Abstract
Intrinsic disorder is believed to contribute to the ability of some proteins to interact with multiple partners which is important for protein functional promiscuity and regulation of the cross-talk between pathways. To better understand the mechanisms of molecular recognition through disordered regions, here, we systematically investigate the coupling between disorder and binding within domain families in a structure interaction network and in terminal and inter-domain linker regions. We showed that the canonical domain–domain interaction model should take into account contributions of N- and C-termini and inter-domain linkers, which may form all or part of the binding interfaces. For the majority of proteins, binding interfaces on domain and terminal regions were predicted to be less disordered than non-interface regions. Analysis of all domain families revealed several exceptions, such as kinases, DNA/RNA binding proteins, certain enzymes, and regulatory proteins, which are candidates for disorder-to-order transitions that can occur upon binding. Domain interfaces that bind single or multiple partners do not exhibit significant difference in disorder content if normalized by the number of interactions. In general, protein families with more diverse interactions exhibit less average disorder over all members of the family. Our results shed light on recent controversies regarding the relationship between disorder and binding of multiple partners at common interfaces. In particular, they support the hypothesis that protein domains with many interacting partners should have a pleiotropic effect on functional pathways and consequently might be more constrained in evolution.
Introduction
Recent computational and experimental studies have revealed that many protein regions lack well-defined structure. These so-called intrinsically disordered proteins (IDPs) have certain properties and functions that distinguish them from proteins with well-defined structures, namely they have specific amino acid composition, propensity for post-translational modifications, and promiscuous binding of different partners. Disorder might be also crucial for providing reduced constraints for alternative splicing and efficient regulation via rapid degradation.1–3
It has been suggested that intrinsic disorder contributes to the ability of some proteins to interact with multiple partners which can be important for protein functional promiscuity, regulation of the cross-talk between pathways, and evolution of new functions.4,5 Both theoretical and experimental studies have suggested that intrinsically disordered proteins are plastic and can adopt different structures upon binding to different partners.6–9 Interactions with multiple partners can be accompanied by disorder-to-order transition or folding upon binding10–14 although disorder may also play an important functional role in protein complexes, especially in homooligomers.15–17 In addition, binding through unfolded or partially unfolded intermediates can provide a kinetic advantage through the “fly-casting” mechanism.18 The binding mechanism, whether binding occurs between folded or unfolded chains, depends on the structural characteristics, interface properties, and degree of minimal frustration of monomers.19,20 Indeed, it has been shown that physicochemical characteristics of interfaces formed by IDPs are on average different from those formed by structured proteins. Namely, they form much larger interfaces with a large number of contacts per residue, exhibit prominent preference for hydrophobic residues and are localized linearly on the primary sequence.17,19,21 A few examples have experimentally demonstrated the coupling between folding and binding,10–14,22,23 and other examples were compiled from the analysis of protein complexes in the Protein Data Bank (PDB).16,21,24–28 Different algorithms have been proposed to predict disordered binding motifs prone to disorder-to-order transition from the protein sequence.26,29–31
The important role of disorder in protein–protein interactions is manifested in the high frequency of disordered proteins in protein–protein interaction networks. Studies of the relationship between disorder content and the degree of a protein in interaction networks showed that some hub proteins are fully or partially disordered, and some structured hub proteins interact with disordered proteins.32–34 Although hub proteins with at least ten interactions seem to be more enriched with disorder compared to proteins with a single interaction,35 the correlation between the disorder of a protein and the number of its partners has been reported to be rather weak.36 In addition, it has been shown that disorder may promote the assembly of large complexes,15 independently of the hubbiness of the protein.37
To gather clearer evidence regarding the relationship between disorder and binding of multiple partners at the same or different interfaces, several studies inspected disorder at binding interfaces. It has been suggested that all hubs might be subdivided into two categories that reflect their different binding and evolutionary properties. Based on co-expression or structural data, one might distinguish “party”38 (or “multi-interface”)39 hubs, which correspond to more evolutionarily conserved proteins binding many protein partners simultaneously, from “date” (or “singlish”) hubs, which correspond to less-conserved proteins forming mutually exclusive, transient interactions. It has been shown that date or singlish hubs might have a higher fraction of disorder than non-hub proteins40,41 while multi-interface hubs have approximately the same disorder content as other proteins.42
To understand the principles of molecular recognition through disordered regions, we performed a rigorous analysis of protein disorder with respect to protein binding and promiscuous binding (or multibinding). The most straightforward way to study these effects would be to invoke the structural interaction networks that provide the data on interaction interfaces and, in particular, interfaces that bind multiple partners (multibinding interfaces). Such an approach using structural networks has been undertaken recently for full chain proteins41 from PDB and the subset of the Saccharomyces cerevisiae proteins confirmed using domain interaction data and binding interfaces inferred from iPfam.42 Our approach is, instead, to explore the full range of observed disorder at the family level, by compiling all binding interfaces of proteins in each family from experimentally-determined structures of protein complexes. Systematically characterizing disorder across domain families helps to avoid the bias caused by over-represented families in protein–protein interaction networks and the large number of interactions between homologous proteins.
