Abstract
The most prevalent type of RNA editing is mediated by ADAR (adenosine deaminase acting on RNA) enzymes, which convert adenosines to inosines (a process known as A→I RNA editing) in double-stranded (ds)RNA substrates. A→I RNA editing was long thought to affect only selected transcripts by altering the proteins they encode. However, genome-wide screening has revealed numerous editing sites within inverted Alu repeats in introns and untranslated regions. Also, recent evidence indicates that A→I RNA editing crosstalks with RNA-interference pathways, which, like A→I RNA editing, involve dsRNAs. A→I RNA editing therefore seems to have additional functions, including the regulation of retrotransposons and gene silencing, which adds a new urgency to the challenges of fully understanding ADAR functions.
An RNA transcript is subjected to various maturation processes, such as 5′ capping, splicing, 3′ processing and polyadenylation, after it is transcribed from the gene. Post-transcriptional processing of primary transcripts is essential to generate mature messenger RNAs that are ready to be translated into proteins1. RNA editing is a post-transcriptional-processing mechanism that results in an RNA sequence that is different from the one encoded by the genome, and thereby contributes to the diversity of gene products. There are different types of RNA-editing mechanism that either add or delete nucleotides, or that change one nucleotide into another2 (BOX 1).
Box 1. Different types of RNA editing.
RNA editing is a post-transcriptional process that changes the nucleotide sequence of an RNA transcript from the DNA sequence encoded by the corresponding gene2. Editing of mRNAs, transfer RNAs and ribosomal RNAs has been reported in bacteria to man. The first example of mRNA editing, which involved the insertion or deletion of many uridine (U) residues, was reported 20 years ago for mRNAs that are encoded by the mitochondrial DNA of trypanosomes. Soon after, other types of RNA editing were discovered, and it became clear that RNA editing is a widespread phenomenon in all three kingdoms of life. In transcripts of the mitochondrial and chloroplast DNAs of plants, for example, the conversion of many cytidine (C) residues to uridine (C→U editing) and the less frequent U→C editing occur, whereas an insertion of guanosine (G) residues occurs in the coding mRNAs of negative-strand RNA viruses. In Physarum polycephalum, different types of RNA editing occur in mitochondrial mRNA and rRNA; insertion of multiple cytidine residues, dinucleotide insertion (CU, GU, UA, AA, UU and GC) and an AAA deletion104,105. C→U editing occurs in the small subunit rRNA in Dictyostelium discoideum mitochondria106.
In mammals, two separate nucleotide-substitution types of RNA editing have been identified. The conversion of a specific cytidine residue to uridine (C→U editing) in apolipoprotein B mRNA is mediated by APOBEC1 cytidine deaminase107. This C→U editing results in the change of a glutamine codon to a translation stop codon and the consequent synthesis of APOB48, a shorter isoform of APOB100, which is translated from the unedited apolipoprotein B mRNA. The second type, adenosine to inosine (A→I) RNA editing, which is the main focus of this review, is the most common type of mammalian RNA editing.
Various nucleotide alterations of tRNA sequences (tRNA editing) are also known. 5′-terminal editing of mitochondrial tRNAs occurs in the amoeboid protist Acanthamoeba castellanii108. A→I editing of tRNAs, which is mediated by ADAT (adenosine deaminase acting on tRNA), occurs in eukaryotes and also in Escherichia coli109. ADAT1 edits A37 (near the anticodon) of tRNAAla, and the heterodimeric ADAT2–ADAT3 complex edits A34 at the wobble position of the anticodon of a subset of tRNAs3–5,110. ADAR genes are thought to have evolved from ADAT genes2–5,110.
The type of RNA editing that is most prevalent in higher eukaryotes converts adenosine (A) residues into inosine (I) in double-stranded (ds)RNAs through the action of ADAR (adenosine deaminase acting on RNA) enzymes3–5. A→I RNA editing of a short dsRNA that has formed between a coding exon and nearby intron sequences can lead to a codon change and an alteration in the protein function. However, it was recently discovered that the most frequent targets of A→I RNA editing seem to be long, but partially double-stranded, RNAs that are formed from inverted Alu repeats and long interspersed element (LINE) repeats located in introns and untranslated regions (UTRs) of mRNAs6–9. Global editing of non-coding RNA might control the expression of genes that harbour these repeat sequences of retrotransposon origin.
Post-transcriptional gene regulation can also occur through RNA interference (RNAi), an evolutionarily conserved phenomenon that involves dsRNA molecules10,11. Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are non-coding RNAs that are generated by a class of RNase III ribonucleases (specifically, Dicer and Drosha). These small RNAs are incorporated into the RNA-induced silencing complex (RISC), which mediates the RNAi process12–16. The idea that the RNAi and A→I RNA editing pathways might compete for a common substrate dsRNA was originally proposed by Bass17. Recent studies showed that precursor RNAs of certain miRNAs indeed undergo A→I RNA editing18–21, and editing seems to regulate the processing and expression of mature miRNAs19. Furthermore, one of the mammalian ADAR-family members sequesters siRNAs, thereby reducing RNAi efficacy22. Last, analysis of ADAR-null Caenorhabditis elegans strains indicates that A→I RNA editing might counteract RNAi silencing of endogenous genes and transgenes23–25.
In this review, I discuss recent findings on new functions of A→I RNA editing in the regulation of non-coding RNAs and on the interplay between RNA editing and RNAi pathways. For comprehensive reviews on A→I RNA editing, see REFS 3–5.
A→I RNA editing by ADARs
Deamination of adenosine to inosine
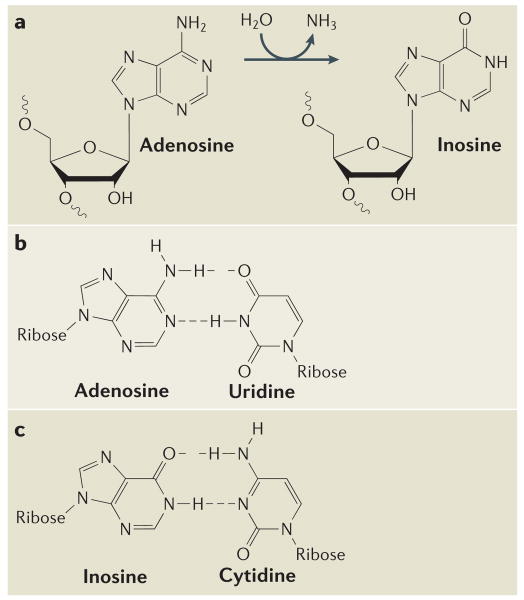
During the A→I RNA editing process, adenosine is converted to inosine by hydrolytic deamination of the adenine base26,27 (FIG. 1a). The translation machinery reads the inosine as if it were guanosine (G) (FIG. 1b), leading to the introduction of missense codons into mRNAs. Reverse transcriptase also reads inosine as guanosine; therefore, A→I RNA editing translates into an A→G change when analysing cDNA sequences.
Figure 1. Deamination of adenosine to inosine by ADAR.
a | A hydrolytic deamination reaction converts adenosine to inosine. b | Adenosine base-pairs with uridine. c | By contrast, inosine base-pairs, as if it were guanosine, in a Watson–Crick-bonding configuration with cytidine.
ADAR genes
The catalytic reaction of A→I RNA editing is mediated by ADAR enzymes (FIG. 2a). ADARs were originally identified in Xenopus laevis eggs and embryos by their dsRNA-unwinding activity28,29. Soon after, however, it was discovered that this activity is in fact a dsRNA-specific adenosine deaminase26,27. The first mammalian ADAR gene, human ADAR1, was cloned following the biochemical purification and microsequencing of the ADAR1 protein30, which then led to the identification of ADAR2 (REFS 31–33) and ADAR3 (REFS 34,35) (FIG. 2a). The enzymatic activity of ADAR1 and ADAR2 has been shown30–33. ADAR3 activity has not yet been shown, although functional domain features are conserved in this family member34,35. Therefore, the function(s) of ADAR3 remains to be established.
