Abstract
Transnitrosylation and denitrosylation are emerging as key post-translational modification events in regulating both normal physiology and a wide spectrum of human diseases. Thioredoxin 1 (Trx1) is a conserved antioxidant that functions as a classic disulfide reductase. It also catalyzes the transnitrosylation or denitrosylation of caspase 3 (Casp3), underscoring its central role in determining Casp3 nitrosylation specificity. However, the mechanisms that regulate Trx1 transnitrosylation and denitrosylation of specific targets are unresolved. Here we used an optimized mass spectrometric method to demonstrate that Trx1 is itself nitrosylated by S-nitrosoglutathione at Cys73 only after the formation of a Cys32-Cys35 disulfide bond upon which the disulfide reductase and denitrosylase activities of Trx1 are attenuated. Following nitrosylation, Trx1 subsequently transnitrosylates Casp3. Overexpression of Trx1C32S/C35S (a mutant Trx1 with both Cys32 and Cys35 replaced by serine to mimic the disulfide reductase-inactive Trx1) in HeLa cells promoted the nitrosylation of specific target proteins. Using a global proteomics approach, we identified 47 novel Trx1 transnitrosylation target protein candidates. From further bioinformatics analysis of this set of nitrosylated peptides, we identified consensus motifs that are likely to be the determinants of Trx1-mediated transnitrosylation specificity. Among these proteins, we confirmed that Trx1 directly transnitrosylates peroxiredoxin 1 at Cys173 and Cys83 and protects it from H2O2-induced overoxidation. Functionally, we found that Cys73-mediated Trx1 transnitrosylation of target proteins is important for protecting HeLa cells from apoptosis. These data demonstrate that the ability of Trx1 to transnitrosylate target proteins is regulated by a crucial stepwise oxidative and nitrosative modification of specific cysteines, suggesting that Trx1, as a master regulator of redox signaling, can modulate target proteins via alternating modalities of reduction and nitrosylation.
Nitric oxide (NO) is an important second messenger for signal transduction in cells. The production of cGMP by guanylyl cyclase, enabled by the binding of NO onto heme, is considered the primary mechanism responsible for the plethora of functions exerted by NO (1). However, S-nitrosylation, the covalent addition of the NO moiety onto cysteine thiols, is increasingly recognized as an important post-translational modification for regulating protein functions (for reviews, see Refs. 2 and 3). S-Nitrosylation is dynamic, reversible, site-specific, and modulated by selected cellular stimuli (4–7). With improved detection sensitivity, an increasing number of S-nitrosylated proteins have been identified by proteomics technologies (5, 8–13). Among the known modified proteins, nitrosylation occurs only on selected cysteines (4, 6, 14–17). Non-enzymatic mechanisms proposed to determine S-nitrosylation specificity include the availability of specific NO donors and protein microenvironments that stabilize the pKa of acidic target cysteines (18). Furthermore, several enzymes, including hemoglobin (19, 20), superoxide dismutase 1 (21, 22), S-nitrosoglutathione reductase (23–25), and protein-disulfide isomerase (26), have been shown to possess either transnitrosylase or denitrosylase activities. However, an enzymatic system that governs site-specific transnitrosylation and denitrosylation, analogous to the kinase/phosphatase paradigm for regulating protein phosphorylation, has remained largely uncharacterized.
Trx11 is an important antioxidant protein with protein reductase activity (27, 28). It has been characterized as an antiapoptotic protein because of its ability to suppress proapoptotic proteins, including apoptosis signal-regulating kinase 1 via disulfide reduction and Casp3 via transnitrosylation of Cys163 (14, 29). Conversely, Trx1 can denitrosylate Casp3 at Cys163, resulting in Casp3 activation (7). Trx1 appears to govern site-specific reversible nitrosylation of selected protein targets (14, 15), but what are the underlying mechanisms that regulate Trx1 transnitrosylation and denitrosylation activities? Are there additional Trx1-mediated transnitrosylation or denitrosylation targets that have not yet been identified? In this study, we used ESI-Q-TOF mass spectrometry (MS) to analyze the nitrosylation of Trx1 and a Casp3 peptide (Casp3p) under different redox conditions. Because of the labile nature of the S–NO bond, direct identification of S-nitrosylated proteins and their specific nitrosylation sites by MS remains challenging (8). A biotin switch method that is based on the derivatization of protein S–NO with a biotinylating agent is typically used for such analyses (8). However, like any indirect method, both false positive and negative identifications have been reported (30). Recently, we developed a method for direct analysis of protein S-nitrosylation by ESI-Q-TOF MS without prior chemical derivatization (31). Here we applied the same technique to determine the regulation of Trx1 by stepwise oxidative and nitrosative modifications of distinct cysteines and its subsequent ability to transnitrosylate target proteins. Nitrosative modification at Cys73 of Trx1 cannot occur without prior attenuation of the Trx1 disulfide reductase and denitrosylase activities via either disulfide bond formation between Cys32 and Cys35 or their mutation to serines. This is a key observation that has never been previously reported. Consequently, we designed a proteomics approach and discovered over 40 putative Trx1 transnitrosylation target proteins. We further characterized the Trx1 transnitrosylation proteome and identified three consensus motifs surrounding the putative Trx1 transnitrosylation sites, suggesting a protein-protein interaction mechanism for determining transnitrosylation specificity.
EXPERIMENTAL PROCEDURES
Reagents
All chemical reagents were purchased from Sigma unless otherwise indicated. ACN and HPLC grade water were obtained from Mallinckrodt Baker. Formic acid was purchased from EMD Chemicals (Merck KGaA). Two-dimensional gel electrophoresis (2DE) materials and SYPRO Ruby stain were purchased from Bio-Rad. The plasmids containing the coding sequences for Trx1 and the Trx1C35S (Cys35 substituted by serine), Trx1C32S/C35S (Cys32 and Cys35 substituted by serines), and Trx1C32S/C35S/C73S (Cys32, Cys35, and Cys73 substituted by serines) mutants were made using the shuttle vector pDC316. FLAG tags were added onto the N termini of the Trx1 sequences for detection purposes. A synthetic human caspase 3 (NCBI gi|16516817) peptide (Casp3p) containing the known nitrosylation site Cys163 (163CRGTELDCGIETD175, m/z 1,409.58) was purchased from AnaSpec (San Jose, CA). The following synthetic peptides containing the putative S-nitrosylated Trx1 (SNO-Trx1) targeting motifs AXC and AC or mutations A to W were purchased from GenScript (Piscataway, NJ): human GAPDH (gi|31645) peptide (146IISNASCTTNCLAPLAK162, m/z 1,720.03) and its mutant, mGAPDH (146IISNWSCTTNCLAPLAK162, m/z 1,835.16), and human tubulin-β (gi|57209813) peptide (280NMMAACDPRHGR291, m/z 1,358.58) and its mutant, mTubulin-β (280NMMWWCDPRHGR291, m/z 1,588.85).
S-Nitrosylation of Casp3p
Cys163 and Cys170 of Casp3p readily formed a disulfide bond in an ambient environment (data not shown). These residues were reduced prior to nitrosylation treatments. Casp3p (25 μg) was dissolved in 50 μl of nitrosylation buffer (NB) containing 10% ACN, 1 mm EDTA (Mediatech, Herndon, VA), and 0.1 mm neocuproine, pH 6.8 and incubated with 2 μl of 50 mm Tris(2-carboxyethyl)phosphine hydrochloride (Pierce) at 37 °C for 60 min to reduce the Cys163-Cys170 disulfide bond. The reduced peptide was desalted using a PepCleanTM C18 spin column (Pierce), and peptides were eluted with 70% ACN and concentrated to ∼25 μl with a SpeedVac. An aliquot of the reduced peptide (1 nmol) was mixed with a 10-fold molar excess of S-nitrosoglutathione (GSNO) in 30 μl of NB at 37 °C for 30 min in the dark and then used directly for MS detection. A Cys163-Cys170 disulfide form of Casp3p was treated in parallel as a negative control and was not found to be nitrosylated.
S-Nitrosylation of Trx1
Recombinant His-tagged human Trx1 (25 μg) in 50 μl of NB was reduced and desalted as described for Casp3p. Either the Cys32-Cys35 reduced form (rTrx1) or the disulfide bond form (oTrx1) of Trx1 (25 μg each) was mixed with a 25-fold molar excess of GSNO in 50 μl of NB at 37 °C for 30 min in the dark. GSNO-treated Trx1 was then used directly for MS detection. To study direct Trx1-mediated transnitrosylation of target proteins, SNO-Trx1 was precipitated with cold acetone at −20 °C for 1 h. The protein pellet was washed four times with cold acetone at −20 °C and dissolved in 30 μl of NB. Non-S-nitrosylated oTrx1, GSNO, and S-nitrosylated BSA (SNO-BSA) were processed in parallel as controls for the evaluation of the specificity of SNO-Trx1-mediated transnitrosylation.
SNO-Trx1-mediated Transnitrosylation of Casp3p and Target Peptides and Proteins
Acetone-precipitated SNO-Trx1 was used to nitrosylate Casp3p or GAPDH and tubulin peptides and their mutants at a 1:1 or 1:10 molar ratio at 37 °C for 30 min in the dark, and nitrosylated peptides were detected by Q-TOF MS as described below. Similarly, acetone-precipitated SNO-Trx1 was used to nitrosylate recombinant human Casp3 protein (Trx1:Casp3, 10:1 molar ratio; BD Pharmingen), recombinant His-tagged human peroxiredoxin 1 protein (Prx1) (Trx1:Prx1, 10:1 molar ratio; Abcam, Cambridge, MA), or HeLa cell protein extract (Trx1:cellular protein, 1:20 mass ratio) at 37 °C for 30 min in the dark. Non-S-nitrosylated oTrx1 and rTrx1 were used as negative controls. GSNO treatment was performed as a positive control. Following treatment, proteins were precipitated with cold acetone and prepared for the biotin switch assay (8) to evaluate their nitrosylation status. Lysozyme (100 μg), which has no free thiols (all eight cysteines form disulfide bonds as confirmed by MS analysis) was used as the protein precipitation carrier. Biotinylated proteins were detected by Western blot as described below.
