Abstract
Background
In insects, like in most invertebrates, olfaction is the principal sensory modality, which provides animals with essential information for survival and reproduction. Odorant receptors are involved in this response, mediating interactions between an individual and its environment, as well as between individuals of the same or different species. The adaptive importance of odorant receptors renders them good candidates for having their variation shaped by natural selection.
Methodology/Principal Findings
We analyzed nucleotide variation in a subset of eight Or genes located on the 3L chromosomal arm of Drosophila melanogaster in a derived population of this species and also in a population of Drosophila pseudoobscura. Some heterogeneity in the silent polymorphism to divergence ratio was detected in the D. melanogaster/D. simulans comparison, with a single gene (Or67b) contributing ∼37% to the test statistic. However, no other signals of a very recent selective event were detected at this gene. In contrast, at the speciation timescale, the MK test uncovered the footprint of positive selection driving the evolution of two of the encoded proteins in both D. melanogaster —OR65c and OR67a —and D. pseudoobscura —OR65b1 and OR67c.
Conclusions
The powerful polymorphism/divergence approach provided evidence for adaptive evolution at a rather high proportion of the Or genes studied after relatively recent speciation events. It did not provide, however, clear evidence for very recent selective events in either D. melanogaster or D. pseudoobscura.
Introduction
Animals can recognise and discriminate chemical signals in the environment, which provides essential information for survival and can profoundly influence their behaviour [1]. In the case of airborne molecules, the recognition starts with their interaction with odorant receptors that reside in the olfactory receptor neurons (ORNs; [2]). These ORNs transmit signals into the Central Nervous System, where they are processed, ultimately leading to behavioural responses.
Odorant receptor (Or) genes encode signal-transduction proteins with seven transmembrane domains. In insects, they are members of a large and rather old multigene family, with orthologs in orders as diverse as Diptera, Homoptera, Hymenoptera and Coleoptera (e.g., [3], [4], [5]). Because olfaction contributes to find food and mates as well as to detect predators, genes involved in olfactory perception are candidates to have evolved by the action of positive natural selection. Indeed, a maximum likelihood analysis of nonsynonymous and synonymous divergence across five species of the melanogaster subgroup with complete genome sequences revealed that the overall evolution of the Or family during the last ∼12 MY was nonneutral [6]. Also, the comparison of Or polymorphism in a specieswide sample of Drosophila simulans and divergence of those from D. melanogaster orthologs provided some evidence of adaptive evolution of OR proteins in the D. simulans lineage [6].
The analysis of polymorphism, unlike that of divergence, can uncover the footprint left on DNA sequences by very recent selective events. Moreover, the analysis of polymorphism and divergence at coding regions constitutes a powerful approach to detect the action of recurrent positive selection driving to fixation amino acid changes after relatively recent speciation events. In an effort to uncover the action of positive selection acting on Or genes at these two timescales, we have analyzed within-population variation in two well characterized species (D. melanogaster and D. pseudoobscura) as well as divergence to a closely related species (D. simulans and D. miranda, respectively) at a subset of eight Or genes —Or63a, Or65a-b-c cluster, Or67a, Or67b, Or67c and Or69a— that were solely chosen for their location on the same chromosomal arm of D. melanogaster (3L or Muller's D element). In D. pseudoobscura, the Or genes studied are located on the XR chromosomal arm, with the exception of genes Or65b2, Or65b4 and Or65b5 that are located on element C (chromosome 3) and gene Or67a on element E (chromosome 2) due to transposition events that predated the X-autosome fusion [7]. Our multilocus analysis of polymorphism and divergence provided no clear indication of very recent action of positive selection on the Or genes studied. It did, however, uncover the footprint of positive selection driving the evolution of a relatively large proportion of the encoded proteins in both the D. melanogaster and D. pseudoobscura lineages.
