Abstract
The bacterial flagellar motor can rotate in both counterclockwise (CCW) and clockwise (CW) directions. It has been shown that the sodium ion-driven chimeric flagellar motor rotates with 26 steps per revolution, which corresponds to the number of FliG subunits that form part of the rotor ring, but the size of the backward step is smaller than the forward one. Here we report that the proton-driven flagellar motor of Salmonella also rotates with 26 steps per revolution but symmetrical in both CCW and CW directions with occasional smaller backward steps in both directions. Occasional shift in the stepping positions is also observed, suggesting the frequent exchange of stators in one of the 11–12 possible anchoring positions around the rotor. These observations indicate that the elementary process of torque generation by the cyclic association/dissociation of the stator with every FliG subunit along the circumference of the rotor is symmetric in CCW and CW rotation even though the structure of FliG is highly asymmetric and suggests a 180° rotation of a FliG domain for the rotor-stator interaction to reverse the direction of rotation.
Keywords: FliG, MotAB stator complex, Salmonella, stepping rotation, switch
Many motile bacteria can move towards more favorable conditions and escape from undesirable ones for their survival by sensing temporal variations of environmental stimuli such as chemical attractants and repellents, light, O2, pH, and temperature. Such behavior is called taxis, which involves sensory reception, signal transduction, and the modulation of the motor function (1). Bacterial motility is also controlled by an intracellular signalling molecule, c-di-GMP, to facilitate biofilm formation, which often causes serious troubles in human life (2–4). Thus, bacterial motility and behavior are fundamental properties for their life cycle, and hence it is of eminent importance to understand their basic mechanisms for the welfare of human society.
Bacteria such as Escherichia coli and Salmonella enterica can swim in liquid environments by rotating helical flagellar filaments with bidirectional rotary motors embedded in the cytoplasmic membrane (1). There are many different types of motor proteins, such as F-actin-myosin and microtubule-kinesin, which are linear motors, and F- and V-ATPases and bacterial flagellar motors, which are rotary motors. Because motor proteins play many important roles in various cellular activities, deep insights into the mechanisms of force or torque generation are essential for the understanding of how cells respond to external stimuli. Interestingly, these biological motors are all unidirectional except for the flagellar motor, putting the flagellar motor in a unique position. The flagellar motor can rotate in both counterclockwise (CCW) and clockwise (CW) directions. Salmonella swims smoothly by CCW motor rotation, but momentary CW rotation causes the cell body to tumble and change their swimming direction. Switching probability can be altered by a number of environmental stimuli, and that is how taxis is achieved (1). Therefore it is also important to understand the mechanism of switching rotational direction.
The flagellar motor of Salmonella converts the energy of proton flow by the electrochemical potential difference across the cytoplasmic membrane into the mechanical work required for its rotation (1, 5, 6). MotA and MotB form the MotA4MotB2 complex within the cytoplasmic membrane (7, 8) and act as the stator that conducts proton (9). A fully functional motor contains at least 11 stator units around the rotor (10). FliF, FliG, FliM, and FliN form the rotor called the MS-C ring; FliF forms the MS ring, which spans through the cytoplasmic membrane with the M ring portion, and the remaining three proteins form the C ring on the cytoplasmic side of the MS ring (11). An electrostatic interaction between the cytoplasmic domain of MotA and the C-terminal domain of FliG is thought to be involved in torque generation (12). The protonation and deprotonation of a highly conserved aspartic acid of MotB is efficiently coupled with torque generation (13, 14).
FliG, FliM, and FliN are also called the switch proteins responsible for switching the direction of motor rotation because certain mutations in these proteins affect the switching between CW and CCW rotation (15). The switching probability is modified by a chemotactic signal transduction pathway (1, 6). The binding of a chemotactic signalling protein, phospho-CheY (CheY-P), to FliM and FliN is thought to induce a cooperative conformational change in the C-terminal FliG domains of the C ring, resulting in a different rotor-stator interaction to allow the motor to spin in the CW direction (16, 17). The switching probability increases linearly with increase in motor torque in both directions, suggesting that the switch senses the load on the stator-rotor interaction as well as the level of CheY-P (18). Recently, it has been reported that the c-di-GMP binding protein YcgR binds to MotA, FliG, and FliM in the presence of c-di-GMP, resulting not only in a reduced motor speed but also in the CCW bias in motor rotation (3, 4). In Rhodobacter sphaeroides, the flagellar motor rotation is unidirectional, but CheY-P acts as a molecular brake to stop motor rotation to change the swimming direction (19).
