Abstract
Surface topography and texture of cell culture substrata can affect the differentiation and growth of adherent cells. The biochemical basis of the transduction of the physical and mechanical signals to cellular responses is not well understood. The lack of a systematic characterization of cell-biomaterial interaction is the major bottleneck. This study demonstrated the use of a novel subcellular fractionation method combined with quantitative MS-based proteomics to enable the robust and high-throughput analysis of proteins at the adherence interface of Madin-Darby canine kidney cells. This method revealed the enrichment of extracellular matrix proteins and membrane and stress fibers proteins at the adherence surface, whereas it shows depletion of extracellular matrix belonging to the cytoplasmic, nucleus, and lateral and apical membranes. The asymmetric distribution of proteins between apical and adherence sides was also profiled. Apart from classical proteins with clear involvement in cell-material interactions, proteins previously not known to be involved in cell attachment were also discovered.
The growth and differentiation of cells in multicellular organisms are regulated by the complex interplay of biochemical and mechanical signals. In the past decades, a plethora of data on the roles of mechanical and structural cues in modulating cellular behaviors has emerged (1–5). It is increasingly evident that cell fates can be changed by engineering the physical properties of the microenvironment to which the cells are exposed (6–8). These observations have inspired the development of functionalized biomaterials that can directly and specifically interact with tissue components, and support or even direct the appropriate cellular activities (9, 10). Although promising progress has been observed in the past few years, several gaps in knowledge in this field have hindered the development of such ”intelligent” biomaterials. In particular, the understanding of the mechanism in which the cell orchestrates physiological and morphological changes by translating mechanical and structural information into biochemical signals is still very limited.
As a standard experimental model, cell lines cultured in vitro as a monolayer over solid substrata are usually used to study the effects of biomaterial surfaces on cellular phenotypes. With this simple model system, ingenious experiments have shown that physical forces applied through the extracellular matrix (ECM)1 can induce changes in cell adhesion molecules and stress-induced ion channels, which then lead to changes in the cytoskeleton and gene expressions (11–13). We term the biochemical structure present at the interface between the substratum and the cellular interior the adherence surface (AS), which is composed of the basal plasma membrane with associated structures such as the ECM on one side and the focal adherence complexes on the other. In monolayer cell culture systems, the AS is the only part of the cells in direct contact with the substratum, and is therefore responsible for the first line of communication between the cells and the biomaterial. It is likely that the AS is the organelle that mediates the communication of mechanical and tectonic signals from the substratum to biochemical transducers in the cells. Given the complexity of this process, it is clear that the understanding of this phenomenon cannot be achieved merely by studying individual biological parts in isolation. It is necessary, therefore, to systematically characterize the biochemical factors that mediate the interactions between cells and materials to yield insights into intracellular signaling processes that are responsible for such cellular responses. Toward this goal, we seek to investigate the biochemical basis of how different biomaterials may impose changes in the composition of the AS of adherent cells.
MS-based proteomics have recently emerged as a standard technique in modern cell biology. Various techniques based on the chemical conjugation of isotopically labeled reporters to proteins or peptides, such as the isobaric tag for relative and absolute quantitation (iTRAQ) and the isotope-coded affinity tags, enable MS-based proteomics to quantify and compare proteome changes between biological samples. As an attractive alternative, stable isotope labeling with amino acids in cell culture (SILAC) is a metabolic labeling technique that enables isotopically encoded cells to be mixed before lysis and fractionation, thus eliminating inherent quantification biases in these steps, and also enables a simpler procedure and more accurate quantitation (14). SILAC MS-based proteomics have recently contributed to organellar proteomes (15, 16), accurate measurement of protein-protein interactions (17), and the characterization of proteome dynamics during cell differentiation (18). The use of MS-based proteomics has enabled the systematic evaluation of proteome changes on the adhesion of cells to substrata of interest. Kantawong et al. (19) applied DIGE and LC-MS/MS to identify proteome changes in cells on surface with nanotopography. Xu et al. (20) investigated proteome differences of human osteoblasts on various nano-sized hydroxyapatite powders with different shapes and chemical compositions using iTRAQ-based two-dimensional LC-MS/MS.
One advantage of proteomics is that it can effectively be combined with subcellular fractionation and allow the comprehensive characterization of the proteins enriched in targeted cellular structures. To yield new insight in molecular interactions in cell-biomaterial interfaces, we aimed to develop a robust protocol for the proteomic characterization of the AS of adherent cells on a biomaterial surface and use it for discovering new cell-biomaterial interface specific biomarkers. Our approach was to develop an isolation technique for AS with high yields and purity for proteomic analysis. The isolated AS on substratum was analyzed by confocal microscopy and Western blotting. SILAC was then used to characterize the fold-enrichment of proteins in the purified AS compared with whole cells and to discover new biomolecules in the cell-biomaterial interface. This study introduces a novel cell-biomaterial interface proteomic procedure, which can be used to identify the AS specific proteome in a high throughput manner and provide a simple and robust method to systematically analyze cell-biomaterial interactions at a molecular level.
EXPERIMENTAL PROCEDURES
Cell Culture
Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection (ATCC number CCL-34) and were cultured in Dulbecco's modified eagle medium (DMEM) high glucose (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), streptomycin (100 μg/mL) (Invitrogen) and penicillin (100 IU/mL) (Invitrogen) at 37 °C and 5% CO2 in air in a humidified incubator. For biochemical experiments, cells were grown on 60-mm tissue culture polystyrene culture dishes (Falcon). For microscopy MDCK cells were grown on glass coverslips (Corning, 2865-18) in six well dishes.
