Abstract
Cellular homeostasis is achieved by the proper balance of regulatory networks that if disrupted can lead to cellular transformation. These cell circuits are fine-tuned and maintained by the coordinated function of proteins and non-coding RNAs (ncRNAs). In addition to the well-characterized protein coding and microRNAs constituents, large ncRNAs are also emerging as important regulatory molecules in tumor-suppressor and oncogenic pathways. Recent studies have revealed mechanistic insight of large ncRNAs regulating key cancer pathways at a transcriptional, post-transcriptional and epigenetic level. Here we synthesize these latest advances within the context of their mechanistic roles in regulating and maintaining cellular equilibrium. We posit that similar to protein-coding genes, large ncRNAs are a newly emerging class of oncogenic and tumor-suppressor genes. Our growing knowledge of the role of large ncRNAs in cellular transformation is pointing towards their potential use as biomarkers and targets for novel therapeutic approaches in the future.
INTRODUCTION
The ultimate cause of cancer is the alteration of the balanced harmony of cellular networks and gene expression programs that maintain cellular homeostasis. Even the slightest perturbation of these pathways can result in cellular transformation. For decades, genetic studies have revealed the mutational alteration of genes that control these critical pathways such as DNA damage response, growth arrest, cell survival or the apoptotic pathway (1).
Genes controlling such balance can be classified into two major groups: tumor-suppressor genes and oncogenes. Tumor-suppressor genes protect cells against deleterious mutations and cellular regulation that could prime transformation. Conversely, genes that initiate the cellular transformation process upon inappropriate activation comprise oncogenes. Recent research points to the need for an expanded definition beyond just protein-coding genes to also include ‘tumor-suppressor non-coding RNAs (ncRNAs)’ and ‘oncogenic ncRNAs’.
Indeed, numerous profiling and characterization studies of microRNAs have identified critical roles for ncRNAs in cancer (2–5). MicroRNA alterations have been involved in the initiation and progression of human cancer. Furthermore, microRNA-expression profiling of human tumors has identified signatures associated with diagnosis, staging, progression, prognosis and response to treatment (4,6–12). Given that microRNAs primarily function as post-transcriptional regulators (13), they can act as tumor suppressor or oncogenes depending on their target genes (14). But in addition to the relatively well-described microRNAs, the growing knowledge of the mammalian non-coding transcriptome is revealing that the genome is also replete with large ncRNAs, which could have a major role in the development and progression of cancer, although their mechanisms of function remain less well understood (15–19).
Besides genetic mutations of tumor suppressor or oncogenes, a great deal of evidence indicates that epigenetic alteration is also a major factor contributing to tumor transformation and cancer (20). Intriguingly, a number of studies suggest that large non-coding RNAs are key components of the epigenetic regulatory networks.
For example, it is now well established that some large ncRNAs such as XIST, HOTAIR, AIR and KCNQ1OT1 interact with chromatin-remodeling complexes targeting them to specific genes to exert their functions (16,21–25). It has been proposed that different ncRNAs may serve as molecular scaffolds for those complexes so they can function in an appropriate spatial and temporal manner (26–28). In support of this hypothesis, a recent study shows that as many as 20% of large intergenic ncRNAs (lincRNAs) expressed in a given cell associate with chromatin-repressive complexes such as polycomb repressive complex 2 (PRC2) and that many of these lincRNAs are bound by multiple chromatin factors. Moreover, depletion of these lincRNAs affects the ability of PRC2 to regulate a specific subset of genes (27,28). More recently, HOTAIR was also discovered to bridge several chromatin-modifying complexes (29). Collectively, these studies point to an emerging theme of large ncRNAs interfacing with chromatin regulation by serving as molecular scaffolds.
It is noteworthy that chromatin-regulatory complexes are linked with the aberrant proliferation of cancer cells. For instance, SUZ12, a subunit of PRC2 complex is overexpressed in colon and breast cancers and EZH2 is upregulated in many tumors, including Hodgkin lymphoma, prostate and breast cancer (30). Moreover, EZH2 expression is associated with poor prognosis and is an indication for the metastatic character of the disease (30). Besides PcG proteins, misregulation of other chromatin complexes is associated with cancer. For example, ∼10% of leukemias bear chromosomal translocations of the trithorax group histone methyltransferase mixed lineage leukemia (31). Collectively, these findings point to an important interplay between ncRNAs and chromatin regulation, which might be relevant for the control of gene expression networks critical for the process of cell transformation. These studies point to the interface of RNA and chromatin representing a new dimension in our understanding of cellular transformation.
Here we synthesize recent advances in our understanding of large ncRNAs in cancer pathways. These include examples with a wide range of molecular mechanisms involved in gene regulation. Although there is a rich literature of the roles of miRNAs and other small RNA pathways in cancer, here we specifically focus on large ncRNAs with particular emphasis on their molecular mechanisms in tumorigenesis.