Results and Discussion
Levels of polymorphism
Table 1 summarizes the estimated levels of nucleotide variation at the Or genes studied in Drosophila melanogaster and D. pseudoobscura. A total of 18.9 and 19.5 kb were analyzed in each of these species, respectively (Table S1). The number of segregating sites was 445 in D. melanogaster and 421 in D. pseudoobscura, with the former species exhibiting a lower overall proportion of polymorphic sites with singletons (31%) than the latter species (62%). In both species, the estimated nonsynonymous nucleotide diversity was almost ten-fold lower than synonymous estimates (Table 1). Estimates of noncoding diversity did not differ significantly from those of synonymous diversity in either D. melanogaster or D. pseudoobscura (Wilcoxon signed-rank test; P = 0.31 and 0.36, respectively), which would seem in contrast with the higher level of constraint at intergenic regions than that at synonymous sites previously observed in D. melanogaster/D. simulans comparisons [8], [9]. Moreover, similarly to previous surveys [10], [11], [12], no significant difference in the level of either noncoding or synonymous polymorphism was detected in D. pseudoobscura between the sex-linked and autosomal genes (Wilcoxon signed-rank test; P = 0.22 and 0.18, respectively). The time elapsed since the X-autosome fusion (8–12 My; [13]) cannot probably account for these results since it would seem sufficient for variation at the newly X-linked arm (XR) to have attained the new equilibrium and therefore for the newly sex-linked genes to exhibit the expected reduction of variation relative to autosomal genes. The previously detected bias in the species sex-ratio toward a higher proportion of females [14] might be one of the factors contributing to the detected similarity.
Table 1. Nucleotide variation in different functional regions of the Or genes.
| S | π | Haplotypes | K | |||||||||||||
| Species | Gene | Na | nc | s | a | Totalb | nc | s | a | Total | No. | Hd | K nc | K s | K a | K a/K s |
| D. melanogaster | Or63a | 12 | 28 | 14 | 6 | 48 (13) | 0.009 | 0.016 | 0.002 | 0.007 | 10 | 0.97 | 0.050 | 0.118 | 0.011 | 0.089 |
| Or65a | 14 | 17 | 17 | 8 | 42 (14) | 0.005 | 0.023 | 0.002 | 0.006 | 13 | 0.99 | 0.028 | 0.110 | 0.023 | 0.208 | |
| Or65b | 13 | 11 | 4 | 6 | 21 (0) | 0.023 | 0.005 | 0.003 | 0.006 | 7 | 0.83 | 0.170 | 0.160 | 0.033 | 0.207 | |
| Or65c | 12 | 6 | 34 | 6 | 47 (24) | 0.014 | 0.039 | 0.002 | 0.011 | 11 | 0.99 | 0.121 | 0.155 | 0.034 | 0.221 | |
| Or67a | 12 | 52 | 17 | 2 | 71 (36) | 0.011 | 0.015 | 0.0004 | 0.007 | 12 | 1 | 0.106 | 0.202 | 0.078 | 0.388 | |
| Or67b | 13 | 20 | 2 | 1 | 23 (4) | 0.009 | 0.004 | 0.0002 | 0.004 | 10 | 0.95 | 0.090 | 0.214 | 0.009 | 0.044 | |
| Or67c | 14 | 34 | 26 | 7 | 67 (34) | 0.006 | 0.022 | 0.002 | 0.006 | 8 | 0.82 | 0.061 | 0.197 | 0.009 | 0.047 | |
| Or69a | 11 | 91 | 23 | 14 | 128 (27) | 0.023 | 0.021 | 0.003 | 0.014 | 11 | 1 | 0.079 | 0.119 | 0.032 | 0.265 | |
| Total | 259 | 137 | 50 | 447 (152) | ||||||||||||