SILAC Labeling of MDCK Cells and Adherence Surface Isolation
Before making lysates, cells were metabolically labeled by culturing for five rounds of cell division in SILAC DMEM medium supplemented with 10% dialyzed fetal calf serum (Biowest, Nuaillé, France) to ensure that all the cellular proteins were labeled until saturation (21). All isotopically labeled amino acids were from Cambridge Isotope Lab (Andover, MA). Cells used for AS isolation were labeled with DMEM containing l-13c6-arginine (r6) and l-4,4,5,5-D4-lysine (K4), whereas cells for making whole cell lysate were labeled cultured in DMEM containing l-arginine (r0) and l-lysine (K0), as shown in Figure 1A.
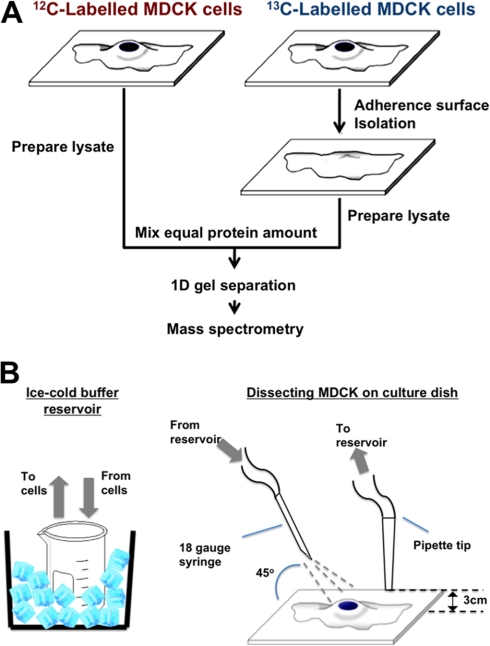
Fig. 1.
Experiment setup and s1‘chematic diagram of machinery used to isolate adherence surface (AS) at cell-biomaterials interface. A, a schematic flow chart of SILAC gel-based proteomics for distinguishing whole cells and AS. B, demonstrates machinery set-up for AS isolation from cells on substrate.
The AS isolation procedure was based on the lysis squirting technique first introduced by Nermut et al. (1989) (22) and a similar technique used by Gates et al. (1993) (23). The method used in this study was modified as follows to increase the yield and reproducibility of isolation and purity. R6K4 labeled cells were seeded on 60-mm noncoated tissue culture dishes with an initial cell density of 1.5 × 103 and cultured for three days to confluence. The cell monolayer was rinsed three times with cold PBS and once with cold hypotonic lysis buffer (2.5 mmol/L Tris-HCl pH 7.4, 1 mmol/L MgCl2, 0.5 mmol/L CaCl2 in Milli-Q water (Millipore, Billerica, MA), and was incubated at 4 °C in this buffer for three minutes. The cell monolayer was then sheared by a pressurized buffer jet at an angle under the equipment setup as shown in Figure 1B, with constant monitoring under a phase contrast microscope (Leica, Microsystems GmbH, Wetzlar, Germany) to ensure complete removal of the apical cell compartments. The pump output was optimized to shear and remove apical cell compartments while leaving the AS on the substratum. The isolated AS on the substratum surface was rinsed with cold 0.5% Triton X-100 in PBS and then three times with cold PBS. For proteomics analysis, the AS retained on the substratum surface was directly harvested in 8-mol/L urea using a cell scraper.
For whole cell lysate, SILAC R0K0-labeled cells was cultured as same as described in preparing AS fractions. Cell monolayer was also subjected to procedures described above (except apical disruption) to ensure fair comparison. Intact cell monolayer was directly lysed and harvested in 8-mol/L urea using a cell scraper.
LC-MS/MS Analysis
The protein concentration of the lysates were determined by the Bradford assay (Pierce). AS fractions and whole cell lysates were mixed at an 1:1 protein amount, followed by concentration with an ultrafiltration cutoff of 10 kDa (Amicon, Millipore Corporation, Billerica, MA). The combined sample was incubated in 10 mmol/L dithithreitol (GE Healthcare) in 8-mol/L urea (USB Corporation, Cleveland, OH) at room temperature for 40 min, followed by incubation in 55 mmol/L iodoacetamide (GE Healthcare) in 8-mol/L urea at room temperature in the dark for another 40 min. The sample was then mixed with SDS loading buffer [0.5 mol/L Tris Cl pH 6,8, 10% glycerol, 2% (w/v) SDS, 0.01% (w/v) bromphenol blue, 5% (w/v) dithiothreitol] and incubated for 10 min at room temperature. Proteins were separated by electrophoresis on a 10% Novex precast gel (Invitrogen). The gel was then fixed and stained with colloidal Coomassie Blue (Invitrogen) according to the manufacturer's instruction. The gel lane was excised into 15 slices. Each gel slice was cut into 1-mm3 pieces, which were subsequently treated with destaining solution (Sigma) according to the manufacturer's instructions. This was followed by incubation in 50 mmol/L ammonium bicarbonate in Milli-Q water (Millipore, Billerica, MA) containing proteomic grade Trypsin (Sigma) (50 ng/gel slice) overnight at 37 °C. The resulting peptides were extracted from each gel slice as described previously (24), separated by HPLC (Dionex, Sunnyvale, CA) on a commercial C18 reverse phase column (inner diameter 75 μm, 5 μm Acclaim pepMap100 medium; Dionex) over an 80-minute gradient (Mobile phase A: 0.1% fluoroacetic acid in 2% ACN in Milli-Q water; mobile phase B: 0.1% fluoroacetic acid in 98% ACN) and then analyzed by a micrOTOF-Q II ESI-Qq-TOF mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany).