Denitrosylation of SNO-Trx1 and Transnitrosylated Target Proteins by rTrx1
Recombinant Trx1 was incubated at 37 °C for 60 min with either 10 mm H2O2 or 10 mm DTT to yield oTrx1 or rTrx1, respectively, and subsequently desalted on a C18 cartridge. Buffer-alone controls had H2O2 and DTT added and were processed in parallel to confirm that these chemicals were removed and had no effect on the following experiments. Ten micrograms of oTrx1 in 100 μl of NB was first treated with 100 μm GSNO for 30 min at 37 °C to produce SNO-Trx1. Excess GSNO was removed by C18 cartridge cleanup. The SNO-Trx1 was mixed with an equimolar amount of oTrx1 or rTrx1 for 30 min at 37 °C and then mixed with 100 μg of lysozyme for acetone precipitation. Pellets were washed in ice-cold acetone, modified by biotin switch assay, and detected by Western blotting. For rTrx1 denitrosylation of transnitrosylated target proteins, HeLa cell proteins (200 μg) were nitrosylated with 10 μg of SNO-Trx1 and then combined with 10 μg of oTrx1 or rTrx1 at 37 °C for 30 min. The proteins were modified by biotin switch assay and detected by Western blotting.
Cell Culture and Molecular Biology
HeLa cells were grown at 37 °C in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere. Cells were transiently transfected with plasmids containing human Trx1, Trx1C35S, Trx1C32S/C35S, and Trx1C32S/C35S/C73S or with the empty pDC316 vector using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Forty-eight hours after transfection, the cells were treated with either 100 μm 1-chloro-2,4-dinitrobenzene (DNCB) for 60 min, H2O2 for 30 min, or corresponding buffer as controls. The treated cells were harvested via centrifugation at 500 × g for 5 min and washed with phosphate-buffered saline (PBS) prior to subsequent analyses.
Determination of Protein S-Nitrosothiol Level
S-Nitrosothiol levels were estimated according to the procedure described by Mannick and Schonhoff (32). Briefly, 500 μg of protein from HeLa cell extract was nitrosylated with either NB, 100 μm GSNO, GSNO plus 25 μg of oTrx1, or GSNO plus 25 μg of rTrx1 at 37 °C for 30 min. Proteins were precipitated with cold acetone, and the pellets were washed four times with cold acetone at −20 °C and then dissolved in 500 μl of NB. Each sample (50 μl) was incubated with 50 μl of 1% sulfanilamide in 0.5 m HCl with 0.2% HgCl2 for 5 min. Subsequently, 100 μl of a solution containing 0.02% N-(1-naphthyl)ethylenediamine dihydrochloride in 0.5 m HCl was added, and the samples were incubated for 5 min at RT. A control reaction was performed without HgCl2. The concentration of azo compounds formed was determined by measuring the absorption at 540 nm and was quantified based on a standard curve generated with serial dilutions of a GSNO stock solution.
Biotin Switch Analysis of Protein Nitrosylation
Nitrosylation is very labile. Although direct detection by MS is possible for synthetic peptides (31), a biotin switch technique (8) was used to identify nitrosylated proteins and their nitrosylation sites (supplemental Fig. S1). In brief, cells were lysed in lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, and 0.1 mm neocuproine) supplemented with a protease inhibitor mixture. After the removal of cell debris from the lysate, the resulting protein concentrations were measured using the bicinchoninic acid (BCA) method (Pierce) and adjusted to 1 μg/μl with lysis buffer. Similarly, recombinant proteins (with or without nitrosylation treatment) re-suspended in HEN buffer (25 mm HEPES, pH 7.7, 1 mm EDTA, and 0.1 mm neocuproine) were supplemented with 100 μg of lysozyme as a carrier for acetone precipitation. Proteins were denatured with 2.5% SDS (Bio-Rad), and free thiols were alkylated using 20 mm methyl methanethiosulfonate (MMTS) (Pierce) with frequent vortexing at 50 °C for 30 min. Excess MMTS was removed by cold acetone precipitation of the proteins. The protein pellets were reconstituted in HEN buffer containing 1% (w/v) SDS and 0.2 mm N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP) (Pierce) with or without 10 mm ascorbate. The reaction mixture was incubated in the dark for 1 h at RT. Excess reagents were removed by cold acetone precipitation of the proteins. The protein pellets were solubilized in non-reducing SDS-PAGE loading buffer (100 mm Tris, pH 6.8, 2% SDS, 15% glycerol, and 0.01% bromphenol blue) for Western blotting or in resuspension buffer (RB; 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, and 0.5% SDS) for immunoprecipitation as described below. For Western blotting, 15 μg of protein was separated using non-reducing SDS-PAGE and transferred onto nitrocellulose membrane. The biotinylated protein was probed with an anti-biotin antibody (1:3,000) (Vector Laboratories, Burlingame, CA) and visualized with enhanced chemiluminescent substrate (PerkinElmer Life Science).
Redox Analysis of Trx1
Analysis of the relative levels of oTrx1 and rTrx1 was performed as described previously (33). Briefly, following the treatment of HeLa cells with 0–200 μm DNCB for 60 min, the cells were washed with PBS, and proteins were extracted with lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, and 1 mm EDTA) supplemented with a protease inhibitor mixture. Separation of oTrx1 from rTrx1 was performed by native gel electrophoresis (15%) followed by Western blotting. Proteins were transferred onto nitrocellulose membrane and detected by anti-Trx1 antibody (1:5,000). The redox status of Trx1 in mouse brain, heart, kidney and lung was analyzed similarly.
Analysis of Prx1 Modifications in Trx1C32S/C35S-overexpressing HeLa Cells
HeLa cells were transiently transfected with either an empty pDC316 vector or Trx1C32S/C35S mutant. Forty-eight hours after transfection, the cells were treated with 0–200 μm H2O2 for 30 min or with the corresponding medium as a control. The treated cells were harvested and washed with PBS. Extracted proteins were modified by biotin switch assay, separated using either non-reducing or reducing SDS-PAGE, and detected by Western blotting. Nitrosylated Prx1 (SNO-Prx1) was detected by anti-Prx1 blotting of proteins enriched with streptavidin beads following the biotin switch assay described above. Prx-SO3H was detected using an anti-Prx-SO3H antibody (Abcam; 1:3,000). To detect the effect of GSNO nitrosylation on Prx1 sensitivity to H2O2-induced overoxidation, HeLa cells were treated with or without 1 mm GSNO for 30 min and then incubated with increasing concentrations of H2O2 up to 200 μm for 30 min. The cells were collected, extracted proteins were separated using either reducing or non-reducing SDS-PAGE, and Prx1 monomer and dimer or Prx-SO3H was detected by Western blotting.
Immunoprecipitation and Detection of S-Nitrosylated Proteins
Proteins modified by the biotin switch assay were precipitated in acetone and dissolved in RB. Protein concentrations were determined by the BCA method. Biotinylated proteins (400 μg) in 200 μl of RB were diluted with 200 μl of PBS and subsequently mixed with 20 μl of streptavidin-agarose beads (Pierce). The mixture was incubated for 1 h at RT with agitation. The beads were washed five times with 1 ml of PBS and incubated with 2× SDS-PAGE loading buffer for 30 min at 37 °C with gentle agitation followed by heating at 100 °C for 5 min. Supernatant proteins were desalted by acetone precipitation and redissolved in 2DE buffer (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm DTT, 0.2% BioLyte, pH 3–10, and 0.01% bromphenol blue) in readiness for 2DE separation. For the IEF step, the proteins were loaded onto 11-cm IPG strips (pH 3–10 non-linear; Bio-Rad). IEF was performed with a Protean IEF cell (Bio-Rad) at 20 °C using the following settings: 12-h rehydration at 50 V, 0.5 h at 250 V, ramped to 8,000 V over 3 h, and held at 8,000 V for 6 h. After IEF, proteins on the IPG strips were reduced with DTT (2%, w/v) for 15 min and then alkylated with iodoacetamide (2.5%, w/v) for 15 min in equilibration buffer (6 m urea, 375 mm Tris-HCl, pH 8.8, 2% SDS, and 20% glycerol). The second dimension separation was performed by 12.5% SDS-PAGE at 120 V. The gels were fixed with 40% methanol and 10% acetic acid for 30 min and stained with SYPRO Ruby overnight. Gel images were acquired on a Typhoon 9400 imager (GE Healthcare). Protein spots in the 2DE gels were excised for trypsin digestion and protein identification by the tandem mass spectrometry methods described below. For Western blot detection of specific nitrosylated proteins, total and avidin-enriched proteins were separated by 12.5% SDS-PAGE and transferred onto nitrocellulose membrane. Membranes were blocked with 5% milk and probed with anti-Prx1 (1:5,000), anti-heat shock protein 90 α/β (HSP90α/β) (sc-7947, Santa Cruz Biotechnology, Santa Cruz, CA; 1:5,000), anti-actin (sc-1616, Santa Cruz Biotechnology; 1:5,000), anti-Trx1 (1:5,000), anti-endothelial nitric-oxide synthase (NOS) (1:2,000), anti-procaspase 3 (1:1,500), anti-thioredoxin reductase (TrxR) (1:2,500), and anti-thioredoxin-interacting protein (Txnip) (1:5,000) antibodies. All antibodies used in this study were from Abcam unless otherwise indicated.
Analysis of Nitrosylated Proteins and Peptides by Mass Spectrometry
Mass measurement of S-nitrosylated proteins and peptides was performed on an API-US Q-TOF tandem mass spectrometer (Waters) equipped with a nano-ESI source (New Objective, Woburn, MA) according to an optimized method developed in our laboratory (31). MS analyses were performed via direct infusion unless stated otherwise. The proteins and peptides were diluted with 5% ACN and 0.1% formic acid prior to MS analyses. The capillary voltage was set to 3 kV. The cone voltage and the collision energy for analyzing S-nitrosylated Casp3p (SNO-Casp3p) and SNO-Trx1 were set to 20 and 4 eV, respectively. The tandem MS (MS/MS) spectrum for SNO-Casp3p was acquired at a collision energy of 32 eV. Argon was used as the collision gas.
