Abstract
Many essential cellular processes depend upon the self-assembly of stable multiprotein entities. The architectures of the vast majority of these protein machines remain unknown because these structures are difficult to obtain by biophysical techniques alone. However, recent progress in defining the architecture of protein complexes has resulted from integrating information from all available biochemical and biophysical sources to generate computational models. Chemical cross-linking is a technique that holds exceptional promise toward achieving this goal by providing distance constraints that reflect the topography of protein complexes. Combined with the available structural data, these constraints can yield three-dimensional models of higher order molecular machines. However, thus far the utility of cross-linking has been thwarted by insufficient yields of cross-linked products and tandem mass spectrometry methods that are unable to unambiguously establish the identity of the covalently labeled peptides and their sites of modification. We report the cross-linking of amino moieties by 1,3-diformyl-5-ethynylbenzene (DEB) with analysis by high resolution electron transfer dissociation. This new reagent coupled with this new energy deposition technique addresses these obstacles by generating cross-linked peptides containing two additional sites of protonation relative to conventional cross-linking reagents. In addition to excellent coverage of sequence ions by electron transfer dissociation, DEB cross-linking produces gas-phase precursor ions in the 4+, 5+, or 6+ charge states that are readily segregated from unmodified and dead-end modified peptides using charge-dependent precursor selection of only quadruply and higher charge state ions. Furthermore, electron transfer induces dissociation of the DEB-peptide bonds to yield diagnostic ion signals that reveal the “molecular ions” of the unmodified peptides. We demonstrate the power of this strategy by cross-linking analysis of the 21-protein, ADP-bound GroEL-GroES chaperonin complex. Twenty-five unique sites of cross-linking were determined.
A wide range of cellular processes are mediated by stable protein complexes that range in subunit size from a few proteins, e.g. the signal recognition particle, to over a hundred, e.g. the spliceosome. Indeed, protein interactions underlie an enormous scope of physiological and pathophysiological processes, encompassing everything from cell cycle regulation to initiation of apoptosis, angiogenesis, and aberrant interactions in cancer.
Presently, the subunit compositions, dynamics, and topographies as well as the overall architectures of most multimeric complexes remain unknown. With a handful of exceptions, most notably the crystal structures of the large and small ribosomal subunits, which were early synchrotron successes determined some 10 years ago (1–3), large molecular machines have proven recalcitrant to high resolution structural analysis. Conversely, a large number of individual cytosolic proteins have been studied at atomic resolution as have a few membrane proteins. However, despite this individual level of detail, our knowledge of structural information on most complexes is inadequate, and new approaches aimed toward unraveling these structures are necessary.
One approach that has proven successful is the generation of computational models that integrate biophysical and biochemical information. For instance, structural data from cryoelectron microscopy of intact complexes and crystallography of individual subunits have been combined with proteomics-based experiments that provide composition and neighbor relationships to model a variety of protein complexes (4).
A promising strategy to further refine the modeling process involves chemical cross-linking in conjunction with modern tandem mass spectrometry. Analysis of covalently cross-linked proteins provides information on subunit interfaces and generates distance constraints that reflect the topography of the complex (5–7). For example, we have previously used such an approach to generate a structure of the bacterial signal recognition particle in complex with its receptor (8). Using cross-link constraints, the modeled structure was in agreement with the x-ray structure deduced later and additionally revealed information on the location of the M-domain that failed to diffract. Taking advantage of recent advances in high resolution mass spectrometry, chemical cross-linking of the 15-protein RNA polymerase II-transcription factor IIF complex has been carried out using the reagent BS3.1 The spatial proximities obtained revealed the locations of yeast transcription factor IIF on the model of the polymerase II surface (9).
However, chemical cross-linking is still not viewed as a robust technique for generating structural information because of the complexities involved in achieving the desired results. These include the low yield of cross-linked products and the complexity of the digested reaction mixture. Moreover, the lack of widespread progress or adoption of cross-linking analysis by the protein biology community rests on the over-reliance by research groups on commercially available chemical reagents that have not been constructed with the ease of successful analysis by mass spectrometry in mind.
Any reagent designed to solve this challenge must take advantage of the power and sensitivity of modern tandem mass spectrometry to provide the sequence and sites of modification for the peptides that constitute the cross-link. The most basic requirement is to experimentally optimize the formation of sequence ion series that will define both peptides and their sites of attachment in a cross-linked species. Both electron transfer dissociation (ETD) (10) and electron capture dissociation (11) energy deposition processes would appear to have a major advantage over the use of collision-induced dissociation (CID) with respect to the analysis of large, highly charged species such as cross-linked peptides. These processes produce extensive, evenly-distributed dissociation along peptide backbones that show less influence to the sequence of the analyte peptide than CID.
To fully exploit this advantage, it is desirable to generate precursor ions with maximal charge state as ETD is known to function optimally with low m/z precursors (12). Furthermore, highly charged analytes can be specifically analyzed in the presence of complex mixtures on the basis of charge state-dependent precursor ion selection in the mass spectrometer (9, 13). From this perspective, commercial cross-linking reagents, which react by acylation of lysine residues, are suboptimal as they remove two potential sites of protonation from any cross-linked species formed.
A number of strategies use labeling with stable isotopes to facilitate identification of cross-linked peptides within complex reaction mixtures on the basis of the unique isotopic signatures bestowed (14–21). However, precursor ion selection based on an isotope pattern is difficult to implement, whereas selection based on charge state is an option routinely available on all tandem mass spectrometers. Hence, isotopic labeling schemes only serve as an aid to correct identification rather than a means to increase the yield of cross-linked peptides that may be selected for MS/MS. Other strategies incorporate a chemical tag into the cross-linking reagent that allows enrichment of labeled peptides (19, 22–25).
However, of the number of chemical cross-linking reagents reported in recent years, nearly all rely on activated esters, such as N-hydroxysuccinimide esters, to acylate the amino moieties of proteins. In addition to decreasing the number of basic sites on the cross-linked peptides, our laboratory has noted that peptides cross-linked in this manner often provide poor sequence information in both CID and ETD MS/MS. Often the product ion spectra favor fragment ions from only one of the two peptides, and sequence ions that contain the site of cross-linking are less frequently observed than unmodified fragments.2 This difficulty has prompted the development of gas-phase cleavable cross-linking reagents (26–32). These reagents dissociate in the mass spectrometer to unmask the now linear component peptides but generally require an additional stage of activation to obtain sequence ions.
Here we describe cross-linking by a novel chemical reagent, 1,3-diformyl-5-ethynylbenzene (DEB), that forms Schiff bases between lysyl ε-amino functions at protein-protein interfaces. These are readily reduced by cyanoborohydride to join proteins through secondary amino linkages. Thus, DEB inserts a rigid backbone spacer between lysyl ε-amino functions while preserving them as sites of protonation that, as noted above, are advantageous for ETD sequence determination. We show the utility of this strategy in studies of the 21-subunit GroEL-GroES chaperonin complex. This complex facilitates protein folding by sequestering polypeptide chains in a compartment formed by a nucleotide-bound, seven-membered homooligomeric GroEL ring and a seven-membered homooligomeric GroES ring. An additional GroEL ring, the trans ring, sits adjacent to and facing the cis (GroES-bound) ring but bears a collapsed conformation, which is unable to bind GroES until ATP hydrolysis and the completion of the catalytic cycle (33–35).
EXPERIMENTAL PROCEDURES
Materials
GroEL and GroES, prepared from overexpression in Escherichia coli, were a generous gift from the Frydman laboratory at Stanford University. Protein concentrations were determined by Bradford assay. Sequencing grade methylated trypsin was ordered from Promega. Synthetic reagents and other chemicals were purchased from Sigma-Aldrich at the highest grade available except for sodium cyanoborohydride (95%), which was ordered from Acros Organics, and formic acid (98%), which was from Fluka. HPLC grade solvents were purchased from Fisher Scientific.