The previous observation of 26 CCW steps per revolution of the flagellar motor (20), which corresponds to the number of FliG subunits that form part of the rotor ring (21), made it clear that the elementary torque generation process is the cyclic association/dissociation of the stator complex MotA/B with every FliG subunit along the circumference of the rotor. The observation of such a fine stepping motion was made possible by reducing the number of functional stators in the sodium ion-driven chimeric flagellar motor and lowering sodium ion motive force to reduce the rotation rate to meet the spatial and temporal resolution of the measurement system (20). It has also been reported that the unidirectional motor of Rhodobacter sphaeroides stops at 27–28 discrete angles, locked in place by a relatively high torque (19). Interestingly, backward CW steps of the sodium ion-driven chimeric flagellar motor were also often observed even in the absence of CheY, but the step size (-10.3°) was slightly smaller than the forward CCW steps (13.8°) (20), suggesting an asymmetric mechanism of bidirectional rotation. Asymmetry in the CW and CCW rotation has also been demonstrated recently with distinct torque-speed curves (22). However, it still remains unclear whether the elementary torque generation process is really asymmetric with different step sizes in the two directions.
In this study, to clarify the elementary process of torque generation of the flagellar motor in both CCW and CW directions, we set up an optical nanophotometry system that has the ability to determine the two-dimensional position of a 100-nanometer (nm) fluorescent bead with nanometer spatial resolution and submillisecond temporal resolution and measured the fine stepping motions of the proton-driven flagellar motor of Salmonella. We partially replaced the wild-type MotA/B complex with the mutant MotA/B(D33N) complex to reduce the number of functional stators incorporated into the motor. We show that the average step size is about 14° in both directions, clearly indicating a symmetric elementary process of bidirectional torque generation.
Results
Spatial Resolution of Nanophotometry.
We have been able to observe the stepping motion of the proton-driven flagellar motor of Salmonella with our fluorescence nanophotometry system at nanometer spatial resolution and submillisecond temporal resolution by labelling the motor with a 100-nm fluorescent bead. The standard deviations of the observed position of 100-nm beads attached on the glass surface show that the accuracy of the bead positioning is 2.2 nm, which corresponds to about 0.8° in the angular positioning (Figs. S1 and S2). But we still needed some other tricks to reduce the rotation rate of the motor to observe the fine steps.
Reduction of the Number of Stators.
Because the involvement of multiple stators for torque generation as well as the high-speed rotation of the motor makes it difficult to observe the stepping motion (20), we tried to reduce the number of functional stators. Because the MotA/B(D33N) complex is known to form a nonfunctional stator with totally impaired proton conductivity around the rotor (23, 24), we coexpressed MotA and MotB(D33N) in wild-type cells to reduce the number of functional stators (Fig. S3). Because the torque at high load is proportional to the number of stators (10, 25, 26), we estimated the number of functional stators in the motor by measuring the rotation rates of 1.0-μm beads attached to partially sheared, sticky filaments of SJW46. Fig. S4A shows the histograms of flagellar rotation rates for cell populations grown with 10 μM or 25 μM IPTG to induce the expression of MotA and MotB(D33N). Under a low induction condition with 10 μM IPTG, the speed histogram showed a majority of data distributed within a range from 20–40 Hz with a small population extended up to 70 Hz, and the entire histogram showed several peaks separated by a roughly equal interval of around 8 Hz (Fig. S4A, middle). The result is similar to what was previously observed for the E. coli motAB double mutant under low induction of MotA and MotB (10). Under a high induction condition with 25 μM IPTG, the histogram showed a major population up to 25 Hz with a few peaks at multiples of 8 Hz (Fig. S4A, lower). Because it has been shown that a single stator unit drives a 1.0-μm bead at about 7 Hz in the proton-driven flagellar motor of E. coli (7) and because our resurrection experiments under high load to measure the step wise increment in rotation rate upon induction of MotA/MotB in a motA/motB null strain of Salmonella showed that the unit increment was 7–8 Hz (Fig. S4B), the result indicates that the number of functional stators was reduced to one or two under the high induction condition. The same unit speed of about 8 Hz obtained by either resurrection or replacing functional stators with nonfunctional ones also assure that the nonfunctional stators do not disturb the rotation behavior and hence the fine stepping motion of the motor we observed.