For protein identification, protein spots of interest from 2DE were excised, reduced by DTT, alkylated with iodoacetamide, digested with trypsin, and desalted with C18 ZipTips (Millipore) as described previously (34). Both MS and MS/MS analyses of the digested peptides were performed on a MALDI-TOF/TOF tandem MS instrument (4800 Proteomics Analyzer, Applied Biosystems, Foster City, CA) with internal mass calibration. Mass spectra (m/z 800–4,000) were acquired in the positive ion mode. Tandem mass spectra of selected ions were acquired in the 1-kV mode. GPS explorer software (v3.5; Applied Biosystems) was used to generate the peak lists for the database search against the human NCBI protein database (October 17, 2007, 119,431 protein entries) using a local Mascot search engine (v2.2). Trypsin was selected as the enzyme with one missed cleavage; carbamidomethyl-modified cysteines and oxidation of methionines were set as variable modifications (biotin-HPDP was not searched as a modification because it was removed via DTT reduction). The mass tolerance for precursor ions was 50 ppm, and that for fragment ions was 0.3 Da. Proteins containing at least two peptides with confidence interval values no less than 95% were considered to be positively identified.
For identification of nitrosylated cysteines, biotinylated proteins were recovered with 0.5% SDS following acetone precipitation of the proteins from the biotin switch assay. The proteins were diluted 10-fold with 50 mm NH4HCO3 and digested with trypsin (1:30 (w/w) enzyme:protein ratio) at 37 °C overnight. The resulting peptides were loaded onto an avidin cartridge (ICAT kit from Applied Biosystems) for enrichment of the biotinylated peptides. After washing the cartridge to remove unmodified peptides with 2 ml of PBS, pH 7.2 and 1 ml of a solution containing 50 mm ammonium bicarbonate and 20% methanol, pH 8.3, the biotinylated peptides were eluted with 30% ACN and 0.4% TFA, dried in a SpeedVac, and resuspended in 2% ACN and 0.1% TFA. For LC/MS/MS analysis, the biotinylated peptides were first separated by Dionex UltiMate® 3000 reversed phase liquid chromatography (capillary PepMap 100 column, 75 μm × 150 mm, 3 μm, 100 Å, C18; Dionex, Sunnyvale, CA). The eluted peptides were analyzed using a Waters API-US Q-TOF MS instrument. MS spectra (m/z 400–1,900) were acquired in the positive ion mode. Argon was used as the collision gas. The collision energy was set from 16 to 60 eV, depending on the precursor ion charge state and mass. MS/MS spectra were acquired in the data-dependent analysis mode in which the three most abundant precursors with two to five charges from each MS survey scan were selected for fragmentation. The peak lists were generated by ProteinLynx (v2.1) into PKL files. Database searches were performed with Mascot (v2.2) against the human NCBI protein database (October 17, 2007, 119,431 protein entries) using the following search parameters. Trypsin was selected as the enzyme with one missed cleavage, the mass tolerance was 100 ppm for MS and 0.6 Da for MS/MS, and MMTS-modified and biotin-HPDP-modified cysteines and methionine oxidation were set as variable modifications. For MS/MS identification of the nitrosylation site of the peptide, we set a Mascot score threshold of at least 34, which corresponded to a confidence interval of 95% or better; the spectra were manually validated for post-translational modifications. The false discovery rate was calculated to be <0.5% according to Peng et al. (35). Proteins and biotinylated peptides belonging to isoforms that are indistinguishable from the MS/MS spectra were grouped together and are presented in supplemental Tables S1 and S2, and the protein isoforms with the most descriptive names and that have been well characterized biochemically are presented in Table I for ease of discussion.
Table I. Putative Trx1 transnitrosylation target proteins and their S-nitrosylation sites (C).
| Accession no. and protein name | m/z | z | Localization of nitrosylation site | Score | Refs. |
|---|---|---|---|---|---|
| Chaperones | |||||
| gi|306890, chaperonin (HSP60) | 986.98 | 2 | 237CEFQDAYVLLSEK250a | 34 | 10, 11, 65 |
| gi|5729877, heat shock protein 70 protein 8 | 658.36 | 2 | 602VCNPIITK609 | 50 | |
| gi|13543666, peptidylprolyl isomerase A (cyclophilin A) | 985.47 | 2 | 56IIPGFMCQGGDFTR69 | 64 | |
| 809.91 | 2 | 155KITIADCGQLE165 | 46 | ||
| Metabolic proteins | |||||
| gi|4503571, enolase 1 | 1,003.00 | 2 | 344VNQIGSVTESIQACK348a | 39 | 5, 10, 65 |
| gi|31645, glyceraldehyde-3-phosphate dehydrogenase | 951.48 | 2 | 235VPTANVSVVDLTCR248a | 106 | 5, 10, 13, 65–73 |
| 1,097.52 | 2 | 146IISNASCTTNCLAPLAK162a | 83 | ||
| gi|4557032, lactate dehydrogenase B | 810.39 | 2 | 159VIGSGCNLDSAR170 | 52 | |
| gi|4505763, phosphoglycerate kinase 1 | 1,070.57 | 2 | 107ACANPAAGSVILLENLR123 | 55 | |
| gi|35505, pyruvate kinase | 955.75 | 3 | 343AEGSDVANAVLDGADCIMLSGETAK367a | 37 | 10 |
| 865.92 | 2 | 44NTGIICTIGPASR56a | 42 | ||
| gi|37267, transketolase | 968.43 | 3 | 115QAFTDVATGSLGQGLGAACGMAYTGK140 | 36 | |
| Structural proteins | |||||
| gi|4503745, filamin 1 | 1,018.84 | 3 | 444CSYQPTMEGVHTVHVTFAGVPIPR467a | 47 | 73 |
| 1,045.52 | 3 | 1247LQVEPAVDTSGVQCYGPGIEGQGVFR1272a | 71 | ||
| gi|32015, α-tubulin | 1,041.13 | 3 | 280AYHEQLSVAEITNACFEPANQMVK303a | 35 | 5, 10, 11, 13, 66, 73 |
| 787.36 | 2 | 339SIQFVDWCPTGFK351a | 51 | ||
| gi|57209813, tubulin, β | 718.80 | 2 | 280NMMAACDPR288a | 35 | 10, 11, 13, 66, 74 |
| gi|37852, vimentin | 931.45 | 2 | 322QVQSLTCEVDALK334a | 48 | 75 |
| Redox | |||||
| gi| 4505591, peroxiredoxin 1 | 926.46 | 3 | 169HGEVCPAGWKPGSDTIKPDVQK190a | 62 | 10, 71, 73 |
| gi|5453549, peroxiredoxin 4 | 925.43 | 3 | 241HGEVCPAGWKPGSETIIPDPAGK263 | 63 | |
| 1,072.83 | 3 | 140SINTEVVACSVDSQFTHLAWINTPR164 | 67 | ||
| Ribosomal proteins | |||||
| gi|7765076, S3 ribosomal protein | 803.52 | 2 | 95GLCAIAQAESLR106a | 38 | 10, 13 |
| gi|4506605, ribosomal protein L23 | 1,171.13 | 2 | 16ISLGLPVGAVINCADNTGAK35 | 39 | |
| gi|34335134, SEC13-like 1 isoform b | 920.79 | 3 | 217DVAWAPSIGLPTSTIASCSQDGR239 | 83 | |
| 640.80 | 2 | 28LATCSSDR35 | 45 | ||
| gi|4759160, small nuclear ribonucleoprotein | 929.10 | 3 | 9VLHEAEGHIVTCETNTGEVYR29 | 100 | |
| gi|31455238, HNRPA2B1 protein | 682.82 | 2 | 35LTDCVVMR42 | 37 | |
| Translation regulators | |||||
| gi|4503483, eukaryotic translation elongation factor 2 | 983.45 | 2 | 581ETVSEESNVLCLSK594a | 60 | 5, 9, 13 |
| gi|4503471, eukaryotic translation elongation factor 1α | 1,122.88 | 3 | 396SGDAAIVDMVPGKPMCVESFSDYPPLGR423a | 71 | 5, 9, 65, 73 |
| 1,131.60 | 3 | 220DGNASGTTLLEALDCILPPTRPTDKPLR247a | 48 | ||
| Others | |||||
| gi|339723, ADP/ATP translocase | 616.82 | 2 | 120GLGDCLVK127a | 34 | 10, 76 |
| gi|56967118, annexin A2 | 1,075.00 | 2 | 100GLGTDEDSLIEIICSR115a | 36 | 9, 71 |
| gi|2521981, α2-HS-glycoprotein | 612.94 | 3 | 99CDSSPDSAEDVRK111 | 38 | |
| gi|741376, cathepsin B | 1,098.03 | 2 | 129GQDHCGIESEVVAGIPR145 | 86 | |
| gi|5453854, poly(rC)-binding protein 1 | 830.38 | 2 | 47INISEGNCPER57a | 40 | 10, 65, 73 |
| gi|188556, migration-inhibitory factor | 708.40 | 2 | 79LLCGLLAER87 | 42 |
a Peptides containing S-nitrosylation sites that have also been previously reported by others. The spectra for the identification of S-nitrosylation sites are shown in supplemental Fig. S10.
Apoptosis Assay
HeLa cells were first transfected with WT Trx1 or mutant plasmids for 24 h in a 6-well plate, divided into 24-well plates for 12 h, and then treated with 5 μg/ml cycloheximide (CHX) and 20 ng/ml tumor necrosis factor-α (TNF-α) for 6 h. Cells were collected for the apoptosis assay using a BD Pharmingen Phycoerythrin-Annexin V Apoptosis Detection kit I (BD Biosciences) and assayed according to the manufacturer's instructions. The cells were washed twice in PBS and incubated at 37 °C with 100 μl of 0.05% trypsin and EDTA until detachment. Subsequently, 1 ml of 10% FBS in PBS was added to stop the trypsinization. The cells were collected into 5-ml FACS flow cytometry tubes via centrifuge for 5 min at 500 × g and washed once with cold PBS. One hundred microliters of the Annexin V binding buffer was added to the cell pellets and stained with 5 μl of phycoerythrin-Annexin V and 7-aminoactinomycin D for 15 min at room temperature in the dark. Four hundred microliters of the binding buffer was added following staining. The stained cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences). Apoptosis levels were determined by calculating the percentage of Annexin V-positive cells.