MASCOT Search and Statistical Analysis
Peak list was generated using Data Analysis, Version 4.0 (Bruker Daltonics). The MS data were searched using the National Center for Biotechnology Information non-redundant protein database for Canis familiaris (01-09-2005 release, 33,651 sequences searched), using the MASCOT search engine, version 2.2 (Matrix Science). Fixed modification was Carbamidomethyl (C) and variable modifications used were oxidation (M), as well as the appropriate SILAC modifications. Trypsin specificity was used, allowing for two missed cleavages, and a mass tolerance of 0.5 Da was used for MS precursors and 0.5 Da for fragment ions. Peptide charges of +2 and +3 were selected. Individual ions with Mascot scores higher than 20 were used (a threshold commonly used for confident protein identification from tandem MS data) (25). Only rank 1 peptides were considered, thus removing duplicate homologous proteins from the results. Under these conditions, the estimated false positive rate is less than 5%, according to a previous analysis (26). SILAC quantitation was performed using the WarpLC software (Bruker Daltonics), which measures the averaged MS peak heights of the isotopic pairs. Proteins with heavy/light isotopic ratios less than 0.01 were discarded as they mostly represented environmental contaminants. Proteins with identification based on only one peptide were also excluded. Only proteins with an S.D. (the variations among the measured peptide ratios for the same protein) of less than 70% of the averaged ratios were accepted for further analysis. Proteins were clustered at 98% sequence similarity, and functional classification of the identified proteins was performed using Proteincenter (Proxeon, Denmark).
Western Blotting
AS and whole cell fractions were harvested and quantified as described above. Proteins were mixed with SDS loading buffer, followed by separation with 10% Novex precast gel (Invitrogen) at 100 V, and transferred to nitrocellulose membrane. The membrane was blocked with 5% (w/v) nonfat milk powder (Nestle) and 0.05% Tween-20 in PBS for 60 min, followed by washing with 0.05% Tween-20 in PBS and incubating with primary antibody in blocking buffer for two hours. Primary antibodies used were anti-fibronectin (Santa Cruz Biotechnology, Inc., CA; sc-9068), anti-transferrin receptor (Santa Cruz, sc-48747), anti-Cadherin (Santa Cruz, sc-10733), anti-tubulin (Santa Cruz, sc-5546), anti-nucleophosmin (Santa Cruz, sc-5564) and anti–nuclear DNA helicase II (NDH II) (Santa Cruz, sc-66997). Secondary antibodies used were horseradish peroxidase-conjugated anti-mouse immunoglobulin G (GE Healthcare) and anti-rabbit immunoglobulin G (GE Healthcare). The membrane was washed and incubated with secondary antibody for 60 min. After washing, the membrane was developed with Amersham Biosciences ECL Plus™ detection kit (GE Healthcare) and was imaged using a Gel Doc with a cool CCD camera (Fujifilm LAS-3000, Fujifilm Corporation, Tokyo, Japan). Image analysis was performed using ImageJ (version 1.42, National Institutes of Health).
Confocal Microscopy
To visualize structures retained on the AS after isolation, AS isolation was performed on cells grown on coverslips. Both AS on substrate and whole cells were fixed for five minutes with 4% freshly prepared paraformaldehyde in PBS at room temperature, washed once with PBS before permeablization in 1% Triton X-100 in PBS for five minutes at room temperature. After permeablization, the cells or AS were washed briefly in PBS and blocked with 50 mmol/L glycine in PBS for 30 min at room temperature. The cells were then incubated with primary antibodies for 60 minutes, washed, and incubated with fluorochrome-conjugated secondary antibodies. For imaging involving organelle-specific fluorescent dyes, live cells were stained with dyes diluted in DMEM at 37 °C, without fixation and permeabilization.
Primary antibodies used were anti-Talin (Santa Cruz, sc-15336) and anti-fibronectin (Santa Cruz, sc-9068). Secondary antibodies used were anti-rabbit-TRITC and anti-rabbit-FITC. Phalloidin-TRITC (Millipore, 90228), lipophilic tracers (Invitrogen, D282), MitoTracker (Invitrogen, M7512) and Hoechst (Sigma) was used to visualize the actin, plasma membrane, mitochondria, and DNA, respectively. The preparations were mounted with fluorescent mounting medium (Dako Denmark A/S Glostrup, Denmark), and examined under a confocal microscope (Leica TCS-SPE).
Atomic Force Microscopy
To reveal the morphology of the AS attaching on the material, cells were grown to 80% confluence and the AS attached to tissue culture polystyrene dish was prepared as previously described, and was then examined under PBS. For comparison, live whole cell were examined in CO2-independent medium (Invitrogen) at room temperature. AFM images were captured with a Veeco BioScope II scanning probe microscope equipped with Silicon Nitride Probes (DNP-10, Veeco Instruments, Inc., Plainview, NY) in fluid contact mode. Images were analyzed by the NanoScopeTM (Veeco).
RESULTS
AS Purification on Substratum
To achieve the purification of AS attached to the biomaterials of interest for subsequent proteomic analysis, several cell disruption procedures were tested. Our initial studies indicated that wet cleaving with nitrocellulose membrane (27) produced a large variability in AS yield (data not shown). The yield was highly sensitive to the wetness of the nitrocellulose membrane, a factor that could not be controlled easily. For the sonication method (28), it was found that cell tended to be either not disrupted or completely detached from the substratum. Lysis squirting methods (29, 30) demonstrated more reproducible and high yields in purifying the AS, however, most of them required the use of pre-coating silica colloidal on the apical side of adhered cells to increase efficiency. This is an extensive experimental perturbation that would likely have interfered with the native cell-biomaterial interaction. In this study, this method was modified to fulfill stringent requirements for cell-biomaterial interface proteomic study.