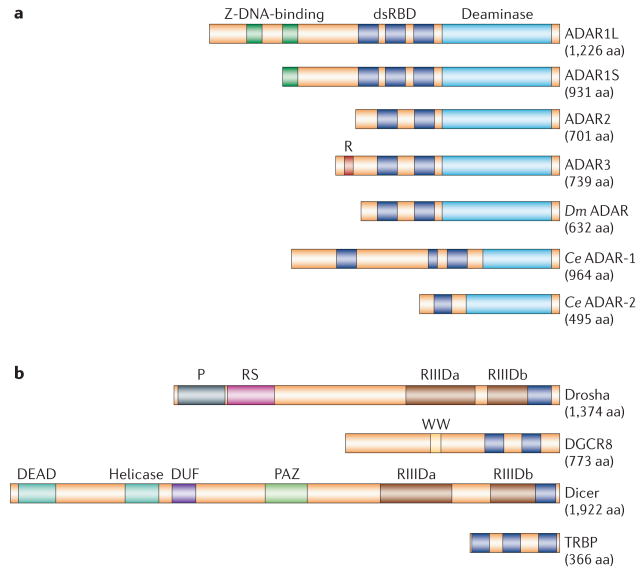
Figure 2. Types of dsRBD-containing protein: ADAR-family proteins and proteins that are required for miRNA biogenesis.
a | Three human ADAR (adenosine deaminase acting on RNA)-family members (ADAR1–3), Drosophila melanogaster (Dm) ADAR and two Caenorhabditis elegans (Ce) proteins, ADAR-1 and ADAR-2, share common functional domains: 2 or 3 repeats of the dsRBD and a catalytic deaminase domain. Certain structural features, such as Z-DNA-binding domains and the Arg-rich (R) domain, are unique to particular ADAR members. Binding of ADAR to double-stranded (ds)RNA substrates is mediated through dsRBDs38, whereas Z-DNA-binding domains might increase the affinity of ADAR1L specifically for short dsRNAs such as siRNAs22. Binding of the R domain to single-stranded RNAs has been reported, but its biological significance is currently unknown35. Two ADAR1 translation products, the isoforms ADAR1L and ADAR1S, result from transcription from different promoters followed by alternative splicing. This leads to translation initiation from the upstream or downstream Met codon41. b | Drosha and Dicer, two RNase III endonuclease family members, are essential for miRNA biogenesis. Drosha and Dicer, as well as cofactors DGCR8 and TRBP, contain one or more dsRBDs. In addition to the catalytic domain RIIID, which is responsible for the RNase III endonucleolytic reaction, unique functional domains, such as the Pro-rich (P) and Arg–Ser-rich (RS) domains, are present in Drosha. By contrast, the DEAD-box RNA helicase, DUF and PAZ domains are present in Dicer. The PAZ domain binds to the 3′ end of miRNAs, whereas the precise role of the DEAD-box RNA helicase domain is unknown. The function of the DUF domain is also unknown. The WW motif of DGCR8 is likely to be involved in protein interactions. Both ADARs and the proteins involved in the miRNA biogenesis pathway bind their dsRNA substrates through dsRBDs. The interaction between dsRNA and dsRBD is not RNA-sequence specific. Therefore, adenosine to inosine (A→I) editing and RNA-interference mechanisms might compete for a common dsRNA substrate, such as primary transcript miRNA (FIGS 6,7). aa, amino acids.
These three ADARs, which were originally identified in human and rodent, are conserved in vertebrates3–5. Only a few ADAR genes have been found in invertebrates. Drosophila melanogaster have only a single ADAR2-like gene, Adar36, whereas C. elegans have two ADAR genes, adar-1 and adar-2 (REF. 24) (FIG. 2a). No ADAR genes have been identified in the genomes of plants, fungi or yeasts.
Domain structure of ADARs
Members of the ADAR family contain common structural features (FIG. 2a). The dsRNA-binding domain (dsRBD; ∼65 amino acids) makes direct contact with the dsRNA37 and is required for dsRNA binding. The C-terminal region of ADAR contains amino-acid residues that are conserved in several cytidine deaminases and are predicted to participate in the formation of the catalytic centre of ADAR30,38. The crystal structure of the catalytic domain of human ADAR2 shows that His394, Glu396 and two Cys residues, Cys451 and Cys516, of ADAR2 are indeed involved in the coordination of a zinc atom and the formation of the catalytic centre39. Most surprisingly, however, the structural studies also revealed the presence of inositol hexakisphosphate (IP6) buried in the enzyme core, but located very close to the catalytic centre. The IP6 molecule could have a crucial role during the deamination reaction39.
ADAR gene expression and regulation
Both ADAR1 and ADAR2 are present in many tissues, whereas ADAR3 is expressed only in the brain30–35. Two isoforms of ADAR1, a full-length ADAR1L and a shorter, N-terminal-truncated ADAR1S, are known40. One of the three promoters that drive the ADAR1 gene is interferon inducible, and the mRNA transcribed from this promoter directs the translation of ADAR1L, initiated from an upstream Met codon41. A substantial increase in ADAR1L expression occurs during experimentally induced inflammation in mice42. Two other ADAR1 mRNAs, transcribed from constitutive promoters, direct the synthesis of ADAR1S, which is initiated from a downstream Met codon due to alternative splicing and skipping of the exon that contains the upstream Met codon (FIG. 2a). ADAR2 expression is regulated by the transcriptional activator cyclic-AMP-response-element binding (CREB) protein43, but the regulatory mechanism for ADAR3 is currently unknown.
ADAR1L is detected mainly in the cytoplasm, whereas ADAR1S localizes in the nucleoplasm and nucleolus40,44,45. ADAR2 localizes predominantly in the nucleolus44,46. The significance of the nucleolar localization of ADAR1S and ADAR2 is not currently clear. The cellular distribution of ADAR1L indicates the localization of its targets, possibly a different class of dsRNA substrate (for example, siRNAs; see below), in the cytoplasm22.
Substrate and editing-site selectivity
Both intermolecular and intramolecular dsRNAs of >20 base pairs (bp) (two turns of the dsRNA helix) can serve as substrates for ADAR47. Many adenosine residues of a long, completely base-paired dsRNA (>100 bp) are edited non-selectively. By contrast, shorts dsRNAs (∼20–30 bp) or a long but partially dsRNA with mismatched bases, bulges and loops (imperfect dsRNAs) are edited selectively; only a few adenosines are specifically chosen, indicating that the secondary structure in ADAR substrates dictates editing-site selectivity48. For example, site-selective A→I RNA editing occurs on an imperfect fold-back dsRNA structure that is formed between the exon sequence around an editing site(s) and a downstream intronic complementary sequence, termed editing-site-complementary sequence (ECS), of glutamate receptor-2 (GluR2) and serotonin (5-HT) receptor-2C (5-HT2CR) pre-mRNAs49,50 (see FIG. 3 and below). The ECS and the dsRNA structure are required for editing3,5,51,52.
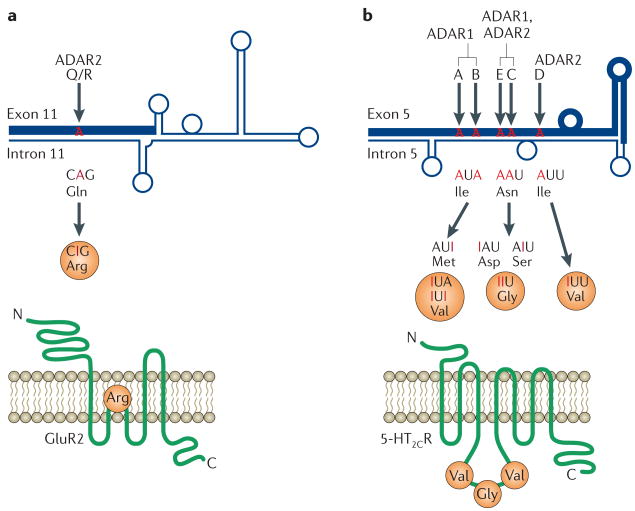
Figure 3. Functional changes by A→I RNA editing of coding sequences.
a | l-glutamate is the predominant excitatory neurotransmitter in vertebrate nervous systems, and the glutamate receptor (GluR) has been implicated in neuronal plasticity and higher functions such as memory and learning51. Adenosine to inosine (A→I) RNA editing of the Gln/Arg (Q/R) site leads to the replacement of a Gln by an Arg residue49,51. Ion-channel receptors that contain the edited GluR2 subunit are impermeable to Ca2+, whereas channels that lack the edited subunit permit influx of Ca2+. Q/R-site editing also regulates the tetramerization and intracellular trafficking of the receptor protein111. b | Serotonin receptors have important roles in physiological and behavioural processes such as circadian rhythms, emotional control and feeding behaviour55,64. G-protein-coupling functions of serotonin (5-HT) receptor-2C (5-HT2CR) are dramatically reduced by A→I RNA editing that occurs at five sites (A, B, C, D and E sites). For example, the potency of the agonist-stimulated G-protein-coupling activity of the fully edited receptor isoform (Val-Gly-Val) is reduced by 20-fold compared with the unedited receptor isoform (Ile-Asn-Ile)50,55. The fold-back double-stranded (ds)RNA structure, which consists of short dsRNA regions, bulges and loops, is formed because of partial complementarity of the exon and intronic editing-site complementary sequence (ECS; which is essential for editing). The thick dark-blue line represents the exon, and the thin dark-blue line represents the intron. Certain sites are exclusively edited only by ADAR1 (adenosine deaminase acting on RNA-1) or ADAR2; ADAR2 edits exclusively the Q/R site of GluR2 subunit and the D site of 5-HT2CR, whereas ADAR1 selectively edits the A and B sites of 5-HT2CR. The molecular mechanism that underlies the editing-site selectivity is not yet completely understood. However, the secondary structure in the fold-back dsRNA substrates, as well as functional interactions between two monomers of ADAR1 or ADAR2, might dictate editing-site selectivity. Several intronic editing sites that have been detected in GluR2 and 5-HT2CR dsRNAs are not shown.
