Abstract
A small 63-residue membrane protein, p7, has essential roles in the infectivity of the Hepatitis C virus (HCV) in humans. This hydrophobic membrane protein forms homo-oligomeric ion channels in bilayers, which can be blocked by known channel blocking compounds. In order to perform structural studies of p7 by NMR spectroscopy, it is necessary to produce milligram quantities of isotopically labeled protein; as is the case for most membrane-associated proteins, this is challenging. We describe the successful expression of full-length p7 and two truncated constructs in E. coli using a fusion partner that directs the over-expressed protein to the inclusion bodies. Following isolation of the fusion proteins by affinity chromatography, they were chemically cleaved with cyanogen bromide. The p7-polypeptides were purified by size exclusion chromatography. Solution NMR two-dimensional HSQC spectra of uniformly 15N-labeled p7-polypeptides in DHPC isotropic micelles are fully resolved, with a single resonance for each amide site. The solid-state NMR spectra of the same polypeptides in magnetically aligned 14-O-PC/6-O-PC bicelles demonstrate their reconstitution into planar phospholipid bilayers.
Introduction
Polypeptides expressed from the Hepatitis C virus (HCV) genome are of interest because the virus infects more than 170 million people (3% of the world’s population). HCV is a member of the Hepacivirus genus of the family Flaviviridae and causes persistent infection in a majority of cases and can lead to chronic hepatitis, cirrhosis and hepatocellular carcinoma.1 A 3,000 amino acid polypeptide is encoded from a single stranded RNA of 9.6 kb. This polypeptide is cleaved proteolytically into 10 mature proteins; 3 structural proteins (E1, E2, core), 6 non-structural proteins (NS2-NS5) and a small 63-residue membrane protein (p7) whose sequence is found between those of the structural and non-structural proteins.2, 3 Because p7 is a membrane protein it is a challenging system for bacterial expression and sample preparation for NMR spectroscopy and other methods of protein structure determination. With key roles in the HCV lifecycle, it is a potential target for antiviral drugs. In this article we describe methods for heterologous expression in E. coli and purification of milligram quantities of isotopically labeled full-length and truncated constructs of p7. High-resolution solution NMR and solid-state NMR spectra demonstrate that the polypeptides are well folded and integrated into micelle and bilayer environments.
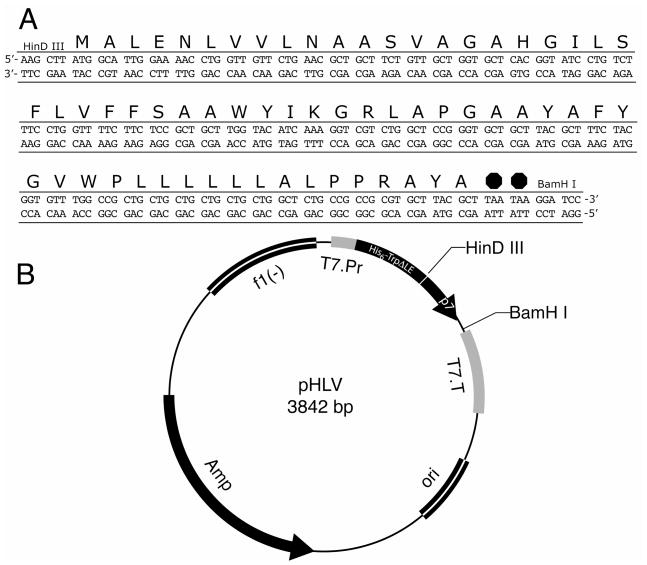
p7 is a viral membrane protein that is found to be in the endoplasmic reticulum of the infected host cell; and since it has been shown to display channel activity, it is classified as a viroporin.4 The amino acid sequence of p7 is shown in Figure 1; also shown are the sequences of two truncated constructs that have also been expressed and purified. The architecture of the protein is generally thought to resemble a ‘hairpin’ with two hydrophobic trans-membrane helices connected by a short 6-residue inter-helical loop that contains highly conserved Arg and Lys residues.5–8 p7 has been shown to form homo-oligomers that function as ion channels in membranes and in lipid bilayers;9–11 the channels can be blocked by amantadine,9 hexamethylene amiloride (HMA),10 and long akyl-chain iminosugar derivatives.11 The ion channels formed by p7 may play an important role in the life cycle of the viruses that encode them, and drugs that block them may affect virus replication as shown with p7 of BVDV.12 p7 is essential for infectivity of HCV, since mutated and deleted versions of p7 were not viable.13
Figure 1.
Amino acid sequences of the three constructs of p7 used in these studies.