MECHANISMS OF CELL TRANSFORMATION AND TUMOR SUPPRESSION BY LARGE ncRNAs
One of the first steps to the identification of ncRNAs relevant to disease is the profiling of their expression across normal and tumor samples. To this end, different profiling strategies have been applied to identify ncRNAs in cancer. Some studies have analyzed the available gene expression data sets in search for tumor-specific ncRNAs (32–34). Other groups have designed tiling arrays covering non-coding sequences of the genome to profile tumor cells (34–36), whereas others have focussed on the identification of ncRNA genes that are differentially methylated in tumors and may thus have a role in cell transformation (37,38). These approaches have led to the identification of several long ncRNAs, whose expression and/or DNA methylation are significantly associated with cancerous tissues. However, this effort has been greatly limited by the incomplete representation of non-coding sequences on DNA microarrays. More recently, this limitation has been overcome by the advances in massively parallel RNA sequencing combined with new computational methods. This has allowed the reconstruction of transcripts that originate the sequence reads (39,40). Application of these methods can result in significant progress in the study of currently poorly annotated ncRNAs, such as lincRNAs, and their splicing isoform diversity.
The studies described above have identified numerous large ncRNAs that exhibit differential expression between normal and tumor states. Although such alterations could be due in some cases to secondary effects of the tumor progression, numerous experimental studies (summarized in Tables 1 and 2) have suggested that ncRNAs play important roles in controlling cellular pathways involved in cellular transformation, thus acting as potential onco- or tumor-suppressor RNAs. A challenging task is to determine how these RNA molecules are able to modulate those pathways. Here, we describe recent studies that have shed new light on the functional and mechanistic roles of large ncRNAs in cancer.
Table 1.
Examples of large ncRNAs potentially oncogenic
| RNA | Organism | Size | Genomic location | Expression | Functional characteristics | Mechanism | References |
|---|---|---|---|---|---|---|---|
| p21 NAT/Bx332409 | Homo sapiens | ≥423 bp | Antisense cdkn1a/p21 | EST sequenced from neuroblastoma | Antisense of Cdnk1a/p21. Negative regulation of Cdnk1a/p21 by epigenetic. silencing | Requirement of Ago1 but not Ago2 for epigenetic silencing of Cdkn1a/p21 | (44) |
| HOTAIR | H. sapiens | 2.2 kb | Intergenic HoxC locus | Distal fibroblasts/metastatic breast tumors | Gene silencing in trans. Metastasis | Interaction with PRC2 and LSD1 complexes and targeting to repressed genes | (22,34) |
| MALAT-1 | H. sapiens | 8.7 kb | Intergenic Chr11 | Overexpression in lung adenocarcinoma, breast, pancreas, colon, prostate and hepatocellular carcinomas | Metastasis | Induction of GAGE6 proto-oncogene transcription by inhibition of PSF repressor | (35,49–51) |
| VL30-1 | Mus musculus | 4.9 kb | Retroelement ncRNA | Ras-mediated transformation of mouse fibroblasts | Induction of Rab23 proto-oncogene transcription by inhibition of PSF repressor | (47,60–62) | |
| ANRIL | H. sapiens | 2.2 kb | Antisense of INK4n/ARF/INK4a and p15/CDKN2B | Upregulated in prostate cancer | Gene silencing of INK4a/ARF/INK4a and p15/CDKN2B | Interaction with CBX7 component of PRC1 complex | (42,43,63–65) |
| H19 | H. sapiens | 2.3 kb | Imprinted H19-Igf2 locus in Chr11 | Bladder, human lung and breast carcinoma, choriocarcinoma | Control of imprinting. Oncogenic or tumor suppressor. Containing microRNA miR-675 | Unknown | (66–70) |
| CUDR | H. sapiens | 2.2 kb | Intergenic Chr19 | Overexpressed in a doxorubicin-resistant human squamous carcinoma subline | May regulate drug sensitivity and promote cellular transformation through resistance to apoptosis | Unknown | (71) |
| Zeb2/Sip1 NAT | H. sapiens | 680 bp | Antisense of Zeb2/Sip1 | Overexpressed in human tumors with low E-cadherin expression | Inhibition of E-cadherin through induction of Zeb2 protein levels | Inhibition of splicing of Zeb2 first exon containing IRES sequence | (46) |
| SRA-1 | H. sapiens | 875 bp | Alternative splicing of SRA gene, loss of coding frame | In breast cancer cells, different balance of coding/non-coding splicing isoforms | Co-activator of steroid receptors and other transcription factors as MyoD. Increased levels of non-coding isoform associated with metastasis | Interaction in ribonucleoprotein complexes with several positive regulators, including SRC-1, p68 and p72, Pus1p and Pus3p, as well as negative regulators, such Sharp and SLIRP to be recruited to promoters of regulated genes | (72–79) |
| PCGEM1 | H. sapiens | 1.6 kb | Intergenic, Chr2 | Upregulated in prostate cancer in African-American patients | Inhibition of apoptosis, promotion of cell growth | Unknown | (80–84) |
| UCA1 | H. sapiens | 1.4 kb | Intergenic, Chr19 | Embryonic development and bladder cancer associated | Increases proliferation, migration, invasion and drug resistance of human bladder cell line | Unknown | (85) |
Table 2.