Here, we integrate analysis of disorder and binding for protein domains, inter-domain linkers, and terminal regions. Such integrative analysis is crucial since previously observed correlations between disorder and hubbiness can be explained by the presence of disordered inter-domain linkers (or terminal regions) in multidomain proteins, examples of which were discussed in a previous review.33 Moreover, the number of interactions and hubbiness depends on the number and characteristics of domains in multidomain proteins and it is not clear how disorder is coupled with binding at the level of individual domains. It has been emphasized previously how important it is to analyze distinct binding modes (not only the number of binding partners) which can give clues about the relationship between network topology and genomic features.39 A large fraction of such binding modes is the result of crystal packing and rigorous filtering should be applied, especially to define multibinding interfaces. Furthermore, different methods of disorder prediction might produce quite different results which may lead to noise, bias, and controversy in understanding the coupling of disorder and binding.
Taking all these into consideration, in this study we applied three independent disorder prediction techniques and ensured biological relevance of interactions with the ultimate goal of trying to reveal mechanisms of molecular recognition through disordered regions. We explore the relationship between disorder and binding using atomic details of protein interactions. In particular, we study interfaces that are reused for binding to different partners, mapping observed binding interfaces to a domain–domain interaction network. Analyzing disorder at the domain family level allows us to measure the relationship between disorder and diversity of interactions for various protein families and identify all domain families with significantly more or less disorder on binding interfaces. We also investigate disorder in terminal and inter-domain linker regions to provide a complete picture of the role of disorder in protein binding.
Results
Propensity for disorder as a function of number of interactions
We define an interaction to be between two domain families with a distinct conserved binding mode in order to measure the variety of interactions rather than the number of interaction partners. Domain–domain interactions were gathered from PDB for families from the Conserved Domain Database,43 as described in Experimental. Disorder was predicted from sequence using the Disopred2,44 FoldUnfold,45 and VSL246,47 algorithms which identified 6%, 11%, and 18%, respectively, of residues in domain footprints as disordered (Table 1). Fraction disorder for each family is the average of disorder over all proteins in the family. Fig. 1 illustrates how fraction disorder on the footprint and binding interface regions depends on the number of domain–domain interactions. As can be seen from this figure, there is a tendency for protein domain families with more interactions to exhibit less disorder on average (with a slight increase in disorder for families with more than 8 interactions). This correlation is rather weak but statistically significant for two disorder prediction methods, Disopred2 and VSL2 (p-value < 0.001), while FoldUnfold does not report significant decrease with the number of interactions.
Table 1.