Motif Analysis
Peptides containing nitrosylated cysteines were subjected to Motif-X analyses (http://motif-x.med.harvard.edu/motif-x.html). The following parameters were used: central character, “C,” width, “13,” occurrences, “5,” significance, “0.001,” background, “ipi. HUMAN.fasta,” and foreground format, “fasta.”
Statistical Analysis
Data are expressed as means ± S.E. Statistical analysis was performed using a two-tailed unpaired Student's t test with Excel. Differences were considered significant for p < 0.05.
RESULTS AND DISCUSSION
Only Cys32-Cys35 Oxidized Trx1 Can Be Nitrosylated at Cys73 in Vitro
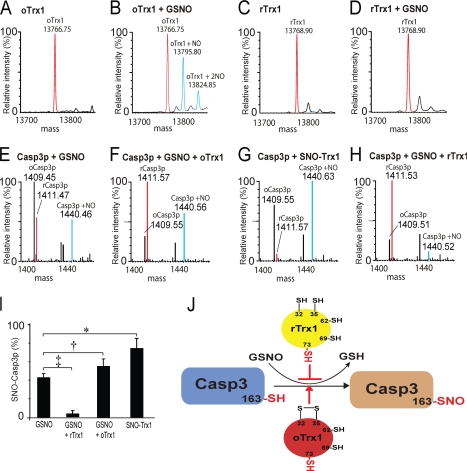
Earlier studies suggested that the redox status of Cys32 and Cys35 may affect the ability of Trx1 to transnitrosylate or denitrosylate target proteins (7). Here we present direct evidence that only after the formation of the Cys32 and Cys35 disulfide bond (forming oTrx1) can oTrx1 be S-nitrosylated in vitro (forming SNO-Trx1). There are five cysteines at residues 32, 35, 62, 69, and 73 in human Trx1 with Cys32 and Cys35 located within the reductive CXXC motif. Under non-reducing conditions, high resolution MS analysis of a recombinant His-tagged human Trx1 (supplemental Fig. S2A) demonstrated that this protein had a mass of 13,766.75 Da, corresponding to Trx1 with three free thiols and one disulfide bond (Fig. 1A). This molecule was readily S-nitrosylated by GSNO, an NO donor, resulting in a product with a mass of 13,795.80 Da (Fig. 1B; 13,766.75 + 29 Da, i.e. oTrx1 + NO). In addition, we also observed lower levels of dinitrosylated Trx1 with a mass of 13,824.85 Da (Fig. 1B; 13,766.75 + 58 Da, i.e. oTrx1 + 2NO). Based on MS/MS analysis of SNO-Trx1, we previously identified the redox status of all five Trx1 cysteines: Cys32 and Cys35 formed a disulfide bond, Cys62 and Cys69 existed as free thiols, and Cys73 was nitrosylated (31). These results were in agreement with observations made by Mitchell and Marletta (14), who demonstrated that Trx1 nitrosylated at Cys73 is the requisite intermediate for Trx1-mediated transnitrosylation of Casp3. For comparison, Trx1 with reduced Cys32 and Cys35 (rTrx1) yielded a mass of 13,768.90 Da (Fig. 1C), corresponding to the addition of ∼2 extra protons to oTrx1. Unexpectedly, we could not detect any SNO-Trx1 MS signal following the incubation of rTrx1 with GSNO (Fig. 1D), an observation contrary to that reported by Hashemy and Holmgren (17). This discrepancy might have arisen through prolonged exposure of the protein solution to air, a subsequent oxidization of rTrx1, and eventual nitrosylation. Alternatively, GSNO can facilitate disulfide formation between Cys32 and Cys35 (36), and it is conceivable that under certain conditions rTrx1 is converted to oTrx1, which becomes nitrosylated. These results suggest that the redox status of Cys32 and Cys35 is critical to nitrosylation of Cys73.
Fig. 1.
S-Nitrosylation of Trx1 and targets of Trx1 transnitrosylation are dependent upon cysteine redox status. Mass spectra of oTrx1 before (A) and after (B) GSNO treatment and of rTrx1 before (C) and after (D) GSNO treatment are shown. All spectra were deconvoluted to singly charged states by MassLynx (v4.1; Waters) for ease of comparison. Red peaks, precursors; blue peaks, nitrosylated; and unlabeled peaks in C and D, sodium adducts. E, spectrum of GSNO-treated Casp3p. An m/z peak at 1,440.46 (labeled blue), corresponding to SNO-Casp3p (1,411.5 + 29 Da, i.e. reduced Casp3p (labeled red) + NO), was observed. F, in the presence of oTrx1, a substantial SNO-Casp3p ion peak was observed. G, direct transnitrosylation of Casp3p by SNO-Trx1. A prominent SNO-Casp3p ion was observed. H, in the presence rTrx1, hardly any SNO-Casp3p was detected as shown by the minor m/z peak at 1,440.52. The disulfide form of Casp3p had a mass of 1,409.46 Da. I, relative percentages of SNO-Casp3p were analyzed using a Student's t test. Data are presented as mean ± S.E.; n = 3. All comparisons were of the SNO-Casp3p ion species relative to all Casp3p ion species within the treatment groups (*, p = 0.0001; †, p = 0.03; ‡, p = 0.002; Student's t test). J, proposed model for Trx1 regulation of Casp3 nitrosylation by Trx1 Cys32 and Cys35 redox status.
There are conflicting reports concerning the S-nitrosylation site(s) on Trx1. For mononitrosylated Trx1, Cys69 was first reported to be nitrosylated based on site-directed mutagenesis in endothelial cells (37); however, this result was not reproducible (17). Direct chemical identification of Cys73 nitrosylation (without genetic manipulation) was reported previously by us (31) and others (14). Through specific cysteine mutagenesis, Hashemy and Holmgren (17) reported the simultaneous nitrosylation of both Cys69 and Cys73 by GSNO using rTrx1 as starting material. Here we report relatively minor amounts of dinitrosylated Trx1, possibly with Cys73 and one other site nitrosylated; this SNO-Trx1 species was not identified because of its low abundance (Fig. 1B) (31). From analysis of crystal structures, Weichsel et al. (16) reported GSNO-mediated nitrosylation of Cys62 at neutral pH and Cys69 at more basic pH. It is likely that experimental conditions for x-ray crystallography, including protein concentration, pH, and buffer composition, may have resulted in these observed nitrosylation sites. Indeed, the same study also reported the dimerization of Trx1 via Cys73, a complex not widely reported in cellular systems. Cys32-SNO is also proposed to serve as a Trx1 denitrosylation intermediate for both GSNO and SNO-proteins (7, 38), but SNO-Cys32 Trx1 as a molecular entity has not been isolated, possibly because of the transient nature of this species.
Transnitrosylation of Casp3p and Protein by SNO-Trx1
To further our investigation, we examined whether the redox status of Cys32 and Cys35 affects Trx1 transnitrosylation of target proteins. Although Casp3 is a well characterized Trx1-mediated transnitrosylation target, impurities from the commercially available sources of Casp3 (supplemental Fig. S2B) made direct MS determination of its nitrosylation status challenging. Therefore, we initially chose to use a synthetic Casp3p containing the known nitrosylation site at Cys163 as a model system for this study, and we subsequently validated the findings with the full-length Casp3 protein. Casp3p had a mass of either 1,411.47 (dithiol) or 1,409.46 Da (disulfide; Fig. 1E). Dithiol Casp3p can be readily nitrosylated with GSNO as demonstrated by the production of an ion with a mass of 1,440.46 Da (Fig. 1E; 1,411.47 + 29 Da, i.e. Casp3p + NO), corresponding to mononitrosylated Casp3p. The presence of oTrx1 slightly enhanced GSNO-mediated S-nitrosylation of Casp3p (Fig. 1F), and significantly, SNO-Trx1 was able to transnitrosylate Casp3p directly (Fig. 1G). On the other hand, rTrx1 dramatically inhibited the S-nitrosylation of Casp3p by GSNO (Fig. 1H). We also confirmed that SNO-Trx1-mediated transnitrosylation of Casp3p was not due to an experimental artifact during sample preparation as SNO-BSA and acetone-precipitated GSNO residual could not transnitrosylate Casp3p (supplemental Fig. S3, A and B). Although there are two cysteines within Casp3p (163CRGTELD170CGIETD), MS/MS analysis found that only Cys163, a known Casp3 nitrosylation site (14), could be nitrosylated (supplemental Fig. S3C). Overall, it appeared that oTrx1 could be readily converted by GSNO into SNO-Trx1, and this not only promoted GSNO nitrosylation of Casp3p but also directly nitrosylated Casp3p by SNO-Trx1 (Fig. 1, I and J). On the other hand, rTrx1 inhibited GSNO nitrosylation of Casp3p (Fig. 1, I and J), consistent with previous reports that the Cys32 free thiol is important for Trx1-mediated denitrosylation of Casp3 (7).
We next validated the observations made using Casp3p with a recombinant human Casp3 protein (supplemental Fig. S2B) and found that neither oTrx1 nor rTrx1 can transnitrosylate Casp3 in the absence of GSNO (supplemental Fig. S4A, lanes 1 and 4). As was the case with Casp3p, rTrx1 inhibited GSNO-mediated nitrosylation of Casp3 (supplemental Fig. S4A, lane 2), whereas both GSNO and SNO-Trx1 directly nitrosylated Casp3 (supplemental Fig. S4A, lanes 3 and 5). In addition to Casp3, we also found that oTrx1 was more effective at promoting GSNO-mediated nitrosylation of cellular protein in vitro than rTrx1 or GSNO alone (supplemental Fig. S4B), suggesting that additional SNO-Trx1-mediated transnitrosylation targets may exist. Multiple nitrosylated protein bands were observed when HeLa cell extract was incubated with SNO-Trx1 alone. When SNO-Trx1 was incubated with an equal molar ratio of oTrx1, its nitrosylation status was preserved, but when combined with rTrx1, it was denitrosylated (supplemental Fig. S5, A and B). Consequently, the level of protein transnitrosylation was preserved in the presence of oTrx1 but was diminished in the presence of rTrx1 (supplemental Fig. S5, C and D). Overall, the extent of Trx1 nitrosylation and the ability of Trx1 to transnitrosylate appear to be regulated by the denitrosylation ability of its reduced form. From these observations, it appears that a Trx1 molecule is unlikely to possess both transnitrosylation and denitrosylation activities simultaneously. A separate molecule of rTrx1 can denitrosylate SNO-Trx1, attenuating the ability of the latter to transnitrosylate target proteins.