Examples of potential tumor-suppressor large ncRNAs
| RNA | Organism | Size | Genomic location | Expression | Functional characteristics | Mechanism | References |
|---|---|---|---|---|---|---|---|
| GAS5 | H. sapiens, M. musculus | Multiple splicing isoforms 0.6–1.8 kb (human) | Intergenic. Some introns encode snoRNAs | Downregulated in breast cancer, upregualted by induced growth arrest | Induces growth arrest and apoptosis | Blocks transcriptional induction by GR by binding to GR DNA binding domain and competing with DNA GRE | (52–54,86–91) |
| lincRNA-p21 | M. musculus | 3.1 kb | Intergenic, upstream of p21/cdkn1a | Induced by p53 upon DNA damage or oncogenic stress | Global gene repression in the p53 transcriptional response inducing cellular apoptosis | Physical interaction with hnRNP-K, targeting genes for transcriptional repression | (36) |
| MEG3 | H. sapiens | Several isoforms 1.6–1.8 kb | Intergenic, Chr 14 | Maternally expressed imprinted gene. Hypermethylated in myeloid leukemia, multiple myeloma and pituitary tumors | Activates p53 activity on specific genes by increasing p53 protein levels by suppressing MDM2 levels. Also inhibits cell proliferation independently of p53 | Unknown | (92–98) |
| ncRNA CCND1/cyclin D1 | H. sapiens | ≥200–300 nt | Transcribed from 5' end of cyclin D1 gene | Induced by DNA damage | Induces repression of cyclin D1 gene | Binding to TLS protein induces TLS allosteric change, allowing interaction with cyclin D1 gene, inhibiting CBP/p300 HAT activity and resulting in cyclin D1 repression | (56) |
Oncogenic ncRNAs
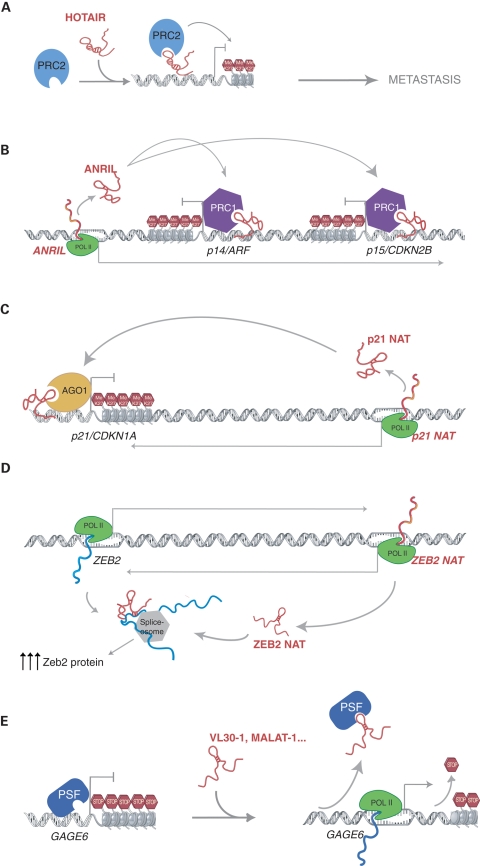
Similar to protein-coding oncogenes, large ncRNAs can also promote cellular pathways that lead to tumorigenesis. One example of such an oncogenic lincRNA is HOTAIR. HOTAIR underscores the importance of understanding the relationship between epigenetic regulation by ncRNAs and cancer. HOTAIR is expressed from the HOXC locus and was initially discovered as a gene repressor of HOXD genes. This repressive action is conferred by the interaction of HOTAIR with the PRC2 complex, imparting PRC2 localization and repression of the HOXD locus (22) (Fig. 1A). A new study has found that HOTAIR is significantly overexpressed in breast tumors (34). Furthermore, HOTAIR expression level in primary breast tumors is a powerful predictor of patient outcomes such as metastasis and death (34). This phenotype seems to be tightly associated with PRC2-dependent gene repression induced by HOTAIR. Enforced expression of HOTAIR results in an altered pattern of H3K27 methylation and increased invasiveness, whereas the depletion of HOTAIR causes the opposite cellular phenotype (34). Collectively, these studies demonstrate how oncogenic lincRNAs can hijack the epigenetic machinery to reshape the epigenetic landscape leading to cancer.
Figure 1.
Mechanisms of gene regulation by oncogenic large ncRNAs. (A) lincRNA HOTAIR recruits PRC2 to specific gene promoters for methylation of lysine 27 of histone 3 (H3K27me), inducing gene repression that leads to breast tumor metastasis. (B) Large ncRNA ANRIL is transcribed antisense of the p14/ARF and p15/CDKN2B genes. ANRIL mediates gene silencing of the locus by interaction and recruitment of CBX7, a component of PRC1 histone 3 lysine 27-methyltransferase complex. (C) p21 NAT ncRNA is transcribed antisense of the p21/CDKN1A gene. This RNA requires Ago1 protein to mediate epigenetic silencing of p21/CDKN1A promoter involving H3K27me. (D) The ncRNA expressed antisense of the Zeb2 gene (Zeb2 NAT) overlaps with the 5′ splice site of one of Zeb2 introns. Zeb2 NAT inhibits the splicing of the intron, which contains an IRES sequence. In this way, Zeb2 protein translation is upregulated. (E) Rab23 proto-oncogene (mouse) and GAGE6 proto-oncogene (human) are repressed by PSF protein. This repression is relieved when VL30-1 ncRNA (mouse) or MALAT-1 and others (human) interact with PSF, displacing it from the promoter.