The modified AS purification method in the current study is shown in Figure 1B. In general, a dual channel peristaltic pump was set to circulate cold lysis buffer between the ice-cooled buffer reservoir and the cell monolayer. Cold lysis buffer was compressed through a 23-gauge syringe, which was clamped into place at a 45° angle to the monolayer, to shear away the apical compartment of cells beneath (Fig. 1). A pipette tip adjacent to the shearing site was kept elevated 3 mm above the cell layer to circulate buffer back to the reservoir, while maintaining the buffer level to just covering the cell layer. The culture dish was moved carefully under the syringe so as to purify all the AS from the surface. The buffer jet pressure, syringe size were optimized so that no precoating was needed.
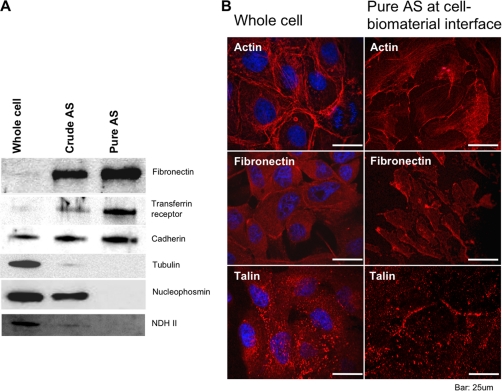
To quantify the purification of AS proteins, we performed immunoblotting using antibodies against proteins with known intracellular distributions. The ECM component fibronectin and the membrane protein transferrin receptor were found to be enriched in the AS when isolated by the standard buffer squirting method (“Crude AS”, Fig 2A), indicating that our protocol could effectively isolate the ECM and AS of MDCK cells. Further characterization of the purity of the crude AS, however, indicated that nuclear proteins nucleophosmin and NDH II, and cytoplasmic protein tubulin were still detectable (Fig 2A). To eliminate the contamination from cytosolic proteins, we introduced an additional step in which the isolated AS (also known as crude AS) was rinsed with 0.5% Triton X-100 (pure AS). Figure 2A suggested that fibronectin was highly enriched in pure AS (33-fold, as judged by densitometry) as compared with the whole cell sample. Transferin receptor protein was highly enriched in pure AS (16-fold), whereas non-membrane markers (Tubulin, nucleophosmin, and NDH II) were undetectable. Interestingly, cadherin is slightly enriched in crude AS and pure AS (2-fold). To further investigate the localization of proteins on pure AS, immunofluorescence (IF) was performed. The IF micrograph shows that actin stress fiber, fibronectin, and talin remained on the AS after isolation. In contrast, Hoechst DNA staining was not detectable on pure AS.
Fig. 2.
Western blottings and immunofluorescence of whole cell and isolated AS for quantifying AS purity. A, shows Immunoblottings on whole cell, crude AS and pure AS fractions against fibronectin, transferrin receptor, cadherin, tubulin,, nucleophosmin and nuclear DNA helicase II. This demonstrates the enrichment and depletion of ECM, membrane, cytoskeleton and nuclear proteins in AS fractions. B, shows immunofluorescence images probing against actin stress fiber, fibronectin and talin, representing cytoskeleton, ECM and focal adhesion, respectively. Whole cell (left) and pure AS (right) can be easily differentiated.
Morphology of the Isolated AS
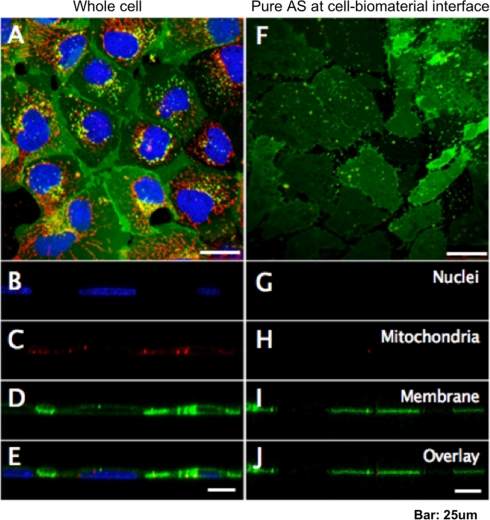
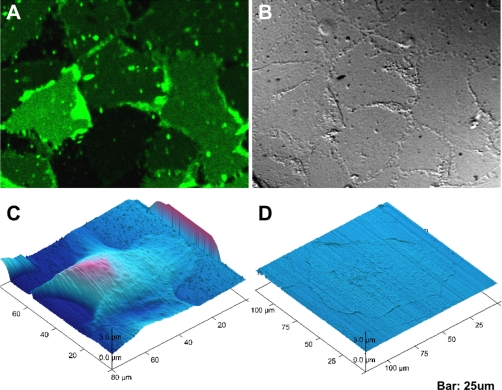
To further visualize the quality of AS isolation, both intact cells and AS were stained with Hoechst, lipophilic Tracers, and MitoTracker, which are live-cell-compatible dyes specific for the cell nucleus, plasma membrane, and mitochondria, respectively. Confocal microscopy of whole cells (Fig. 3A) indicated positive staining of all probes. Transverse XZ sections (Fig. 3B to 3E) demonstrated that nuclei and mitochondria were enclosed by the plasma membrane. Isolated AS showed only positive plasma membrane staining (Fig. 3F). Remarkably, the transverse confocal sections (Fig. 3G to 3J) showed only thin layers of isolated AS on the substratum, whereas the nuclei and mitochondria were undetectable. The size of the lipophilic tracer-labeled structures was comparable to the size of intact cells, suggesting these structures represented intact pieces of AS and not cell debris. Figure 4A, B shows the differential interference contrast and fluorescence images of the pure AS, in which the AS appeared as a lawn of membranes of similar size, indicating that the isolation procedure yielded a nearly homogenous preparation of AS. To inspect the morphology of the isolated AS at a higher resolution, we performed atomic force microscopy (AFM) on unfixed adherent cells and AS, using an AFM system that could operate on wet biological samples. AFM images (Fig. 4C, D) indicated that the height of an intact MDCK cell was approximately 5 μm, consistent with the size of a typical adherent mammalian cell, while the AS was only less than 1 μm thick. The isolated AS was still completely attached to the substratum, but was devoid of any discernible intracellular structures, suggesting a vast majority of structures that were associated with apical portion of the cell had been removed, and only structures in close contact with the biomaterial remained.