NMR spectroscopy has been long used to study the structure of larger globular proteins. Recently, it has been applied to studies of membrane proteins of increasing size, including proteins with multiple transmembrane helices. It provides an opportunity to study these proteins in lipid environments that resemble the native cell membrane. Solution NMR enables studies of the protein in micelles and isotropic bicelles, while solid-state NMR can be used to study the protein in phospholipid bilayers. These studies require tens of milligrams of isotopically labeled polypeptides. In practice, this requires that the protein be heterologously expressed in bacteria that can be grown on defined isotope-containing media.
The over expression of membrane proteins is often hindered by their lethality to the host bacteria due to overloading of the cell membrane. To overcome this problem the polypeptide sequence of interest is expressed as a fusion protein, which directs it to inclusion bodies where it is sequestered away from the cell membrane.14–16 Following isolation and solubilization, the fusion partner is cleaved from the target protein, and purified by chromatography.
Intrinsic membrane proteins are, by their hydrophobic nature, insoluble in water without the presence of denaturants, detergents, or phospholipids, and at high concentrations they can form aggregates. Care must be taken during protein isolation and purification, as well as during sample preparation to ensure that the protein is properly folded.17–20 Membrane proteins with multiple transmembrane segments, once cleaved from their fusion partner, can be reconstituted into lipid environments where they assume their native conformations. This typically requires the complete removal of denaturant or detergent at the conclusion of the purification.
The preparation of milligram quantities of purified isotopically labeled p7 protein is an essential first step for structure determination by NMR spectroscopy. By screening a number of detergents and lipids for sample preparation, conditions can be identified that yield well resolved NMR spectra.21–24
Materials and Methods
Cloning
Synthesized oligonucleotides (Integrated DNA Technologies, www.idtdna.com) corresponding to the full-length sequence of p7 from the J4 genotype of HCV subtype 1b25 were used to make amplification products that could be ligated into the pHLV expression vector containing the TrpΔLE fusion protein (Figure 2).14, 26 The forward primer included a restriction site for Hind III endonuclease, a methionine codon for cyanogen bromide cleavage of the protein and five additional 5′ bases to increase the efficiency of the restriction digestion. The reverse primer included two stop codons at the end of the p7 coding region, a BamHI restriction site and four extra 5′ bases. PCR was performed using Pfu DNA polymerase (Stratagene, www.stratagene.com) and the following temperature schedule in a thermocycler (Hybaid PCRSprint, Thermo Scientific, www.thermo.com); 1 minute at 94°C, 1 minute at 60°C and 2 minutes at 72°C. After the completion of 30 cycles the reaction was incubated at 4°C for 10 minutes.
Figure 2.
Sequence of the expression vector of the p7 constructs. The DNA and protein sequence of the full-length protein are shown above the vector map. The pHLV plasmid containing the His tagged TrpΔLE fusion partner was used by inserting the p7 DNA sequence into the vector between the HinDIII and BamHI restriction sites. The DNA insert included an N-terminal methionine for cleavage and two C-terminal stop codons.
Amplified PCR products and supplied plasmids were digested with the restriction enzymes Hind III and BamHI for 1 hour at 37°C. The restricted insert was purified using PCR cleanup kit (Qiagen, www.qiagen.com) and the restricted vector products were separated on a 1.5% agarose gel. The bands corresponding to the digested plasmid were cut out and the DNA was purified using a gel extraction kit (Qiagen).
The insert was ligated into the plasmid using T4 ligase (New England Biolabs, www.neb.com). The reaction mixture was kept at room temperature for 2 hours. The reactions were transformed into DH5α cells (Invitrogen, www.invitrogen.com) and plated on LB/carbenicillin agar media. The plates were incubated overnight at 37°C. Healthy colonies were picked and grown overnight in 5 ml of LB/carbenicillin at 37°C in the shaker. The plasmids from the cells were purified using a mini-prep kit (Qiagen) and confirmed by sequencing (Eton Bioscience, www.etonbio.com).
Plasmids for the p7TM1 and p7TM2 constructs were prepared by priming the regions of the full-length p7 vector with oligonucleotides that amplified the desired sequence with the restriction sites described above. The amplified products were ligated into the restricted vector as described for the full-length construct. Colonies were grown for each of the truncated sequences and were confirmed by sequencing.
Expression
Plasmids with the correct DNA sequence were transformed into BL21(DE3) (Invitrogen) cells for growth on defined media. Transformation cells were plated on LB medium supplemented with carbenicillin and placed in an incubator at 37°C overnight. A cell stock solution was made by inoculating 5 ml of LB/carbenicillin with a single colony and allowing it to grow to an optical density at 600 nm (OD600) of 0.4. Sterilized glycerol was added to a final concentration of 7%. Fifty microliters of the cells were then aliquoted into 1.5 ml centrifuge tubes and placed in the −80°C freezer to be used for expression growths.