Furthermore, some editing sites are preferentially edited only by ADAR1 or ADAR2 (FIG. 3), indicating a significant difference in their RNA–substrate interactions, possibly through their dsRBDs (different numbers and spacing between different dsRBDs)53. The distinctive site selectivity of ADAR1 and ADAR2 could also be mediated through functional interactions between the two monomers of ADAR1 or ADAR2, as such interactions possibly position specific adenosine residues relative to the catalytic centre of ADAR53,54.
Physiological significance of editing
Editing sites found in protein-coding regions
A limited number of targets (∼30 genes), such as mammalian GluR49 and 5-HT2CR55 as well as potassium channel Kv1.1 (REF. 56) and D. melanogaster Na+-channel57 gene transcripts, have been identified that are subjected to A→I RNA editing in their coding sequences51,52,56. In addition to cellular genes, transcripts of certain viruses, such as hepatitis delta virus, are also edited58.
Most often, RNA editing of protein-coding genes alters and diversifies the functions of the respective proteins, as shown by the two most studied examples (FIG. 3). Seeburg and colleagues identified a total of eight A→I RNA editing sites in the coding regions of receptors for several GluR subunits49,51. Among the eight editing sites, the Gln/Arg (Q/R) site located in the channel-pore-loop domain of the GluR2 subunit has the most important role in ion-channel function; editing of this single site makes the tetrameric channel protein impermeable to Ca2+ (FIG. 3a). Emeson and colleagues discovered a total of five A→I RNA editing sites located in the second intracellular loop or G-protein-coupling domain of 5-HT2CR55. Combinatorial editing of the five sites results in changes in three codons, Ile, Asn and Ile, to possibly six different amino-acid residues, resulting in the expression of up to 24 receptor isoforms with altered G-protein-coupling functions. For example, the ligand (5-HT) responsiveness of the receptor that has been fully edited at all five sites is reduced by 20-fold compared with that of the unedited receptor (FIG. 3b).
RNA-editing deficiencies
The inactivation of ADAR-gene-family members has significant physiological consequences, reported as phenotypic alterations of ADAR-gene mutants created in various species. Flies with a homozygous deletion in the Adar gene exhibit brain-related changes such as a lack of coordinated locomotion and age-dependent neurodegeneration36. Strains of C. elegans that contain homozygous deletions of both adar-1 and adar-2 display defective chemotaxis24. Mice with a homozygous Adar2-null mutation die several weeks after birth. These mice experience repeated episodes of epileptic seizures that originate from excess influx of Ca2+ and consequent neuronal death caused by under-editing of GluR2 pre-mRNA at the Q/R site59, which is a major target of ADAR2 (FIG. 3a). Last, the inactivation of ADAR1 leads to an embryonic lethal phenotype that is caused by defective erythropoiesis and widespread apoptosis60–62.
Human diseases or pathophysiologies can also be caused by dysfunction of the A→I RNA editing mechanism63,64. Heterozygosity for the ADAR1-gene functional-null mutation results in dyschromatosis symmetrica hereditaria, a human pigmentary genodermatosis of autosomal-dominant inheritance65. RNA-editing deficiencies also underlie disorders of the central nervous system. Under-editing of the Q/R site of GluR2 pre-mRNA (FIG. 3a) has been proposed to be responsible for the death of sporadic amyotrophic lateral sclerosis (ALS) motor neurons66, as well as apoptotic death of ischaemic neurons during ischaemia caused by cardiac arrest and disruption of the blood flow to the brain43. Last, RNA editing of 5-HT2CR might have some causative relevance to neuropsychiatric disorders, such as depression, as the editing pattern of 5-HT2CR mRNA (FIG. 3b) is significantly altered in the prefrontal cortex of suicide victims64,67,68.
Global editing of non-coding RNAs
The initial identification of physiologically important editing target genes, such as GluR2, and the consequent alterations of protein functions has fascinated many investigators. However, the number of genes that have been identified as editing targets has been far lower than that predicted by the amount of inosine that can be detected in rat brain poly(A)+ RNA69. This led to global searches for A→I editing sites in coding and non-coding regions.
Bioinformatics screening for A→I RNA editing sites
Several groups have recently developed a systematic, computational analysis method for the genome-wide identification of new A→I RNA editing sites6–9. Reverse transcriptase recognizes inosine as if it were guanosine (FIG. 1b). Therefore, an A→I RNA editing site can be identified when a cDNA sequence or an expressed sequence tag (EST) and the corresponding genome sequence are aligned, given that guanosine residues reverse-transcribed from inosines are detected in place of gene-encoded adenosines (FIG. 4a). The screening strategy consists of an algorithm to align a cluster of A→G mismatches in cDNAs or ESTs to the genome sequence and to assemble them into clusters that contain complete or partial genes in the dsRNA regions (as predicted by the presence of complementary sequences in a limited distance through a computer-assisted programme). This is followed by the elimination of single nucleotide polymorphisms (SNPs) and the evaluation of data quality. With this technique, a much larger than expected number of human A→I RNA editing sites has been identified6–9. Most surprisingly, almost all of these new sites that were identified in the human transcriptome (∼15,000 sites, mapped in ∼2,000 different genes) reside in non-coding regions that consist of inversely oriented repetitive elements (FIG. 4b), mostly Alu repeats (∼90%) and some LINE repeats (∼10%), representing ∼13% and ∼21% of the human genome, respectively.
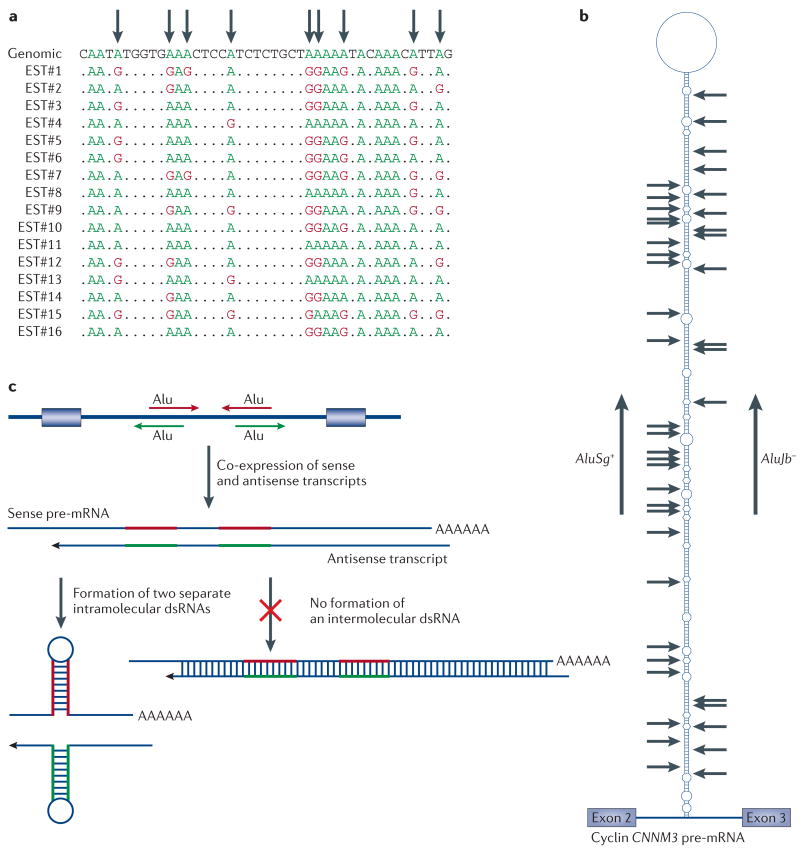
Figure 4. Extensive A→I RNA editing of non-coding repeat sequences.
a | A typical alignment of genomic and expressed sequence tag (EST) cDNA sequences is shown. Adenosine to inosine (A→I) RNA editing sites that have been identified as A→ guanosine (G) changes are marked by arrows. b | Detection of A→I RNA editing sites (arrows) in intron 2 of the cyclin CNNM3 pre-mRNA. Both bioinformatics screening and PCR after reverse transcription of RNA (RT-PCR) experiments have identified numerous extensively edited sites (some sites are 80–90% edited) in the double-stranded (ds)RNA structure that contains two inversely oriented Alu-subfamily members, AluSg+ and AluJb− (REF. 76). c | Scarcity of an intermolecular RNA duplex with sense and antisense transcript pair. Co-expression of CNNM3 pre-mRNAs and antisense transcripts in NT2-N neurons was discovered while analysing RT-PCR products76. As with CNNM3 pre-mRNA, extensive A→I RNA editing of these antisense transcripts was limited to the Alu-repeat sequences (shown in red and green), which indicated that sense and antisense strand RNAs formed two separate intramolecular dsRNAs instead of a completely complementary, long intermolecular dsRNA76. Together, it seems that in vivo formation of intermolecular RNA duplexes of sense and antisense transcripts is very rare, if it occurs at all.