| Average | 0.013 | 0.018 | 0.002 | 0.008 | 0.944 | 0.088 | 0.159 | 0.029 | 0.184 | |||||||
| D. pseudoobscura | Or63a | 8 | 39 | 9 | 12 | 60 (35) | 0.039 | 0.011 | 0.005 | 0.014 | 8 | 1 | 0.082 | 0.065 | 0.011 | 0.173 |
| Or65b1 | 8 | 21 | 22 | 5 | 48 (26) | 0.034 | 0.028 | 0.002 | 0.015 | 8 | 1 | 0.063 | 0.058 | 0.024 | 0.411 | |
| Or65b2 | 7 | 7 | 12 | 7 | 26 (16) | 0.011 | 0.017 | 0.002 | 0.007 | 7 | 1 | - | - | - | - | |
| Or65b4 | 6 | 13 | 14 | 17 | 44 (36) | 0.019 | 0.020 | 0.007 | 0.012 | 6 | 1 | 0.048 | 0.061 | 0.017 | 0.285 | |
| Or65b5 | 7 | 26 | 21 | 11 | 58 (27) | 0.013 | 0.029 | 0.005 | 0.012 | 7 | 1 | 0.053 | 0.115 | 0.021 | 0.183 | |
| Or67a | 8 | 22 | 32 | 12 | 66 (42) | 0.036 | 0.039 | 0.004 | 0.015 | 8 | 1 | 0.078 | 0.077 | 0.015 | 0.199 | |
| Or67b | 8 | 7 | 4 | 1 | 12 (6) | 0.003 | 0.004 | 0.0003 | 0.002 | 7 | 0.96 | 0.021 | 0.030 | 0.005 | 0.180 | |
| Or67c | 8 | 47 | 24 | 2 | 73 (43) | 0.011 | 0.030 | 0.001 | 0.009 | 8 | 1 | 0.028 | 0.040 | 0.005 | 0.121 | |
| Or69a | 8 | 20 | 7 | 7 | 34 (30) | 0.002 | 0.003 | 0.001 | 0.002 | 8 | 1 | 0.033 | 0.038 | 0.008 | 0.224 | |
| Total | 202 | 145 | 74 | 421 (261) | ||||||||||||
| Average | 0.019 | 0.020 | 0.003 | 0.010 | 0.996 | 0.051 | 0.060 | 0.013 | 0.222 | |||||||
S, no. of segregating sites; π, nucleotide diversity; K, nucleotide divergence; nc, noncoding; s, synonymous; a, nonsynonymous; Hd, haplotype diversity.
Number of lines sequenced.
Number of singletons are given in parentheses.
There is evidence of recombination in the history of all genes studied in both D. melanogaster and D. pseudoobscura (i.e., R m≥1), with the exception of gene Or67b in the latter species (Table S2). As expected from recombination rates based on genetic map distances, the overall degree of genetic association between polymorphisms (as summarized by the Z nS statistic; Table S2) was generally higher in D. melanogaster (from 0.20 to 0.66) than in D. pseudoobscura (from 0.14 to 0.53).
No clear indication of very recent adaptive substitutions
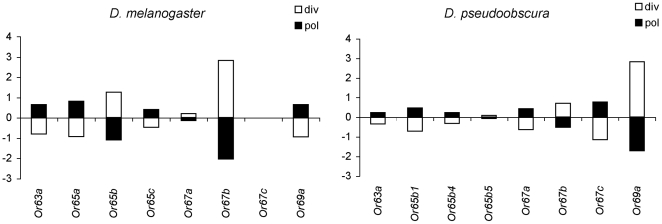
Multilocus HKA tests were performed using silent polymorphism (in D. melanogaster and D. pseudoobscura) and divergence (between D. melanogaster and D. simulans, and between D. pseudoobscura and D. miranda, respectively; Fig. 1). Only in the D. melanogaster/D. simulans comparison, the low probability associated to the test statistic (χ2 = 13.27; P = 0.07) pointed to a possible decoupling between levels of polymorphism and divergence across genes. In this comparison, a single gene exhibiting a local reduction in polymorphism —Or67b— contributed 36.6% to the test statistic. However, no clear signature of a recent selective sweep was detected in the pattern of polymorphism at this gene using either summary statistics based on the frequency spectrum (Tajima's D and normalized Fay and Wu's H [16], [17], [18]; see below) or the Kim and Stephan test [15], which also considers the spatial distribution of variation (results not shown).