A starter culture was prepared by adding 10 ul of the cell stock to 5 ml of LB and carbenicillin. The culture was put in the shaker for 2 hours at 270 rpm and 37°C. One milliliter of the starter culture was added to 500 ml of sterile M9 media (7.5 mM ammonium sulfate, 39 mM glucose, 0.04 mM disodium hydrogen phosphate, 0.02 mM potassium dihydrogen phosphate, 0.009 mM sodium chloride, 1 mM and 1% LB in a 2 L baffled flask. The flask was placed in the shaker for 16 hours. The culture was then transferred to 4.5 liters of M9 media in a batch/continuous bioreactor (New Brunswick Scientific, Bioflo, www.nbsc.com). The pH was maintained at 7.0 by the addition of 10 N NaOH and the set temperature was regulated at 37°C. When the cells reached an OD600 of 0.8, protein expression was induced by the addition of 0.5 mM isopropyl-beta-D-thiogalactopyranoside (IPTG). The temperature was lowered to 25°C and the cells were allowed to grow for 18 hours. The cells were harvested by transferring them to 1 L polycarbonate centrifuge bottles (Beckman) and spinning at 7,000 rpm for 25 minutes at 4°C (Beckman Coulter Avanti J-20XP Centrifuge, Beckman JLA-8.1000 rotor, www.beckmancoulter.com).
Purification
Thirty milliliters of Resuspension Buffer I (1 mM EDTA, 50 mM Tris pH 8.0, 1 mM NaN3, 15% (v/v) glycerol) were added to the pelleted cells from each liter of culture media. The cell pellet was disrupted using a spatula, and the slurry was transferred to a 50 ml polypropylene centrifuge tube (Beckman). The resuspended cells were lysed by sonication (Fisher Scientific 550 Sonic Dismembrator, www.fishersci.com) on ice for 4 minutes. The disrupted cells were centrifuged at 17,000 rpm for 25 minutes at 4°C (Beckman JA-25.50 rotor). The supernatant was removed and the pellet containing the membrane fraction and inclusion bodies was resuspended in 30 ml of Resuspension II Buffer (1 mM EDTA, 50 mM Tris pH 8.0, 1 mM NaN3, 1% (w/v) deoxycholic acid, 1% IGEPAL CA-630). The suspension was sonicated and centrifuged as described above. The supernatant was removed and the pellet, which contained the isolated inclusion bodies, was broken up in 30 ml of Binding Buffer (6 M guanidine hydrochloride, 0.5 M sodium chloride, 20 mM Tris pH 8.0, 5 mM imidazole) and stored at 4°C overnight.
Partially dissolved inclusion bodies were sonicated on ice as described above and centrifuged at 12,000 rpm to remove any particulates that remained. The protein solution was loaded onto a Ni-NTA (Qiagen) column (30 ml) that had been charged with 3 column volumes of 0.1 M NiSO4 and equilibrated with 2 column volumes of Binding Buffer. After the protein was loaded, non-specific bound proteins were washed off of the column using 2 column volumes of Wash Buffer (6 M guanidine hydrochloride, 0.5 M sodium chloride, 20 mM Tris pH 8.0, 50 mM imidazole). Purified fusion protein was eluted from the column using 45 ml of Elution Buffer (6 M guanidine hydrochloride, 0.5 M sodium chloride, 20 mM Tris pH 8.0, 500 mM imidazole). The flow-through was collected throughout the purification procedure and the samples were analyzed by SDS-PAGE. The eluted fusion protein was transferred to a 10 kDa dialysis bag (Spectra/Por, www.spectrapor.com) and dialyzed in 4 L of ddH2O with constant water changes. After removal of guanidine the protein was lyophilized overnight (Freezone 4.5, Labconco, www.labconco.com).
The fusion partner was cleaved from the target protein by treatment with cyanogen bromide. Approximately 10 mg/ml of the fusion protein was dissolved in 70% formic acid, and 100 mg/ml cyanogen bromide was added to the solution in the lyophilizing vessel. The reaction vessel was covered in foil and incubated for 3 hours at room temperature with constant swirling. The reaction was stopped by adding an equal volume of 1 N NaOH, diluted 4-fold with ddH2O and transferred to a 1 kDa dialysis bag. The cleavage products were dialyzed in ddH2O with continuous water changes until the solution became neutral. The solution was then transferred back to the reaction vessel and placed on the lyophilizer overnight.
The dry protein powder was dissolved in 10% SDS (10 mg/ml) and placed in a bath sonicator for 15 minutes. The protein concentration was diluted in half with FPLC Running Buffer (20 mM sodium phosphate, 4 mM SDS, 1 mM EDTA, 1 mM sodium azide, pH 8.2) and loaded onto a Sephacryl column (HiPrep 26/60 Sephacryl S-200, GE Healthcare, www1.gelifesciences.com) for size exclusion FPLC. The elution profile was monitored by the absorbance at 280 nm and the fractions containing protein were collected and analyzed by SDS-PAGE. The fractions containing pure p7 were transferred to a dialysis bag with a 1 kDa size cutoff, and the detergent was removed by exhaustive dialysis. Precipitated protein was collected by centrifugation and lyophilized under vacuum overnight.