Synthesis of DEB
Generally the procedure of Bhagwat et al. (36) was followed for the preparation of 1,3-dihydroxymethyl-5-ethynylbenzene from diethyl-5-hydroxyisophthalate with the exception that the trimethylsilyl (TMS) group was not removed. Oxidation to the dialdehyde was effected by the Dess-Martin procedure. Briefly, 0.7 mmol of 1,3-dihydroxymethyl-5-trimethylsilyl-ethynylbenzene was stirred with 1.8 mmol of Dess-Martin periodinane in CH2Cl2 at 0 °C. After 1 h, the ice bath was removed, and the mixture was stirred for an additional 2 h. The mixture was washed with aqueous sodium thiosulfate and then with aqueous sodium bicarbonate and dried, and the residue was purified by flash chromatography on silica gel using a solvent system consisting of 15% ethyl acetate, 84.9% hexane, 0.1% formic acid to yield the TMS-protected diformyl compound.
The TMS group was then removed by dissolving the 1,3-diformyl-5-trimethylsilylethynylbenzene in 5:1 MeOH:THF and adding a 3 molar excess of 2 n K2CO3 solution dropwise. Saturated ammonium chloride solution was added, and the product was extracted into ethyl acetate, which was dried and removed under vacuum. The crude product was purified by flash chromatography on silica gel using a mobile phase of 17% ethyl acetate, 82.9% hexane, 0.1% formic acid. The final product was characterized by 1H NMR (400 MHz, CDCl3) δ: 10.09 (s, 2H), 8.35 (t, J = 1.53 Hz, 1H), 8.23 (d, J = 1.54 Hz, 2H), 3.27 (s, 1H); and high resolution MS (ESI): m/z [M + H]+ calculated for [C10H7O2]+, 159.0441; observed, 159.0440.
Cross-linking Reactions
GroEL and GroES were exchanged into buffer A (50 mm HEPES, 20 mm Mg(OAc)2, 10 mm KCl, pH 8.0) by using 5- and 30-kDa cutoff Ultrafree MC centrifugal concentrators (Millipore). Cross-linking reactions contained 33 μg of GroEL and 17 μg of GroES in a final volume of 50 μl of buffer A (final protein concentrations, 11 and 33 μm for GroEL and GroES, respectively). Cross-linking reactions also contained 1 mm adenosine diphosphate (ADP) (from 10× stock in buffer A) and 2.5 mm DEB (from 40× stock in DMSO). Reactions were equilibrated to 37 °C, and reduction of Schiff base adducts (cross-linking) was initiated by the addition of NaCNBH3 to 20 mm final concentration (from 50× stock in 0.01 n NaOH). Reactions were incubated at 37 °C for 1 h and terminated by acetone precipitation. 200 μl of ice cold acetone was added, and the samples were placed on dry ice for 2 h. The protein was recovered by centrifugation for 20 min at 15,000 × g and then washed twice with ice-cold acetone. Residual acetone was removed on a SpeedVac.
The acetone-precipitated pellets were then dissolved in Laemmli sample buffer, separated by SDS-PAGE, and visualized with Coomassie Blue staining. The region of the gel corresponding to molecular mass greater than 54 kDa was divided into four gel bands, which were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin overnight at 37 °C. Peptides were extracted from the gel slices using two aliquots of 5% formic acid, 50% acetonitrile solution with 30 min of vortexing and sonication. Extracts were evaporated to dryness, desalted on C18 OMIX solid phase extraction tips (Varian). Approximately 0.5% of each sample was injected for LC-MS/MS analysis.
The DEB molecule was designed with an aryl alkyne moiety to allow click chemistry-based enrichment. However, for simple systems, enrichment of cross-linked peptides by SDS-PAGE isolation of high molecular weight protein species followed by in-gel tryptic digestion and charge state-dependent selection of ETD precursor ions was found to be an excellent method of isolating cross-linked species. We will present the results from the bioconjugate enrichments and depletions of cross-linked reaction mixtures elsewhere.3
Mass Spectrometry
Tryptic digests of cross-linked proteins were separated by reverse phase chromatography using a Waters Nanoacquity ultraperformance LC system equipped with a 100-μm-inner diameter × 100-mm column packed with 1.7-μm-diameter, 300-Å-pore size C18 particles (Waters). Peptide mixtures were separated using 90-min gradients of 2–30% solvent B (solvent A, 0.1% formic acid in H2O; solvent B: 0.1% formic acid in acetonitrile) at a flow rate of 400 nl/min.
Eluting peptides were analyzed on an ESI LTQ-Orbitrap XL with an ETD module installed (Thermo Scientific). Precursor spectra were measured in the Orbitrap analyzer by averaging two microscans of 100-ms maximal duration with an automatic gain control setting of 2 × 106. Components observed in quadruply charged or higher charge states were selected for ETD analysis. High resolution ETD product ion spectra were measured in the Orbitrap averaging two microscans of 500-ms maximal duration and an automatic gain control setting of 2 × 105. The minimal signal required for product ion selection was 10,000. ETD activation times were varied between 50 and 200 ms, and supplemental activation was turned on. The precursor isolation window was set to 5 m/z units. In some experiments, ETD or CID of the same precursors was simultaneously measured in the linear ion trap.
Data Analysis
Separate ETD and CID peak lists were generated using the in-house package PAVA (version, July 28, 2009) (37) such that optimal interrogation of ETD-specific fragmentation and sequence correlation was possible (38). Putative cross-linked peptides were identified by searching for arbitrary mass modifications greater than 400 Da on Lys residues or at protein N termini using Protein Prospector version 5.6. These searches were performed with parent mass tolerance of 100 ppm and a fragment tolerance of 20 ppm. Carbamidomethylcysteine was searched as a constant modification. Variable modifications included methionine oxidation, loss of initiator methionine, protein N-terminal acetylation, and peptide N-terminal glutamine cyclization to pyroglutamate. Additionally, type 0 (“dead-end”) DEB modifications (C10H6O1) and type 0 DEB modifications in which the free aldehyde is reduced to an alcohol (C10H8O1) were searched as variable modifications on lysine residues and protein N termini. We use the nomenclature: type 0, 1, and 2 to refer to dead-end, intrapeptide, and interpeptide modifications, respectively (39). Mass modifications of any integer value between 400 and 5000 Da on lysine residues or protein N termini were searched as variable modifications. These searches look for mass modifications over a range of integers plus a mass defect based on the averagine mass (40). Thus, the elemental formula of the hypothetical species in the mass modification searches is unknown, necessitating a precursor mass tolerance larger than the instrumental limitation. No more than two variable modifications were allowed on any given peptide. Up to three missed tryptic cleavages were allowed, and variable modifications that modified lysine residues were not counted as one of these three missed cleavages.
Mass modification searches were performed against a restricted database consisting of only GroEL and GroES. An earlier search of the data against the Swiss-Prot (version, December 15, 2009) database (513,877 entries) had established these as the only major components in the sample (the other results were contaminating cytosolic E. coli proteins identified by four or fewer peptides). This search used the same parameters as above except that the precursor mass tolerance was set to 20 ppm, and mass modifications were turned off.
As a default setting, Batch-Tag in Protein Prospector only considers the 20 most intense peaks in each half of the mass range of any given MS/MS spectrum to search a total of 40 peaks. However, putative cross-linked spectra, e.g. those that were identified as bearing an arbitrary modification greater than 400 Da, were re-searched using the 100 most intense product ion signals in the peak list. The top 100 scoring peptides in this search were examined for complementarity. That is, this list was examined for pairs of peptides whose mass values combined with the mass of the DEB cross-linker bridge (C10H6) equal the mass of the selected precursor M + H (with a tolerance of 15 ppm). This search was performed using a developmental version of Protein Prospector, version 5.3.1xl. The developmental cross-linking version of Protein Prospector assigns a peptide score to each individual peptide hit as well as an overall score to the cross-linked peptide. The complementarity search features will be added to the public version of Protein Prospector (http://prospector.ucsf.edu) with the next major release.