Figure 1. Multilocus HKA.
Summary of a multilocus HKA test, which compares polymorphism within D. melanogaster and D. pseudoobscura to divergence from D. simulans and D. miranda, respectively. Solid bars represent contributions to the overall χ2 test statistic caused by polymorphism levels at each locus; open bars represent contributions caused by divergence. Positive values indicate an excess of polymorphism or divergence relative to neutral expectations. Likewise, negative values indicate a defect relative to expectation.
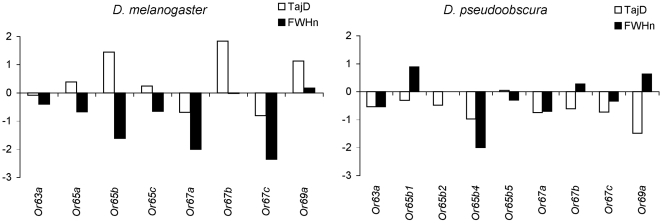
The frequency distribution of nucleotide variants was investigated using Tajima's D and normalized Fay and Wu's H (Fig. 2; [16], [17], [18]). In D. melanogaster, the estimated D values varied widely across genes whereas the H estimates were generally negative (Fig. 2). The estimated values did not depart from neutral expectations either under stationarity or under the bottleneck scenario proposed for derived European populations ([19], [20], [21]; results not shown).
Figure 2. Summary statistics.
Tajima's D and normalized Fay and Wu's H statistics for D. melanogaster and D. pseudoobscura.
In D. pseudoobscura, a general skew toward negative values of both Tajima's D and Fay and Wu's H was observed, which resulted in average negative values for both statistics (−0.648 and −0.265, respectively). A similar observation concerning the folded frequency spectrum (i.e., Tajima's D statistic) was previously reported in this species and led the authors to consider a scenario of population expansion as the most plausible explanation for the detected pattern [11], [22].
Evidence for adaptive evolution of ORs in the D. melanogaster and D. pseudoobscura lineages
The MK test that was performed for each gene separately yielded highly significant results for genes Or65c and Or67a in the D. melanogaster/D. simulans comparison, and for genes Or65b1 and Or67c in the D. pseudoobscura/D. miranda comparison (Table 2). In all these cases, an excess of fixed nonsynonymous changes was detected. When correcting for multiple testing (using the stringent sequential Bonferroni correction; [23]), the tests remained significant for the same four genes. When applying the MK test to the pooled set of genes, highly significant results were obtained in both comparisons (D. melanogaster/D. simulans and D. pseudoobscura/D. miranda), indicating a general trend toward an excess of fixed nonsynonymous changes. In all cases, the removal of singleton polymorphisms did not affect the results (results not shown), which together with the below-one values of the neutrality index [24] for all four genes (0.16 and 0.08 for Or65c and Or67a and 0.15 and 0.06 for Or65b1 and Or67c, respectively) suggests that these genes exhibited indeed a significant excess of nonsynonymous fixed mutations. Moreover, in the D. melanogaster comparison, the polarized MK test (using D. yakuba as the outgroup) revealed a significant excess of fixed nonsynonymous mutations at genes Or65c and Or67a in the D. melanogaster lineage (results not shown). Little is known about the specific functions of the encoded receptors in each species except that in D. melanogaster the receptors encoded by genes of the Or65 cluster seem to have pheromones as ligands [25] whereas genes Or67a and Or67c are known to respond strongly to a broad range of food odours [26].