On the basis of this analysis, it is predicted that >85% of pre-mRNAs are possibly edited, with the vast majority being targeted in introns (∼90%) and the rest in UTRs6. A similar screening strategy that is restricted to coding regions resulted in the identification of only a few editing target genes70,71. Together, these results indicate that the most common targets of ADARs are the non-coding sequences of transcriptomes and that protein re-coding as a result of A→I RNA editing is rare.
Editing of repeat RNAs in non-primate species
If global editing of non-coding Alu repeats in the human transcriptome has some biological significance, one might expect that the same is true in other organisms. Alu repeats are short interspersed elements (SINEs) that are unique to primates. However, SINE elements that are considered to have a common evolutionary origin with Alu repeats do exist in other organisms. Therefore, computational analyses have been carried out to search for A→I RNA editing sites in mouse EST databases8,72. The editing level in SINEs in mouse is at least an order of magnitude lower compared with Alu repeats in humans8,72. This substantial reduction in frequency might be explained by the differences in repeat length (∼300 bp versus ∼150 bp for human Alu and mouse SINE, respectively) and higher sequence homogeneity among human Alu repeats compared with mouse SINEs8,72. Screening for A→I RNA editing sites in rat, chicken and fly transcriptomes showed that non-coding repeat sequences are major targets of ADARs, but the editing frequency is again much lower than that observed in human transcriptomes72. So, although there is variability in the editing frequency of different organisms, A→I RNA editing of non-coding, repetitive RNA sequences seems to be a widespread phenomenon in the animal kingdom.
Editing of non-coding antisense transcripts
Global transcriptome analysis has shown that a large fraction of the genome produces transcripts from both sense and antisense strands (70%). Most sense and antisense transcript pairs are coordinately expressed, which indicates that antisense transcription might contribute to the control of sense transcripts73,74. However, it is unknown how frequently mammalian sense and antisense transcripts form into intermolecular dsRNAs. Because A→I RNA editing occurs only on dsRNA, the global examination of editing sites for sense and antisense transcripts could provide useful information on the in vivo formation of intermolecular RNA duplexes that consist of sense and antisense transcript pairs (FIG. 4c).
Recent bioinformatics studies of human EST databases for sense and antisense RNA pairs indicate that A→I RNA editing is restricted to intramolecular RNA duplexes that consist of inversely oriented repeat sequences of either sense or antisense RNA. However, A→I RNA editing is not detected in the regions outside of repetitive sequences75. PCR after reverse transcription of RNA (RT-PCR) and sequencing analysis of sense and antisense cyclin CNNM3 RNAs derived from an intronic region that contains two inverted Alu repeats confirmed that both sense and antisense RNAs are extensively edited, but only in their intramolecular fold-back dsRNA structures76 (FIG. 4b). No editing was detected outside of the Alu sequences, which indicates that the formation of an intermolecular sense–antisense RNA duplex does not occur76 (FIG. 4c). Interestingly, analysis of an equimolar mixture of sense and antisense CNNM3 RNAs that were edited in vitro by recombinant ADAR1 and ADAR2 indicate again that A→I RNA editing is restricted to the intramolecular fold-back structure, which indicates that inversely oriented Alu repeats predominantly form an intramolecular dsRNA and that their interaction with ADARs might prevent the formation of intermolecular RNA duplexes76.
Implications of repetitive RNA editing
What are the implications of global A→I RNA editing of non-coding, repetitive sequences for the control of gene expression (FIG. 5a)? The A→I sequence changes that are introduced in pre-mRNAs seem to be recognized by the splicing machinery. Furthermore, several cellular activities seem to specifically recognize and function on inosine-containing RNA (I-RNA) or dsRNA (I-dsRNA).
Figure 5. Possible regulatory functions for non-coding RNA editing.
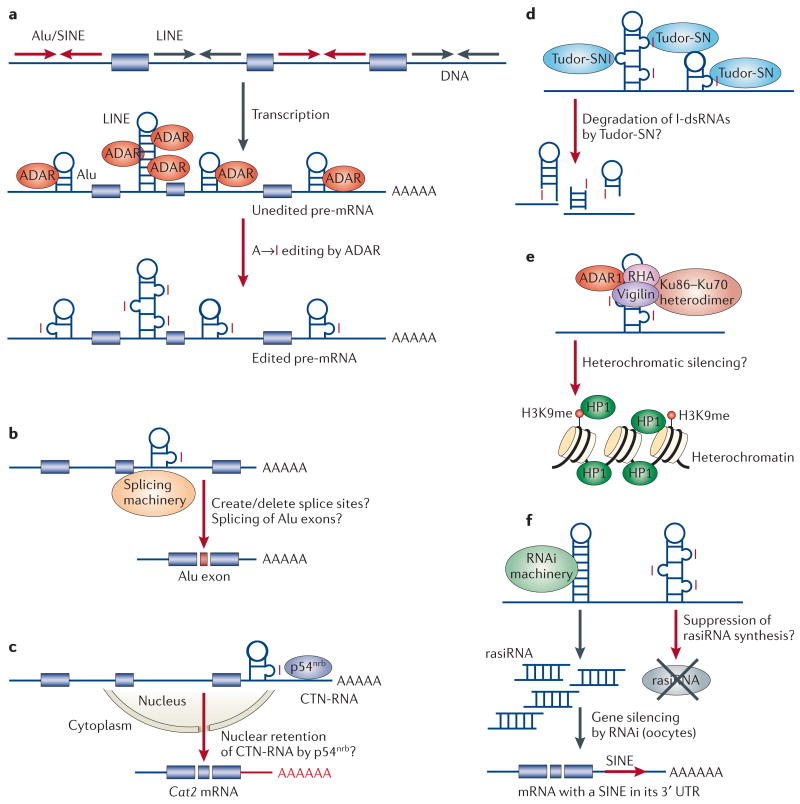
a | Extensive adenosine to inosine (A→I) editing of an RNA-duplex structure that consists of inverted Alu or LINE repeats. The inverted Alu or LINE repeats in introns and untranslated regions (UTRs) form intramolecular RNA duplexes genome wide, which are then subjected to A→I RNA editing by ADAR (adenosine deaminase acting on RNA). b | An inosine is interpreted by the splicing machinery as a guanosine. Therefore, splice sites might be created or deleted due to A→I editing of intronic Alu fold-back double-stranded (ds)RNAs, leading to the inclusion or exclusion of Alu exons6. c | A→I editing of a SINE fold-back dsRNA present in the 3′ UTR of CTN-RNA and its binding to p54nrb might be involved in the regulatory mechanism that retains this RNA in nuclear speckles81. When cells are placed under stress, CTN-RNA is cleaved and de novo polyadenylated at an alternative site to release the protein-coding Cat2 mRNA, which is then translated into cationic amino-acid transporter-2 protein81. The factors involved in the cleavage and de novo polyadenylation mechanisms are unknown. d | Tudor staphylococcal nuclease (Tudor-SN), an RNA-induced silencing complex (RISC)-associated component that lacks an assigned function in the RNA interference (RNAi) mechanism, has recently been identified as a potential inosine-containing dsRNA (I-dsRNA)-specific ribonuclease83. A→I editing of pre-mRNAs containing Alu or LINE fold-back dsRNA structures might be degraded by Tudor-SN, which, in turn, might control the expression levels of genes harbouring repeat sequences. e | The possibility that A→I RNA editing is involved in the heterochromatic silencing mechanism has been indicated by findings of Vigilin–ADAR1 complex formation and binding of Vigilin to inosine-containing RNAs85. Vigilin is an RNA-binding protein localized both in the nucleus and cytoplasm. The Drosophila melanogaster homologue of Vigilin, DDP1, has been known to have a role in heterochromatic gene silencing. The heterochromatic silencing process modifies the chromatin structure through various mechanisms, including histone H3 Lys9 methylation (H3K9me) and HP1 binding, which might eventually lead to methylation of cytosines in DNA (see recent reviews on heterochromatic silencing86,87,91). HP1, heterochromatin protein-1; RHA, RNA helicase A. f | In somatic cells and tissues, A→I editing of Alu or LINE fold-back dsRNAs might suppress the generation of rasiRNAs and therefore RNAi-mediated silencing in trans of genes that harbour the Alu or LINE sequence in UTRs. In mouse oocytes, rasiRNAs are generated92, possibly due to the absence of A→I editing. Modified with permission from REF. 112 © (2004) MacMillan Magazines Ltd.