In addition to intergenic large ncRNAs such as HOTAIR, global transcriptome analysis shows that up to 70% of protein-coding transcripts have antisense partners, and the perturbation of the antisense RNA can alter the expression of the sense gene (41). Some of these genes encode tumor-suppressor proteins that can become epigenetically silenced by the expression of the antisense ncRNA. Thus, misregulation of these antisense ncRNAs could lead to cellular transformation.
Indeed, the antisense ncRNA ANRIL controls expression in the INK4A/ARF locus comprising the tumor-suppressor genes INK4n/ARF/INK4a, p16/CDKN2A and p15/CDKN2B, which regulate cell cycle progression and senescence. ANRIL is transcribed antisense to the INK4n/ARF/INK4a promoter and overlaps with two exons of p15/CDKN2B. Independent studies have shown that overexpression of ANRIL in prostate cancer results in the silencing of INK4n/ARF/INK4a and p15/CDKN2B by heterochromatin formation (42,43). ANRIL interacts with CBX7, a component of the polycomb repressive complex 1 (PRC1), resulting in the targeting of this complex to the chromatin and the establishment of repressive epigenetic marks (43) (Fig. 1B). Another example of a tumor-suppressor gene that is epigenetically silenced by an antisense RNA is the cell cycle regulator p21/CDKN1A. In this case, the silencing mechanism requires the component of the RNAi pathway Ago-1 (44) (Fig. 1C). Thus, perhaps similar to ANRIL, the p21 antisense ncRNA may also be upregulated in cancer rendering p21 inert, leading to cellular transformation.
In addition to the regulation of tumor-suppressor pathways by epigenetic silencing shown by previous examples, some antisense transcripts can also fine tune gene expression at the post-transcriptional level. E-cadherin is a gene correlated with gastric, breast, colorectal, thyroid and ovarian cancers. Its loss of function is thought to contribute to progression in cancer by increasing proliferation, invasion and/or metastasis (45). A strong association has been demonstrated between the expression of a particular natural antisense transcript (NAT) and human tumors with low E-cadherin expression (46). NAT overlaps with an intronic 5' splice site of the Zeb2 gene and prevents its splicing. The retained intron contains an internal ribosome entry site (IRES) necessary for the increased translation of Zeb2 protein, which can subsequently function as a transcriptional repressor of E-cadherin (46) (Fig. 1D). Collectively, these studies provide strong impetus for further investigation of antisense ncRNAs in cancer pathways.
Other ncRNAs can induce the expression of the proto-oncogene Rab23 resulting in transformation and metastasis of skin fibroblasts (47). These ncRNAs act as inhibitory molecules, by complexing with the DNA and RNA-binding PSF (polypyrimidine tract-binding protein-associated splicing factor) protein to block its function as a transcriptional repressor, resulting in aberrant Rab23 expression. Interestingly, the interaction of ncRNAs with PSF is conserved between mouse and human, although species-specific ncRNAs are involved. In the mouse VL30-1 RNA, a member of the VL30 family of mouse retroelement ncRNAs mediates this mechanism (48), whereas in human five different RNAs can interact with PSF to induce the expression of the proto-oncogene GAGE6. These include the retroelements L1PA16 and MER11C, the mitochondrial gene HN encoding the humanin peptide and the ncRNA MALAT-1 (49) (Fig. 1E). Interestingly, many studies have identified the large ncRNA MALAT-1 as a tumor marker that is overexpressed in many different tumor types (35,50,51). However, it remains to be determined whether MALAT-1 acts exclusively through inhibition of the tumor-suppressor PSF.
Collectively, these studies point to the possibility of ‘oncogenic large ncRNAs’ that upon misregulation could either silence tumor-suppressor genes or induce the expression of oncogenes priming the cell for transformation.
Tumor-suppressor ncRNAs
Tumor-suppressor ncRNAs could phenotypically affect cells by promoting tumor-suppressor pathways, and when their function is compromised, cells are prone to develop cancer. In support of this notion, a few new studies have elucidated several examples (Table 2) of ‘tumor-suppressor large ncRNAs’.