Fig. 3.
Morphology of isolated AS comparing to whole cell visualized with confocol microscopy. Fluorescence confocol micrographs on MDCK whole cells (A) and adherence surface (F). Nucleous, mitochondria and plasma membrane are shown as blue, red and green, respectively. Horizontal sections of (A) are shown in (B–E), and similarly (G–J) are correspond to (F).
Fig. 4.
Morphology of isolated AS comparing to whole cell visualized by AFM and bright field (DIC). Fluorescence image of AS on substratum (A) is compared side by side with its differential interference contrast (DIC) image (B). AFM micrographs (C) shows the contour of whole cells while (D) shows contour of pure AS.
Proteomic Analysis of the Isolated AS at Cell-Biomaterial Interface
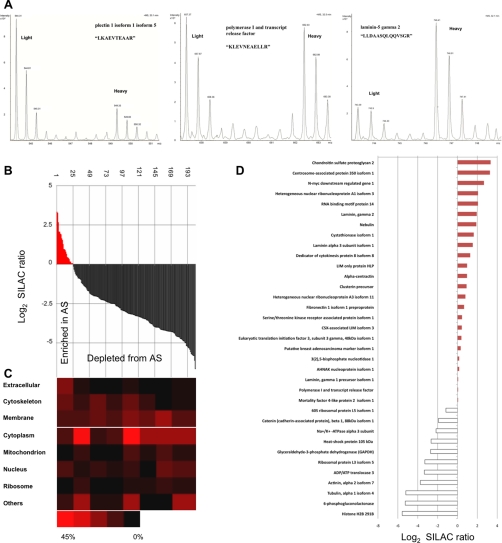
After establishing a robust protocol for AS isolation, we proceeded to characterize the proteins enriched in the AS preparation by LC/MS/MS proteomics with the aid of SILAC. Proteins from the AS isolated from SILAC “heavy” R6 K4–labeled cells were mixed with an equal amount of whole cell lysate prepared from SILAC “light” R0 K0–labeled cells. Mixed lysate was separated by one-dimensional gel, in-gel digested, and analyzed with LC-MS/MS. There were a total 3,478 unique peptides identified (supplemental Table 1). The intensity ratio between each heavy isotope-labeled peptide and its light isotope counterpart could be measured. Figure 5A shows examples of peptides with heavy/light (H/L) isotopic ratios of less than 1, close to 1, and more than 1. Protein average SILAC ratio (H/L) was calculated by averaging its associating peptide ratios. For each protein, the average SILAC ratio therefore indicated the extent of its enrichment in the isolated AS preparation. Among the 204 proteins with reliable quantitation (supplemental Table 1), 24 (12%) had SILAC ratios of more than 1, i.e. they were enriched in AS. The remaining 180 (88%) proteins with ratios <1 were considered depleted from AS. These 204 proteins were classified according to their Gene Ontology (GO) “cellular component” annotations (Fig. 5B,C). This classification shows that the proportion of AS-associated proteins (ECMs, membrane, cytoskeletons) is highly enriched for proteins with a ratio of more than 1 and decreased for those with a ratio of less than 1. The case is reversed for non-AS associated proteins such as those from nucleus, ribosome, mitochondrion and cytoplasm. This finding is consistent with the immunoblotting and IF data in which the isolated AS retained cytoskeletons, ECMs, and membrane proteins, whereas most of intracellular organelles were removed.
Fig. 5.
Proteomics analysis of AS proteins isolated from cell-biomaterial interface. A, shows SILAC pairs of three peptides in MS spectrums, which belong to plectin isoform 1 isoform 5, polymerase I and transcript release factor and laminin-5 gramma 2, representing “Depleted from AS” (left), “Equally distributed within cell” (Middle) and “Enriched in AS” (right). B, shows normalized H/L ratio of all identified & quantified proteins (204) in descending order, AS enriched proteins (normalized ratio > 0) are in red (24), AS depleted are in black (180). Heat map (C) is complemented to (B), showing proteins subcellular origin, according to GO, at different normalized ratio. D, shows noteworthy proteins and its normalized ratio.
Among the proteins enriched in the AS at cell-biomaterial interface were some of the classic ECM proteins, e.g. laminin, and focal adhesion components, e.g. fibronectin (Fig. 5D). Proteins known to regulate cell adhesion or motility were also identified. For example, the chondroitin sulfate proteoglycan, nebulin, was found to be enriched 10 and 3.7 times in the AS preparation, respectively. In contrast, catenin delta-1 (CTNND1), Na+/K+ ATPase 2 (ATP1A2) were all depleted from the AS. Apart from proteins with clear involvement in adhesion and cell-biomaterial interaction that were found at cell-biomaterial interface, centrosome-associated protein 350, heterogeneous nuclear ribonucleoprotein A1, A3, and RNA binding motif protein 14, which were not confirmed its role at cell-biomaterials interaction, were also found enriched in the AS at the cell-biomaterials interface.