All of the hits from this complementarity search were then validated by manual annotation of the product ion spectra. Furthermore, the charge state and monoisotopic mass determination of the precursors were validated manually.
RESULTS
Complementary Mass Modification Searching
As described above, the ability of Protein Prospector to search for arbitrarily sized mass modifications on any lysine residue (40) was used to generate a list of putative cross-linked peptide species. From 8263 product ion spectra, 843 were found to match a tryptic peptide bearing a mass modification greater than 400 Da on an internal lysine or at the protein N terminus. A novel search strategy was then used to identify bona fide cross-linked peptides. This process involved re-searching a putatively cross-linked spectrum against all possible tryptic peptides that could be formed from the GroES-GroEL proteins, again allowing for arbitrary value mass modifications between 400 and 5000 Da, and examining the list of the top 100 resulting hits for complementarity. As noted already above, complementarity is defined as a peptide, P, whose mass modification value matches the mass of a second peptide, Q, plus the mass of the DEB bridge (C10H6) and vice versa. Although the mass modification search to the individual peptide requires sacrificing accurate mass on account of not knowing the elemental formula of the modification (typically, we use 100-ppm MS tolerance for Orbitrap data), the second search, which looks for complementarity, reintroduces mass tolerance that matches the limits of the instrumentation (15 ppm for Orbitrap data).
High Resolution ETD Analysis of Cross-linked Peptides
Based on the complementarity criterion, of the 843 putative spectra of cross-linked peptides, it was possible to assign 388 (46%) bona fide cross-linked peptides. However, this list was highly redundant as the same precursor was generally selected for dissociation multiple times and because the same cross-linked residues were observed in slightly different analogs with respect to methionine oxidations or missed cleavages. Finally the number of cross-linked peptides was reduced to 25 non-redundant lysine-lysine or lysine-protein N-terminal cross-links as shown in Table I.
Table I. Cross-linked peptide pairs from GroEL-GroES identified by complementary mass modification searching.
| No. | m/z | z | Errora | XL scoreb | %Fragc | Peptide Pd | P + He | P + H + XLe | Peptide Qd | Q + He | Q + H + XLe |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 448.995 | 4 | 3.02 | 40.3 | 100.0 | DVK(771.451)FGNDAR | 1021.506 | 1148.560 | VK(1146.550)MLR | 646.407 | 773.461 |
| 2 | 512.489 | 5 | 3.33 | 44.0 | 100.0 | VTLGPK(1730.907)GR | 827.510 | 954.564 | SFGAPTITK(952.557)DGVSVAR | 1605.860 | 1732.914 |
| 3 | 466.060 | 5 | 8.69 | 30.3 | 100.0 | VTLGPK(1498.762)GR | 827.510 | 954.564 | AVAAGMNPMDLK(952.569)R | 1373.703 | 1500.757 |
| 4 | 602.125 | 5 | 3.63 | 50.9 | 82.6 | NVVLDK(1416.706)SFGAPTITK | 1589.890 | 1716.944 | VGAATEVEMK(1714.939)EK | 1291.656 | 1418.710 |
| 5 | 518.278 | 6 | 9.89 | 50.4 | 97.9 | SFGAPTITK(1498.772)DGVSVAR | 1605.860 | 1732.914 | AVAAGMNPMDLK(1730.929)R | 1373.703 | 1500.757 |
| 6 | 819.853 | 5 | 3.67 | 51.4 | 65.5 | GYLSPYFINK(855.511)PETGAVELESPFILLADKK | 3239.724 | 3366.778 | K(3364.777)ISNIR | 730.457 | 857.511 |
| 7 | 709.071 | 6 | 7.68 | 85.2 | 68.8 | GYLSPYFINK(1009.664)PETGAVELESPFILLADKK | 3239.724 | 3366.778 | GIVK(3364.795)VAAVK | 884.593 | 1011.647 |
| 8 | 776.911 | 6 | 2.62 | 46.2 | 69.4 | GYLSPYFINK(1416.707)PETGAVELESPFILLADKK | 3239.724 | 3366.778 | VGAATEVEMK(3364.775)EK | 1291.656 | 1418.710 |
| 9 | 624.010 | 6 | 2.09 | 65.2 | 90.4 | GYLSPYFINK(499.297)PETGAVELESPFILLADKK | 3239.724 | 3366.778 | K(3364.770)AR | 374.251 | 501.305 |
| 10 | 435.779 | 4 | 2.87 | 28.4 | 95.8 | K(1009.636)ISNIR | 730.457 | 857.511 | GIVK(855.501)VAAVK | 884.593 | 1011.647 |
| 11 | 511.546 | 4 | 6.23 | 32.9 | 96.4 | K(1312.704)ISNIR | 730.457 | 857.511 | VAAVK(855.508)APGFGDR | 1187.653 | 1314.707 |
| 12 | 515.041 | 4 | 2.59 | 11.7 | 71.4 | K(1326.687)ISNIR | 730.457 | 857.511 | ATLEDLGQAK(855.490)R | 1201.654 | 1328.708 |
| 13 | 537.545 | 4 | 2.80 | 18.4 | 63.3 | K(1416.701)ISNIR | 730.457 | 857.511 | VGAATEVEMK(855.502)EK | 1291.656 | 1418.710 |
| 14 | 524.877 | 5 | 4.63 | 40.4 | 97.5 | VAAVK(1432.702)APGFGDR | 1187.653 | 1314.707 | VGAATEVEM(oxidation)K(1312.704)EK | 1307.651 | 1434.705 |
| 15 | 666.161 | 5 | 4.67 | 63.4 | 86.2 | VVINK(927.496)DTTTIIDGVGEEAAIQGR | 2399.278 | 2526.332 | EK(2524.332)LQER | 802.442 | 929.496 |
| 16 | 582.317 | 6 | 8.29 | 59.3 | 98.2 | QQIEEATSDYDREK(1250.803)LQER | 2238.063 | 2365.118 | VAK(2363.131)LAGGVAVIK | 1125.735 | 1252.790 |
| 17 | 382.223 | 5 | 4.42 | 35.8 | 95.0 | 739.4819-M(oxidation)NIRPLHDR | 1167.605 | 1294.659 | VIVK(1292.652)R | 614.435 | 741.489 |
| 18 | 406.017 | 5 | 2.08 | 46.7 | 95.5 | 858.4519-M(oxidation)NIRPLHDR | 1167.605 | 1294.659 | K(1292.648)EVETK | 733.409 | 860.463 |
| 19 | 613.931 | 5 | 2.91 | 78.1 | 91.7 | 1914.018-MNIRPLHDR | 1151.610 | 1278.664 | EVETK(1276.658)SAGGIVLTGSAAAK | 1788.970 | 1916.024 |
| 20 | 568.709 | 5 | 5.47 | 59.2 | 93.2 | 1671.909-M(oxidation)NIRPLHDR | 1167.605 | 1294.659 | SAGGIVLTGSAAAK(1292.659)STR | 1546.855 | 1673.909 |
| 21 | 368.976 | 4 | 0.14 | 19.8 | 93.8 | VIVK(858.447)R | 614.435 | 741.489 | K(739.473)EVETK | 733.409 | 860.463 |
| 22 | 506.496 | 5 | 2.50 | 15.0 | 47.6 | VIVK(1914.015)R | 614.435 | 741.489 | EVETK(739.480)SAGGIVLTGSAAAK | 1788.970 | 1916.024 |
| 23 | 572.338 | 4 | 1.09 | 9.4 | 44.7 | VIVK(1671.896)R | 614.435 | 741.489 | SAGGIVLTGSAAAK(739.476)STR | 1546.855 | 1673.909 |
| 24 | 513.087 | 5 | 0.36 | 38.1 | 100.0 | RK(1671.894)EVETK | 889.510 | 1016.564 | SAGGIVLTGSAAAK(1014.550)STR | 1546.855 | 1673.909 |
| 25 | 703.178 | 5 | 5.81 | 32.9 | 80.7 | SAGGIVLTGSAAAK(1965.008)STR | 1546.855 | 1673.909 | VGDIVIFNDGYGVK(1671.914)SEK | 1839.949 | 1967.003 |
a Mass error in ppm. The theoretical M + H is given by: P + Q + XL where P and Q are the unprotonated monoisotopic mass values of the individual peptides and XL represents the DEB fragment and charge bearing proton, C10H7.