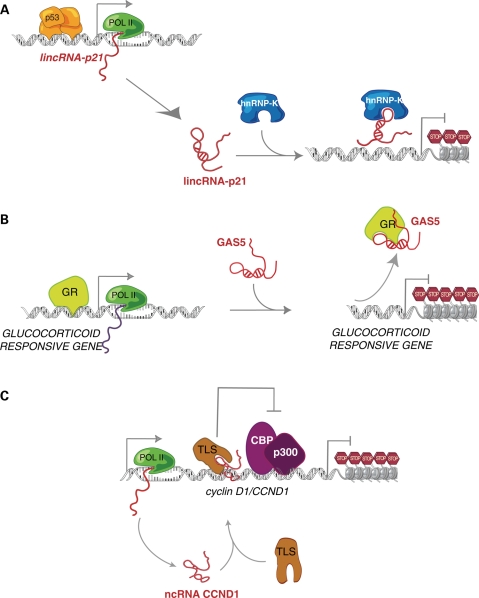
For example, recent studies identified numerous lincRNAs that are induced by the p53 tumor-suppressor pathway (17,36). When cells are subjected to stress, the transcription factor p53 initiates a tumor-suppressor program that involves the expression and repression of many genes. Surprisingly, among the genes specifically induced by p53, there are many lincRNAs. In particular, one of these lincRNAs, named lincRNA-p21 is directly induced by p53 to play a critical role in the p53 transcriptional response. LincRNA-p21 is required for the global repression of genes that interfere with p53 function regulating cellular apoptosis. Interestingly, lincRNA-p21 can mediate gene repression by physically interacting with the protein hnRNP-K, allowing its localization to promoters of genes to be repressed in a p53-dependent manner (36) (Fig. 2A). This study underlines the importance of well-tuned regulation of lincRNAs to orchestrate transcriptional programs that maintain cellular homeostasis.
Figure 2.
Mechanisms of gene regulation by tumor-suppressor large ncRNAs. (A) lincRNA-p21 expression is directly induced by p53. Then, lincRNA-p21 specifically interacts with hnRNP-K for localization to gene promoters for repression. (B) GAS5 mimics the conformation of DNA GREs, binding to GR. In this manner, GR loses the ability to activate transcription of target genes. (C) DNA damage induces the expression of ncRNA CCND1 from the 5′ of cyclin D1/CCND1 gene. ncRNA CCND1 interacts with the TLS protein, inducing a conformational change in TLS that allows its binding to the cyclin D1 promoter. This causes inhibition of cyclin D1 gene expression by blocking of CBP and p300 HAT activity.
GAS5 (growth arrest-specific 5) represents another example of a large ncRNA that regulates the expression of a critical subset of genes with tumor suppressive consequences. GAS5 is induced under starvation conditions being highly expressed in cells whose growth is arrested (52,53). GAS5 functions by outcompeting the DNA-binding sites of the glucocorticoid receptor (GR), thus reducing cell metabolism (54). Specifically, the GAS5 RNA conformation mimics that of the gluticorticoid responsive element (GRE) DNA, blocking the ability of GR to bind gene promoters to induce their transcription (54) (Fig. 2B). Interestingly, GAS5 has also been observed to be downregulated in breast cancer, perhaps to keep cancer cells active even under low nutrient conditions (52,53).
Another tumor-suppressor ncRNA is involved in the regulation of cyclin D1/CCND1 gene expression. Cyclin D1 is a cell cycle regulator frequently mutated, amplified and overexpressed in a variety of tumors (55). When cells are subjected to DNA damage, ncRNAs are expressed from the 5' regulatory regions of cyclin D1 gene, thereby mediating its transcriptional repression. Indeed, these ncRNAs interact with the TLS protein, inducing its allosteric modification. This conformational change allows the association of TLS to the cyclin D1 promoter, which inhibits transcriptional induction by histone acetyltransferases such as CBP and p300 (56) (Fig. 2C).
These studies show that tumor-suppressor ncRNAs can be rapidly induced by cellular stress to regulate gene expression. Possibly, RNA molecules, due to their quick turn over rate, are ideal effectors when a rapid response is required to protect cells from external insults.
FUTURE PERSPECTIVES
The studies reviewed herein contribute to the growing evidence of the important roles of large ncRNAs in cancer, both by regulating tumor-suppressor and oncogenetic pathways. Thus, some ncRNAs play a critical role in maintaining cellular homeostasis and, when we have a deeper understanding of their roles in cancer, they can be used as diagnostic tools in conjunction with protein-coding genes.
An intriguing common theme is emerging of large ncRNAs forming ribonucleic–protein complexes that impart key regulatory functions in cellular circuits. We have discussed HOTAIR, lincRNA-p21 ANRIL or MALAT-1 among others that share a common functionality of forming RNA–protein complexes with chromatin regulatory factors. However, a higher-resolution understanding of cancer will require a comprehensive identification of large ncRNAs misregulated across a spectrum of cancer types and their associated protein complexes. Both biochemical approaches combined with in vivo studies will be required to fully understand the mechanistic and phenotypic roles of large ncRNAs in cancer.