Table 2. McDonald and Kreitman tests.
| Species | Gene | FS | FNS | PS | PNS | P-valuea |
| D. melanogaster | Or63a | 28 | 8 | 14 | 6 | 0.54 |
| Or65a | 24 | 22 | 17 | 8 | 0.22 | |
| Or65b | 37 | 28 | 4 | 6 | 0.50 | |
| Or65c | 28 | 30 | 34 | 6 | <0.001*** | |
| Or67a | 47 | 67 | 17 | 2 | <0.001*** | |
| Or67b | 52 | 9 | 2 | 1 | 0.40 | |
| Or67c | 36 | 6 | 26 | 7 | 0.54 | |
| Or69a | 44 | 47 | 23 | 14 | 0.18 | |
| TOTAL | 296 | 217 | 137 | 50 | <0.001*** | |
| D. pseudoobscura | Or63a | 15 | 6 | 9 | 12 | 0.12 |
| Or65b1 | 12 | 18 | 22 | 5 | 0.003** | |
| Or65b4 | 7 | 7 | 14 | 17 | 1.00 | |
| Or65b5 | 24 | 16 | 21 | 11 | 0.81 | |
| Or67a | 12 | 8 | 32 | 12 | 0.39 | |
| Or67b | 8 | 5 | 4 | 1 | 0.61 | |
| Or67c | 3 | 4 | 24 | 2 | 0.011** | |
| Or69a | 18 | 13 | 7 | 7 | 0.75 | |
| TOTAL | 99 | 77 | 133 | 67 | 0.044* |
FS, fixed synonymous substitutions; FNS, fixed nonsynonymous substitutions; PS, polymorphic synonymous substitutions; PNS, polymorphic nonsynonymous substitutions.
*P<0.05;
**P<0.01;
***P<0.001.
Two-tailed Fisher's exact test.
In both D. melanogaster and D. pseudoobscura, two of the eight Or genes studied exhibited the footprint of protein adaptive evolution. The estimated proportion (0.25 in both lineages) is based on a small number of genes and does not differ significantly from that estimated (0.1) in a genomewide study, which included a larger number of Or genes (20) that were partially sequenced in a sample including both African and cosmopolitan lines of D. melanogaster [27]. This relatively high proportion would seem consistent with diverse observations in D. melanogaster. Indeed, in this species the expression of some chemoreceptor genes is highly sexually dimorphic and frequently sexually antagonistic, and the extent of transcriptional responses to changing conditions is heterogeneous among the chemoreceptor repertoire [28]. Moreover, some of the encoded proteins have indeed pheromones as ligands and they might either signal the presence of inappropriate mating partners or contribute to the identification of conspecific partners [29]. Other odorant receptors exhibit a strong response to food odours and might serve to signal food sources in the environment. The challenges imposed by changing environmental conditions, such as those often associated with speciation and species range expansions, might thus trigger the adaptive evolution of ORs and also promote adaptive regulatory changes in the chemoreceptor genes. However, the proportion of Or genes under positive selection detected in both our study and the genomewide study [27], as well as that of Gr genes (2 out of 20) in the latter study, do not differ significantly from the proportion of non-chemosensory genes (29 out of 379; [27]). A similar result was obtained when chemosensory (Or and Gr) genes in D. simulans [6] were compared to a genomewide sample of non-chemosensory genes [30]. In Drosophila, adaptive protein evolution at the speciation timescale —as evidenced by the polymorphism to divergence comparison in D. melanogaster, D. simulans and D. pseudoobscura— would thus seem as pervasive among ORs as among the rest of proteins.
Materials and Methods
Drosophila strains
Fourteen isochromosomal lines for the third chromosome of Drosophila melanogaster obtained from a natural population of Sant Sadurní d'Anoia (Spain; [31]), and 13 highly inbred lines of D. pseudoobscura from a natural population of Davis (USA; kindly provided by C. Segarra) were used for the analysis of polymorphism. Highly inbred lines obtained by ten generations of sib-mating were also used for the analysis of divergence: one line each of D. simulans (Mozambique; [32]) and D. miranda (kindly provided by C. Segarra).
DNA extraction, amplification and sequencing
DNA was extracted from i) one single individual per inbred line (a male in the case of D. pseudoobscura and D. miranda); and ii) ten individuals per isochromosomal line, using either a modification of protocol 48 in Ashburner [33] or the PUREGENE DNA Purification kit (Gentra Systems, Inc.) for DNA extraction of a single fly.