Stepping Rotation.
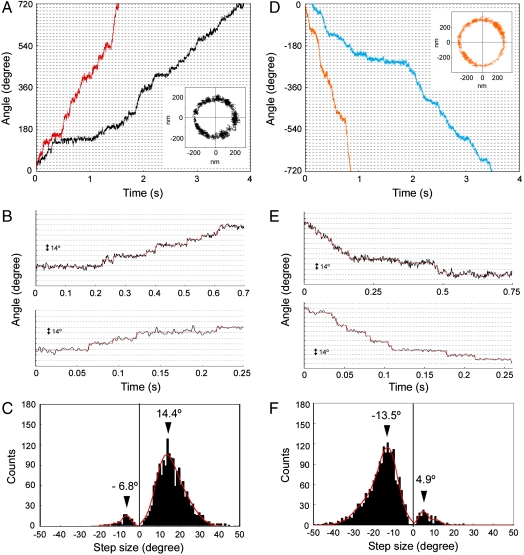
A much smaller bead, such as 100-nm bead, for low-load measurements is preferable for the observation of fine steps since a high-load probe is likely to show a low compliance, smearing the rotational movement of the motor by elasticity and flexibility of the axial structures, such as the hook and filament (6). It has been shown, however, that the motor speed near zero load is independent of the stator number (26, 27). To reduce the rotation rate further under low load, we reduced the intracellular pH without changing the proton motive force (pmf) significantly by lowering the external pH in the presence of 20 mM potassium benzoate (28, 29). We carried out bead assays at an external pH of 6.0 in the presence of 20 mM potassium benzoate and 25 μM IPTG. Because we used a high-intensity dye laser as a light source for nanophotometry, cumulative photo damage may have further reduced the rotation rate by reducing pmf as previously described (20). Fig. 1A shows a temporal record of the rotation angles of 100-nm fluorescent beads attached to the partially sheared sticky filament of MM3076iC, which rotates exclusively CCW due to the deletion of chemotaxis genes. Stepping motions can be clearly seen with motors rotating CCW at speeds less than 2 Hz (Fig. 1B and Fig. S5A). The step size histogram shows a peak at 14.4° (Fig. 1C). The rotational fluctuation of the beads attached to the paralyzed motors of a Salmonella motA motB double null mutant was about 2.5° (Fig. S2). Therefore, we conclude that the step size of the proton-driven motor rotating CCW is about 14°, which is in good agreement with previous results with the sodium ion-driven chimeric motor (20).
Fig. 1.
Stepping rotation of the Salmonella motor in CCW and CW directions. (A), (B) Angular positions of the beads around the motor axis plotted against the time. (A) The beads attached to the flagellar filaments of MM3076iC (exclusively rotates CCW), and (D) those of SJW46 (rotates CCW and CW). The increase and the decrease in the angle correspond to CCW and CW rotation, respectively. Different colors correspond to the rotation of different motors of different cells. The bead positions were determined from images sampled at a frame rate of 2.4 kHz, and epifluorescence excitation was done using a dye laser (≈20 mW). A two-dimensional trajectory of a bead corresponding to one of the two angular curves is shown in the inset of (A), (D), respectively. (B), (E) Selected parts of the angular plots in (A), (D), respectively. Red lines are steps determined by a step-finding algorithm. (C), (F) Histograms of step angles obtained from CCW and CW rotation, respectively. Cells were grown with 25 μM IPTG for 4 h. All the measurements were carried out at an external pH 6.0 in the presence of 20 mM potassium benzoate at 23 °C.