A key goal for future progress is to identify large ncRNAs that could potentially serve as biomarkers for specific disease states. A clear advantage in the diagnostic use of ncRNA detection versus that of protein-coding RNAs is that in the former the RNA itself is the effector molecule, thus its expression levels may be a better indicator of the intrinsic characteristics of the tumor. Indeed, microRNA expression profiling has been successfully used for cancer classification, reflecting the developmental lineage and differentiation state of the tumors (10–12). In the near future, the great technological advance and decrease in cost of parallel massive sequencing will allow the profiling of the entire transcriptome of every type of tumor, including small and large ncRNA molecules, allowing the most powerful and informative diagnosis. In fact, the application of the new genomic technologies to the profiling of multiple cancers is already a reality. The tremendous amount of data generated by these projects present great possibilities for prognosis and therapeutic application. This has called for the creation of the International Cancer Genome Consortium (ICGC) that will coordinate the international effort to systematically study more than 25 000 cancer genomes at the genomic, epigenomic and transcriptomic levels (57). We can easily predict that in the next few years a complete catalogue of the large ncRNA expression as well as the genetic mutations, amplifications and deletions in non-coding regions associated with different types of tumors will be available.
Besides the imminent use of our knowledge of cancer-associated large ncRNAs for diagnosis, therapeutic applications may be possible in a more distant future. The progress in the use of RNAi-mediated gene silencing for the treatment of different diseases is encouraging and could be applied to selectively silence oncogenic ncRNAs. Gene therapy could also be applied for the delivery to specific cells of tumor-suppressor large ncRNAs for the treatment of cancer. However, many technical challenges have to be overcome for a wider use of therapeutic RNAi and gene therapy, including the development of reliable delivery systems, dosage regimes and techniques to ameliorate RNAi off target effects (58,59). When the technical limitations are overcome, ncRNAs may be ideal targets for therapy due to their high turnover rate as well as their direct and specific regulatory functions. Predictably, therapeutic targeting of ncRNAs will carry fewer negative effects than those of protein-coding genes, given that they function regulating specific facets of their protein interacting partners.
In summary, overwhelming evidence reveals that large ncRNAs are molecules that keep in perfect tune the balance of gene expression networks, and discordance in their function results in homeostatic imbalance, ultimately causing cellular transformation. Large ncRNAs are shedding new light on our understanding of these cancer pathways and may represent a ‘missing link’ in cancer.
FUNDING
J.L.R. is a Damon Runyon-Rachleff, Searle and Smith Family Foundation Scholar and Richard Merkin awardee. M.H. is supported by the NIH Directors New Innovator Award (Grant number 1DP2OD00667-01), Smith Family Foundation, the Damon Runyon Cancer Foundation and Searle Scholar Program.
ACKNOWLEDGEMENTS
We would like to thank Moran Cabili-Kalmar, Magdalena J. Koziol, Sabine Loewer and Bárbara Tazón-Vega for critical comments on the manuscript and Sigrid Hart (Broad Institute) for illustration support.
Conflict of Interest statement. None declared.
References
- 1.Cowin P.A., Anglesio M., Etemadmoghadam D., Bowtell D.D. Profiling the cancer genome. Annu. Rev. Genomics Hum. Genet. 2010 doi: 10.1146/annurev-genom-082509-141536. Epub ahead of print 2 July 2010. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Pekarsky Y., Croce C.M. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best Pract. Res. Clin. Haematol. 2007;20:425–437. doi: 10.1016/j.beha.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 4.Ruan K., Fang X., Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Calin G.A., Croce C.M. Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes. Blood. 2009;114:4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho W.C. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Volinia S., Galasso M., Costinean S., Tagliavini L., Gamberoni G., Drusco A., Marchesini J., Mascellani N., Sana M.E., Abu Jarour R., et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson P., Lu J., Zhang H., Shai A., Chun M.G., Wang Y., Libutti S.K., Nakakura E.K., Golub T.R., Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Bishop J.A., Benjamin H., Cholakh H., Chajut A., Clark D.P., Westra W.H. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin. Cancer Res. 2010;16:610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 12.Swanton C., Caldas C. Molecular classification of solid tumours: towards pathway-driven therapeutics. Br. J. Cancer. 2009;100:1517–1522. doi: 10.1038/sj.bjc.6605031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carninci P. Non-coding RNA transcription: turning on neighbours. Nat. Cell Biol. 2008;10:1023–1024. doi: 10.1038/ncb0908-1023. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Song X., Glass C.K., Rosenfeld M.G. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb. Perspect. Biol. 2010 doi: 10.1101/cshperspect.a003756. Epub ahead of print 23 June 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R., Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 22.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammad F., Mondal T., Guseva N., Pandey G.K., Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–2499. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 26.Zappulla D.C., Cech T.R. RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 2006;71:217–224. doi: 10.1101/sqb.2006.71.011. [DOI] [PubMed] [Google Scholar]