Mass Spectrometry
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) spectrometry was performed on a Voyager DE-STR instrument (Applied Biosystems, www.appliedbiosystems.com). Purified protein was dissolved in 50% acetonitrile and 0.1% trifluoroacetic acid and mixed 1:1 with a matrix solution of 3,5-dimethoxy-4-hydroxycinnamic acid dissolved in 30% acetonitrile and 0.1% trifluoroacetic acid.
NMR Sample Preparation and Spectroscopy
For solution NMR samples, the dry protein powder was dissolved in a 0.4 M 1,2-dihexyl-1-sn-glycero-3-phosphocholine (DHPC) (Avanti, avantilipids.com) solution with vortexing. The final concentrations in the samples for solution-state NMR were 0.5 mM protein, 125 mM DHPC and 10% D2O. The pH of the sample was adjusted to 4.0 with the addition of a small amount of 0.1 N NaOH. The sample was transferred to a 5 mm × 180 mm NMR tube.
Solution NMR spectra were acquired on a Bruker DMX 600 MHz spectrometer equipped with a triple-resonance probe with three-axis pulsed field gradients and a deuterium lock channel. All of the experiments were performed at 50°C using a 1.5 sec recycle delay. A 15N-1H fast Heteronuclear Single Quantum Cohererance (fHSQC)27 pulse sequence was used with 1024 points in t2 and 256 points in t1. The spectra were referenced to water at 4.70 ppm.
Solid-state samples were prepared by dissolving 3 mg of dried protein in 9.5 mg of 1,2-di-O-hexyl-sn-glycero-3-phosphocholine (6-O-PC) (Avanti) in 100 ul of water with vortexing. The dissolved protein/lipid mixture was added to 46.5 mg of 1,2-Di-O-tetradecyl-sn-glycero-3-phosphocholine (14-O-PC) (Avanti) in 100 ul of water to make a bicelle sample that was 28% lipid (wt/vol) and had a q value (long chain/short chain) of 3.2. The pH of the sample was adjusted to 4.0 using a small amount of 0.1 N NaOH. Bicelles were formed by putting the sample through several heating/cooling/vortexing cycles as described previously.28 The sample was transferred to a 5 mm × 15 mm flat-bottomed NMR tube (New Era, www.newera-spectro.com), capped with a rubber stopper and sealed with Teflon tape.
Solid-state NMR experiments were performed on a Bruker Avance 700 MHz spectrometer equipped with a home-built 1H/15N double resonance probe with a 5 mm solenoid coil with a strip shield.29 Aligned bicelle samples were measured at 42°C. Spectra were obtained using MOIST30 cross-polarization and SPINAL1631, 32 modulated decoupling with 512 t1 points and 2048 scans. The chemical shifts were referenced to external 15N- ammonium sulfate at 26.8 ppm for 15N, and the 1H resonance of water at 4.7 ppm.
The NMR data was processed using NMRPipe33 and the figures were prepared using Sparky (Goddard and Kneller, SPARKY 3, University of California San Francisco).
Results
In order to optimize conditions, primed oligonucleotides for the sequence of p7 of the J4 genotype were inserted into several different fusion systems for the expression in BL21(DE3) cells. Due to the lethality of overexpressed membrane proteins to the bacteria, growth of the bacteria to relatively high cell densities was only possible when the p7 constructs were directed to inclusion bodies in the cells. This eliminated the possibilities of using approaches where the protein was expressed directly, without a fusion protein, or directed to the membrane in the cell. Initially the N-terminal ketosteroid isomerase (KSI) fusion system, which directs the expressed protein to inclusion bodies, showed some promise but difficulties encountered in the isolation of the p7 protein led to the exploration of other vector systems. The TrpΔLE fusion protein, which also directs the protein to inclusion bodies, was found to be a suitable expression system.
The proteins expressed also contain a mutation at residue 27. A single cysteine was replaced with a serine. When working with hydrophobic proteins at high concentrations the presence of a cysteine can lead to undesirable disulfide bonds even in the presence of reducing agent. To eliminate this complication the mutation was made.