DISCUSSION
The objective of this study was to develop a protocol that allows for the proteomic characterization of the adherence surface, together with the associated ECM, of mammalian cells adhered to a biomaterial (substratum) in monolayer cell culture. We believe that the methodology developed in this work can offer a simple and convenient technique for the biochemical investigation of the interface between cells and biomaterials. Although we developed this techniques based on a well-characterized mammalian cell line MDCK cultured on a standard plastic surface, this protocol can readily be adopted on a wide range of cell lines on different substrata.
Several methods have been described for the isolation of the AS of mammalian cells using techniques such as wet-cleaving (22, 27), sonication (28), controlled lysis squirting (22, 31–33), isolation on poly(l-lysine)-coated beads (34, 35), and sandwiching (22, 36). Most of these protocols, however, were developed for the microscopy studies of the inner face of the plasma membrane, and the yields of the isolated AS tended to be low and variable. We have now modified one of these approaches to achieve a large-scale isolation of AS with a reproducible yield and purity.
The Western blot results suggested that the rinsing step after the AS isolation procedure with detergent (0.5% Triton X-100 in PBS) significantly reduced the contamination with non-membrane associated proteins. The contamination in the crude AS was likely caused by protein-protein associations between AS proteins and intracellular proteins. The detergent rinsing decreased such binding and reduced contamination. Such improvement successfully increased the enrichment of ECM and membrane markers. The 33-fold enrichment of fibronectin in pure AS was considerably higher than a similar attempt by Gates et al. which only yielded a 13-fold enrichment, which implies we obtained a more specific isolation of AS proteins making them easier to be detected by MS. Interestingly, the enrichment of cadherin, which is involved in tight junction formation with neighboring cells, was only 2-fold. A possible reason for this is that the cells in the monolayer were sheared close to basal fraction, during which the tight junction in the lateral fraction was removed along with the apical fraction, resulting in only a small enrichment of cadherin being detected. The IF micrographs also showed that focal adhesion complex was retained on pure AS, which was represented by the positive staining of talin. Furthermore, actin stress fiber, which associated with focal adhesion complex, was also retained on pure AS. This suggests that the AS isolation enriched not only ECMs and AS proteins, but also the associated cytoskeletons. ECMs, AS proteins, and cytoskeletons are all active players in cell-biomaterial interactions, and evidence indicates that our method purified AS proteins on the substratum.
Our fluorescence and AFM images suggested that the isolation procedure removed most of the cytosol components and apical membranes of attached cells, and retained intact AS attached to the substrate. AS patches under bright-field images were similar to those reported in previous literatures (23, 27, 29, 33). By comparison of those published protocols, our method allowed a greater yield in AS proteins preparation, had less experimental perturbations, and was more efficient. Thus, this enables extensive proteomics study of interface between cells and materials surfaces.
In our SILAC LC/MS/MS experiment, a total 204 proteins were quantified with confidence (supplemental Table 1). The SILAC ratio indicated that 24 of these highly expressed at the AS at the cell-biomaterial interface, whereas the remainder were depleted from the interface. Some of the classic ECM and focal adhesion components, such as fibronectin and laminin (Fig. 5E), were among these proteins enriched in the isolated AS at the cell-biomaterial interface While several of these AS markers were also detected through IF and Western blotting, the AS proteins enrichment protocol was cross-validated. Remarkably, we have identified all three chains of laminin in the isolated AS, indicating that this protocol provides a fast and convenient system for isolating and studying the ECM. Other proteins known to function in the regulation of cell adhesion or motility were also identified. For example, chondroitin sulfate proteoglycan was found to be enriched 10 times in AS preparation. This protein is a hyaluronic acid -binding protein and interacts with fibronectin, which plays a role in intercellular signaling and in connecting cells with the extracellular matrix (37). Nebulin (3.7 times enrichment) is involved in maintaining the structural integrity of sarcomeres and the membrane system associated with the myofibrils, and plays a role in regulating actin filament length (38, 39). Filamin-binding LIM protein (1.4 times) serves as an anchoring site for cell-ECM adhesion proteins and filamin-containing actin filaments. It plays a role in cell spreading and motility, and participates in the regulation of filamin-mediated cross-linking and stabilization and assembly of actin filaments (40, 41). The enrichment of membrane protein is inferior to the enrichment of ECMs and cytoskeleton proteins, the result was expected as the AS fraction should only constitute less than a half of the total plasma membrane (ventral, lateral, and apical). The use of Triton X-100 washing step, although necessary to eliminate some intracellular contaminants, may lead to the sacrifice of some membrane proteins. Taken together, our AS proteomic study has revealed a list of proteins with the expected identity signature of that of the biological interface. Our experiment also indicated certain membrane proteins preferentially located away from AS. Catenin delta-1 (CTNND1), Na+/K+ ATPase 3 (ATP1A3) were all depleted from the AS. It is possible these proteins are expressed more on the apical membrane instead. For example, Na+/K+ ATPase tends to accumulate at apical side (in contact with medium) to regulate cellular ion concentration (42). This gives evidence of asymmetric organization of membrane proteins in cells adhered on surface. This also validated the AS proteins enrichment protocol.