- 27.Koziol M.J., Rinn J.L. RNA traffic control of chromatin complexes. Curr. Opin. Genet. Dev. 20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D., Thomas K., Presser A., Bernstein B.E., van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon J.A., Lange C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Albert M., Helin K. Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 21:209–220. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Sang X., Zhao H., Lu X., Mao Y., Miao R., Yang H., Yang Y., Huang J., Zhong S. Prediction and identification of tumor-specific noncoding RNAs from human UniGene. Med. Oncol. 2009;27:894–898. doi: 10.1007/s12032-009-9302-0. [DOI] [PubMed] [Google Scholar]
- 33.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 34.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez D.S., Hoage T.R., Pritchett J.R., Ducharme-Smith A.L., Halling M.L., Ganapathiraju S.C., Streng P.S., Smith D.I. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum. Mol. Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 36.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M., et al. A large intergenic non-coding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum. Mol. Genet. 2007;16(Spec no. 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 38.Cheung H.H., Lee T.L., Davis A.J., Taft D.H., Rennert O.M., Chan W.Y. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br. J. Cancer. 2010;102:419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., Fan L., Koziol M.J., Gnirke A., Nusbaum C., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 42.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap K.L., Li S., Munoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris K.V., Santoso S., Turner A.M., Pastori C., Hawkins P.G. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berx G., van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb. Perspect. Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beltran M., Puig I., Pena C., Garcia J.M., Alvarez A.B., Pena R., Bonilla F., de Herreros A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song X., Wang B., Bromberg M., Hu Z., Konigsberg W., Garen A. Retroviral-mediated transmission of a mouse VL30 RNA to human melanoma cells promotes metastasis in an immunodeficient mouse model. Proc. Natl Acad. Sci. USA. 2002;99:6269–6273. doi: 10.1073/pnas.092112199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G., Cui Y., Zhang G., Garen A., Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc Natl Acad Sci USA. 2009;106:16794–16798. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L., Feng T., Lian Y., Zhang G., Garen A., Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl Acad. Sci. USA. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji P., Diederichs S., Wang W., Boing S., Metzger R., Schneider P.M., Tidow N., Brandt B., Buerger H., Bulk E., et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 51.Lin R., Maeda S., Liu C., Karin M., Edgington T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 52.Coccia E.M., Cicala C., Charlesworth A., Ciccarelli C., Rossi G.B., Philipson L., Sorrentino V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell. Biol. 1992;12:3514–3521. doi: 10.1128/mcb.12.8.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 54.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diehl J.A. Cycling to cancer with cyclin D1. Cancer Biol. Ther. 2002;1:226–231. doi: 10.4161/cbt.72. [DOI] [PubMed] [Google Scholar]
- 56.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Lu Z., Wientjes M.G., Au J.L. Delivery of siRNA Therapeutics: Barriers and Carriers. AAPS J. 2010 doi: 10.1208/s12248-010-9210-4. Epub ahead of print 15 June 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owen R.D., Bortner D.M., Ostrowski M.C. ras oncogene activation of a VL30 transcriptional element is linked to transformation. Mol. Cell. Biol. 1990;10:1–9. doi: 10.1128/mcb.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song X., Sun Y., Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl Acad. Sci. USA. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song X., Sui A., Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc. Natl Acad. Sci. USA. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cunnington M.S., Santibanez Koref M., Mayosi B.M., Burn J., Keavney B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guastadisegni M.C., Lonoce A., Impera L., Albano F., D'Addabbo P., Caruso S., Vasta I., Panagopoulos I., Leszl A., Basso G., et al. Bone marrow ectopic expression of a non-coding RNA in childhood T-cell acute lymphoblastic leukemia with a novel t(2;11)(q11.2;p15.1) translocation. Mol. Cancer. 2008;7:80. doi: 10.1186/1476-4598-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pasmant E., Laurendeau I., Heron D., Vidaud M., Vidaud D., Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 66.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gabory A., Ripoche M.A., Yoshimizu T., Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet. Genome Res. 2006;113:188–193. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 68.Lottin S., Vercoutter-Edouart A.S., Adriaenssens E., Czeszak X., Lemoine J., Roudbaraki M., Coll J., Hondermarck H., Dugimont T., Curgy J.J. Thioredoxin post-transcriptional regulation by H19 provides a new function to mRNA-like non-coding RNA. Oncogene. 2002;21:1625–1631. doi: 10.1038/sj.onc.1205233. [DOI] [PubMed] [Google Scholar]
- 69.Matouk I.J., DeGroot N., Mezan S., Ayesh S., Abu-lail R., Hochberg A., Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sotomaru Y., Katsuzawa Y., Hatada I., Obata Y., Sasaki H., Kono T. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J. Biol. Chem. 2002;277:12474–12478. doi: 10.1074/jbc.M109212200. [DOI] [PubMed] [Google Scholar]