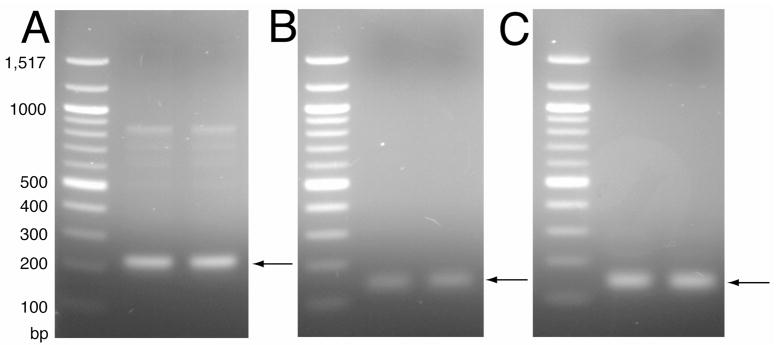
The amplified oligonucleotides were separated by electrophoresis on a 1.5% agarose gel (Figure 3A) and ligated into the vector downstream of the fusion protein that has an N-terminal His6 tag using the restriction sites Hind III and BamHI. A single methionine residue engineered into the sequences separated the TrpΔLE and p7 proteins as a site for cleavage by cyanogen bromide. For plasmids containing the truncated sequences p7TM1 and p7TM2, oligonucleotides were designed to be used as primers with the original oligonucleotides to amplify the sequences corresponding to the N-terminal 37 and the C-terminal 39 amino acids with the same restriction sites. These constructs were designed for studies of the individual transmembrane segments. The amplified products (Figure 3B and 3C) were separated on an agarose gel and ligated into the original vector in the same manner as the full-length construct. Ligated plasmids were transformed into DH5α cells and grown on LB/Carb plates. Several colonies were sent for sequencing and plasmids with the correct sequence were transformed into BL21(DE3) competent cells for expression and isotopic labeling in defined growth media.
Figure 3.
Agarose gel of PCR products for p7 construct insert DNA. The 1.5% gel shows the amplification of the sequences for (A) p7, (B) p7TM1 and (C) p7TM2. Lane 1 for each of the gels is the base pair size marker 100 Ladder (New England Biolabs).
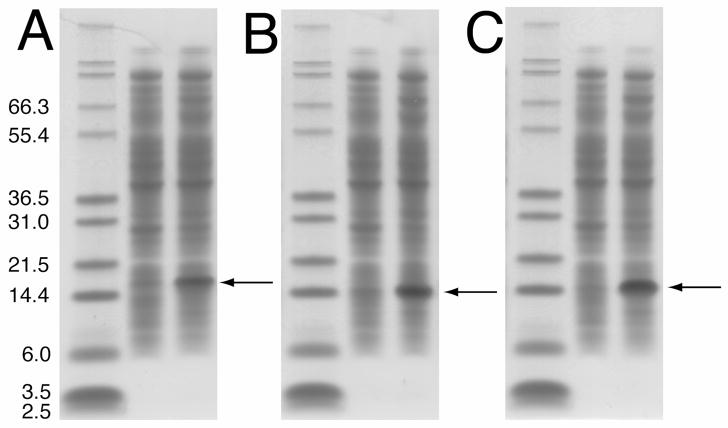
For the initial trials, the cells were grown in LB media to evaluate protein expression. These experiments indicated that the fusion protein was in fact expressed at a high level in the rich media. For the preparation of isotopically labeled proteins, the cells were grown in M9 minimal media containing 15N-labeled ammonium sulfate as the sole nitrogen source and expression was induced by the addition of IPTG. The use of a bioflo fermenter allowed the regulation of pH, temperature and oxygen during the growth. The constant monitoring of these conditions allowed the cell density to reach an OD600 of >4, almost double what was observed for the same cells grown in baffled flasks. The fusion proteins for all three constructs were highly expressed in the minimal media, as shown by SDS-PAGE (Figure 4). The prominent bands, which were not present prior to induction, in the three gels at ~21 kDa (Fig 4A), p7TM1 at ~18 kDa (Fig 4B) and p7TM2 at ~19 kDa (Fig 4C) correspond to the three fusion proteins.
Figure 4.
SDS-PAGE gel showing the expression of each of the three p7 protein constructs. The gels for (A) p7, (B) p7TM1 and (C) p7TM2 show the expression level of the fusion protein before and 4 hours after induction with IPTG. The arrow indicates the band representing the over-expressed fusion protein in each case.
The fusion proteins were isolated from the inclusion bodies by a two-step lysis. The first step utilized a Tris and glycerol buffer and sonication to remove the soluble proteins from the cell lysate. The second step removed the membrane fraction of the lysate by using a buffer that contains detergents to solubilize it during sonication. The fusion proteins in the inclusion bodies, pelleted by centrifugation, were denatured in 6 M guanidine buffer with sonication prior to being added to the nickel column for isolation by affinity chromatography. The bound protein was washed with Binding Buffer and a wash buffer containing 50 mM imidazole to remove non-specifically bound proteins. The fusion protein was then eluted in a buffer containing 500 mM imadazole. The flow through, wash and elution samples were analyzed by SDS-PAGE, and the majority of the fusion protein was found to be in the elution. The eluted protein was dialyzed against water, which immediately resulted in its precipitation. After overnight dialysis the protein was transferred to a flask, frozen and put on the lyophilizer overnight.