Modulating splicing sites?
An inosine is interpreted by the splicing machinery as a guanosine. A→I RNA editing could therefore create or delete splice donor and acceptor sites. For example, a highly conserved canonical 5′-splice site dinucleotide recognition sequence, GU (AU→IU = GU), or a 3′-splice acceptor site, AG (AA→AI = AG), can be created by editing5. Self-editing of the intronic dsRNA sequence of ADAR2 pre-mRNA indeed results in the creation of an alternative 3′-splice acceptor site and the suppression of ADAR2 expression77. Also, a number of genes (for example, ADAR2b) that contain internal protein-coding Alu exons have been reported32,33,78,79. It is possible that some of these Alu exons are generated by the creation of splice sites following A→I RNA editing of Alu fold-back dsRNA (FIG. 5b). Several examples of exclusion and inclusion of the Alu exon due to editing of the Alu fold-back dsRNA sequence have been identified through the analysis of human cDNA sequences6 (FIG. 5b). A→I RNA editing might therefore affect alternative splicing, perhaps more than currently noted, of introns that contain Alu fold-back dsRNA.
Nuclear retention?
Affinity chromatography using I-RNA (that is, synthetic RNA that contains many inosines in place of guanosines) led to the identification of p54nrb (REF. 80). p54nrb is a nuclear localized multifunctional protein that interacts with splicing factor PSF and matrin-3 (a nuclear matrix protein), and it has been proposed to have a role in the mechanism for trapping extensively edited polyoma virus RNAs in the nucleus80. Previously, it was not known whether p54nrb regulated the nuclear retention of any cellular RNAs that contain many inosines as a result of A→I RNA editing. However, it now seems that A→I RNA editing of a long dsRNA formed on inverted repeats of SINEs that are present in the 3′ UTR of CTN-RNA and its binding to p54nrb might be involved in the regulatory mechanism that retains this RNA in nuclear speckles (also known as interchromatin granule clusters)81. Under stress, CTN-RNA is post-transcriptionally cleaved and de novo polyadenylated at an alternative site to produce protein-coding Cat2 mRNA, which is then translated into cationic amino-acid transporter-2 proteins81. The factors involved in the cleavage and de novo polyadenylation mechanisms are unknown (FIG. 5c).
Degradation?
A ribonuclease activity that specifically cleaves I-dsRNA has been reported82. Preferential cleavage by this ribonuclease occurs on both RNA strands of a dsRNA that contains multiple I·U base pairs82. The ribonuclease is specific to I-dsRNAs; dsRNAs that contain Watson–Crick base pairs, or dsRNAs that contain G·U base pairs in place of I·U base pairs, are not cleaved. Interestingly, Tudor staphylococcal nuclease (Tudor-SN), a RISC-associated component that lacks an assigned function in the RNAi mechanism16, has recently been identified as a potential I-dsRNA-specific ribonuclease, or at least as an essential cofactor of the activity83. Although Tudor-SN localizes to the cytoplasm of X. laevis oocytes83, its cellular distribution in somatic cells remains to be established84. A→I RNA editing of Alu or LINE fold-back dsRNA structures might therefore lead to the degradation of pre-mRNAs by Tudor-SN, which, in turn, might control the expression levels of genes that harbour repeat sequences (FIG. 5d).
Heterochromatic silencing?
The possible involvement of A→I RNA editing in the heterochromatic silencing mechanism has been proposed following the identification of Vigilin as another cellular factor that binds to I-RNAs85. Vigilin is found in complexes that contain ADAR1, the Ku86–Ku70 heterodimer (DNA-binding proteins that are involved in the DNA-repair mechanism) and RNA helicase A (RHA). Vigilin localizes to heterochromatin, and the D. melanogaster homologue of Vigilin, DDP1, is essential for heterochromatic gene silencing in flies. RHA has been suggested to have various functions such as unwinding a dsRNA structure formed around the exon–intron of D. melanogaster Na+-channel gene, which is also one of the A→I RNA editing targets57. The Vigilin–ADAR1–Ku-heterodimer–RHA complex recruits the DNA-dependent protein kinase PKcs enzyme, which phosphorylates a set of targets including heterochromatin protein-1 (HP1). HP1 has a major role in the chromatin-silencing mechanism85 (see also recent reviews on heterochromatic silencing86,87). Although the findings described above are suggestive, the significance of Vigilin–ADAR1 complex formation and binding of I-RNAs to Vigilin, as well as their relation to the heterochromatic silencing mechanism, remain to be established (FIG. 5e).
Suppression of rasiRNA?
The fold-back dsRNAs of C. elegans and D. melanogaster retrotransposons are processed into siRNA-like molecules — rasiRNAs, also known as repeat-associated siRNAs — in germline cells. rasiRNAs are proposed to constrain the expression of retro elements and protect the genome integrity of eggs and early embryos by the RNAi-mediated heterochromatic silencing mechanism86,88,89. The details of how rasiRNAs activate the mechanism are unknown.
Are rasiRNAs generated and are they involved in a similar RNAi-mediated silencing mechanism in mammalian cells (see reviews on RNAi-mediated heterochromatic gene silencing86,90,91)? Numerous rasiRNAs have been recently identified in mouse eggs and early embryos, which shows that fold-back dsRNAs of mammalian retrotransposon sequences can be processed to rasiRNAs92. Furthermore, rasiRNAs are reported to degrade a reporter target mRNA that contains the repetitive element in the 3′ UTR when they are injected into mouse oocytes. This indicates that retrotransposons are suppressed through the RNAi pathway in mouse oocytes92. Because A→I RNA editing alters the fold-back dsRNA structure, processing of rasiRNAs might be affected by editing and therefore by ADAR expression levels (FIG. 5f). For example, the generation of rasi-RNAs might be suppressed through A→I RNA editing of the fold-back dsRNA in somatic cells and tissues so that mRNAs that harbour repetitive elements in their UTRs are not silenced in trans (FIG. 5f). In support of this hypothesis, A→I RNA editing of repeat RNAs occurs only at low levels in ovaries and testes8. Furthermore, the nuclear versus cytoplasmic localization and activation of ADARs are regulated during maturation of oocytes and early embryos of X. laevis93.
Crosstalk between RNA editing and RNAi
In parallel with the recent findings on editing of non-coding repeat RNAs, a line of evidence has been accumulating that A→I RNA editing and RNAi pathways frequently interact, revealing another new function of editing that also affects global expression of many genes.
Suppression of RNAi by A→I RNA editing
RNAi, like A→I RNA editing, is a process that functions on viral and cellular dsRNAs14,16. Many proteins that are involved in the RNAi mechanism, such as Dicer, Drosha, DGCR8 and TRBP, contain dsRBDs, as do ADARs (FIG. 2b). Multiple adenosines of a long dsRNA can be deaminated by ADAR, whereas the RNase III-like ribonuclease Dicer processes long dsRNAs to 19–21 bp siRNAs (FIG. 6). Subsequently, AGO2 nuclease, a component of RISC, degrades cognate mRNAs through the siRNA-guided RNAi mechanism14,16.
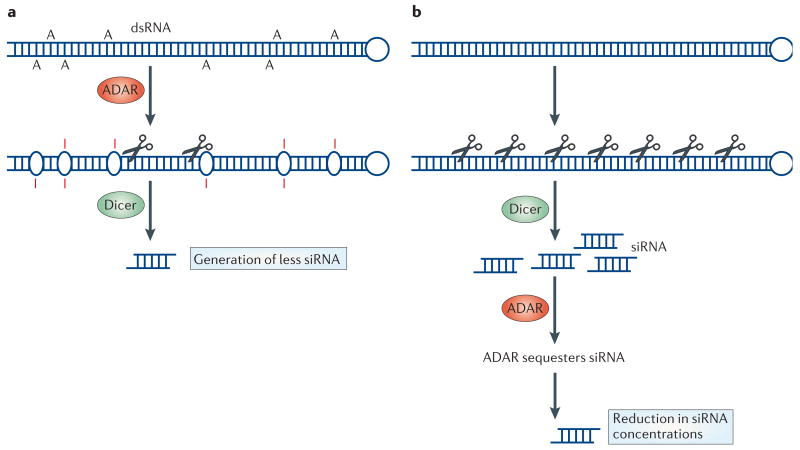
Figure 6. Interaction between RNA editing and RNA-interference pathways.