The isolated AS proteome contains many proteins previously not known to be involved in cell adhesion or cell-biomaterial interaction. For example, we found that centrosome-associated protein 350 was highly enriched in the isolated AS. Centrosome-associated protein 350 is a crucial protein that is involved in mitosis by playing an essential role in centriole growth and in anchoring microtubules to the centrosomes maintaining the integrity of the microtubule network. Its enrichment in the AS suggests a previously unknown role in the regulation of cell proliferation on biomaterial surface. Moreover, a specific subset of RNA-binding proteins such as heterogeneous nuclear ribonucleoprotein A1, A3, and RNA binding motif protein 14 were found enriched in AS. This is consistent with the recent discovery of the association of RNA-binding proteins with the “spreading initiation centers,” a novel structure important for the attachment of cells to the substratum (43). This study has shed new light on previously unavailable information on the cell-biomaterial interface. This study also determined that by simple adjustments of buffer jet pressure, syringe size, and confluence of cells, the AS of various adherent cell lines (e.g. MC3T3-E1, pre-osteoblast) on different substrata could be purified (data not shown). This implies that our novel protocol is versatile and can be extensively applied to the detailed investigation of cell-biomaterial molecular interactions. Thus, it should benefit the development of functionalized biomaterials.
CONCLUSION
This study demonstrated that the AS isolation successfully enriched low abundance AS proteins of cells adhered on a solid substrate and suppressed the detection of non-related protein from other cellular compartments. Our study also suggested that the AS of various adherent cell lines cultured on different substrata could be purified by simple adjustments of buffer jet pressure, syringe size, and confluence of cells. We believe this AS enrichment procedure can unlock a way for a high throughput and system-oriented identification and quantification of low abundance AS proteins on substrates, thereby enabling new opportunities for studying cell-biomaterial interactions. This breakthrough allows a more systematic description of the cell-biomaterial interface at a molecular level. In addition, it enables the analysis of proteome differences at the interface between cells and different engineered biomaterial surfaces.
Supplementary Material
Footnotes
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ECM
- extracellular matrix
- AS
- adherence surface
- MDCK
- Madin-Darby canine kidney
- iTRAQ
- isobaric tag for relative and absolute quantitation
- SILAC
- stable isotope labeling with amino acids in cell culture
- DMEM
- Dulbecco's modified Eagle's medium
- R6
- l-13c6-arginine
- k4
- l-4,4,5-D4-lysine
- R0
- l-arginine
- k0
- l-lysine
- NDH II
- nuclear DNA helicase II
- IF
- immunofluorescence
- AFM
- atomic force microscopy.
REFERENCES
- 1.Klein E. A., Yin L., Kothapalli D., Castagnino P., Byfield F. J., Xu T., Levental I., Hawthorne E., Janmey P. A., Assoian R. K. (2009) Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr. Biol. 19, 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito T., Hayashi H., Kameyama T., Hishida M., Nagai K., Teraoka K., Kato K. (2010) Suppressed proliferation of mouse osteoblast-like cells by a rough-surfaced substrate leads to low differentiation and mineralization. Materials Sci. Engineer. C 30, 1–7 [Google Scholar]
- 3.Bigerelle M., Anselme K., NoÎl B., Ruderman I., Hardouin P., Iost A. (2002) Improvement in the morphology of Ti-based surfaces: a new process to increase in vitro human osteoblast response. Biomaterials 23, 1563–1577 [DOI] [PubMed] [Google Scholar]
- 4.Mendonca G., Mendonca D. B., Simoes L. G., Araujo A. L., Leite E. R., Duarte W. R., Aragao F. J., Cooper L. F. (2009) The effects of implant surface nanoscale features on osteoblast-specific gene expression. Biomaterials 30, 4053–4062 [DOI] [PubMed] [Google Scholar]
- 5.Lam M. T., Huang Y.-C., Birla R. K., Takayama S. (2009) Microfeature guided skeletal muscle tissue engineering for highly organized 3-dimensional free-standing constructs. Biomaterials 30, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 6.Lutolf M. P., Gilbert P. M., Blau H. M. (2009) Designing materials to direct stem-cell fate. Nature 462, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan B. P., Hui T. Y., Yeung C. W., Li J., Mo I., Chan G. C. F. (2007) Self-assembled collagen-human mesenchymal stem cell microspheres for regenerative medicine. Biomaterials 28, 4652–4666 [DOI] [PubMed] [Google Scholar]
- 8.Liao S., Chan C. K., Ramakrishna S. (2008) Stem cells and biomimetic materials strategies for tissue engineering. Materials Sci. Engineer. C 28, 1189–1202 [Google Scholar]
- 9.Williams D. F. (2008) On the mechanisms of biocompatibility. Biomaterials 29, 2941–2953 [DOI] [PubMed] [Google Scholar]
- 10.Williams D. F. (2009) On the nature of biomaterials. Biomaterials 30, 5897–5909 [DOI] [PubMed] [Google Scholar]
- 11.Ingber D. E.Tensegrity-based mechanosensing from macro to micro. Prog. Biophysics Mol. Biol. 97, 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamenovic D., Wang N., Ingber D. E., Michael R. K.Cellular tensegrity models and cell-substrate interactions. In: King M. R. (ed). Principles of Cellular Engineering. Burlington, MA: Academic Press; 2006: 81–101 [Google Scholar]
- 13.Chen C. S., Alonso J. L., Ostuni E., Whitesides G. M., Ingber D. E. (2003) Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 307, 355–361 [DOI] [PubMed] [Google Scholar]