Transnitrosylation of Cellular Proteins by Mutant Trx1 with Attenuated Reductase Activity
To identify potential Trx1-mediated transnitrosylation target proteins in vivo, we overexpressed a Trx1 mutant in HeLa cells with both Cys32 and Cys35 replaced by Ser (Trx1C32S/C35S). Although structurally similar to rTrx1, this mutant mimics the function of oTrx1 in which its protein reductase and denitrosylation activities are attenuated (39). Furthermore, Trx1C32S/C35S may be nitrosylated to form SNO-Trx1C32S/C35S, resulting in the transnitrosylation of target proteins. HeLa cells were chosen because they are known to express sufficient endothelial NOS to supply endogenous NO donors (40, 41). Overexpression of WT Trx1 and mutants did not alter endothelial NOS levels (supplemental Fig. S6). Surprisingly, without the addition of exogenous NO donors, the levels of nitrosylated proteins were dramatically elevated in Trx1C32S/C35S-expressing cells compared with vector-, Trx1-, or Trx1C35S-expressing cells (Fig. 2, A and B). Protein nitrosylation differences among individual cell groups were amplified by the addition of DNCB, a TrxR inhibitor (Fig. 2B). Expression of Trx1C32S/C35S/C73S triple mutant resulted in decreased nitrosylation of most but not all protein compared with Trx1C32S/C35S-expressing cells (Fig. 2C), suggesting that Cys73 in SNO-Trx1C32S/C35S plays a prominent role in mediating protein transnitrosylation. However, alternative mechanisms are also possible. The overall structure of Trx1C32S/C35S is similar to that of WT (15, 42, 43), and it can behave as a competitive inhibitor of TrxR binding to Trx1 (44), resulting in an accumulation of endogenous oTrx1. In the presence of NO donors, oTrx1 becomes nitrosylated at Cys73 and facilitates transnitrosylation of downstream target proteins. As expected, we observed an increase in the amount of oTrx1 in DNCB-treated HeLa cells (supplemental Fig. S7A) that correlated with increased protein nitrosylation (supplemental Fig. S7B). oTrx1 can be detected in mouse tissues (supplemental Fig. S7C), suggesting that SNO-Trx1 may play an important physiological role in these tissues. In contrast to previous reports of increased protein nitrosylation in endothelial cells overexpressing WT Trx1 (37, 45), no appreciable increase in protein nitrosylation was observed in HeLa cells overexpressing WT Trx1 (Fig. 2B). However, DNCB treatment did elevate protein nitrosylation (Fig. 2B), presumably due to an increased level of oTrx1 (supplemental Fig. S7A). Interestingly, lower protein nitrosylation levels were observed in DNCB-treated Trx1C35S-expressing cells (Fig. 2B). These results suggest that without a redox-active thiol at amino acid 35 with which to form a disulfide bond the free thiol of Cys32 may either inhibit protein nitrosylation by other NO donors or denitrosylate proteins (7). However, the precise mechanisms and function for maintaining Cys32 as a free thiol in this mutant need to be investigated.
Fig. 2.
Increased protein nitrosylation in Trx1C32S/C35S-expressing HeLa cells. A, Trx1C32S/C35S expression resulted in increased protein nitrosylation levels determined by Western blotting of biotinylated proteins. An ascorbate control was used to confirm nitrosylation detection specificity. B, DNCB treatment further enhanced Trx1C32S/C35S-related increases in protein nitrosylation. Trx1C35S-expressing cells contained lower levels of nitrosylated proteins compared with cells subjected to other treatments. All comparisons were made with vector (Vec)-transfected cells (*, p = 0.006; †, p = 0.004; ‡, p = 0.007; §, p = 0.009). C, Trx1C32S/C35S/C73S expression resulted in decreased protein nitrosylation levels compared with Trx1C32S/C35S-expressing cells as determined by Western blotting of biotinylated proteins (**, p = 0.006; Student's t test). Values are the mean ± standard error (S.E.) for experiments performed in triplicate.
The data presented suggest that Trx1-mediated transnitrosylation and reduction may be functionally divergent modalities for regulating target proteins. In support of this notion, only rTrx1 serves as a reductase (27) or a denitrosylase (7); it is resistant to S-nitrosylation and does not function as a transnitrosylase. On the other hand, S-nitrosylation hinders Trx1 reductase activities (17), an observation that may well reflect a requisite for Cys32-Cys35 disulfide bond formation prior to nitrosylation of Trx1. Detailed structural analyses of the Cys32/Cys35 dithiol form following its S-nitrosylation point to the formation of a Cys32-Cys35 disulfide bond within nitrosylated Trx1 (16, 17). It would appear that only the reductase-inactive form of Trx1 can become nitrosylated and function as a transnitrosylase. In contrast, when Cys32/Cys35 are reduced, Trx1 denitrosylates target proteins and GSNO with concomitant generation of nitrooxyl (HNO) (46), a molecular species described as a product of S-nitrosothiols (47). As mentioned earlier, Trx1C32S/C35S structurally resembles rTrx-1 rather than oTrx1 (15). Therefore, the ability of Trx1C32S/C35S to transnitrosylate target proteins is probably due to a lack of free thiols to carry out reductase and denitrosylase functions in the catalytic site rather than an enhanced association with target proteins.
Identification of Putative Trx1 Transnitrosylation Targets
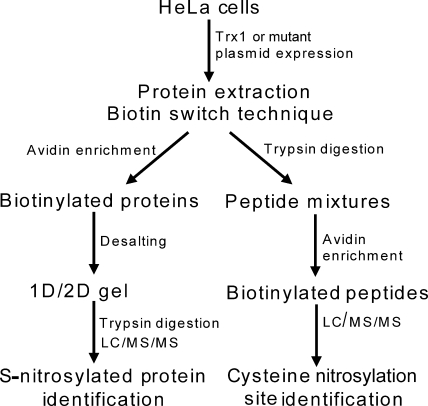
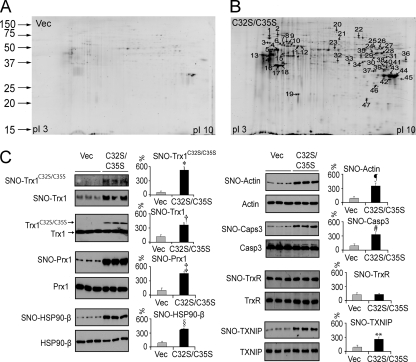
To identify cellular Trx1-mediated transnitrosylation target proteins, we used the biotin switch technique (8) to convert nitrosylated cysteines into biotinylated cysteines (supplemental Fig. S1), and then we affinity-purified the biotinylated proteins with streptavidin-agarose beads (Fig. 3). We recovered the proteins and separated them by 2DE (Fig. 4, A and B). Substantially more nitrosylated proteins were isolated from Trx1C32S/C35S-expressing cells compared with control cells (Fig. 4, A and B). Using MS/MS methods, 33 proteins in 47 gel spots were identified (Figs. 3 and 4B and supplemental Table S1). In addition, the nitrosylation sites within 36 peptides from 28 proteins were also identified using avidin enrichment of the tryptic peptides obtained from biotin switch-processed soluble proteins (Table I). These approaches brought the total number of putative Trx1-mediated transnitrosylation targets identified to 47, including over half of the proteins reported previously either to be nitrosylated or to contain the same nitrosylation sites as reported here (Table I and supplemental Table S2), validating our approach. To confirm the proteomics findings, we verified the increase in the nitrosylation levels of Prx1, HSP90β, and actin in Trx1C32S/C35S-overexpressing cells by Western blotting (Fig. 4C). Although Casp3 was not abundant enough to be detected by MS, we were able to verify by Western blot that it was significantly more nitrosylated in Trx1C32S/C35S-expressing cells (Fig. 4C). As expected, SNO-Trx1C32S/C35S was observed in Trx1C32S/C35S-expressing cells (Fig. 4C). Although the expression level of Trx1C32S/C35S was only about 10% relative to the endogenous Trx1 level (Fig. 4C), Trx1C32S/C35S expression resulted in an over 2-fold increase in SNO-Trx1 compared with control cells (Fig. 4C), suggesting that both SNO-Trx1C32S/C35S and SNO-Trx1 may be responsible for the increased protein nitrosylation levels observed. Recently, Txnip, a cellular inhibitor of Trx1 reductase activity, was shown to promote cellular protein nitrosylation (48). Although the authors attributed this observation to the inhibition of Trx1 Cys32/Cys35 dithiol-mediated denitrosylase activity by Txnip, it is unclear whether the transnitrosylase activity of Trx1 can be enhanced by Txnip. Curiously, although Txnip expression was unaffected by Trx1C32S/C35S expression, its nitrosylation levels were elevated (Fig. 4C), suggesting a possible novel cross-talk between Trx1 and Txnip via transnitrosylation, a hypothesis that warrants more detailed future study. By comparison, TrxR expression and nitrosylation levels were not dramatically altered in Trx1C32S/C35S-expressing HeLa cells (Fig. 4C), suggesting that Trx1C32S/C35S-mediated transnitrosylation is not due to down-regulation of TrxR expression or nitrosylation.
Fig. 3.
Work flow for identification of putative Trx1 transnitrosylation targets and identification of S-nitrosylation sites.
Fig. 4.