Two ways of interaction between RNA editing and RNA-interference pathways have been proposed. a | The introduction of many inosine·uridine (I·U) mismatched base pairs and the alteration of the double-stranded (ds)RNA structure by ADAR (adenosine deaminase acting on RNA) leads to the generation of fewer siRNAs by Dicer, because such dsRNAs that contain many I·U mismatched base pairs become resistant to Dicer cleavage95. b | Also, a fraction of already processed siRNAs might be sequestered by certain ADAR-gene-family members, reducing the effective siRNA concentration. For example, cytoplasmic ADAR1L binds siRNA tightly. Gene silencing by siRNA is significantly more effective in the absence of ADAR1, which indicates that ADAR1L is a cellular factor that limits siRNA potency in mammalian cells by decreasing the effective siRNA concentration and its incorporation into the RNA-induced silencing complex (RISC)22.
In general, dsRNA-binding proteins lack sequence specificity in the strict sense94. Therefore, it has been speculated that the A→I RNA editing mechanism might interact with the RNAi pathway by competing for shared dsRNA substrates and reducing RNAi efficacy17. The dsRNA that is extensively edited in vitro by ADAR indeed becomes resistant to Dicer, resulting in the generation of less siRNA and reduced RNAi95 (FIG. 6a). Dicer is thought to distinguish dsRNAs that contain I·U wobble base pairs from dsRNAs that contain only Watson–Crick base pairs95.
Strains of C. elegans that contain homozygous deletions of both adar-1 and adar-2 genes (FIG. 2a) display defective chemotaxis24. These phenotypic alterations, however, can be reverted in C. elegans strains that have an RNAi deficiency, indicating that ADAR-null worm phenotypes are RNAi dependent23. Expression of a gene that is involved in the chemotaxis mechanism (‘chemotaxis gene’) might be under control of the balance between A→I RNA editing and RNAi on dsRNA derived from the chemotaxis gene (FIG. 6a). It is assumed that overly enhanced RNAi effects and suppression of the chemotaxis gene result in ADAR-null worm phenotypes, but details of this RNA editing and RNAi pathway interaction remain to be established.
In addition, studies on the expression of transgenes in ADAR-null worms indicate that A→I RNA editing of dsRNAs that are derived from inverted repeats of transgenes seems to prevent silencing of the transgenes by RNAi in C. elegans25 (FIG. 6a). The results indicate once again the antagonistic effects of ADAR in vivo on RNAi that control the invasion of transgenes, viral infection and activities of transposons11,14,16. This type of transgene silencing (co-suppression), as well as silencing of viral RNAs through RNAi, is efficient in plants and fungi that lack ADAR genes and the A→I RNA editing system11,14,16,96. In these organisms, RNAi seems to be the sole defence mechanism against invasion of transgenes and viral infection. The A→I RNA editing system might have evolved to counteract RNAi in organisms in which more advanced immune systems developed.
Suppression of siRNA by ADAR1L
In the studies described above, long dsRNA was proposed to be the target of ADAR23,25. And, Dicer and ADAR are thought to compete for long dsRNA substrates (FIG.6a). In addition, the function of siRNAs, which have already been processed from the long dsRNA by Dicer, might be quenched in mammalian cells (FIG. 6b). Certain viral and cellular factors function as suppressors of RNAi. For example, ERI-1 is a 3′→5′ exonuclease that affects the efficacy of the endogenous RNAi mechanism by specifically degrading siRNAs97. By contrast, a 19-kDa protein (p19) homodimer synthesized by tombusvirus binds tightly and specifically to siRNAs, thereby suppressing the host plant defence RNAi mechanism98,99. Cytoplasmic ADAR1L has also been reported to bind siRNA tightly22. Gene silencing by siRNA is significantly more effective in mouse fibroblasts that are homozygous for an Adar1-null mutation than in wild-type cells22. These findings implicate ADAR1L as a cellular factor that limits siRNA potency in mammalian cells, as does p19, by decreasing the effective siRNA concentration and its incorporation into RISC (FIG. 6b)22. Eri1 and Adar1 gene expression is induced in mice that have been injected with high doses of non-specific siRNA100, which indicates the involvement of ADAR1 and ERI1 in a cellular feedback mechanism in response to siRNA. The endogenous siRNAs or siRNA-like molecules that are regulated through binding of ADAR1L (for example, the rasiRNAs described above and in FIG. 5f) remain to be identified.
Editing of miRNA precursor sequences
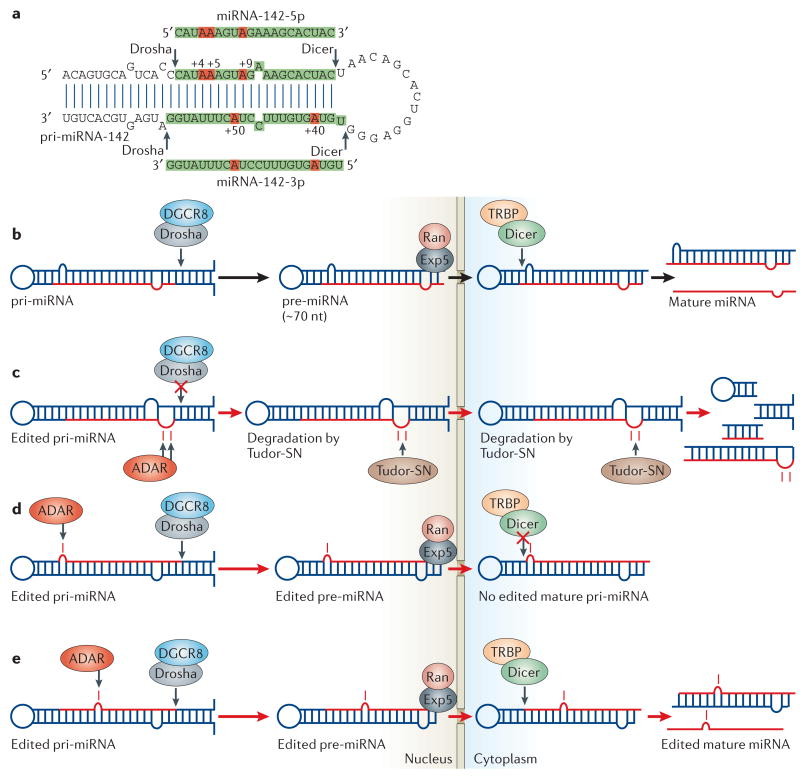
Numerous cellular and viral small non-coding RNAs, which are known as miRNAs, have been discovered12–16. These small RNA molecules function through a mechanism that is similar to siRNA-mediated RNAi13,15. Although miRNA is single stranded, it is generated from a long primary transcript (pri-miRNA) that consists of an imperfect short dsRNA region and a loop (FIG. 7a). Nuclear Drosha, together with the dsRNA-binding protein DGCR8 (FIG. 2b), cleaves pri-miRNAs, releasing 60–70-nucleotide intermediate precursors (pre-miRNAs). Recognition of correctly processed pre-miRNAs and their nuclear export is carried out by exportin-5 and RanGTP. Cytoplasmic Dicer, together with the dsRNA-binding protein TRBP (FIG. 2b), then processes the pre-miRNAs into 20–22-nucleotide siRNA-like duplexes (FIG. 7b)13,15. One or both strands of the duplex might serve as the mature miRNA. Following their incorporation into RISC, miRNAs block the translation of partially complementary targets that are located in the 3′ UTR of specific mRNAs or they guide the degradation of target mRNAs, as do siRNAs12–16. Any dsRNAs that are recognized by the RNAi mechanism are also potential targets for A→I RNA editing, and the possibility that pri-miRNAs might be edited by ADAR has been pointed out previously61.
Figure 7. Regulation of microRNA processing and expression by RNA editing.
a | Adenosine to inosine (A→I) editing sites of pri-miRNA-142. The region to be processed into mature sense and antisense strand miRNA-142 (5p and 3p, respectively) is highlighted in green. Five major editing sites are indicated by an A in red. The 5′ end of the mature miRNA-142-5p sequence is numbered as +1. Editing of the +4 and the +5 sites inhibits cleavage by the Drosha–DGCR8 complex19. Modified with permission from REF. 19 © (2006) MacMillan Magazines Ltd. b | The Drosha-DGCR8 complex cleaves pri-miRNAs in the nucleus, producing ∼70-nucleotide pre-miRNA intermediates, which are exported by exportin-5 and RanGTP into the cytoplasm. The Dicer–TRBP complex executes the second cleavage, generating mature miRNAs. c | Drosha cleavage of pri- to pre-miRNA is suppressed by A→I editing of certain sites, such as the +4 and +5 sites of pri-miRNA-142. Also, highly edited pri-miRNA-142 is degraded by Tudor staphylococcal nuclease (Tudor-SN)19. d | A→I editing of certain sites might lead to inhibition of the Dicer–TRBP cleavage step. e | Editing of pri-miRNAs at certain sites might lead to the expression of ‘edited mature miRNAs’ (for example, Kaposi-sarcoma-associated virus miRNA, miRNA-K12-10b)20.