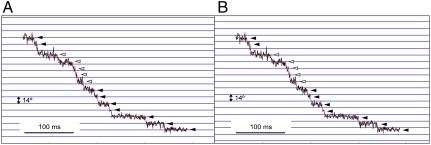
Although the motor of MM3076iC rotated mostly CCW, backward steps were also occasionally observed as was shown previously (20), but the step sizes were much smaller (6.8°) than those observed in the previous result (10.9°) (20) (Fig. 1C). When the motor abruptly switches the rotational direction momentarily, the observed step size can be smaller than the actual one due to an elastic response of the hook and filament (6). However, when the backward step is prolonged enough for the distortion in the elastic elements of the flagellum to relax, the step size should be the same as that of the forward one. We actually observed backward steps of normal size occasionally (Fig. 2). Therefore, the backward steps of smaller sizes are presumably due to the artefact of measurement and analysis.
Fig. 2.
Full 14° backward steps observed with motors of two different cells. (A) MM3076iC and (B) SJW46 motors. Arrowheads indicate switching events. The upward and downward displacements correspond to CCW and CW rotation, respectively.
To observe steps in CW rotation, we attached the 100-nm fluorescent bead to the filament of SJW46, whose motor rotates in both CCW and CW directions. We carried out bead assays at external pH 6.0 in the presence of 20 mM benzoate not only to reduce the rotation rate but also because chemoreceptors sense a reduced cytoplasmic pH and this increases the probability of CW-biased rotation of the motor (30, 31). The motor extensively rotated in CW direction (Fig. 1D) and showed stepping motions (Fig. 1E and Fig. S5B). The peak of the step size histogram was found at 13.5° (Fig. 1F), which is almost the same as that of CCW rotation considering the relatively wide distribution. Small backward steps were also observed (Fig. 1F). These results indicate that the elementary processes involved in torque generation are equivalent in both CCW and CW directions, suggesting a symmetric mechanism of torque generation.
We also observed occasional shifts in the angular step positions of regular 14° interval (Fig. 3). The sets of consecutive steps labeled with solid and open arrowheads both have a step size of 14°, but those labeled with open arrowheads are shifted by about 10° relative to those labeled with solid arrowheads. The shifts in the step position are probably caused by an exchange of an active stator unit with a new one anchored at a different position, which is not related with the previous one by the 26-fold rotational symmetry of the rotor, but is presumably one of the 11–12 possible positions for the stators to be anchored around the rotor. This result indicates that the stator anchoring positions around the rotor are not related with the rotor structure. In the stepping rotation record shown in Fig. 3, the five steps labeled with open arrowheads in the second phase is shifted by about 10° from the first phase but the remaining seven steps labeled with solid arrowheads in the third phase are back in the same original position as the first phase, suggesting that the active stator in the first phase did not detach from the rotor completely and became active again after the other one that was active in the second phase became inactive. Such an exchange of active stators may be a frequent event in the process of torque generation.
Fig. 3.
Occasional shifts in the step positions of regular 14° interval. A set of horizontal lines in (B) is shifted from that in (A) by 9° to indicate the amount of shift in the second phase labeled with open arrowheads. Solid and open arrowheads indicate the step positions matching the horizontal lines in (A), (B), respectively.
Discussion
The fine stepping motion of the flagellar motor with 26 steps per revolution has been observed only with a sodium ion-driven chimeric motor (20). The present observation of the 26 steps per revolution with the proton-driven flagellar motor indicates that the torque generation mechanisms are similar between the two types of flagellar motors regardless of the power source. Although bidirectional flagellar motor rotation had been thought to be symmetric (32), two recent observations suggested asymmetry in the mechanism of torque generation. One showed different step sizes in the forward and backward rotation (20); the other showed distinctly different torque-speed curves in CW and CCW rotation (22). Our present observations clearly show that at least the elementary process of torque generation is symmetric in the bidirectional motor rotation. Occasional shift in the angular positions of 26 steps per revolution and its restoration to the original ones suggest that the stator anchoring positions are not related with the rotor structure and that even the stators anchored around the rotor become temporarily inactive without leaving the motor. These observations will add some useful constraints to distinguish specific mechanisms of motor rotation.