Average fraction disorder (as a percentage) present in domain footprint (F) and interaction interface (I), according to the Disopred2, FoldUnfold, and VSL2 prediction methods. Data are shown for all families in the dataset and selected subsets
| Family description | Number of families | Disopred2 |
FoldUnfold |
VSL2 |
|||
|---|---|---|---|---|---|---|---|
| F | I | F | I | F | I | ||
| All families | 1364 | 6.1 | 4.4 | 11.4 | 9.6 | 18.2 | 15.7 |
| 1DDIa | 611 | 7.7 | 5.8 | 13.5 | 11.8 | 21.5 | 18.7 |
| >1DDI | 753 | 4.7 | 3.4 | 9.8 | 7.9 | 15.6 | 13.2 |
| >8DDI | 61 | 3.1 | 2.1 | 9.4 | 7.8 | 13.6 | 11.4 |
| Promiscuous | 108 | 4.1 | 4.1 | 10.3 | 8.7 | 16.6 | 14.8 |
| Signaling | 49 | 11.3 | 8.4 | 8.6 | 6.3 | 24.4 | 20.4 |
Domain–domain interaction.
Fig. 1.
Average fraction disorder per domain family plotted against number of domain–domain interactions. Disorder is measured over domain footprint (unmarked line) and interface region (line with symbol) for three disorder prediction methods.
Although there is a tendency towards less disorder as the number of interactions grows, we observe diversity of fraction disorder among different members of domain families. For example, 24% of families predicted by Disopred2 and FoldUnfold and 10% of families predicted by VSL2 exhibit quite large variation in disorder content (where the ratio between mean value and standard deviation of fraction disorder within the family is greater than one). Large variation in disorder might indicate that disorder is not conserved and is non-functional in these cases. Another possibility is that different family members might have specific interaction partners which employ various disordered regions (either structured interfaces for binding disordered proteins or disordered interfaces binding structured proteins). This scenario has been observed in several cases outlined previously.42,48 Indeed, we observe a correlation between diversity in disorder content within the domain family and the number of interactions.
We also observe that promiscuous domain families (we analyzed 108 non-redundant domain families corresponding to 215 domains from this study), defined from the independent study49 based purely on domain architecture analysis, have slightly less disorder on all domain regions compared to the overall dataset, but this difference is significant only for the footprint region for disorder predicted with Disopred2 (p-value < 0.03) (Table 1). Our finding is consistent with the previous observation that promiscuous domains recombine with many other domains in evolution (by definition), suggesting a large number of interaction partners.
Signaling proteins were previously found to have significantly greater disorder than proteins with other functions50 and the kinase family, in particular, was enriched among single-interface hub proteins.42 We identified 49 potential signaling families in our dataset as the families where a domain is annotated with the “signal transducer activity” function or the “signal transduction” biological process according to the curated Gene Ontology Annotation.51 We find that the number of interaction partners does not differ significantly between signaling and non-signaling domains yet the fraction disorder in signaling domains is significantly higher than in non-signaling domains according to the Disopred2 and VSL2 algorithms (Table 1). This is consistent with the previous study of the role of disorder in domain–domain interaction networks of S. cerevisiae, where the authors showed that multibinding domains (“singlish” according to their terminology) enriched with signaling and kinase functions have a higher fraction of Disopred2 predicted disorder.39,42
It has been shown that a much larger fraction of eukaryotic proteins contain long disordered regions compared to bacterial and archaeal proteins.44 Indeed, we observe that families containing only eukaryotic proteins (416 families) have 1.6–2.5 times as much disorder on domain footprint and interface regions compared to families that contain only prokaryotic proteins (543 families) according to the VSL2 and Disopred2 methods. (FoldUnfold reported a slight increase in disorder in prokaryotic families.) Our data set is well-balanced between eukaryotes and prokaryotes with 46% of domains from eukaryotic proteins, 43% from bacteria, and 6% from archaea, and the number of domain–domain interactions is distributed similarly for eukaryote-only families and prokaryote-only families (though statistically distinct according to the t-test). These suggest that the taxonomic source of the proteins in our data set does not overly influence our overall findings regarding the relationship between interaction and disorder.