b Protein Prospector XL score for cross-link match.
c Percent fragmentation is defined as the number of c- and z-ions observed divided by the number searched.
d The mass modifications reported by Protein Prospector are given in parentheses following the site of modification. Modifications to the protein N-terminus precede the peptide sequence. The modification on P is equal to Q + XL − H.
e Mass values of the diagnostic fragment ions.
Table I reports the individual peptide sequences, P and Q with the arbitrary mass modifications matched by Protein Prospector along with the overall XL score for the combination of peptides, and the fragmentation percentage observed for each match. Fragmentation percentage is defined as the number of sequence unique c- and z•-ions that were observed for a peptide sequence divided by the number of possible unique fragment ions (12). Most of the reported cross-linked peptides in this study produced excellent ETD sequence fragment ion coverage of both halves of the cross-linked peptides (see Figs. 1 and 2 and supplemental figures). We observed 18 of 25 spectra that were matched by over 80% of the possible sequence ions, whereas 23 of 25 were matched by over 50%.
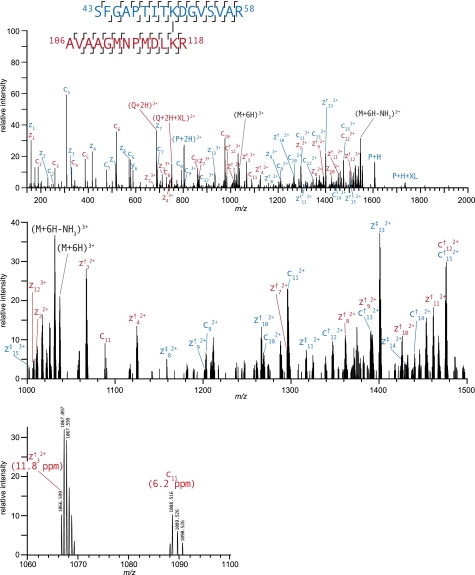
Fig. 1.
High resolution ETD product ion spectrum of m/z 518.2786+ corresponding to intersubunit DEB cross-link between Lys51 and Lys117 on GroEL. The dagger symbol marks c+1 and z+1 ions (c† and z†). The double dagger denotes c+2 and z+2 ions (c‡ and z‡). Diagnostic fragment ions, P + H and Q + H, match the M + H value of individual, unmodified peptides. The second panel shows a zoomed in view of the region from m/z 1000 to 1500, and the third panel is further zoomed in to show the product ions at isotopic resolution.
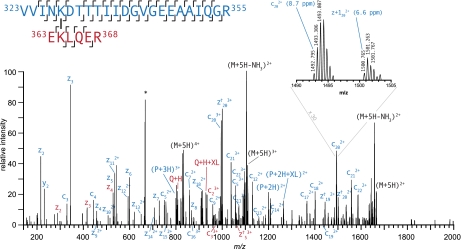
Fig. 2.
High resolution ETD product ion spectrum of m/z 666.1615+ corresponding to intersubunit DEB cross-link between Lys327 and Lys364 of GroEL. The Q + H and Q + H + XL signals confirm the identity of the peptide EKLQER. The asterisk (*) denotes contaminating signals that are within the precursor selection window. The dagger symbol marks c+1 and z+1 ions (c† and z†). High resolution product ion spectra enable confirmation of fragment ion charge state.
As would be expected from peptide fragmentation induced by electron transfer, all of the fragment ions observed result from dissociation of a single bond. Thus, c- and z•-ions, which contained the modified lysine residues, were observed with the expected mass shift intact. That is, these c- and z•-ions from peptide P were shifted such that they accounted for the intact mass of peptide Q plus the cross-linked bridge (C10H6). Often, these fragment ions were observed as doubly or triply charged signals, which were readily distinguished and assigned unambiguously as the ETD fragments were measured at high mass resolution and high mass measurement accuracy in the Orbitrap analyzer (see Figs. 1 and 2). In contrast, it should be noted that the same product ion spectra measured in the linear ion trap lacked sufficient mass resolution to precisely determine the charge state of multiply charged fragment ion signals.
In addition to the generally high coverage of sequence ions, another considerable advantage of DEB cross-links is that under ETD cross-linked peptides dissociate the carbon-nitrogen bond between the benzylic position of DEB and the reductively alkylated amine. This process produces fragment ions that correspond to each of the peptides that make up the cross-link as if they were unmodified (P + H and Q + H) as well as the same peptides bearing a tag from the DEB molecule (P + H + XL and Q + H + XL). Fig. 3 illustrates these diagnostic fragment ions and suggests a likely mechanism of their formation. This fragmentation pathway was found to be present among all lysine-lysine-cross-linked spectra, and these dissociations were found to be extremely useful in confirming the identity of putative cross-links. However, this dissociation pathway was generally less prevalent from modifications located at the N terminus of the protein. The presence of these diagnostic fragments was particularly helpful in assignments in which one of the peptides was short and identified by only a handful of c- and z•-ions as shown in Fig. 2. Despite the low score of this spectrum matched to the peptide EK(2524.332)LQER, the identity of this species is confirmed by the Q + H and Q + H + XL ion signals at 802.440 and 929.491 m/z, respectively.
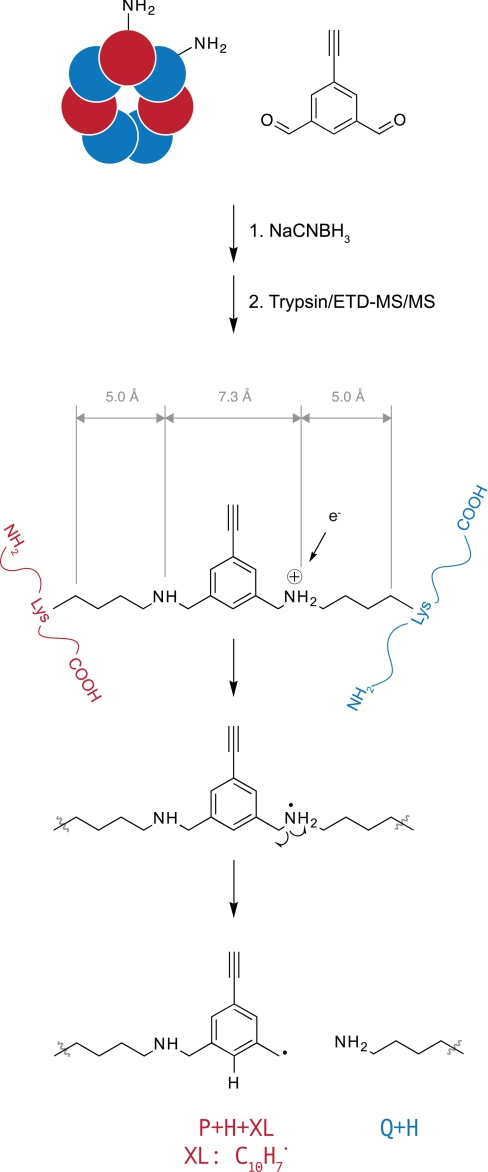
Fig. 3.