Amplification and sequencing primers were designed based on the D. melanogaster and D. pseudoobscura genome sequences using program Oligo 4 (Molecular Biology Insights, Inc.). In general, amplification primers were designed to be conserved between species. Sequencing primers were species-specific and spaced on average 500 nucleotides. The purification step was a modification of the protocol described in Dean et al. [34]. Sequencing products were ethanol precipitated and later separated on automatic sequencers ABI 377 or ABI 3700 (ABI Applied Biosystems). All sequences were obtained on both strands. The sequences reported in this article are deposited in the EMBL sequence database library under accession numbers EU274289, EU128651 and FR669264 – FR669446.
Sequence Analysis
For newly generated sequences, consensus sequences were obtained using the SeqMan program of the DNASTAR Lasergene software package [35]. Or genes from D. yakuba were downloaded from the Comparative Assembly Freeze 1 (CAF1), according to the GLEANR Annotation in the AAAWiki website (http://rana.lbl.gov/drosophila/; [36]). Sequences were aligned using the MegAlign program of the DNASTAR Lasergene software package [35] or the BioEdit program [37].
The MacClade program [38] was used to edit the DNA alignments for further analysis. Most analyses of polymorphism and divergence were performed using the DnaSP program [39]. The normalized Fay and Wu's H statistic [18] was calculated with a program kindly provided by S. E. Ramos-Onsins.
The level of DNA polymorphism was estimated as the per-site nucleotide diversity (π: [40]), and nucleotide divergence between species as K, the number of per-site substitutions corrected according to Jukes and Cantor [41]. The minimum number of recombination events (R m) was calculated according to Hudson and Kaplan [42]. The Z nS statistic [43] was used to quantify the overall genetic association (linkage disequilibrium) between polymorphic sites.
Four tests were used in order to detect the footprint left by recent selective events on the level and pattern of polymorphism: the Hudson-Kreitman-Aguadé test (HKA test: [44]), the Tajima's D [16] and the normalized Fay and Wu's H [17], [18] tests, and the maximum likelihood Kim and Stephan test [15]. The multilocus HKA test was conducted using program HKA (distributed by Jody Hey through http://lifesci.rutgers.edu/~heylab). Moreover, the McDonald and Kreitman test (MK test; [45]), which compares the ratios of nonsynonymous to synonymous polymorphic and fixed changes was used to detect the footprint left by recurrent positive selection acting at the protein level after speciation.
Supporting Information
Number of nucleotide positions.
(0.01 MB PDF)
Genetic association.
(0.01 MB PDF)
Acknowledgments
We thank D. Orengo and C. Segarra for providing the isochromosomal D. melanogaster strains and the highly inbred D. pseudoobscura and D. miranda lines, respectively; S. E. Ramos-Onsins for sharing his unpublished program; and Servei de Genòmica at Serveis Científico-Tècnics from Universitat de Barcelona for automatic sequencing facilities.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: I.C.C. was supported by predoctoral fellowship SFRH/BD/11107/2002 from Fundacao para a Ciencia e a Tecnologia, Portugal. This work was supported by grants BFU2004-02253 and BFU2007-63229 from Comision Interdepartamental de Ciencia y Tecnologia, Spain, and 2005SGR-00166 from Comissio Interdepartamental de Recerca i Innovacio Tecnologica, Generalitat de Catalunya, Spain, to M.A. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- 2.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 3.Krieger J, Klink O, Mohl C, Raming K, Breer H. A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189:519–526. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 4.Jones WD, Nguyen TA, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr Biol. 2005;15:R119–121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhou JJ, Vieira FG, He XL, Smadja C, Liu R, et al. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Molecular Biology. 2010;19(Suppl 2):113–122. doi: 10.1111/j.1365-2583.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- 6.McBride CS, Arguello JR, O'Meara BC. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics. 2007;177:1395–1416. doi: 10.1534/genetics.107.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conceição IC, Aguadé M. High Incidence of Interchromosomal Transpositions in the Evolutionary History of a Subset of Or Genes in Drosophila. J Mol Evol. 2008 doi: 10.1007/s00239-008-9071-y. [DOI] [PubMed] [Google Scholar]