Analysis of disorder in connection to domain binding
To study the coupling between disorder and binding, we analyzed the preference of disordered regions to be located on binding interfaces. We would like to reiterate that disorder on interface refers to the sequence-based prediction of disorder implying that the interface region would probably be disordered in unbound state, but does not exclude the possibility that the interface might undergo disorder-to-order transition upon binding. We found that for the majority of domain families the binding interface region is predicted to contain less disorder than the footprint (Table 1; Fig. 1), and the mean values of fraction disorder for the footprint and interface regions are statistically significantly different from each other (t-test p-value < 0.0001). Restricting this analysis to domains with at least 5 or 10 disordered residues in the footprint produced the same result.
Mapping the disordered regions on the common reference frame allowed us to analyze the tendency of disordered regions to be located on multibinding interfaces for each individual domain family. Multibinding interface is defined as those positions that participate in interactions with at least two different non-redundant domain families (see Methods). Table 2 lists the 23 families with statistically significant bias (p-value < 0.05 using the binomial test) toward/against disorder on different regions observed using all three disorder prediction methods. As can be seen from this table, the first thirteen families have a significant bias towards disorder on interface and multibinding interface regions. These families include kinases, DNA/RNA binding proteins, enzymes, and regulatory proteins. The second ten families comprise mostly enzymes with disorder located on regions other than interfaces, which points to the possible role of this disorder in allosteric regulation, post-translational modifications or substrate selectivity, rather than direct involvement in the binding of other protein partners. A more comprehensive list of families with statistically significant bias towards/against disorder on different regions is illustrated in Fig. S1 and listed in Table S1 of the ESI.†
Table 2.
Families with significant bias of disorder towards footprint, interface, or multibinding interface over all three prediction methods, and the number interactions. The first 13 families exhibit bias towards interface or multi-binding interface over the full domain footprint, while the remaining 10 families exhibit bias on footprint over interface
| Family name | Structure rep | Domaina | Intsb | Biasc |
|---|---|---|---|---|
| Phosphoglucose isomerase | 1HOXA | PRK00179 | 1 | I |
| P-loop NTPase | 2HYIC | cd00079 | 7 | I,M |
| PLAT (polycystin-1, lipoxygenase, α-toxin)/LH2 (lipoxygenase homology 2) | 1W52X | cd01759 | 2 | I |
| Somatotropin hormone | 1KF9D | pfam00103 | 2 | I,M |
| GHMP kinases N-terminal | 1WUUA | pfam00288 | 1 | I |
| Peptidase family M41 | 2DHRB | pfam01434 | 1 | I |
| DNA polymerase III β subunit, C-terminal | 1MMIB | pfam02768 | 2 | I |
| L-Rhamnose isomerase (RhaA) | 1BXBB | PRK12677 | 3 | I |
| Ntn hydrolase | 1JD22 | cd03754 | 13 | M |
| Sm and Sm-like proteins | 1JRIA | PRK00737 | 3 | M |
| Copper amine oxidase | 1W2ZC | pfam01179 | 9 | M |
| Tetracyclin repressor, C-terminal all-α | 2NS7C | pfam02909 | 3 | M |
| TIM phosphate binding | 1O95A | cd02929 | 33 | M |
| Carbonic anhydrase | 1T75A | PRK10437 | 3 | F |
| Chorismate binding enzyme | 1K0EA | pfam00425 | 4 | F |
| Eukaryotic aspartyl protease | 1M4HA | pfam00026 | 2 | F |
| Myo-inositol-1-phosphate synthase | 1U1IC | pfam01658 | 2 | F |
| Ribulose bisphosphate carboxylase large chain, N-terminal | 1GK8A | pfam02788 | 5 | F |
| 3-Oxoacyl-[acyl-carrier-protein (ACP)] synthase III C-terminal | 2D3MA | pfam02797 | 4 | F |
| ATP synthase α/β family, β-barrel | 1OHHA | pfam02874 | 3 | F |
| Myotubularin-related | 1ZVRA | pfam06602 | 1 | F |
| IRSp53/MIM homology | 1Y2OA | pfam08397 | 1 | F |
| Protein tyrosine phosphatase | 1XXVA | cd00047 | 6 | F |
CDD accession.
Number of interactions.
Bias type: interface (I), multi-binding interface (M), footprint (F).