2DE analysis of biotinylated proteins from vector (Vec)- (A) and Trx1C32S/C35S (B)-transfected HeLa cells is shown. Nitrosylated proteins were biotinylated, enriched and eluted from streptavidin beads, desalted with ice-cold acetone, dissolved in a 2DE rehydration buffer, and analyzed by 2DE. Gels were stained with SYPRO Ruby dyes, and the protein spots whose densities were increased in Trx1C32S/C35S-transfected cells were excised for trypsin digestion and protein identification by tandem MS (supplemental Table SI). C, proteins recovered from streptavidin beads were analyzed by Western blotting with individual antibodies specific for the detection of nitrosylated Trx1, Prx1, HSP90β, actin, Casp3, Txnip, and TrxR. Values are the mean ± S.E. for experiments performed in triplicate (*, p = 0.0006; †, p = 0.001; ‡, p = 0.0001; §, p = 0.0007;  , p = 0.001; #, p = 0.0009; **, p = 0.002; Student's t test). Non-streptavidin-enriched proteins were analyzed in parallel to detect proteins regardless of their nitrosylation status.
, p = 0.001; #, p = 0.0009; **, p = 0.002; Student's t test). Non-streptavidin-enriched proteins were analyzed in parallel to detect proteins regardless of their nitrosylation status.
Because the specificity of Trx1-mediated transnitrosylation is likely to rely on specific protein-protein interactions between SNO-Trx1 and the target proteins, we also performed a primary sequence motif analysis of the peptides whose nitrosylation sites were identified in this study. Although the number of protein sequences was small, we were able to identify three independent motifs (Fig. 5). The first motif is AXC (where X is any amino acid and C is nitrosylated cysteine), found in eight peptides (Fig. 5A and Table I). Similarly, other motifs, CXXXXA and AC, are found in six peptides (Fig. 5, B and C, and Table I). We compared the ability of SNO-Trx1 to transnitrosylate peptides containing the AXC or AC motifs with mutants containing tryptophan instead of consensus alanine and found that motif recognition may indeed be important (Fig. 5D). However, because the number of peptides found in this study is relatively small, the appearance of potential motifs could be coincidental. These observations suggest a sequence motif-specific mechanism for Trx1-mediated transnitrosylation of target proteins, a hypothesis that warrants further study.
Fig. 5.
Putative Trx1-mediated transnitrosylation target motifs. The motifs were identified using Motif-X (http://motif-x.med.harvard.edu/motif-x.html) analyses of the nitrosylated peptides identified in Trx1C32S/C35S-expressing HeLa cells (see “Experimental Procedures” and Table I). Three putative motifs were identified as AXC (A), CXXXXA (B), and AC (C) where X is any amino acid and C (in red) is nitrosylated cysteine. Casp3 (gi|16516817; see sequence in supplemental Fig. S2B) has a Trx1 trans- and denitrosylation site at Cys163; it is aligned in motif AC. D, SNO-Trx1 transnitrosylates target peptides in the AXC motif, and mutation of the motif from AXC to WXC attenuates transnitrosylation. In mTubulin peptide, both Ala283 and Ala284 were changed to the bulkier amino acid Trp because each represents a consensus Ala within a transnitrosylation target motif. Values are the mean ± S.E. for experiments performed in triplicate (*, p = 0.002; **, p = 0.0035; Student's t test). Acc., accession.
Transnitrosylation of Prx1 by Either Mutant Trx1 or SNO-Trx1 in Vitro
To verify the functional significance of Trx1-mediated transnitrosylation, we further examined the effects of Trx1 transnitrosylation of Prx1, a classic 2-Cys Prx that converts its peroxidatic Cys52–SH thiol (CysP) (supplemental Fig. S2C) to a sulfenic acid CysP-SOH during reduction of peroxides (for reviews, see Refs. 49 and 50). Prx1 uses a recovery cycle that involves the formation of a disulfide bond between CP-SOH and a resolving Cys173 (CysR) on a separate Prx1 peptide that can then be reduced by rTrx1 to regenerate the active Prx1 CysP-SH (49). Overoxidation of CysP-OH to CysP-SO2H may be reversed by an ATP-dependent sulfiredoxin (Srx) (51). However, further oxidation to CysP-SO3H results in the inactivation of Prx1 (52, 53). We observed that the in vivo nitrosylation levels of Prx1 (forming SNO-Prx1) were dramatically elevated in Trx1C32S/C35S-overexpressing HeLa cells (Table I and Fig. 4C), suggesting that it may be regulated by Trx1-mediated transnitrosylation, a function that has not been described previously. However, it is possible that Trx1C32S/C35S may promote the nitrosylation of Prx1 indirectly via an intermediate S-nitrosylated species, such as metal-mediated dinitrosyliron complexes described by Lancaster and co-workers (54). Therefore, although the association of Prx1 and WT Trx1 has been widely reported (55), we confirmed that Prx1 is associated with Trx1C32S/C35S (supplemental Fig. S8A), and Prx1 could be directly nitrosylated by SNO-Trx1 in vitro (Fig. 6A). In addition, two SNO-Trx1 transnitrosylation sites in Prx1 were identified by MS/MS analysis, including CysR in 169HEGVC*PAGWKPGSDTIKPDVQK190 (where C* is nitrosylated cysteine; Fig. 6B), which was also found in Trx1C32S/C35S-overexpressing cells in vivo (Table I). CysR appears to be sensitive to cellular redox environments as it has also been reported to undergo glutathionylation (56). In addition, Cys83, a key residue for the maintenance of the human Prx1 dimer-dimer interface (57), was also nitrosylated (supplemental Fig. S8B). By comparison, the CysP was not substantially nitrosylated by SNO-Trx1 in vitro or in Trx1C32S/C35S-expressing cells (supplemental Fig. S8C).
Fig. 6.
SNO-Trx1 transnitrosylation of Prx1. A, both SNO-Trx1 and GSNO directly transnitrosylated Prx1 in vitro. B, MS/MS spectrum of Prx1 169HEGVC*PAGWKPGSDTIKPDVQK190 (m/z 2,349.2). Cys173 (C*, biotinylated) was nitrosylated. C, SNO-Prx1 levels were higher in Trx1C32S/C35S-expressing cells, and the differences were amplified following H2O2 treatment. D and E, Western blots of Prx1 species after separation on non-reducing gels. Overexpression of Trx1C32S/C35S rendered Prx1 more resistant to H2O2-induced dimer to monomer conversion (D), which correlated with the reduction of Prx-SO3H levels (E). Prx1 expression was not affected by either Trx1C32S/C35S expression or H2O2 treatment. F, statistical evaluation of E. Comparisons were made for Prx1 dimer, monomer, and Prx-SO3H between Trx1C32S/C35S-overexpressing cells and control cells. Values are the mean ± S.E. for experiments performed in triplicate (*, p = 0.0007; †, p = 0.006; ‡, p = 0.0001; Student's t test). Full-size blots are shown in supplemental Fig. S9B. Vec, vector.
Transnitrosylation Protects Prx1 from H2O2-induced Dimer Disruption and Sulfonylation
Drapier and co-workers (58) have shown that NO donors attenuate the overoxidation of Prx1 in macrophages, which the authors attributed to NO induction of Srx. We found that H2O2 treatment increased nitrosylation of proteins in both vector- and Trx1C32S/C35S-expressing cells, albeit more so in the latter cells (Fig. 6C). Increasing H2O2 concentrations up to 200 μm resulted in the disruption of the disulfide-linked Prx1 dimer, although less disruption was observed in Trx1C32S/C35S-expressing cells (Fig. 6D). Coincidentally, H2O2-induced Prx1-SO3H levels were significantly attenuated in Trx1C32S/C35S-overexpressing cells (Fig. 6, E and F). One could argue that the protective effect of Trx1C32S/C35S on Prx1 may be attributed to effects other than transnitrosylation. Therefore, we evaluated whether the addition of GSNO to cells could also protect Prx1 from H2O2-induced dimer disruption and sulfonylation in HeLa cells. Indeed, we confirmed that nitrosylation protected Prx1 from overoxidation in HeLa cells (supplemental Fig. S9). The mechanism of how Trx1 transnitrosylation of CysR and Cys83 protects Prx1 from overoxidation is unknown. However, the C-terminal domain near CysR has been shown to regulate the susceptibility of Prx to overoxidation (59, 60). Therefore, S-nitrosylation of CysR may be a means for maintaining an overoxidation-resistant structure, perhaps by enhancing the ability of CysR to form disulfide bonds with CysP during the peroxidase recovery cycle, a function that has also been suggested for the S-glutathionylation of CysR (56).
Although nitrosylation appears to inhibit Prx peroxidase activity (61), Trx1-mediated transnitrosylation may ultimately be a cellular defense mechanism to preserve Prx1 from overoxidation (Fig. 7). Within physiological environments, the Trx1/Prx1/Srx system can adequately handle reactive oxygen species (ROS; Fig. 7A). However, during prolonged ROS exposure, the TrxR/Trx1 system may be overextended, resulting in the accumulation of oTrx1. Prx1 may become overoxidized to form Prx1-SO2H, which can be repaired by Srx (Fig. 7B). However, Prx1 becomes overoxidized to Prx-SO3H and is inactivated as a peroxidase with escalating ROS levels (62). In contrast, in the presence of NO donors and oTrx1, SNO-Trx1 was produced, and this transnitrosylated Prx1 (Fig. 7C), preventing Prx1 overoxidation. Once cellular ROS levels are reduced, specific denitrosylase can denitrosylate Prx1 and restore its peroxidase function.
Fig. 7.
Proposed function for Trx1-mediated transnitrosylation of Prx1. A, Prx1 peroxidase cycle. Peroxides oxidize Prx1-SPH into Prx1-SPOH (I), which can then form a disulfide bond with the SRH in another Prx1 molecule to form a covalent dimer (II); this dimer can then be reduced by Trx1 (III). B, overoxidation and Srx repair cycle. Prx1-SPOH can be further oxidized to Prx1-SPO2H (IV), which can either be repaired by Srx (V) or terminally oxidized to Prx1-SPO3H (VI), resulting in inactivation of its peroxidase activity. C, nitrosylation cycle. During periods of elevated ROS, oTrx1 can accumulate and be nitrosylated by NO donors into SNO-Trx1 (VII), which in turn transnitrosylates Prx1 into SNO-Prx1 (VIII). SNO-Prx1 may become reactivated by denitrosylases (IX).