The purified fusion proteins were dissolved in 70% formic acid and cleaved by cyanogen bromide at the methionine residue separating the p7 construct and the TrpΔLE fusion partner. After cleavage, concentrated NaOH was added to neutralize the solution. After exhaustive dialysis in water the solution was transferred to the lyophilizer. The cleaved proteins were clearly visible on SDS-PAGE and the efficiency of the cleavage was found to be > 95%.
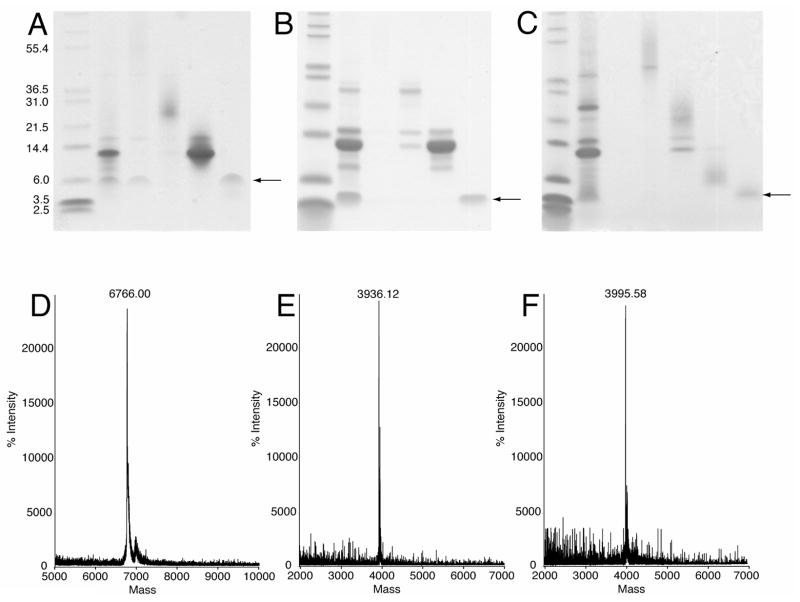
The final purification step was performed by size-exclusion FPLC in sodium phosphate buffer containing 4 mM SDS. The cleavage products eluted from the column in four major peaks. Each of these peaks was analyzed by SDS-PAGE (Figure 5). Three of the peaks corresponded to the expected cleavage products of uncleaved TrpΔLE-p7, TrpΔLE and p7. In addition, the fourth peak also containing p7 eluted at the void volume (~150 mL) and may be an oligomeric form of the protein. The three constructs of p7 stain rather poorly by Coomassie relative to the other proteins in the mixture. Nonetheless, a band of pure protein can be observed for each of the constructs (Figures 5A–C). To confirm that these bands were indeed p7, mass spectrometry was performed. All three spectra showed a single protein with the correct mass for the construct (Figures 5D–F). The elution fractions found to contain pure p7 were combined for dialysis against water. Once the protein was exhaustively dialyzed it was transferred to the lyophilizer and dried. Approximately 2 milligrams of purified protein was isolated from each liter of growth on minimal media.
Figure 5.
SDS-PAGE gel and Mass Spectroscopy of the final purification of the p7 protein constructs. The 12% Tris-Tricine gel shows the elution products of the size exclusion chromatography. The arrow indicates the position of the pure protein for (A) p7, (B) p7TM1 and (C) p7TM2. Below each of the gels is the corresponding mass spectrum for (D) p7, (E) p7TM1 and (F) p7TM2, each with the correct mass.
The initial step in the NMR studies is to obtain well-resolved solution NMR spectra of protein-containing micelles. By counting the appropriate number of resonances, this assures that the correct sequence was expressed, and that if the resonances have approximately the same lineshapes and linewidths that the protein is not severely aggregated or misfolded24. In our experience, high quality solution NMR spectra are a prerequiste for the preparation of well-behaved protein-containing phospholipid bilayer samples for solid-state NMR studies. In order to confirm the purity and correct folding of the p7 proteins, 1H-15N HSQC spectra were recorded under standard solution NMR conditions. Dried protein was dissolved in DHPC (0.4 M) in water. The protein/lipid mixture was vortexed until it became a clear solution and was diluted with deionized water and deuterium oxide. The final sample contained 125 mM DHPC and 10% D2O. The pH of the sample was adjusted to 4.0 by adding a small amount of 0.1 N NaOH. The spectra were obtained at 323 K on a spectrometer with a 1H resonance frequency of 600 MHz.