We also subdivided all multibinding domain families with more than two different interacting partners into two categories: 130 domains with multibinding interface greater than 50% of full interface to represent families that reuse the same interface for different partners, and 156 domains with multibinding interface less than 10% of full interface to represent families with little or no overlap in their interfaces with different binding partners. We call these groups “mb50” and “mb10” respectively. These definitions were chosen to provide a sizable data set, comprising 9.5% and 11.4% of all families, respectively, with a buffer to reduce false positives. It should be mentioned that our definition of multibinding interface (with more than 50% overlap) is different from the singlish interface used in the previous studies.39,42 The latter was defined using mutually exclusive interfaces with overlap between interfaces of different partners of at least one residue. We explicitly make sure that the same interface region can bind different domain partners and therefore use a conservative threshold of 50% overlap.
Although there is a certain tendency for multibinding interfaces to contain less disorder, we believe this is the result of the dependence of fraction disorder on the number of interactions as was shown in Fig. 1 (mb50 group has many more interactions compared to the mb10 group with average 2.2 interactions for families in the mb10 group compared to 9.3 interactions for the mb50 group). Overall we conclude that there is no significant difference between these two groups of domains and the whole dataset if normalized by the number of interactions (Table 3). Further, we found that families tend to interact with other families of similar fraction of multibinding interface, and this holds true if we exclude homodimer domain–domain interactions. On average, the partners of families in the mb50 group have multibinding interface that is 47% of the full interface, compared to 15% for families in the mb10 group. This result is consistent with the previous study obtained on the full protein yeast interaction network which hypothesized that interaction between singlish interfaces is caused by the cascading property of these interactions and their involvement in signaling pathways.42
Table 3.
Average percent disorder for domains, grouped by number of interactions, on footprint and interface regions
| (A) Multibinding interface <10% | |||||||
|---|---|---|---|---|---|---|---|
| DDIa | Number of families | Disopred2 |
FoldUnfold |
VSL2 |
|||
| Fb | Ic | F | I | F | I | ||
| All | 156 | 5.1 | 3.7 | 9.1 | 6.6 | 17.7 | 14.8 |
| 2 | 125 | 5.3 | 4.1 | 8.5 | 6.1 | 18.1 | 15.3 |
| 3 | 26 | 4.8 | 2.3 | 8.7 | 5.6 | 15.9 | 11.9 |
| >4 | 5 | 3.5 | 1.9 | 20.0 | 16.4 | 16.7 | 15.6 |
| (B) Multibinding interface >50% | |||||||
| DDI | Number of families | Disopred2 |
FoldUnfold |
VSL2 |
|||
| F | I | F | I | F | I | ||
| All | 130 | 3.8 | 2.9 | 9.6 | 8.1 | 13.5 | 11.7 |
| 2 | 11 | 5.9 | 1.7 | 11.7 | 9.8 | 14.3 | 10.2 |
| 3 | 6 | 5.3 | 4.2 | 15.0 | 11.8 | 13.0 | 9.5 |
| 4 | 15 | 5.6 | 3.8 | 12.3 | 11.3 | 12.6 | 10.8 |
| 5 | 13 | 4.6 | 3.8 | 7.6 | 4.0 | 14.5 | 12.1 |
| 6 | 18 | 4.3 | 4.7 | 7.9 | 5.9 | 14.5 | 13.4 |
| 7 | 13 | 2.8 | 3.1 | 8.5 | 9.5 | 15.7 | 16.7 |
| 8 | 8 | 0.9 | 0.5 | 6.4 | 6.3 | 6.5 | 5.5 |
| >8 | 46 | 3.0 | 2.1 | 9.7 | 8.1 | 13.6 | 11.5 |
Number of domain–domain interactions.
Footprint.
Interface.