The bidirectional force generation by the flagellar motor is unique among the motor proteins. The members of F-actin-myosin and microtubule-kinesin families are all unidirectional (33, 34). Interestingly, some of them move in the opposite directions by modifications in the neck regions of myosin and kinesin family proteins, while the step sizes are determined by the subunit repeat distances of F-actin and microtubule structures (33, 34). It is thought that the direction of myosin and kinesin motions are determined by biased thermal motions of the head domains of myosin and kinesin, which are presumably determined by the conformational bias of their neck regions. In contrast, in flagellar motor rotation, the rotor ring protein FliG, which determines the step size, is also responsible for switching rotational direction. The crystal structures of FliG show highly asymmetric nature (35, 36), and the charged residues on the surface of its C-terminal domain are presumed to line up with their partner residues on the cytoplasmic domain of MotA for an electrostatic interaction between these two domains in a torque generation step (12). To achieve a symmetric elementary process of torque generation in both CCW and CW rotation, the C-terminal domain of FliG may need to go through a 180° rotation to reorient its charged residues in the two opposite directions. Such a conformational change albeit may seem quite drastic would be possible because the C-terminal domain in the crystal structure is relatively isolated from the rest of the molecule (35, 36). The binding of CheY-P to FliM and FliN could trigger such a conformational change of FliG because FliM subunits are in direct contact with FliG subunits in the C ring structure (5). Although the structure of the cytoplasmic domain of the MotA/B complex is not yet available, it is unlikely that the cytoplasmic domain flips its orientation by 180° for torque generation in the opposite direction because the domain consists of loops and terminal chains that are tightly anchored to the transmembrane domain of the complex that is fixed in a unique orientation in the membrane. That the stator unit is composed of four MotA and two MotB molecules with a twofold symmetry axis perpendicular to the membrane may be to present two distinct surface areas for interactions with the C-terminal domain of FliG in the two opposite orientations for bidirectional torque generation. The distinctly different torque-speed curves in the two directions of rotation (22) may be due to different kinetic parameters involved in the torque generation cycle, and it may imply that the rotor-stator interactions are achieved in structurally distinct manners. Although the 180° rotation of the FliG domain is merely a speculation, it may explain the distinct torque-speed curves.
Materials and Methods
Bacterial Strains, Plasmids, Transduction, DNA Manipulation, and Media.
Salmonella strains SJW46 (fliC(Δ204–292)) (37) and MM3076iC (Δ(cheA-cheZ), fliC (Δ204–292)) (29) were used in this study. SJW46 has intact motors and chemotaxis system, and hence the motor can rotate both CCW and CW, while MM3076iC has intact motors but lacks the chemotaxis system, and hence the motor rotates exclusively CCW. A plasmid, pNSK9-D33N, which encodes MotA and MotB(D33N) on pTrc99A (Pharmacia) was transformed into both cells to reduce the number of functional stators. To carry out resurrection experiments, we used a plasmid, pYC20, which encodes MotA and MotB on pBAD24 (24). DNA manipulation was carried out as previously described (38). Luria broth and motility medium were also prepared as previously described (28).
Setup of an Optical System and Bead Assays.
An optical system was built on an inverted fluorescence microscope (IX-70, Olympus) with a cooled, back-thinned, electron-multiplying charge-coupled device (EMCCD) camera (iXon, Andor), and set on a vibration-free table (Newport). Fluorescent dye-absorbed 100-nm beads (FluoSpheres® “orange,” excitation at 540 nm, emission at 560 nm; Invitrogen) were attached to the sticky filaments and were excited by either green light from an ultra-high-pressure mercury lump (HBO103W/2, Osram) or the 546 nm emission line from a dye laser (599 standing wave dye laser, Coherent) through a 100× oil immersion objective lens (PlanApo, NA 1.4, Olympus). The cube filter set 41007 (Chroma) was used for fluorescence microscopy. The single bead image was projected onto the EMCCD at a magnification of 500×. To bring the bead image onto the EMCCD camera, the specimens placed on an ultrastable stage (Ikeda-Rika) were manipulated by a two-dimensional piezo-micromanipulator (P-734, PI).
Bead assays with 1.0 μm polystyrene beads (Invitrogen) and 0.1 μm fluorescent beads were carried out as previously described (14, 29).
Correction for Elliptic Rotational Trajectory and Step Analysis.