Trx1-mediated Transnitrosylation Protects HeLa Cells from Apoptosis
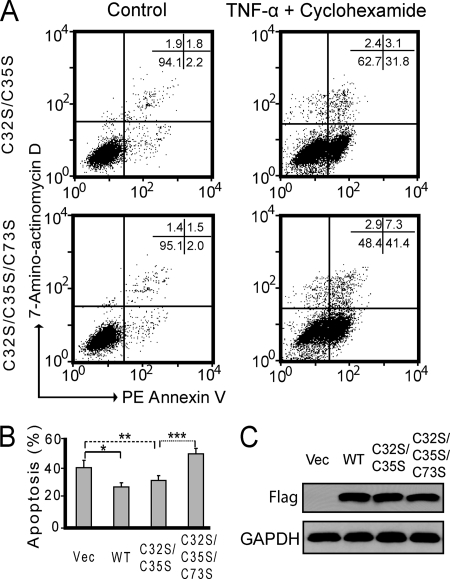
As demonstrated by previous studies, overexpression of WT Trx1 can inhibit apoptosis induced by TNF-α in endothelial cells, whereas expression of TrxC32S/C35S shows only a partial inhibition of the apoptosis (37), a function that is further diminished with NOS inhibition. To determine whether Trx1 Cys73-mediated transnitrosylation of proteins plays a role in protecting cells against apoptosis, we compared the degree of apoptosis in HeLa cells expressing WT Trx1, Trx1C32S/C35S, and Trx1C32S/C35S/C73S mutants treated with TNF-α and CHX (Fig. 8, A and B). Comparable expression of all plasmids was verified by Western blotting (Fig. 8C). As expected, Trx1 significantly inhibited apoptosis (p = 0.004; Fig. 8B), whereas Trx1C32S/C35S showed a degree of apoptosis inhibition comparable with that of WT Trx1 (p = 0.04; Fig. 8B). By comparison, Trx1C32S/C35S/C73S did not protect the cells from apoptosis compared with vector-expressing cells (Fig. 8B). Trx1C32S/C35S/C73S-expressing cells had significantly elevated apoptosis compared with Trx1C32S/C35-expressing cells (p = 0.002; Fig. 8, A and B), suggesting that Cys73-mediated transnitrosylation (Fig. 2C) is important for protecting HeLa cells from TNF-α- and CHX-induced apoptosis. Therefore, Trx1-mediated transnitrosylation and disulfide reduction (2, 15) are both important for protecting cells from apoptosis.
Fig. 8.
Trx1-mediated transnitrosylation protects HeLa cells from apoptosis. HeLa cells overexpressing WT Trx1, Trx1C32S/C35S, and Trx1C32S/C35S/C73S were treated with 5 μg/ml CHX and 20 ng/ml TNF-α for 6 h. The apoptosis assay by flow cytometry is described under “Experimental Procedures.” All cell types showed an increase in apoptosis when treated with TNF-α and CHX (A and B). Overexpression of WT Trx1 significantly inhibited apoptosis (*, p = 0.005) (B). Trx1C32S/C35S/C73S-overexpressing cells had significantly higher cellular apoptosis compared with Trx1C32S/C35S-overexpressing cells (***, p = 0.002) (A and B) and inversely correlated with protein nitrosylation levels in these cells (Fig. 2C). Values are the mean ± S.E.; n = 4; *, p = 0.005; **, p = 0.04; ***, p = 0.002. Expression of WT Trx1 and mutants was confirmed by Western blotting using an anti-FLAG antibody (C). PE, phycoerythrin; Vec, vector.
The transnitrosylation activity of SNO-Trx1 clearly has pathological/pharmacological relevancy. The role of Trx-mediated transnitrosylation following the chemical inhibition of Trx/TrxR during cancer therapy (63, 64) or when the Trx system becomes overwhelmed by cellular oxidative stress during events such as ischemia reperfusion needs to be investigated. Although it is assumed that TrxR can maintain Trx1 in its reduced status, we were able to detect substantial levels of oTrx1 in several tissues, including lung and kidney (supplemental Fig. S7C). In oxidatively active tissues transnitrosylation-active Trx1 may be important for both normal cell physiology and the stress defense response. Therefore, our findings are important both for the understanding of oxidative stress-related pathologies and for therapeutics targeting transnitrosylation.
Conclusion
In this study, we have delineated a redox-based mechanism that distinguishes Trx1-mediated transnitrosylation from its denitrosylation function and identified putative consensus sequence motifs among Trx1 transnitrosylation targets. Trx1 normally protects proteins via its reductase activity. Within highly oxidative environments, oTrx1 accumulates, becomes S-nitrosylated, and offers an alternative modality of protein regulation via transnitrosylation. Trx1-mediated transnitrosylation of specific proteins may serve two purposes: (a) temporarily preventing proteins from acquiring irreversible oxidative modifications when the cellular antioxidant systems can no longer reduce the proteins (17) and (b) serving as a signaling mechanism for altering protein function, contributing to the cellular stress response.
Supplementary Material
Acknowledgments
We appreciate the suggestions from Drs. Andrew Harris, Annie Beuve, Raymond Birge, and Carolyn Suzuki during the preparation of this manuscript.
* This work was supported, in whole or in part, by National Institutes of Health Grant NS046593 (to H. L. for support of a University of Medicine and Dentistry of New Jersey (UMDNJ) Neuroproteomics Core Facility) and Grants AG23039 and HL91469 (to J. S.). This work was also supported by a UMDNJ Foundation grant (to H. L. and J. S.).
 This article contains supplemental Tables S1 and S2 and Figs. S1–S10.
This article contains supplemental Tables S1 and S2 and Figs. S1–S10.
1 The abbreviations used are:
- Trx
- thioredoxin
- 2DE
- two-dimensional gel electrophoresis
- BCA
- bicinchoninic acid
- biotin-HPDP
- N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)propionamide
- Casp3
- caspase 3
- Casp3p
- caspase 3 peptide
- CHX
- cycloheximide
- DNCB
- 1-chloro-2,4-dinitrobenzene
- GSNO
- S-nitrosoglutathione
- HSP
- heat shock protein
- MMTS
- methyl methanethiosulfonate
- NB
- nitrosylation buffer
- oTrx1
- disulfide bond form of Trx1
- Prx
- peroxiredoxin
- RB
- resuspension buffer
- rTrx1
- reduced form of Trx1
- SNO-BSA
- S-nitrosylated BSA
- SNO-Casp3p
- S-nitrosylated Casp3p
- SNO-Prx1
- nitrosylated Prx1
- SNO-Trx1
- S-nitrosylated Trx1
- Srx
- sulfiredoxin
- TrxR
- thioredoxin reductase
- Txnip
- thioredoxin-interacting protein
- ROS
- reactive oxygen species.
REFERENCES
- 1.Arnold W. P., Mittal C. K., Katsuki S., Murad F. (1977) Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. U.S.A. 74, 3203–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mannick J. B. (2007) Regulation of apoptosis by protein S-nitrosylation. Amino Acids 32, 523–526 [DOI] [PubMed] [Google Scholar]
- 3.Foster M. W., Hess D. T., Stamler J. S. (2009) Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J., Xin C., Eu J. P., Stamler J. S., Meissner G. (2001) Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. U.S.A. 98, 11158–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao C., Guo H., Wei J., Mi Z., Wai P. Y., Kuo P. C. (2005) Identification of S-nitrosylated proteins in endotoxin-stimulated RAW264.7 murine macrophages. Nitric Oxide 12, 121–126 [DOI] [PubMed] [Google Scholar]
- 6.Sayed N., Baskaran P., Ma X., van den Akker F., Beuve A. (2007) Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 104, 12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benhar M., Forrester M. T., Hess D. T., Stamler J. S. (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaffrey S. R., Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 9.Greco T. M., Hodara R., Parastatidis I., Heijnen H. F., Dennehy M. K., Liebler D. C., Ischiropoulos H. (2006) Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. U.S.A. 103, 7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao G., Derakhshan B., Shi L., Campagne F., Gross S. S. (2006) SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. U.S.A. 103, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefièvre L., Chen Y., Conner S. J., Scott J. L., Publicover S. J., Ford W. C., Barratt C. L. (2007) Human spermatozoa contain multiple targets for protein S-nitrosylation: an alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics 7, 3066–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torta F., Usuelli V., Malgaroli A., Bachi A. (2008) Proteomic analysis of protein S-nitrosylation. Proteomics 8, 4484–4494 [DOI] [PubMed] [Google Scholar]
- 13.Paige J. S., Xu G., Stancevic B., Jaffrey S. R. (2008) Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 15, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell D. A., Marletta M. A. (2005) Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 1, 154–158 [DOI] [PubMed] [Google Scholar]
- 15.Mitchell D. A., Morton S. U., Fernhoff N. B., Marletta M. A. (2007) Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl. Acad. Sci. U.S.A. 104, 11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weichsel A., Brailey J. L., Montfort W. R. (2007) Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry 46, 1219–1227 [DOI] [PubMed] [Google Scholar]
- 17.Hashemy S. I., Holmgren A. (2008) Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J. Biol. Chem. 283, 21890–21898 [DOI] [PubMed] [Google Scholar]
- 18.Nedospasov A., Rafikov R., Beda N., Nudler E. (2000) An autocatalytic mechanism of protein nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 97, 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pezacki J. P., Pelling A., Kluger R. (2000) S-Nitrosylation of cross-linked hemoglobins at β-cysteine-93: stabilized hemoglobins as nitric oxide sources. J. Am. Chem. Soc. 122, 10734–10735 [Google Scholar]
- 20.McMahon T. J., Doctor A. (2006) Extrapulmonary effects of inhaled nitric oxide: role of reversible S-nitrosylation of erythrocytic hemoglobin. Proc. Am. Thorac. Soc. 3, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jourd'heuil D., Laroux F. S., Miles A. M., Wink D. A., Grisham M. B. (1999) Effect of superoxide dismutase on the stability of S-nitrosothiols. Arch. Biochem. Biophys. 361, 323–330 [DOI] [PubMed] [Google Scholar]
- 22.Johnson M. A., Macdonald T. L., Mannick J. B., Conaway M. R., Gaston B. (2001) Accelerated S-nitrosothiol breakdown by amyotrophic lateral sclerosis mutant copper,zinc-superoxide dismutase. J. Biol. Chem. 276, 39872–39878 [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Yan Y., Zeng M., Zhang J., Hanes M. A., Ahearn G., McMahon T. J., Dickfeld T., Marshall H. E., Que L. G., Stamler J. S. (2004) Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 [DOI] [PubMed] [Google Scholar]