Analysis of disorder on chain terminal regions and domain linkers
To understand the effect of disorder outside domain footprints, in inter-domain linker regions and terminal protein regions, we considered the interactions between full protein chains, that is, chains containing domains from our previous domain dataset. Disorder in regards to binding at interdomain and terminal regions has not been explicitly addressed in the previous studies. Conserved Domain Database (CDD) domain footprints were used to partition each chain into domain, inter-domain (linker), and N- and C-terminal regions. Redundant sequences were clustered as described in the previous section, and all calculated values of region sizes and disorder counts were averaged over each group of non-redundant sequences. Altogether we gathered interactions for 35 812 chains from 4615 non-redundant clusters.
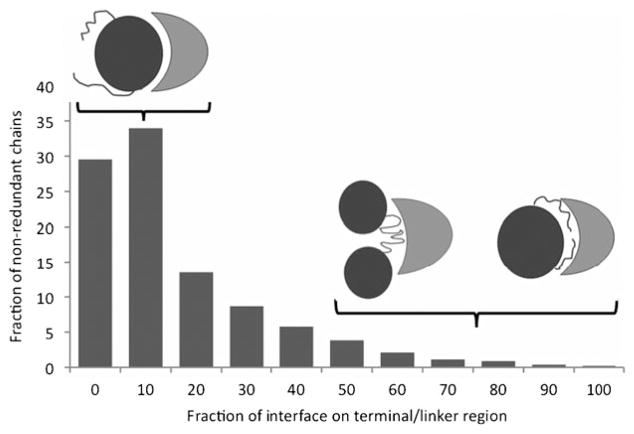
Table 4 and Fig. 2 and 3 show how often terminal and domain linker regions contribute to the formation of the interfaces and whether these interfaces are disordered. Values presented in this table were averaged over all non-redundant chains. First, one can see that N- and C-terminal regions are often located on interfaces. The interface occupies 19–23% of these regions (and conversely, these regions on average occupy ~12% of the interface). Terminal regions participate in protein interactions more often than inter-domain linkers. Statistical analysis shows that in 10% of chain non-redundant groups, terminal and inter-domain linkers have a higher propensity to form interfaces than domain footprints (p-value < 0.05 from the binomial test) while 30% of chain non-redundant groups have interfaces preferentially located on domain footprints. For the remaining cases (60%), there is no significant tendency for the interface to be located exclusively on different regions and terminal, inter-domain linkers, and domain footprints may partially form an interface.
Table 4.
The number of chain non-redundant clusters, region sizes, and interface fraction (fraction of a given region that is on interface) for full-chain, protein–protein interactions
| Region | Chains containing region | Fraction of chain | Interface fraction |
|---|---|---|---|
| Whole chain | 4615 | 100 | 27.9 |
| N-terminal | 4068 | 8.0 | 18.7 |
| C-terminal | 3935 | 7.8 | 22.9 |
| Inter-domain | 1482 | 7.5 | 15.8 |
| Domain | 4615 | 83.9 | 29.2 |
Fig. 2.
Histogram of the fraction of interface on terminal and linker regions for all non-redundant chain groups. Each bin contains all chains with fraction interface up to the bin label, in increments of 10%.
Fig. 3.
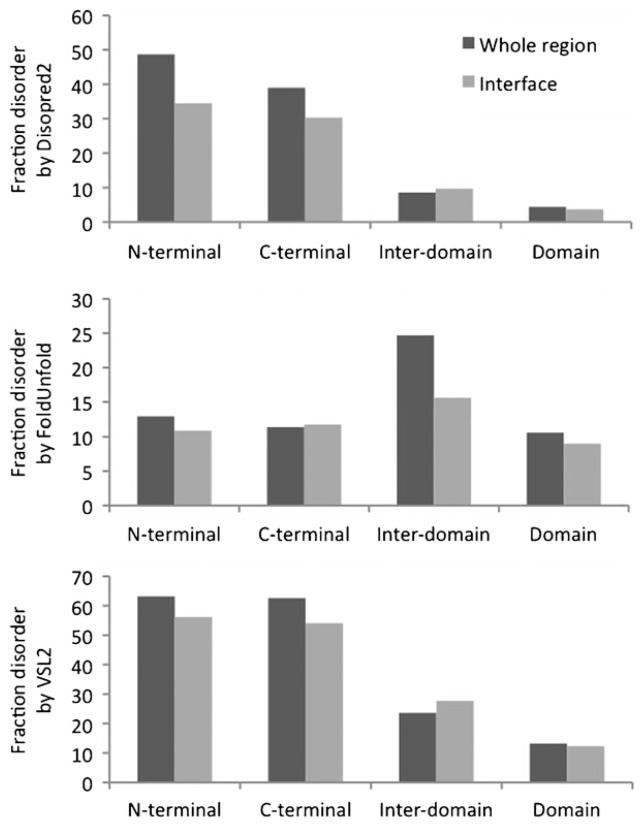
Fraction disorder on terminal, inter-domain, and domain regions for three disorder prediction methods.