Recent studies showed that certain miRNA precursors are indeed edited by ADAR18–21. A systematic survey of human pri-miRNA sequences identified A→I RNA editing sites in ∼6% of all pri-miRNAs examined21. However, this could be a low estimate21, and in vitro editing studies of randomly selected pri-miRNAs predict that as many as 50% of all pri-miRNAs might have specific A→I RNA editing sites19. The editing of miRNA precursors could have important implications for their processing, as well as the expression and the functions of mature miRNAs. A→I RNA editing alters the fold-back dsRNA structure of miRNA precursors; this might affect their subsequent processing and export steps.
Recent studies have revealed that the editing of two specific sites of pri-miRNA-142 (+4 and +5 sites in FIG. 7a) completely suppresses its cleavage by the Drosha–DGCR8 complex19. Also, Tudor-SN promotes the degradation of highly edited pri-miRNA-142 (REF. 19) (FIG. 7c). As expected, mature miRNA-142 expression levels are substantially increased in Adar1-null or Adar2-null mutant mice19. Although this is yet to be shown, A→I RNA editing of certain pri-miRNAs at specific sites is expected to suppress pre-miRNA export from the nucleus by exportin-5 and RanGTP, and the cleavage of pre-miRNA to mature miRNA by the Dicer–TRBP complex (FIG. 7d). In the pri-miRNA-142 studies, editing of certain sites, such as the +40 site (FIG. 7a), did not affect cleavage by Drosha or Dicer19. So, the structural changes of certain miRNA precursors that are caused by editing at a few selected sites might be tolerated. This implies that editing of certain pri-miRNAs might result in the expression of edited mature miRNAs, depending on the location of the editing site(s). Indeed, the expression of a Kaposi-sarcoma-associated virus miRNA (miRNA-K12-10b) that was edited at position 2 (+2 site) has been reported20. Edited miRNA can silence a set of target genes that are different from those silenced by the unedited miRNA, especially if an editing site is located in the ‘seed sequence’; that is, the 5′ half (+2 to +8) of the miRNA sequence that is important for pairing with the target mRNA12,13 (FIG. 7e). Alternatively, editing might affect the selection of the ‘effective’ miRNA strand that is loaded onto RISC and guides it to the target mRNA. The selection of the ‘effective’ strand depends on the local stability of the sense–antisense miRNA duplex12,13,16. A→I RNA editing is expected to affect the local stability of the duplex.
Concluding remarks and outlook
ADARs were originally discovered as a mysterious dsRNA-unwinding activity, but they were soon identified as enzymes that are involved in A→I RNA editing, which is essential for the re-coding of important mammalian genes. The roles of ADAR genes and A→I RNA editing, however, need to be redefined, as we have now realized that non-coding, repetitive RNAs are their most frequent targets. Furthermore, recent findings all point to an intimate interplay between A→I RNA editing and RNAi. Indeed, we are just beginning to grasp the magnitude of the biological significance of A→I RNA editing, with many questions remaining to be answered.
The RNAi machinery is functional in ADAR-null worms and is therefore independent of A→I RNA editing25. Furthermore, ADAR genes are missing in plant, fungi and yeast genomes, whereas these species do have RNAi. Has A→I RNA editing evolved specifically to tune and regulate RNAi in the animal kingdom, possibly along with the expansion of repeat elements in the genome? Although SINEs are edited genome wide in different species, the editing frequency of primate-specific Alu repeats is substantially higher (30–40 fold) than that of mouse SINE repeats. Does this mean that A→I RNA editing is less important in non-primate species, even though all three ADAR genes remain conserved among vertebrates? Alternatively, does this mean that the biologically most important dsRNA targets for A→I RNA editing have yet to be discovered? A large new class of small RNAs (∼26–30 nucleotides) in complex with PIWI-family proteins (piRNAs, PIWI-interacting RNAs) has been reported in mammalian testes92,101–103. It would be interesting to determine whether A→I RNA editing is at all involved in the suppression of piRNA biogenesis.
The inactivation of Adar1 leads to an embryonic lethal phenotype, which is caused by widespread apoptosis60–62. It seems that the editing of currently unknown target dsRNA(s) protects developing embryos from massive apoptosis. Perhaps by addressing the questions mentioned above and by achieving a better understanding of the interaction between A→I RNA editing and RNAi pathways, we might uncover the mechanism that underlies the phenotype of Adar1-null mutant mouse embryos.
Acknowledgments
I am grateful to J. M. Gott, H. H. Kazazian, J. M. Murray and also members of my laboratory, especially L. Valente and Y. Kawahara, for their comments and suggestions. This work was supported in part by grants from the US National Institutes of Health, Juvenile Diabetes Research Foundation and the Commonwealth Universal Research Enhancement Program of the Pennsylvania Department of Health.
- ADAR
An adenosine deaminase that catalyses an RNA-editing reaction whereby an adenosine is converted to an inosine.
- Alu repeat
A dispersed, moderately repetitive DNA sequence found in the human genome with ∼1.4 million copies. The sequence is ∼300 base pairs long. The name Alu comes from the restriction endonuclease (AluI) that cleaves the sequence.
- LINE
A long interspersed element (LINE) sequence that is typically used for non-long terminal repeat retrotransposons.
- Non-coding RNA
RNA that is transcribed from DNA, but that is not translated into protein. Introns, 5′ and 3′ untranslated regions of mRNA, antisense transcripts (RNAs transcribed from the antisense strand of DNA), siRNA, miRNA, RNAs transcribed from repetitive sequences, tRNA, rRNA, small nuclear (sn)RNA and small nucleolar (sno)RNA are all non-coding RNAs.
- Retrotransposon
A mobile genetic element; its DNA is transcribed into RNA, which is reverse-transcribed into DNA and then is inserted into a new location in the genome.
- RNA interference (RNAi)
A post-transcriptional gene-silencing process in which double-stranded (ds)RNA triggers the degradation of homologous mRNA. Degradation of the target mRNA is induced by siRNAs that are derived from long dsRNA.
- Small interfering RNA (siRNA)
A small (19–23 base pair) non-coding double-stranded (ds)RNA that is processed from a longer dsRNA. Such non-coding RNAs hybridize with mRNA targets, and confer target specificity to the silencing complexes in which they reside.
- microRNA (miRNA)
A small (19–23 nucleotide) single-stranded RNA that is processed from a precursor that consists of a short double-stranded (ds)RNA region, internal loops or bulges, and a loop. miRNAs have an essential role in suppressing translation or in the degradation of a target mRNA by the miRNA-mediated RNA-interference mechanism.
- RNase III family
A group of double-stranded (ds)RNA-specific endonucleases that cleave dsRNA into short fragments with a 3′ overhang and a recessed 5′ phosphate on each strand. Drosha and Dicer, which are essential for RNA interference, belong to this family.
- RNA-induced silencing complex (RISC)
This complex, which contains siRNAs and protein factors, such as AGO2, mediates the degradation of target mRNAs with high sequence complementarity to the siRNA. A similar complex that contains miRNA instead of siRNA (miRISC) suppresses the translation of target mRNAs with partial complementarity to the miRNA.
- Deamination
The chemical process that replaces a primary amino group by a hydroxyl group, resulting in the conversion of one nucleoside to another.
- Double-stranded RNA-binding domain (dsRBD)
This compact (∼65 amino acids) domain with an α–β–β–β–α structure makes direct contact with the dsRNA. Proteins that function on dsRNAs contain a single or multiple dsRBDs.
- Inositol hexakisphosphate (IP6)
A phospholipid that is widely distributed throughout the animal kingdom and is affiliated with a wide-ranging array of important physiological activities.
- Z-DNA
A left-handed DNA form that is different from the A and B forms and that is believed to be involved in specific biological functions.
- Expressed sequence tag (EST)
A single-pass, short read of complementary DNA that is generated from a transcribed region of the genome.
- Single nucleotide polymorphism (SNP)
Typically a bi-allelic base-pair substitution, which is the most common form of genetic polymorphism.
- SINE
Short interspersed, repetitive sequences, such as Alu elements, generated by retrotransposition.