Because the bead trajectories are mostly in elliptic shape due to the tilt of the rotational axes of the motors located on various positions of nearly cylindrical cell surface, the angular positions of the rotation were determined by correcting for the elliptic deformation by the least-squares fitting of an ellipse to each bead trajectory.
Step analysis was carried out with a minor modification of the procedure described by Sowa et al. (20). First, an entire episode of trajectory was divided into two intervals, A and B, at a point that gives the best least-squares fit to a single-step function. Then, the same procedure was applied to each interval, and this procedure was repeated until the standard deviation became a preset value estimated from the overall standard deviation of the data. If a step angle was smaller than the standard deviation of the intervals on the both side, the step was removed, and the two adjacent intervals were concatenated.
Supplementary Material
Acknowledgments.
We thank R.M. Berry and Y. Sowa for helpful discussion and comments and F. Oosawa and M. Macnab for continued support and encouragement. S.N. was a research fellow of the Japan Society for the Promotion of Science. This work has been supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S.N., T.M., and K.N.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007448107/-/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 3.Boehm A, et al. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:1–10. doi: 10.1016/j.cell.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a "backstop brake" mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr Opin Struct Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 7.Braun TF, Blair DF. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry. 2001;40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 8.Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 9.Blair DF, Berg HC. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 10.Reid SW, et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 14.Che Y-S, et al. Suppressor analysis of the MotB(D33E) mutation to probe the bacterial flagellar motor dynamics coupled with proton translocation. J Bacteriol. 2008;190:6660–6667. doi: 10.1128/JB.00503-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi S, et al. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyer CM, Vartanian AS, Zhou H, Dahlquist FW. A molecular mechanism of bacterial flagellar motor switching. J Mol Biol. 2009;388:71–84. doi: 10.1016/j.jmb.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarker MK, Paul K, Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci USA. 2010;107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan J, Fahrner KA, Berg HC. Switching of the bacterial flagellar motor near zero load. J Mol Biol. 2009;390:394–400. doi: 10.1016/j.jmb.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pilizota T, et al. A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor. Proc Natl Acad Sci USA. 2009;106:11582–11587. doi: 10.1073/pnas.0813164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sowa Y, et al. Direct observation of steps in rotation of the bacterial flagellar motor. Nature. 2005;437:916–919. doi: 10.1038/nature04003. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, Yonekura K, Namba K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol. 2004;337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 22.Yuan J, Farner KA, Turner L, Berg HC. Asymmetry in the clockwise and counterclockwise rotation of the bacterial flagellar motor. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1007333107. 1007333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J, et al. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto YV, Che Y-S, Minamino T, Namba K. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS lett. 2010;584:1268–1272. doi: 10.1016/j.febslet.2010.02.051. [DOI] [PubMed] [Google Scholar]
- 25.Block SM, Berg HC. Successive incorporation of force-generating units in the bacterial rotary motor. Nature. 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 26.Ryu WS, Berry RM, Berg HC. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature. 2000;403:444–447. doi: 10.1038/35000233. [DOI] [PubMed] [Google Scholar]
- 27.Yuan J, Berg HC. Resurrection of the flagellar rotary motor near zero load. Proc Natl Acad Sci USA. 2008;105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamino T, et al. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J Bacteriol. 2003;185:1190–1194. doi: 10.1128/JB.185.4.1190-1194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura S, et al. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J Mol Biol. 2009;386:332–338. doi: 10.1016/j.jmb.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 30.Kihara M, Macnab RM. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Repaske DR, Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981;145:1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berry RM, Turner L, Berg HC. Mechanical limits of bacterial flagellar motors probed by electrorotation. Biophys J. 1995;69:280–286. doi: 10.1016/S0006-3495(95)79900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 34.Endow SA, Higuchi H. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature. 2000;406:913–916. doi: 10.1038/35022617. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd SA, Whitby FG, Blair DF, Hill CP. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 36.Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshioka K, Aizawa S-I, Yamaguchi S. Flagellar filament structure and cell motility of Salmonella typhimurium mutants lacking part of the outer domain of flagellin. J Bacteriol. 1995;177:1090–1093. doi: 10.1128/jb.177.4.1090-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J Mol Biol. 2004;343:457–466. doi: 10.1016/j.jmb.2004.08.067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.