U., P. Wu, M. Rothe, M. Brasseur, and S. L. McKnight. 1995. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity 2:689-697. [DOI] [PubMed] [Google Scholar]
- 55.Skinnider, B. F., A. J. Elia, R. D. Gascoyne, B. Patterson, L. Trumper, U. Kapp, and T. W. Mak. 2002. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 99:618-626. [DOI] [PubMed] [Google Scholar]
- 56.Smerz-Bertling, C., and A. Duschl. 1995. Both interleukin 4 and interleukin 13 induce tyrosine phosphorylation of the 140-kDa subunit of the interleukin 4 receptor. J. Biol. Chem. 270:966-970. [DOI] [PubMed] [Google Scholar]
- 57.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 58.Swain, S. L., A. D. Weinberg, M. English, and G. Huston. 1990. IL-4 directs the development of Th2-like helper effectors. J. Immunol. 145:3796-3806. [PubMed] [Google Scholar]
- 59.Takeda, K., T. Tanaka, W. Shi, M. Matsumoto, M. Minami, S. Kashiwamura, K. Nakanishi, N. Yoshida, T. Kishimoto, and S. Akira. 1996. Essential role of Stat6 in IL-4 signalling. Nature 380:627-630. [DOI] [PubMed] [Google Scholar]
- 60.Takeshita, T., H. Asao, K. Ohtani, N. Ishii, S. Kumaki, N. Tanaka, H. Munakata, M. Nakamura, and K. Sugamura. 1992. Cloning of the gamma chain of the human IL-2 receptor. Science 257:379-382. [DOI] [PubMed] [Google Scholar]
- 61.Thornhill, M. H., S. M. Wellicome, D. L. Mahiouz, J. S. Lanchbury, U. Kyan-Aung, and D. O. Haskard. 1991. Tumor necrosis factor combines with IL-4 or IFN-gamma to selectively enhance endothelial cell adhesiveness for T cells. The contribution of vascular cell adhesion molecule-1-dependent and -independent binding mechanisms. J. Immunol. 146:592-598. [PubMed] [Google Scholar]
- 62.Verma, S. C., S. Borah, and E. S. Robertson. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348-10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, H. W., M. W. Trotter, D. Lagos, D. Bourboulia, S. Henderson, T. Makinen, S. Elliman, A. M. Flanagan, K. Alitalo, and C. Boshoff. 2004. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat. Genet. 36:687-693. [DOI] [PubMed] [Google Scholar]
- 64.Wilkinson, P. C., and L. N. Islam. 1989. Recombinant IL-4 and IFN-gamma activate locomotor capacity in human B lymphocytes. Immunology 67:237-243. [PMC free article] [PubMed] [Google Scholar]
- 65.Wurster, A. L., V. L. Rodgers, M. F. White, T. L. Rothstein, and M. J. Grusby. 2002. Interleukin-4-mediated protection of primary B cells from apoptosis through Stat6-dependent up-regulation of Bcl-xL. J. Biol. Chem. 277:27169-27175. [DOI] [PubMed] [Google Scholar]
- 66.Yoshimoto, T., and W. E. Paul. 1994. CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J. Exp. Med. 179:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng, Y., X. Zhang, Z. Huang, L. Cheng, S. Yao, D. Qin, X. Chen, Q. Tang, Z. Lv, L. Zhang, and C. Lu. 2007. Intracellular Tat of human immunodeficiency virus type 1 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus: role of JAK/STAT signaling. J. Virol. 81:2401-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, M., Y. Zhou, C. Xie, F. Zhou, Y. Chen, G. Han, and W. J. Zhang. 2006. STAT6 specific shRNA inhibits proliferation and induces apoptosis in colon cancer HT-29 cells. Cancer Lett. 243:38-46. [DOI] [PubMed] [Google Scholar]