- Nuclear speckle
An irregularly shaped nuclear organelle that can be visualized by immunofluorescence microscopy using anti-splicing-factor antibodies. Usually, ∼25–50 speckles are present in the interphase mammalian nucleus, and they are thought to constitute storage and/or assembly sites for certain splicing factors.
- rasiRNA (repeat-associated siRNA)
siRNA derived from repetitive sequences such as Alu or LINE retrotransposon elements or centromeric repeat sequences.
- Wobble base pair
Non-G · C, A · U pairing, such as the thermodynamically less stable G · U, I · U pairing. Wobble base pairs, like Watson–Crick pairs, participate in forming helical regions in RNA folding.
- piRNA
An siRNA-like, small non-coding RNA (26–30 nucleotides) that was identified as an RNA component that is complexed with Piwi-family proteins in testes.
Footnotes
Competing interests statement: The author declares no competing financial interests.
References
- 1.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 3.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keegan LP, Leroy A, Sproul D, O'Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]; An up-to-date review on A→I editing and ADAR genes.
- 6.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim DD, et al. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nature Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]; References 6–9 report a genome-wide screening strategy, leading to the identification of numerous A→I editing sites in non-coding Alu repeat RNAs.
- 10.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 14.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 16.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 17.Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 18.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nature Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that A→I editing of a miRNA-142 precursor suppresses its processing by Drosha–DGCR8 and also that the highly edited precursor RNAs are degraded by Tudor-SN.
- 20.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nature Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 21.Blow MJ, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W, et al. ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem. 2005;280:3946–3953. doi: 10.1074/jbc.M407876200. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows that ADAR1L functions as an RNAi suppressor by sequestering siRNAs.
- 23.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]; Shows the RNAi dependence of ADAR-null worm phenotypes.
- 24.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]; Shows, for the first time, that A→I editing prevents RNAi-mediated transgene silencing, which implies an interaction between RNAi and RNA-editing pathways.
- 26.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 27.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 29.Rebagliati MR, Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 30.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melcher T, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 32.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerber A, O'Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 34.Melcher T, et al. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 35.Chen CX, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 37.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 39.Macbeth MR, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawakubo K, Samuel CE. Human RNA-specific adenosine deaminase (ADAR1) gene specifies transcripts that initiate from a constitutively active alternative promoter. Gene. 2000;258:165–172. doi: 10.1016/s0378-1119(00)00368-1. [DOI] [PubMed] [Google Scholar]
- 42.Yang JH, et al. Widespread inosine-containing mRNA in lymphocytes regulated by ADAR1 in response to inflammation. Immunology. 2003;109:15–23. doi: 10.1046/j.1365-2567.2003.01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng PL, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Desterro JM, et al. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 45.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol Cell Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishikura K, et al. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 49.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, et al. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- 51.Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr Opin Neurobiol. 2003;13:279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- 52.Reenan RA. The RNA world meets behavior: A→I pre-mRNA editing in animals. Trends Genet. 2001;17:53–56. doi: 10.1016/s0168-9525(00)02169-7. [DOI] [PubMed] [Google Scholar]
- 53.Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14:345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Cho DS, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J Biol Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 55.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 56.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 57.Reenan RA, Hanrahan CJ, Ganetzky B. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 58.Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 59.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 60.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 62.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 63.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]; An up-to-date review on human diseases caused by defective A→I editing.
- 64.Schmauss C. Regulation of serotonin 2C receptor pre-mRNA editing by serotonin. Int Rev Neurobiol. 2005;63:83–100. doi: 10.1016/S0074-7742(05)63004-8. [DOI] [PubMed] [Google Scholar]
- 65.Miyamura Y, et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet. 2003;73:693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 67.Gurevich I, et al. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 68.Niswender CM, et al. RNA editing of the human serotonin 5-HT2C receptor. Alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 69.Paul MS, Bass BL. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levanon EY, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clutterbuck DR, Leroy A, O'Connell MA, Semple CA. A bioinformatic screen for novel A–I RNA editing sites reveals recoding editing in BC10. Bioinformatics. 2005;21:2590–2595. doi: 10.1093/bioinformatics/bti411. [DOI] [PubMed] [Google Scholar]; References 70 and 71 report that A→I editing of protein-coding regions is exceptionally rare, as demonstrated by a genome-wide screening strategy.
- 72.Eisenberg E, et al. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense–antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 75.Neeman Y, Dahary D, Levanon EY, Sorek R, Eisenberg E. Is there any sense in antisense editing? Trends Genet. 2005;21:544–547. doi: 10.1016/j.tig.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 76.Kawahara Y, Nishikura K. Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense–antisense duplex formation. FEBS Lett. 2006;580:2301–2305. doi: 10.1016/j.febslet.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 75 and 76 show that antisense RNA is extensively edited, but only in regions containing an inverted Alu repeat dsRNA, showing that the formation of sense–antisense intermolecular dsRNAs is very rare.
- 77.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 78.Sorek R, et al. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in Alu exons. Mol Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 79.Dagan T, Sorek R, Sharon E, Ast G, Graur D. AluGene: a database of Alu elements incorporated within protein-coding genes. Nucleic Acids Res. 2004;32:D489–D492. doi: 10.1093/nar/gkh132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 81.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]; Reports that A→I editing of SINE repeats located in the 3′ UTR might regulate nuclear retention and the release of cationic amino-acid transporter-2 mRNAs.
- 82.Scadden AD, Smith CW. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nature Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]; Reports that Tudor-SN, previously identified as a RISC-associated protein, is a ribonuclease specific for inosine-containing dsRNAs, revealing a mechanistic connection between RNAi and RNA-editing pathways.
- 84.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein–Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]; Vigilin in complex with ADAR1 binds to inosine-containing RNAs, revealing a possible role for A→I editing in the heterochomatic gene-silencing mechanism.
- 86.Martienssen RA, Zaratiegui M, Goto DB. RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe. Trends Genet. 2005;21:450–456. doi: 10.1016/j.tig.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 87.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006 doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 88.Aravin AA, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 89.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 90.Aravin A, Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 91.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nature Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 92.Watanabe T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saccomanno L, Bass BL. The cytoplasm of Xenopus oocytes contains a factor that protects double-stranded RNA from adenosine-to-inosine modification. Mol Cell Biol. 1994;14:5425–5432. doi: 10.1128/mcb.14.8.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 95.Scadden AD, Smith CW. RNAi is antagonized by A→I hyper-editing. EMBO Rep. 2001;2:1107–1111. doi: 10.1093/embo-reports/kve244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vance V, Vaucheret H. RNA silencing in plants-defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 97.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 98.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 99.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hong J, et al. High doses of siRNAs induce eri-1 and adar-1 gene expression and reduce the efficiency of RNA interference in the mouse. Biochem J. 2005;390:675–679. doi: 10.1042/BJ20050647. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports the induction of ADAR-1 and ERI-1, an siRNA-specific ribonuclease and an RNAi suppressor, respectively, by high concentrations of siRNA. This indicates the presence of a feedback mechanism.
- 101.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 102.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 103.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 104.Byrne EM, Stout A, Gott JM. Editing site recognition and nucleotide insertion are separable processes in Physarum mitochondria. EMBO J. 2002;21:6154–6161. doi: 10.1093/emboj/cdf610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gott JM, Parimi N, Bundschuh R. Discovery of new genes and deletion editing in Physarum mitochondria enabled by a novel algorithm for finding edited mRNAs. Nucleic Acids Res. 2005;33:5063–5072. doi: 10.1093/nar/gki820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Barth C, Greferath U, Kotsifas M, Fisher PR. Polycistronic transcription and editing of the mitochondrial small subunit (SSU) ribosomal RNA in Dictyostelium discoideum. Curr Genet. 1999;36:55–61. doi: 10.1007/s002940050472. [DOI] [PubMed] [Google Scholar]
- 107.Navaratnam N, Sarwar R. An overview of cytidine deaminases. Int J Hematol. 2006;83:195–200. doi: 10.1532/IJH97.06032. [DOI] [PubMed] [Google Scholar]
- 108.Lohan AJ, Gray MW. Methods for analysis of mitochondrial tRNA editing in Acanthamoeba castellanii. Methods Mol Biol. 2004;265:315–331. doi: 10.1385/1-59259-775-0:315. [DOI] [PubMed] [Google Scholar]
- 109.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 111.Greger IH, Khatri L, Kong X, Ziff EB. AMPA receptor tetramerization is mediated by Q/R editing. Neuron. 2003;40:763–774. doi: 10.1016/s0896-6273(03)00668-8. [DOI] [PubMed] [Google Scholar]
- 112.Nishikura K. Editing the message from A to I. Nature Biotechnol. 2004;22:962–963. doi: 10.1038/nbt0804-962. [DOI] [PubMed] [Google Scholar]