Structure of DEB-cross-linked peptides after reductive amination of protein complex with sodium cyanoborohydride and tryptic digestion. DEB spans an interlysine distance of 7.3 Å measured between ε-amines and 17.3 Å measured between the β-carbons. Electron transfer to protonated secondary amine linkages initiates dissociation of this bond to generate diagnostic P + H and Q + H + XL fragment ions, which match the molecular ion of the individual peptides and were observed in all cross-linked spectra.
In contrast, P + H and Q + H + XL ions were never observed in CID product ion spectra. Although CID in the linear ion trap did generate b- and y-ion fragment series, these series were less extensive than the corresponding c- and z•-ions from ETD and were often dominated by a few intense fragment ion signals. Because charge reduction takes place during ETD but not CID, b- and y-ions had higher charge states than corresponding c- and z•-ions. Because DEB cross-linking generates highly charged precursor ions, a sizeable fraction of the b- and y-ions were triply charged or higher. Most search algorithms do not consider fragment ions to be greater than doubly charged. Thus, product ion spectra should be measured at high resolution and searched with an algorithm that considers fragment ions in higher charge states to analyze DEB-cross-linked samples by CID.
GroEL-GroES Cross-linking
Because of the homooligomeric nature of the GroEL complex, a lysine-lysine-cross-linked peptide pair identified by mass spectrometry can logically originate from at least 14 different topologically distinct subunit pairings. Each of the 25 identified cross-linked peptide pairs was examined with reference to all possible pairings on the crystal structure of the GroEL-GroES-ADP complex (Protein Data Bank code 1PF9 (33)). Distances were measured between ε-amines as well as β-carbons of the cross-linked lysine pairs. The rigid DEB molecule spans an interamine distance of 7.3 Å (Fig. 3). However, to account for rotation of the lysine side chains relative to their location in the crystal structure, the interlysine β-carbon distances were also measured. Because the distance of the lysine side chain from β-carbon to ε-amine is 5 Å, a distance constraint of 17.3 Å can be placed on this measurement.
Of the 25 cross-links, 21 were between two lysine residues, whereas the other four were between the α-amino group of GroES and a lysine. Of the 21 lysine-lysine cross-links obtained from these experiments, 20 fit within the 17.3-Å constraint for at least one possible subunit pairing of matched residues. The twenty-first, Lys13-Lys20 on GroES, exceeds this constraint by 3.0 Å. Table II reports interlysine β-carbon-cross-linked distances that are less than 22 Å from both the capped cis and lidless trans conformations of GroEL and GroES (Fig. 4). At most, one configuration in the cis and one pairing in the trans conformation were consistent with the 17.3-Å distance constraint. In a number of cases, the cross-link could arise exclusively from the cis or the trans conformation. Each of the links detected in this study was visually inspected for feasibility to ensure that there were no obvious clashes with the known atomic resolution structure of the protein (see Fig. 5).
Table II. Cross-linking sites and measured distances of GroEL-GroES cross-links.
| No. | Peptide 1a | Residue 1 | Protein 1 | Peptide 2a | Residue 2 | Protein 2 | Configurationb | Nϵ-Nϵc | Cβ-Cβd | Configurationb | Nϵ-Nϵc | Cβ-Cβd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | DVK*FGNDAR | 7 | GroEL | VK*MLR | 15 | GroEL | cis intra | 5.7 | 6.6 | trans intra | 5.3 | 6.3 |
| 2 | VTLGPK*GR | 34 | GroEL | SFGAPTITK*DGVSVAR | 51 | GroEL | cis intra | 12.6 | 11.2 | trans intra | 11.3 | 9.1 |
| 3 | VTLGPK*GR | 34 | GroEL | AVAAGMNPMDLK*R | 117 | GroEL | cis inter (A-G) | 16.3 | 11.6 | trans inter (H-I) | 12.1 | 10.4 |
| 4 | NVVLDK*SFGAPTITK | 42 | GroEL | VGAATEVEMK*EK | 390 | GroEL | cis intra | 6.1 | 11.0 | trans intra | 15.4 | 20.8 |
| 5 | SFGAPTITK*DGVSVAR | 51 | GroEL | AVAAGMNPMDLK*R | 117 | GroEL | cis inter (A-G) | 11.8 | 11.8 | trans inter (H-I) | 6.3 | 13.2 |
| 6 | GYLSPYFINK*PETGAVELESPFILLADKK | 207 | GroEL | K*ISNIR | 226 | GroEL | cis | N.M.e | N.M. | trans inter (H-I) | 7.2 | 13.9 |
| 7 | GYLSPYFINK*PETGAVELESPFILLADKK | 207 | GroEL | GIVK*VAAVK | 272 | GroEL | cis intra | 7.1 | 12.4 | trans intra | 11.5 | 12.5 |
| 8 | GYLSPYFINK*PETGAVELESPFILLADKK | 207 | GroEL | VGAATEVEMK*EK | 390 | GroEL | cis | N.M. | N.M. | trans intra | 7.7 | 11.0 |
| 9 | GYLSPYFINK*PETGAVELESPFILLADKK | 207 | GroEL | K*AR | 393 | GroEL | cis | N.M. | N.M. | trans intra | 6.4 | 11.9 |
| 10 | K*ISNIR | 226 | GroEL | GIVK*VAAVK | 272 | GroEL | cis | N.M. | N.M. | trans inter (H-N) | 11.8 | 10.5 |
| 11 | K*ISNIR | 226 | GroEL | VAAVK*APGFGDR | 277 | GroEL | cis intra | 9.5 | 9.1 | trans intra | 12.7 | 9.7 |
| 12 | K*ISNIR | 226 | GroEL | ATLEDLGQAK*R | 321 | GroEL | cis | N.M. | N.M. | trans inter (H-N) | 14.9 | 15.7 |
| 13 | K*ISNIR | 226 | GroEL | VGAATEVEMK*EK | 390 | GroEL | cis | N.M. | N.M. | trans inter (H-N) | 10.3 | 15.6 |
| 14 | VAAVK*APGFGDR | 277 | GroEL | VGAATEVEMK*EK | 390 | GroEL | cis | N.M. | N.M. | trans inter (H-N) | 8.7 | 13.6 |
| 15 | VVINK*DTTTIIDGVGEEAAIQGR | 327 | GroEL | EK*LQER | 364 | GroEL | cis inter (A-G) | 8.2 | 15.8 | trans | N.M. | N.M. |
| 16 | QQIEEATSDYDREK*LQER | 364 | GroEL | VAK*LAGGVAVIK | 371 | GroEL | cis intra | 13.2 | 10.6 | trans intra | 8.9 | 11.6 |
| 17 | M*NIRPLHDR | 1 | GroES | VIVK*R | 13 | GroES | intra | 14.2 | 15.0 | |||
| 18 | M*NIRPLHDR | 1 | GroES | RK*EVETK | 15 | GroES | Inter (O-U) | 12.4 | 15.2 | |||
| 19 | M*NIRPLHDR | 1 | GroES | EVETK*SAGGIVLTGSAAAK | 20 | GroES | Inter (O-U) | 22.7 | 18.9 | |||
| 20 | M*NIRPLHDR | 1 | GroES | SAGGIVLTGSAAAK*STR | 34 | GroES | Inter (O-U) | 24.5 | 21.5 | |||
| 21 | VIVK*R | 13 | GroES | K*EVETK | 15 | GroES | Intra | 9.2 | 7.8 | |||
| 22 | VIVK*R | 13 | GroES | EVETK*SAGGIVLTGSAAAK | 20 | GroES | Intra | 19.9 | 20.3 | |||
| 23 | VIVK*R | 13 | GroES | SAGGIVLTGSAAAK*STR | 34 | GroES | Intra | 13.9 | 13.2 | |||
| 24 | K*EVETK | 15 | GroES | SAGGIVLTGSAAAK*STR | 34 | GroES | Intra | 15.1 | 9.8 | |||
| 25 | SAGGIVLTGSAAAK*STR | 34 | GroES | VGDIVIFNDGYGVK*SEK | 74 | GroES | Intra | 15.1 | 15.0 | Inter (O-P) | 16.6 | 13.2 |
a Peptide sequence with the site of DEB attachment denoted by the asterisk (*).