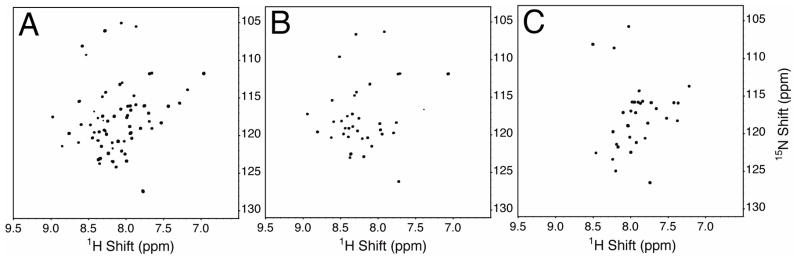
The two-dimensional HSQC spectra in Figure 6 are well resolved and each has the appropriate number of resonances, one for each amide in the protein. Notably, there are the correct number of resonances in the glycine region (15N chemical shifts < 110 ppm) are also present in each of the spectra. There are 5 glycines in full-length p7 (Figure 6A) and 3 in each of the single TM sequences (Figures 6B and 6C). The expected number of peaks corresponding to side-chain and tryptophan indole amide groups are also present in this spectra (not shown in the displayed region). Further comparison of the spectra reveals that the resonances of the individual TM proteins overlap with the corresponding resonances in the spectrum of the full-length protein.
Figure 6.
Two-dimensional 1H-15N HSQC solution NMR spectra of uniformly 15N labeled p7 constructs in DHPC micelles at pH 4.0 and 323 K. (A) Full-length p7. (B) The C-terminal truncated construct p7TM1. (C) The N-terminal construct p7TM2. Each of the spectra is fully resolved with the correct number of amide resonances for the expressed and purified polypeptide.
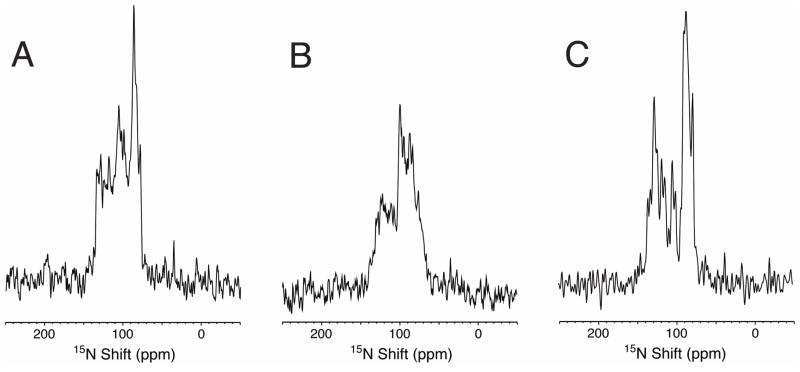
To determine if the proteins could be reconstituted into phospholipid bilayers for solid-state NMR experiments, the proteins were incorporated into bicelles made with 14-O-PC and 6-O-PC with a q of 3.2. The protein dissolved easily in the short-chain lipid and the bicelles were formed after a few cycles of heating and cooling as demonstrated by a clear mixture and correct phase transition. The signal intensity from 70 to 120 ppm in the one-dimensional 15N solid-state NMR spectra indicates that all three of the p7 constructs are well aligned along with the phospholipids in these samples (Figure 7).
Figure 7.
One-dimensional solid-state 15N NMRspectra of the three p7 constructs in magnetically aligned 14-O-PC/6-O-PC (q=3.2) bicelles at 315 K. (A) Full-length p7. (B) The C-terminal truncated construct p7TM1. (C) The N-terminal construct p7TM2. The partially resolved resonance intensity between 70 ppm and 120 ppm is consistent with trans-membrane helices that are well aligned in the phospholipid bilayers.
Discussion
Membrane proteins are challenging systems for experimental studies because of their hydrophobic nature. In general, this has little to do with the size of the polypeptide, and p7, although a small 63-residue protein, has required extensive development to establish the protocols and conditions described here and result in the high-resolution NMR spectra presented in Figures 6 and 7.
The characteristics of the solution NMR spectra of protein-containing micelles and of the solid results indicate that the protein/lipid samples have excellent homogeneity and that these samples are suitable for further studies by solution and solid-state NMR. Moreover, since NMR spectroscopy places stringent demands on the quality of the samples, these same procedures can be adapted to prepare samples for other methods of protein structure determination. The further characterization of p7 is an essential step in the development of drugs targeted to this unique protein in the HCV genome.
Acknowledgments
We thank Yanwen Mai for assistance with the preparation of the protein samples. This research was supported by grants from the National Institutes of Health and a gift from Gilead Sciences. It utilized the Biomedical Technology Resource for NMR Molecular Imaging of Proteins at the University of California, San Diego, which is supported by grant P41EB002031.