- 8.Halligan DL, Eyre-Walker A, Andolfatto P, Keightley PD. Patterns of Evolutionary Constraints in Intronic and Intergenic DNA of Drosophila. Genome Res. 2004;14:273–279. doi: 10.1101/gr.1329204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacevic M, Schaeffer SW. Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics. 2000;156:155–172. doi: 10.1093/genetics/156.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- 12.Gallach M, Arnau V, Marin I. Global patterns of sequence evolution in Drosophila. BMC Genomics. 2007;8:408. doi: 10.1186/1471-2164-8-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobzhansky TG. New York: Columbia University Press.; 1970. Genetics of the evolutionary process. p. ix, 505 p. p. [Google Scholar]
- 14.Pascual M, Mestres F, Serra L. Sex-ratio in natural and experimental populations of Drosophila subobscura from North America. Journal of Zoological Systematics & Evolutionary Research. 2004;42:33–37. [Google Scholar]
- 15.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–777. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glinka S, Ometto L, Mousset S, Stephan W, De Lorenzo D. Demography and Natural Selection Have Shaped Genetic Variation in Drosophila melanogaster: A Multi-locus Approach. Genetics. 2003;165:1269–1278. doi: 10.1093/genetics/165.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orengo DJ, Aguadé M. Detecting the footprint of positive selection in a european population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics. 2004;167:1759–1766. doi: 10.1534/genetics.104.028969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutter S, Li H, Beisswanger S, De Lorenzo D, Stephan W. Distinctly different sex ratios in African and European populations of Drosophila melanogaster inferred from chromosomewide single nucleotide polymorphism data. Genetics. 2007;177:469–480. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamblin MT, Aquadro CF. DNA sequence variation and the recombinational landscape in Drosophila pseudoobscura. A study of the second chromosome. Genetics. 1999;153:859–869. doi: 10.1093/genetics/153.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm S. A simple sequential rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- 24.Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- 25.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 26.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro JA, Huang W, Zhang C, Hubisz MJ, Lu J, et al. Adaptive genic evolution in the Drosophila genomes. Proc Natl Acad Sci U S A. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S, Stone EA, Mackay TF, Anholt RR. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. 2009;5:e1000681. doi: 10.1371/journal.pgen.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Goes van Naters W, Carlson JR. Receptors and neurons for fly odors in Drosophila. Curr Biol. 2007;17:606–612. doi: 10.1016/j.cub.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biology. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orengo DJ, Aguade M. Detecting the footprint of positive selection in a european population of Drosophila melanogaster: multilocus pattern of variation and distance to coding regions. Genetics. 2004;167:1759–1766. doi: 10.1534/genetics.104.028969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozas J, Gullaud M, Blandin G, Aguadé M. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics. 2001;158:1147–1155. doi: 10.1093/genetics/158.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M. New York: Cold Spring Harbor Laboratory Press; 1989. Drosophila: A laboratory handbook.476 [Google Scholar]
- 34.Dean MD, Ballard KJ, Glass A, Ballard JWO. Influence of Two Wolbachia Strains on Population Structure of East African Drosophila simulans. Genetics. 2003;165:1959–1969. doi: 10.1093/genetics/165.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 36.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 37.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 38.Maddison WP, Maddison DR. MacClade 4: Analysis of Phylogeny and Character Evolution: Sinauer Associates, Inc. 2000 doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- 39.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 40.Nei M. New York: Columbia University Press; 1987. Molecular Evolutionary Genetics. [Google Scholar]
- 41.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic Press; 1969. pp. 21–123. [Google Scholar]
- 42.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of nucleotide positions.
(0.01 MB PDF)
Genetic association.
(0.01 MB PDF)