b Configuration used for measurement. Cross-link can be either intersubunit or intrasubunit on either the GroEL cis or trans ring or GroES. For intersubunit matches, the subunit designation of residue 1 and residue 2 used for the measurement are given in parentheses. These refer to Protein Data Bank code 1PF9. A to G, GroEL cis ring; H to N, GroEL trans ring; O to U, GroES.
c Distance in Å between the ϵ-amines of cross-linked lysines (or α-amines for protein N-terminal links).
d Distance in Å between the β-carbons of cross-linked lysines (or α-amines for protein N-terminal links).
e N.M. denotes “no match” <22 Å.
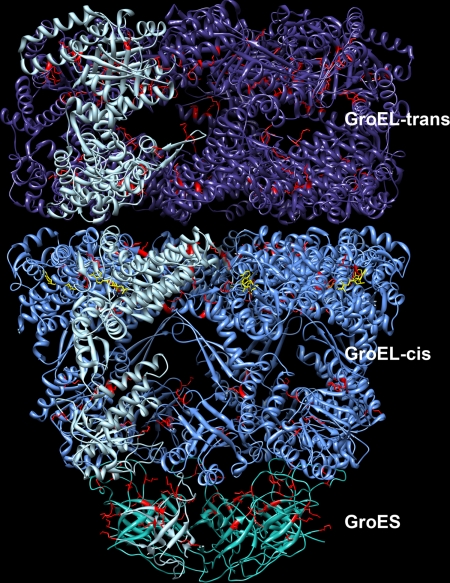
Fig. 4.
Structure of ADP-bound GroEL-GroES complex. A single subunit of each of the three heptameric rings is colored light blue to show the monomer structure. ADP is colored yellow. Residues detected to be cross-linked are highlighted in red.
Fig. 5.
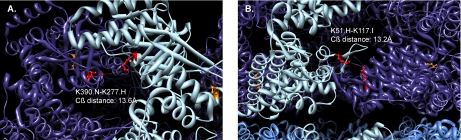
Detail of intersubunit cross-links on GroEL trans ring between Lys277(H) and Lys390(I) (A) and Lys51(H) and Lys117(I) (B). Amino acid residues that constitute acceptable cross-linking configurations are highlighted in red. Orange amino acid residues illustrate alternate, intrasubunit cross-linking configurations that were ruled out because they violated distance constraints and steric requirements.
For cross-links between the α-amine of the N terminus and a lysine side chain, the α-amine-β-carbon distance is expected to fit a constraint of 12.3 Å. However, none of the four cross-links to the GroES N-terminal methionine fit this constraint despite the high confidence in these spectral assignments. Instead the α-amine-β-carbon distances of these linkages spanned 15.0–21.5 Å. As already noted, the one lysine-lysine linkage that exceeded the 17.3-Å constraint was also on the N terminus of GroES (Lys13-Lys20). Hence, our results provide evidence that the N-terminal domain of GroES is somewhat flexible in solution and thus able to form the cross-linked peptides observed.
DISCUSSION
In this work, we report the development of a powerful electron transfer-based mass spectrometric strategy for the elucidation of the precise topography of large protein complexes using a newly designed bifunctional chemical cross-linking reagent, DEB. We show that this reagent samples the presence of free α- and ε-amino functions under physiological conditions within a proximity of ∼7 Å by formation of Schiff bases. Subsequent reduction forms secondary amine linkages that are stable during proteolytic liberation of the cross-linked peptides formed from the protein complex. In addition, this reduction introduces two additional sites of protonation that are advantageous for both (a) increasing the overall charge density of the species during electrospray ionization and (b) favoring the formation of peptide sequence ion series under electron transfer energy deposition conditions. Hence, these spectra contain the sequences and sites of attachment of both peptides participating in the cross-link as well as signals that represent molecular ion mass values corresponding to the individual molecular weights of both peptides. As will be discussed below, these increased charge states are of further analytical advantage in that they facilitate gas-phase “isolation” of cross-linked peptides in complicated reaction mixtures by enabling charge-dependent selection of quadruply charged and higher precursor ion signals during ETD analysis.
The E. coli GroEL-GroES complex was chosen as a multiprotein complex on which to evaluate the effectiveness of our cross-linking methodology. The homooligomeric GroEL complex forms a structure consisting of two stacked heptameric rings. ATP binds to a nucleotide binding site at the equatorial domain of GroEL that catalyzes a conformational extension and counterclockwise twist of the GroEL apical domain. This facilitates binding of the GroES lid, another heptameric homooligomeric ring. The GroES-bound GroEL cis complex encompasses a hydrophobic cavity that sequesters substrate proteins, allowing them to fold properly. Physiologically, hydrolysis of the γ-phosphate of ATP at the GroEL nucleotide binding site drives the catalytic cycle, which consists of coordinated cycling of the cis and trans GroEL rings between extended GroES-bound and collapsed conformers. ADP binding, on the other hand, creates a stable, asymmetric complex in which the cis GroEL ring is extended and capped by the GroES ring, whereas the trans ring is collapsed and open (see Fig. 4) (33–35, 41).
Fig. 4 shows the x-ray structure of the GroEL-GroES-ADP structure with residues established as part of a cross-linked peptide pair highlighted in red. All of the intersubunit cross-links were determined to come from adjacent subunits in one of the rings. This result is consistent with the fact that the vast majority of the contacting protein-protein surface area is present in these regions and that there are few lysine residues in the inter-ring-contacting regions. This is also consistent with results reported from an earlier cross-linking study of GroEL-GroES that found that inter-ring cross-links were formed at a much lower rate (42).
The homooligomeric nature of the GroEL-GroES has complicated our analysis somewhat because the cross-linked species identified by mass spectrometry can originate from a number of different conformational pairings. However, all the cross-links we observed fit the distance imposed by the DEB geometry to at most one pair of residues in the cis and one pair in the trans complex, and many fit exclusively to only one conformation. As a measure of reliability, we determined the lysine-lysine β-carbon distances between every possible pairing of lysine residues on the complex. For each lysine pair, it was determined whether at least one possible subunit pairing matched the distance constraint of 17.3 Å. Of 1130 possible pairings between all possible lysine side chains, 155 fit the distance constraint for at least one possible subunit configuration. Thus, the likelihood of a single cross-link matching randomly is 13.7%. Therefore, it is highly unlikely that 20 of 21 of our reported lysine-lysine cross-links meet this requirement by chance alone.
As mentioned, the four links to the N terminus of GroES as well as the Lys13-Lys20 GroES cross-link require exceeding the expected geometric constraints somewhat. Amino acid residues 13–32 are known to constitute the GroES “mobile loop,” which sits at the contact region between GroEL and GroES. Thus, the cross-linking results are consistent with the findings of NMR spectroscopy and crystallography with respect to the conformational flexibility of this region (41, 43, 44).