- 71.Tsang W.P., Wong T.W., Cheung A.H., Co C.N., Kwok T.T. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;13:890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooper C., Guo J., Yan Y., Chooniedass-Kothari S., Hube F., Hamedani M.K., Murphy L.C., Myal Y., Leygue E. Increasing the relative expression of endogenous non-coding steroid receptor RNA activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37:4518–4531. doi: 10.1093/nar/gkp441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Colley S.M., Leedman P.J. SRA and its binding partners: an expanding role for RNA-binding coregulators in nuclear receptor-mediated gene regulation. Crit. Rev. Biochem. Mol. Biol. 2009;44:25–33. doi: 10.1080/10409230802661719. [DOI] [PubMed] [Google Scholar]
- 74.Xu B., Yang W.H., Gerin I., Hu C.D., Hammer G.D., Koenig R.J. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol. Cell. Biol. 2009;29:1719–1734. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colley S.M., Iyer K.R., Leedman P.J. The RNA coregulator SRA, its binding proteins and nuclear receptor signaling activity. IUBMB Life. 2008;60:159–164. doi: 10.1002/iub.22. [DOI] [PubMed] [Google Scholar]
- 76.Caretti G., Lei E.P., Sartorelli V. The DEAD-box p68/p72 proteins and the noncoding RNA steroid receptor activator SRA: eclectic regulators of disparate biological functions. Cell Cycle. 2007;6:1172–1176. doi: 10.4161/cc.6.10.4228. [DOI] [PubMed] [Google Scholar]
- 77.Caretti G., Schiltz R.L., Dilworth F.J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F.V., Hoffman E.P., Tapscott S.J., Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 78.Hube F., Guo J., Chooniedass-Kothari S., Cooper C., Hamedani M.K., Dibrov A.A., Blanchard A.A., Wang X., Deng G., Myal Y., et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 79.Lanz R.B., McKenna N.J., Onate S.A., Albrecht U., Wong J., Tsai S.Y., Tsai M.J., O'Malley B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 80.Ifere G.O., Ananaba G.A. Prostate cancer gene expression marker 1 (PCGEM1): a patented prostate- specific non-coding gene and regulator of prostate cancer progression. Recent Pat. DNA Gene Seq. 2009;3:151–163. doi: 10.2174/187221509789318360. [DOI] [PubMed] [Google Scholar]
- 81.Bialkowska-Hobrzanska H., Driman D.K., Fletcher R., Harry V., Razvi H. Expression of human telomerase reverse transcriptase, Survivin, DD3 and PCGEM1 messenger RNA in archival prostate carcinoma tissue. Can. J. Urol. 2006;13:2967–2974. [PubMed] [Google Scholar]
- 82.Fu X., Ravindranath L., Tran N., Petrovics G., Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–141. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 83.Petrovics G., Zhang W., Makarem M., Street J.P., Connelly R., Sun L., Sesterhenn I.A., Srikantan V., Moul J.W., Srivastava S. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–611. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 84.Srikantan V., Zou Z., Petrovics G., Xu L., Augustus M., Davis L., Livezey J.R., Connell T., Sesterhenn I.A., Yoshino K., et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc. Natl Acad. Sci. USA. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang F., Li X., Xie X., Zhao L., Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 86.Fleming J.V., Fontanier N., Harries D.N., Rees W.D. The growth arrest genes gas5, gas6 and CHOP-10 (gadd153) are expressed in the mouse preimplantation embryo. Mol. Reprod. Dev. 1997;48:310–316. doi: 10.1002/(SICI)1098-2795(199711)48:3<310::AID-MRD2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 87.Meier I., Fellini L., Jakovcevski M., Schachner M., Morellini F. Expression of the snoRNA host gene gas5 in the hippocampus is upregulated by age and psychogenic stress and correlates with reduced novelty-induced behavior in C57BL/6 mice. Hippocampus. 2010;20:1027–1036. doi: 10.1002/hipo.20701. [DOI] [PubMed] [Google Scholar]
- 88.Mourtada-Maarabouni M., Hedge V.L., Kirkham L., Farzaneh F., Williams G.T. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J. Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 89.Muller A.J., Chatterjee S., Teresky A., Levine A.J. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine chromosome 1. Mamm. Genome. 1998;9:773–774. doi: 10.1007/s003359900862. [DOI] [PubMed] [Google Scholar]
- 90.Nakamura Y., Takahashi N., Kakegawa E., Yoshida K., Ito Y., Kayano H., Niitsu N., Jinnai I., Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet. Cytogenet. 2008;182:144–149. doi: 10.1016/j.cancergencyto.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 91.Smith C.M., Steitz J.A. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5'-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X., Rice K., Wang Y., Chen W., Zhong Y., Nakayama Y., Zhou Y., Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2009;151:939–947. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Benetatos L., Hatzimichael E., Dasoula A., Dranitsaris G., Tsiara S., Syrrou M., Georgiou I., Bourantas K.L. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk. Res. 2009;34:148–153. doi: 10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 94.Benetatos L., Dasoula A., Hatzimichael E., Georgiou I., Syrrou M., Bourantas K.L. Promoter hypermethylation of the MEG3 (DLK1/MEG3) imprinted gene in multiple myeloma. Clin. Lymphoma Myeloma. 2008;8:171–175. doi: 10.3816/CLM.2008.n.021. [DOI] [PubMed] [Google Scholar]
- 95.Gejman R., Batista D.L., Zhong Y., Zhou Y., Zhang X., Swearingen B., Stratakis C.A., Hedley-Whyte E.T., Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab. 2008;93:4119–4125. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 97.Zhao J., Dahle D., Zhou Y., Zhang X., Klibanski A. Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors. J. Clin. Endocrinol. Metab. 2005;90:2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- 98.Zhang X., Zhou Y., Mehta K.R., Danila D.C., Scolavino S., Johnson S.R., Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab. 2003;88:5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]