References
- 1.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ, et al. Proc Natl Acad Sci U S A. 1991;88(6):2451–5. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin C, Lindenbach BD, Pragai BM, McCourt DW, Rice CM. J Virol. 1994;68(8):5063–73. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selby MJ, Glazer E, Masiarz F, Houghton M. Virology. 1994;204(1):114–22. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez ME, Carrasco L. FEBS Lett. 2003;552(1):28–34. doi: 10.1016/s0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 5.Carrere-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. J Virol. 2002;76(8):3720–30. doi: 10.1128/JVI.76.8.3720-3730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. J Gen Virol. 2004;85(Pt 2):451–61. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 7.Luik P, Chew C, Aittoniemi J, Chang J, Wentworth P, Jr, Dwek RA, Biggin PC, Venien-Bryan C, Zitzmann N. Proc Natl Acad Sci U S A. 2009;106(31):12712–6. doi: 10.1073/pnas.0905966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patargias G, Zitzmann N, Dwek R, Fischer WB. J Med Chem. 2006;49(2):648–55. doi: 10.1021/jm050721e. [DOI] [PubMed] [Google Scholar]
- 9.Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. FEBS Lett. 2003;535(1–3):34–8. doi: 10.1016/s0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 10.Pavlovic D, Neville DC, Argaud O, Blumberg B, Dwek RA, Fischer WB, Zitzmann N. Proc Natl Acad Sci U S A. 2003;100(10):6104–8. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Premkumar A, Wilson L, Ewart GD, Gage PW. FEBS Lett. 2004;557(1–3):99–103. doi: 10.1016/s0014-5793(03)01453-4. [DOI] [PubMed] [Google Scholar]
- 12.Harada T, Tautz N, Thiel HJ. J Virol. 2000;74(20):9498–9506. doi: 10.1128/jvi.74.20.9498-9506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai A, Claire MS, Faulk K, Govindarajan S, Emerson SU, Purcell RH, Bukh J. PNAS. 2003;100(20):11646–11651. doi: 10.1073/pnas.1834545100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estephan R, Englander J, Arshava B, Samples KL, Becker JM, Naider F. Biochemistry. 2005;44(35):11795–810. doi: 10.1021/bi0507231. [DOI] [PubMed] [Google Scholar]
- 15.Thai K, Choi J, Franzin CM, Marassi FM. Protein Sci. 2005;14(4):948–55. doi: 10.1110/ps.041244305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma D, Liu Z, Li L, Tang P, Xu Y. Biochemistry. 2005;44(24):8790–800. doi: 10.1021/bi050256n. [DOI] [PubMed] [Google Scholar]
- 17.Kiefer H. Biochim Biophys Acta. 2003;1610(1):57–62. doi: 10.1016/s0005-2736(02)00717-4. [DOI] [PubMed] [Google Scholar]
- 18.Tian C, Tobler K, Lamb RA, Pinto LH, Cross TA. Biochemistry. 2002;41(37):11294–300. doi: 10.1021/bi025695q. [DOI] [PubMed] [Google Scholar]
- 19.Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ. Protein Sci. 2002;11(3):546–57. doi: 10.1110/ps.37302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogl H, Kosemund K, Kuhlbrandt W, Collinson I. FEBS Lett. 1998;432(1–2):21–6. doi: 10.1016/s0014-5793(98)00825-4. [DOI] [PubMed] [Google Scholar]
- 21.Vinogradova O, Sonnichsen F, Sanders CR., 2nd J Biomol NMR. 1998;11(4):381–6. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 22.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J Biomol NMR. 2004;28(1):43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 23.Page RC, Moore JD, Nguyen HB, Sharma M, Chase R, Gao FP, Mobley CK, Sanders CR, Ma L, Sonnichsen FD, Lee S, Howell SC, Opella SJ, Cross TA. J Struct Funct Genomics. 2006;7(1):51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 24.McDonnell PA, Opella SJ. J Magn Reson. 1993;102:120–125. [Google Scholar]
- 25.Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. Virology. 1998;244(1):161–72. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 26.Miozzari GF, Yanofsky C. J Bacteriol. 1978;133(3):1457–66. doi: 10.1128/jb.133.3.1457-1466.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 28.De Angelis AA, Opella SJ. Nat Protoc. 2007;2(10):2332–8. doi: 10.1038/nprot.2007.329. [DOI] [PubMed] [Google Scholar]
- 29.Wu CH, Grant CV, Cook GA, Park SH, Opella SJ. J Magn Reson. 2009 doi: 10.1016/j.jmr.2009.06.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levitt MH, Suter D, Ernst RR. J Chem Phys. 1986;84(8):4243. [Google Scholar]
- 31.Fung BM, Khitrin AK, Ermolaev K. J Magn Reson. 2000;142(1):97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 32.Sinha N, Grant CV, Wu CH, De Angelis AA, Howell SC, Opella SJ. Journal of Magnetic Resonance. 2005;177(2):197–202. doi: 10.1016/j.jmr.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]