Several of the cross-links identified could only have originated from sampling exclusively the capped cis GroEL conformation or the lidless trans conformation. For instance, the cross-link between Lys277 and Lys390 of GroEL fits well between Lys277 on subunit H and Lys390 on subunit N of the trans complex with a Cβ-Cβ distance of 13.6 Å as illustrated in Fig. 5. However, the best fit on the cis complex gives a Cβ-Cβ distance between Lys277 (chain A) and Lys390 (chain A) of 33.3 Å. Furthermore, the intrasubunit links on both the trans and cis conformations are sterically bad fits, which further validates Lys277(H)-Lys390(N) or an equivalent configuration on the trans ring as the cross-linked subunit match.
Several research groups have introduced cross-linking reagents that insert a low energy gas-phase, infrared multiphoton dissociation, or UV light-cleavable bond (26–29, 45). This simplifies cross-linking analysis by producing fragment ions that provide the molecular ion of the individual peptides. However, these strategies tend not to simultaneously produce sequence ions that would identify the peptide. This leaves the identity of the peptides to be inferred by precursor mass alone (46, 47) or requires non-standard instrumentation that permits multiple stages of dissociation (29). With electron transfer dissociation, DEB cross-linking not only produces sequence ions efficiently but also releases the individual molecular ions at equivalent intensity. These species are invaluable in confirming the correct peptide assignment.
Discrimination of cross-linked species by charge state-dependent precursor selection is now an established technique (9, 13). Unmodified and type 0 modified peptides are typically doubly or triply charged, whereas peptides cross-linked by acylating agents (typically N-hydroxysuccinimide esters) are mostly triply or quadruply charged. The charge state shift is due to the additional sites of protonation introduced by the α-amine and C-terminal tryptic residue of the extra peptide. Cross-linking with DEB introduces a further two sites of protonation and results in cross-linked peptides that have on average five charges. We found no cross-linked peptides in our initial experiments that were less than quadruply charged and thus implemented this as a requirement for precursor selection. In comparison, cross-linking with BS3 produces a significant percentage of triply charged cross-linked peptides (9) (see the supplemental chart). Hence, charge state discrimination is more effective with DEB as it is possible to reject a much greater portion of the highly abundant unmodified and dead-end modified species. We expect the efficacy of strong cation exchange to isolate cross-linked species to also be enhanced.
The relatively small size and rigid structure of the DEB molecule leads to increased resolution in the structural inferences made possible via this cross-linking analysis. A recent in silico analysis of 54 protein complexes with solved crystal structures demonstrated that, for the purpose of macromolecular structure modeling based on cross-link-derived distance constraints, the quality of the model depends on the number of cross-links as well as the maximal distance defined by those cross-links. It is desirable to have large numbers of shorter distance restraints (7). However, these factors are in conflict with each other as there will be fewer possible cross-links derived from shorter length cross-linkers.
DEB spans a lysine ε-amino to lysine ε-amino function distance of 7.3 Å. Measured from the β-carbons of the modified lysine residues to account for flexibility in the side chain orientations the distance is 17.3 Å. Other recent studies use the popular, commercially available cross-linkers disuccinimidyl suberate and BS3, which produce identical cross-linked bridges with inter ε-amine distances of 11.4 Å (9, 13). Therefore, DEB-derived distance constraints provide greater structural resolution for modeling purposes than disuccinimidyl suberate- or BS3-derived constraints. Our study found 20 lysine-lysine-cross-linked peptides within the strict restraints of the DEB geometry with one additional cross-link that is likely correct if we allow 3 Å for loop flexibility. Additionally, four cross-links were identified between lysine residues on the GroES mobile loop and the α-amine of GroES.
CONCLUSIONS
It is without doubt that more effective cross-linking methods coupled with optimized electron transfer or electron capture ion optical systems will play a critical role in providing distance constraints that, together with cryoelectron microscopy of a protein complex and x-ray structures of individual subunits, will help solve the structure of macromolecular complexes. Many protein complexes are recalcitrant to crystallography, whereas cryoelectron microscopy often does not provide sufficient resolution (∼4 Å) to determine the orientation and arrangement of the individual subunits. Furthermore, there is growing interest in mapping protein interaction networks as it is now understood that almost all physiological phenomena involve protein-protein interactions (48–51). Understanding not only the identity of interacting proteins but also mapping the structural domains involved in interaction is required to understand how proteins cooperate to carry out cellular processes and will aid in designing new therapeutics, both antibody- and small molecule-based, that target this vast and emerging class of therapeutic targets (52).
By producing cross-linked peptide pairs that are bound through positively charged secondary amines, DEB in conjunction with high resolution ETD mass spectrometry will accelerate cross-link analysis of protein machines by producing robust fragmentation, diagnostic fragments, and increased precursor charge state that benefit charge-based selection schemes. Thus, easily obtained, high quality structural constraints will provide the missing link in our ability to connect individual proteins with their functional partners through exploitation of the tools of computational biology.
Supplementary Material
Acknowledgments
We are grateful to Erik Miller at Stanford University for the gift of GroEL and GroES and to Danica Fujimori at the University of California, San Francisco for providing space and facilities in which to perform chemical synthesis. We thank Giselle Knudsen and Robert Chalkley for review of the manuscript and Peter Baker for Protein Prospector support. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
* This work was supported, in whole or in part, by National Institutes of Health Grant P41RR001614 from the Biomedical Research Technology Program of the National Center for Research Resources (to the Bio-Organic Biomedical Mass Spectrometry Resource at the University of California, San Francisco).
 This article contains supplemental figures and a chart.
This article contains supplemental figures and a chart.
2 M. Trnka, unpublished results.
3 M. J. Trnka and A. L. Burlingame, manuscript in preparation.
1 The abbreviations used are:
- BS3
- bis(sulfosuccinimidyl) suberate
- DEB
- 1,3-diformyl-5-ethynylbenzene
- ETD
- electron transfer dissociation.