- 14.Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 15.Lam Y. W., Lamond A. I., Mann M., Andersen J. S. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam Y. W., Evans V. C., Heesom K. J., Lamond A. I., Matthews D. A. (2010) Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell. Proteomics 9, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vermeulen M., Selbach M. (2009) Quantitative proteomics: a tool to assess cell differentiation. Curr. Opin. Cell Biol. 21, 761–766 [DOI] [PubMed] [Google Scholar]
- 19.Kantawong F., Burchmore R., Wilkinson C. D. W., Oreffo R. O. C., Dalby M. J. (2009) Differential in-gel electrophoresis (DIGE) analysis of human bone marrow osteoprogenitor cell contact guidance. Acta Biomaterialia 5, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 20.Xu J. L., Khor K. A., Sui J. J., Zhang J. H., Chen W. N. (2009) Protein expression profiles in osteoblasts in response to differentially shaped hydroxyapatite nanoparticles. Biomaterials 30, 5385–5391 [DOI] [PubMed] [Google Scholar]
- 21.Ong Se, Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 22.Nermut M. V. (1989) Strategy and tactics in electron microscopy of cell surfaces. Electron. Microsc. Rev. 2, 171–196 [DOI] [PubMed] [Google Scholar]
- 23.Gates R. E., Hanks S. K., King L. E., Jr. (1993) Focal-adhesion components are enriched in ventral membranes isolated from transformed keratinocytes in culture. Biochem. J. 289 (Pt 1), 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 25.Bindschedler L. V., Palmblad M., Cramer R. (2008) Hydroponic isotope labelling of entire plants (HILEP) for quantitative plant proteomics; an oxidative stress case study. Phytochemistry 69, 1962–1972 [DOI] [PubMed] [Google Scholar]
- 26.Rudnick P. A., Wang Y., Evans E., Lee C. S., Balgley B. M. (2005) Large scale analysis of MASCOT results using a mass accuracy-based threshold (MATH) effectively improves data interpretation. J. Proteome Res. 4, 1353–1360 [DOI] [PubMed] [Google Scholar]
- 27.Feltkamp C. A., Pijnenburg M. A., Roos E. (1991) Organization of talin and vinculin in adhesion plaques of wet-cleaved chicken embryo fibroblasts. J. Cell Sci. 100 (Pt 3), 579–587 [DOI] [PubMed] [Google Scholar]
- 28.Drees F., Reilein A., Nelson W. J. (2005) Cell-adhesion assays: fabrication of an E-cadherin substratum and isolation of lateral and Basal membrane patches. Methods Mol. Biol. 294, 303–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaney L. K., Jacobson B. S. (1983) Coating cells with colloidal silica for high yield isolation of plasma membrane sheets and identification of transmembrane proteins. J. Biol. Chem. 258, 10062–10072 [PubMed] [Google Scholar]
- 30.Goode R. J., Simpson R. J. (2009) Purification of basolateral integral membrane proteins by cationic colloidal silica-based apical membrane subtraction. Methods Mol. Biol. 528, 177–187 [DOI] [PubMed] [Google Scholar]
- 31.Lang R. D., Nermut M. V., Williams L. D. (1981) Ultrastructure of sheep erythrocyte plasma membranes and cytoskeletons bound to solid supports. J. Cell Sci. 49, 383–399 [DOI] [PubMed] [Google Scholar]
- 32.Nicol A., Nermut M. V., Doeinck A., Robenek H., Wiegand C., Jockusch B. M. (1987) Labeling of structural elements at the ventral plasma membrane of fibroblasts with the immunogold technique. J. Histochem. Cytochem. 35, 499–506 [DOI] [PubMed] [Google Scholar]
- 33.Cattelino A., Albertinazzi C., Bossi M., Critchley D. R., de Curtis I. (1999) A cell-free system to study regulation of focal adhesions and of the connected actin cytoskeleton. Mol. Biol. Cell 10, 373–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen C. M., Kalish D. I., Jacobson B. S., Branton D. (1977) Membrane isolation on polylysine-coated beads. Plasma membrane from HeLa cells. J. Cell Biol. 75, 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobson B. S., Branton D. (1977) Plasma membrane: rapid isolation and exposure of the cytoplasmic surface by use of positively charged beads. Science 195, 302–304 [DOI] [PubMed] [Google Scholar]
- 36.Aggeler J., Werb Z. (1982) Initial events during phagocytosis by macrophages viewed from outside and inside the cell: membrane-particle interactions and clathrin. J. Cell Biol. 94, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmermann D. R., Ruoslahti E. (1989) Multiple domains of the large fibroblast proteoglycan, versican. EMBO J. 8, 2975–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labeit S., Kolmerer B. (1995) The complete primary structure of human nebulin and its correlation to muscle structure. J. Mol. Biol. 248, 308–315 [DOI] [PubMed] [Google Scholar]
- 39.Ma K., Wang K. (2002) Interaction of nebulin SH3 domain with titin PEVK and myopalladin: implications for the signaling and assembly role of titin and nebulin. FEBS Lett. 532, 273–278 [DOI] [PubMed] [Google Scholar]
- 40.Tu Y., Wu S., Shi X., Chen K., Wu C. (2003) Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell 113, 37–47 [DOI] [PubMed] [Google Scholar]
- 41.Takafuta T., Saeki M., Fujimoto T., Fujimura K., Shapiro S. (2003) A new member of the LIM protein family binds to filamin B and localizes at stress fibers. J. Biol. Chem. 278, 12175–12181 [DOI] [PubMed] [Google Scholar]
- 42.Jaitovich A. A., Bertorello A. M. (2006) Na+, K+-ATPase: An Indispensable IOCn Pumping-Signaling Mechanism Across Mammalian Cell Membranes, Seminars in Nephrology 26, 386–392 [DOI] [PubMed] [Google Scholar]
- 43.de Hoog C. L., Foster L. J., Mann M. (2004) RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117, 649–662 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.