- 24.Que L. G., Liu L., Yan Y., Whitehead G. S., Gavett S. H., Schwartz D. A., Stamler J. S. (2005) Protection from experimental asthma by an endogenous bronchodilator. Science 308, 1618–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lima B., Lam G. K., Xie L., Diesen D. L., Villamizar N., Nienaber J., Messina E., Bowles D., Kontos C. D., Hare J. M., Stamler J. S., Rockman H. A. (2009) Endogenous S-nitrosothiols protect against myocardial injury. Proc. Natl. Acad. Sci. U.S.A. 106, 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zai A., Rudd M. A., Scribner A. W., Loscalzo J. (1999) Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J. Clin. Investig. 103, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore E. C. (1967) A thioredoxin–thioredoxin reductase system from rat tumor. Biochem. Biophys. Res. Commun. 29, 264–268 [DOI] [PubMed] [Google Scholar]
- 28.Fu C., Wu C., Liu T., Ago T., Zhai P., Sadoshima J., Li H. (2009) Elucidation of thioredoxin target protein networks in mouse. Mol. Cell. Proteomics 8, 1674–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang B., Chen C. (2006) An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic. Biol. Med. 41, 562–567 [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Liu T., Wu C., Li H. (2008) A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannick J. B., Schonhoff C. M. (2006) Analysis of protein S-nitrosylation. Curr. Protoc. Protein Sci. 46, 14.6.1–14.6.22 [DOI] [PubMed] [Google Scholar]
- 33.Watson W. H., Pohl J., Montfort W. R., Stuchlik O., Reed M. S., Powis G., Jones D. P. (2003) Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J. Biol. Chem. 278, 33408–33415 [DOI] [PubMed] [Google Scholar]
- 34.Das A., Li H., Liu T., Bellofatto V. (2006) Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150, 201–210 [DOI] [PubMed] [Google Scholar]
- 35.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 36.Nikitovic D., Holmgren A. (1996) S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 37.Haendeler J., Hoffmann J., Tischler V., Berk B. C., Zeiher A. M., Dimmeler S. (2002) Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat. Cell Biol. 4, 743–749 [DOI] [PubMed] [Google Scholar]
- 38.Sengupta R., Ryter S. W., Zuckerbraun B. S., Tzeng E., Billiar T. R., Stoyanovsky D. A. (2007) Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry 46, 8472–8483 [DOI] [PubMed] [Google Scholar]
- 39.Hirota K., Matsui M., Iwata S., Nishiyama A., Mori K., Yodoi J. (1997) AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl. Acad. Sci. U.S.A. 94, 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nisoli E., Clementi E., Paolucci C., Cozzi V., Tonello C., Sciorati C., Bracale R., Valerio A., Francolini M., Moncada S., Carruba M. O. (2003) Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science 299, 896–899 [DOI] [PubMed] [Google Scholar]
- 41.Bulotta S., Ierardi M. V., Maiuolo J., Cattaneo M. G., Cerullo A., Vicentini L. M., Borgese N. (2009) Basal nitric oxide release attenuates cell migration of HeLa and endothelial cells. Biochem. Biophys. Res. Commun. 386, 744–749 [DOI] [PubMed] [Google Scholar]
- 42.Dyson H. J., Jeng M. F., Model P., Holmgren A. (1994) Characterization by 1H NMR of a C32S,C35S double mutant of Escherichia coli thioredoxin confirms its resemblance to the reduced wild-type protein. FEBS Lett. 339, 11–17 [DOI] [PubMed] [Google Scholar]
- 43.Weichsel A., Gasdaska J. R., Powis G., Montfort W. R. (1996) Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer. Structure 4, 735–751 [DOI] [PubMed] [Google Scholar]
- 44.Oblong J. E., Berggren M., Gasdaska P. Y., Powis G. (1994) Site-directed mutagenesis of active site cysteines in human thioredoxin produces competitive inhibitors of human thioredoxin reductase and elimination of mitogenic properties of thioredoxin. J. Biol. Chem. 269, 11714–11720 [PubMed] [Google Scholar]
- 45.Haendeler J., Hoffmann J., Zeiher A. M., Dimmeler S. (2004) Antioxidant effects of statins via S-nitrosylation and activation of thioredoxin in endothelial cells: a novel vasculoprotective function of statins. Circulation 110, 856–861 [DOI] [PubMed] [Google Scholar]
- 46.Stoyanovsky D. A., Tyurina Y. Y., Tyurin V. A., Anand D., Mandavia D. N., Gius D., Ivanova J., Pitt B., Billiar T. R., Kagan V. E. (2005) Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J. Am. Chem. Soc. 127, 15815–15823 [DOI] [PubMed] [Google Scholar]
- 47.Arnelle D. R., Stamler J. S. (1995) NO+, NO, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 318, 279–285 [DOI] [PubMed] [Google Scholar]
- 48.Forrester M. T., Seth D., Hausladen A., Eyler C. E., Foster M. W., Matsumoto A., Benhar M., Marshall H. E., Stamler J. S. (2009) Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J. Biol. Chem. 284, 36160–36166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 50.Hall A., Karplus P. A., Poole L. B. (2009) Typical 2-Cys peroxiredoxins—structures, mechanisms and functions. FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Biteau B., Labarre J., Toledano M. B. (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 52.Wagner E., Luche S., Penna L., Chevallet M., Van Dorsselaer A., Leize-Wagner E., Rabilloud T. (2002) A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem. J. 366, 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem. 277, 38029–38036 [DOI] [PubMed] [Google Scholar]
- 54.Bosworth C. A., Toledo J. C., Jr., Zmijewski J. W., Li Q., Lancaster J. R., Jr. (2009) Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 106, 4671–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vignols F., Bréhélin C., Surdin-Kerjan Y., Thomas D., Meyer Y. (2005) A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 16729–16734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manevich Y., Feinstein S. I., Fisher A. B. (2004) Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U.S.A. 101, 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee W., Choi K. S., Riddell J., Ip C., Ghosh D., Park J. H., Park Y. M. (2007) Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J. Biol. Chem. 282, 22011–22022 [DOI] [PubMed] [Google Scholar]
- 58.Diet A., Abbas K., Bouton C., Guillon B., Tomasello F., Fourquet S., Toledano M. B., Drapier J. C. (2007) Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J. Biol. Chem. 282, 36199–36205 [DOI] [PubMed] [Google Scholar]
- 59.Koo K. H., Lee S., Jeong S. Y., Kim E. T., Kim H. J., Kim K., Song K., Chae H. Z. (2002) Regulation of thioredoxin peroxidase activity by C-terminal truncation. Arch. Biochem. Biophys. 397, 312–318 [DOI] [PubMed] [Google Scholar]
- 60.Sayed A. A., Williams D. L. (2004) Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J. Biol. Chem. 279, 26159–26166 [DOI] [PubMed] [Google Scholar]
- 61.Fang J., Nakamura T., Cho D. H., Gu Z., Lipton S. A. (2007) S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 104, 18742–18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neumann C. A., Cao J., Manevich Y. (2009) Peroxiredoxin 1 and its role in cell signaling. Cell Cycle 8, 4072–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo M. H., Xu X. M., Carlson B. A., Patterson A. D., Gladyshev V. N., Hatfield D. L. (2007) Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS One 2, e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.López-Sánchez L. M., Corrales F. J., López-Pedrera C., Aranda E., Rodríguez-Ariza A. (2010) Pharmacological impairment of S-nitrosoglutathione or thioredoxin reductases augments protein S-nitrosation in human hepatocarcinoma cells. Anticancer Res. 30, 415–421 [PubMed] [Google Scholar]
- 65.López-Sánchez L. M., Corrales F. J., González R., Ferrín G., Muñoz-Castañeda J. R., Ranchal I., Hidalgo A. B., Briceño J., López-Cillero P., Gómez M. A., De La Mata M., Muntané J., Rodríguez-Ariza A. (2008) Alteration of S-nitrosothiol homeostasis and targets for protein S-nitrosation in human hepatocytes. Proteomics 8, 4709–4720 [DOI] [PubMed] [Google Scholar]
- 66.Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 67.Molina y Vedia L., McDonald B., Reep B., Brüne B., Di Silvio M., Billiar T. R., Lapetina E. G. (1992) Nitric oxide-induced S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase inhibits enzymatic activity and increases endogenous ADP-ribosylation. J. Biol. Chem. 267, 24929–24932 [PubMed] [Google Scholar]
- 68.Padgett C. M., Whorton A. R. (1995) S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am. J. Physiol. Cell Physiol. 269, C739–C749 [DOI] [PubMed] [Google Scholar]
- 69.Mohr S., Stamler J. S., Brüne B. (1996) Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J. Biol. Chem. 271, 4209–4214 [DOI] [PubMed] [Google Scholar]
- 70.Mohr S., Hallak H., de Boitte A., Lapetina E. G., Brüne B. (1999) Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 274, 9427–9430 [DOI] [PubMed] [Google Scholar]
- 71.Martínez-Ruiz A., Lamas S. (2004) Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch. Biochem. Biophys. 423, 192–199 [DOI] [PubMed] [Google Scholar]
- 72.Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 73.Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang B., Chen S. C., Wang D. L. (2009) Shear flow increases S-nitrosylation of proteins in endothelial cells. Cardiovasc. Res. 83, 536–546 [DOI] [PubMed] [Google Scholar]
- 75.Wadham C., Parker A., Wang L., Xia P. (2007) High glucose attenuates protein S-nitrosylation in endothelial cells: role of oxidative stress. Diabetes 56, 2715–2721 [DOI] [PubMed] [Google Scholar]
- 76.Vieira H. L., Belzacq A. S., Haouzi D., Bernassola F., Cohen I., Jacotot E., Ferri K. F., El Hamel C., Bartle L. M., Melino G., Brenner C., Goldmacher V., Kroemer G. (2001) The adenine nucleotide translocator: a target of nitric oxide, peroxynitrite, and 4-hydroxynonenal. Oncogene 20, 4305–4316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.