REFERENCES
- 1.Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., Yonath A. (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 [DOI] [PubMed] [Google Scholar]
- 2.Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Structure of the 30S ribosomal subunit. Nature 407, 327–339 [DOI] [PubMed] [Google Scholar]
- 3.Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 A Resolution. Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 4.Robinson C. V., Sali A., Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982 [DOI] [PubMed] [Google Scholar]
- 5.Sinz A. (2006) Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 25, 663–682 [DOI] [PubMed] [Google Scholar]
- 6.Jin Lee Y. (2008) Mass spectrometric analysis of cross-linking sites for the structure of proteins and protein complexes. Mol. Biosyst. 4, 816–823 [DOI] [PubMed] [Google Scholar]
- 7.Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu F., Shan S. O., Moustakas D. T., Alber F., Egea P. F., Stroud R. M., Walter P., Burlingame A. L. (2004) Unraveling the interface of signal recognition particle and its receptor by using chemical cross-linking and tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 16454–16459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zubarev R. A. (2004) Electron-capture dissociation tandem mass spectrometry. Curr. Opin. Biotechnol. 15, 12–16 [DOI] [PubMed] [Google Scholar]
- 12.Good D. M., Wirtala M., McAlister G. C., Coon J. J. (2007) Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 6, 1942–1951 [DOI] [PubMed] [Google Scholar]
- 13.Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller D. R., Schindler P., Towbin H., Wirth U., Voshol H., Hoving S., Steinmetz M. O. (2001) Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal. Chem. 73, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 15.Back J. W., Notenboom V., de Koning L. J., Muijsers A. O., Sixma T. K., de Koster C. G., de Jong L. (2002) Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal. Chem. 74, 4417–4422 [DOI] [PubMed] [Google Scholar]
- 16.Pearson K. M., Pannell L. K., Fales H. M. (2002) Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 16, 149–159 [DOI] [PubMed] [Google Scholar]
- 17.Collins C. J., Schilling B., Young M., Dollinger G., Guy R. K. (2003) Isotopically labeled crosslinking reagents: resolution of mass degeneracy in the identification of crosslinked peptides. Bioorg. Med. Chem. Lett. 13, 4023–4026 [DOI] [PubMed] [Google Scholar]
- 18.Petrotchenko E. V., Olkhovik V. K., Borchers C. H. (2005) Isotopically coded cleavable cross-linker for studying protein-protein interaction and protein complexes. Mol. Cell. Proteomics 4, 1167–1179 [DOI] [PubMed] [Google Scholar]
- 19.Chu F., Mahrus S., Craik C. S., Burlingame A. L. (2006) Isotope-coded and affinity-tagged cross-linking (ICATXL): an efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 128, 10362–10363 [DOI] [PubMed] [Google Scholar]
- 20.Seebacher J., Mallick P., Zhang N., Eddes J. S., Aebersold R., Gelb M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 [DOI] [PubMed] [Google Scholar]
- 21.Petrotchenko E. V., Serpa J. J., Borchers C. H. (July9, 2010) An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics. Mol. Cell. Proteomics 10.1074/mcp.M110.001420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury S. M., Du X., Toliæ N., Wu S., Moore R. J., Mayer M. U., Smith R. D., Adkins J. N. (2009) Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 81, 5524–5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nessen M. A., Kramer G., Back J., Baskin J. M., Smeenk L. E., de Koning L. J., van Maarseveen J. H., de Jong L., Bertozzi C. R., Hiemstra H., de Koster C. G. (2009) Selective enrichment of azide-containing peptides from complex mixtures. J. Proteome Res. 8, 3702–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan F., Che F. Y., Rykunov D., Nieves E., Fiser A., Weiss L. M., Hogue Angeletti R. (2009) Nonprotein based enrichment method to analyze peptide cross-linking in protein complexes. Anal. Chem. 81, 7149–7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vellucci D., Kao A., Kaake R. M., Rychnovsky S. D., Huang L. (2010) Selective enrichment and identification of azide-tagged cross-linked peptides using chemical ligation and mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 1432–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury S. M., Munske G. R., Tang X., Bruce J. E. (2006) Collisionally activated dissociation and electron capture dissociation of several mass spectrometry-identifiable chemical cross-linkers. Anal. Chem. 78, 8183–8193 [DOI] [PubMed] [Google Scholar]
- 27.Lu Y., Tanasova M., Borhan B., Reid G. E. (2008) Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal. Chem. 80, 9279–9287 [DOI] [PubMed] [Google Scholar]
- 28.Gardner M. W., Vasicek L. A., Shabbir S., Anslyn E. V., Brodbelt J. S. (2008) Chromogenic cross-linker for the characterization of protein structure by infrared multiphoton dissociation mass spectrometry. Anal. Chem. 80, 4807–4819 [DOI] [PubMed] [Google Scholar]
- 29.Yang L., Tang X., Weisbrod C. R., Munske G. R., Eng J. K., von Haller P. D., Kaiser N. K., Bruce J. E. (2010) A photocleavable and mass spectrometry identifiable cross-Linker for protein interaction studies. Anal. Chem. 82, 3556–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gardner M. W., Brodbelt J. S. (2010) Preferential cleavage of N-N hydrazone bonds for sequencing bis-arylhydrazone conjugated peptides by electron transfer dissociation. Anal. Chem. 82, 5751–5759 [DOI] [PubMed] [Google Scholar]
- 31.Kao A., Chiu C., Vellucci D., Yang Y., Patel V. R., Guan S., Randall A., Baldi P., Rychnovsky S. D., Huang L. (August24, 2010) Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10.1074/mcp.M110.002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderblom E. J., Bobay B. G., Cavanagh J., Goshe M. B. (2007) Tandem mass spectrometry acquisition approaches to enhance identification of protein-protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. Rapid Commun. Mass Spectrom. 21, 3395–3408 [DOI] [PubMed] [Google Scholar]
- 33.Chaudhry C., Farr G. W., Todd M. J., Rye H. S., Brunger A. T., Adams P. D., Horwich A. L., Sigler P. B. (2003) Role of the [gamma]-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics. EMBO J. 22, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyagi N. K., Fenton W. A., Horwich A. L. (2009) GroEL/GroES cycling: ATP binds to an open ring before substrate protein favoring protein binding and production of the native state. Proc. Natl. Acad. Sci. U.S.A. 106, 20264–20269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishna K. A., Rao G. V., Rao K. R. (2007) Chaperonin GroEL: structure and reaction cycle. Curr. Protein Pept. Sci. 8, 418–425 [DOI] [PubMed] [Google Scholar]
- 36.Bhagwat S. S., Roland D. M., Main A. J., Gude C., Grim K., Goldstein R., Cohen D. S., Dotson R., Mathis J., Lee W. (1992) Thromboxane receptor antagonism combined with thromboxane synthase inhibition. 7. Pyridinylalkyl-substituted arylsulfonylamino arylalkanoic acids. Bioorg. Med. Chem. Lett. 2, 1623–1626 [Google Scholar]
- 37.Lynn A. J., Chalkley R. J., Baker P. R., Medzihradszky K. F., Guan S., Burlingame A. L. (2008) The effect of peaklist generation software on database search results, in 56th ASMS Conference, Denver, June 1–5, 2008, American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 38.Baker P. R., Medzihradszky K. F., Chalkley R. J. (2010) Improving software performance for peptide ETD data analysis by implementation of charge-state and sequence-dependent scoring. Mol. Cell. Proteomics 9, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schilling B., Row R. H., Gibson B. W., Guo X., Young M. M. (2003) MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 [DOI] [PubMed] [Google Scholar]
- 40.Chu F., Baker P. R., Burlingame A. L., Chalkley R. J. (2010) Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol. Cell. Proteomics 9, 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z., Horwich A. L., Sigler P. B. (1997) The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741–750 [DOI] [PubMed] [Google Scholar]
- 42.Azem A., Weiss C., Goloubinoff P. (1998) Structural analysis of GroE chaperonin complexes using chemical cross-linking. Methods Enzymol. 290, 253–268 [DOI] [PubMed] [Google Scholar]
- 43.Shewmaker F., Maskos K., Simmerling C., Landry S. J. (2001) The disordered mobile loop of GroES folds into a defined β-hairpin upon binding GroEL. J. Biol. Chem. 276, 31257–31264 [DOI] [PubMed] [Google Scholar]
- 44.Fiaux J., Bertelsen E. B., Horwich A. L., Wüthrich K. (2002) NMR analysis of a 900K GroEL-GroES complex. Nature 418, 207–211 [DOI] [PubMed] [Google Scholar]
- 45.Tang X., Bruce J. E. (2010) A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. Mol. Biosyst. 6, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson G. A., Tolic N., Tang X., Zheng C., Bruce J. E. (2007) Informatics strategies for large-scale novel cross-linking analysis. J. Proteome Res. 6, 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Tang X., Munske G. R., Tolic N., Anderson G. A., Bruce J. E. (2009) Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol. Cell. Proteomics 8, 409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 49.Hutchins J. R., Toyoda Y., Hegemann B., Poser I., Hériché J. K., Sykora M. M., Augsburg M., Hudecz O., Buschhorn B. A., Bulkescher J., Conrad C., Comartin D., Schleiffer A., Sarov M., Pozniakovsky A., Slabicki M. M., Schloissnig S., Steinmacher I., Leuschner M., Ssykor A., Lawo S., Pelletier L., Stark H., Nasmyth K., Ellenberg J., Durbin R., Buchholz F., Mechtler K., Hyman A. A., Peters J. M. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott J. D., Pawson T. (2009) Cell signaling in space and time: where proteins come together and when they're apart. Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells J. A., McClendon C. L. (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.