Further, we observe that the fraction of predicted disorder on interfaces is significantly higher for terminal and inter-domain regions compared to the domain footprints. Indeed, it is well known that terminal regions are more flexible and more disordered than core domain regions. Similarly to domain regions reported earlier, we also show that interfaces formed by terminal regions are predicted to be less disordered compared to the terminal regions which do not form the interface (t-test and exact Fisher test p-values ≪ 0.0001; this holds true for Disopred2 and VSL2 methods but not for FoldUnfold). The interface within inter-domain linkers is predicted to be as disordered as non-interface regions.
Discussion
The relationship between disorder and binding is not very well understood. Disorder-to-order transition might be important for uncoupling binding affinity from specificity, to provide kinetic advantages through fly-casting mechanisms,18 and might contribute to interactions with multiple partners. In the present study we used atomic details of structure interaction networks based on protein complexes to analyze this coupling at the level of protein domains, terminal regions, and inter-domain linkers. It should be noted that the PDB is biased toward stable obligatory complexes and our findings may not capture all properties of more transient protein interactions.
First, we found that binding interfaces on domain, N- and C-terminal regions are predicted to be less disordered than non-interface regions (this observation does not hold for inter-domain linkers). Moreover, we found that average fraction disorder of a domain family diminishes with the number of interactions of its members. Indeed, protein domains or regions which interact with many other partners in general should have a pleiotropic effect on functional pathways and as a consequence should be constrained in evolution according to the classic Fisher’s hypothesis.52 Indeed, it has been shown previously that proteins and protein regions involved in interactions are more evolutionary conserved (see ref. 53 and references within). Our observation is also congruent with the recent studies by Kim et al. which showed that binding interfaces are less disordered compared to the rest of the proteins from the yeast structural interaction network.42 It should be mentioned that the significant amount of disorder on the full chain proteins used in the later study (including highly disordered terminal and inter-domain linker regions) could account for the difference in predicted disorder between the interface and the rest of the protein. Our analysis of binding interfaces on domain footprints stratifies this result even further.
Disorder-to-order transition on binding interfaces may be expected if the interface is predicted to be disordered and therefore might be disordered in the unbound state (the interface is ordered by definition in complexes since it is defined from residue contacts). Thus our results might imply that in many of PDB proteins, disorder-to-order transition upon binding is not directly seen on interfaces. In our previous study focusing on experimentally determined disorder in proteins in bound and unbound states, we found that disorder-to-order transition occurred directly on binding interfaces in only 40% of cases.16 However, we should mention that in the present work we analyze partially disordered proteins, not completely disordered proteins, and such disorder-to-order transitions which are seen, for example, in MoRFs24 might not be seen in our study or in previous studies based on structural interaction networks.
There is no question that disorder plays an important role in binding of proteins with certain functions and we do observe significant bias of disorder on interfaces and putative disorder-to-order transition for kinases, DNA/RNA binding, certain enzymes and regulatory proteins. At the same time, disorder can contribute to binding indirectly through allosteric regulation and post-translational modifications. Moreover, it has been shown that disorder flanking structured binding motifs suppresses their toxic aggregation and allows certain flexibility necessary for reversible binding with high selectivity.54 According to another hypothesis, the ordered hubs might interact with disordered partners in a cascade fashion.33,42
We did not find any significant difference in disorder on domain interfaces that bind single partners (putative obligatory interactions) and domain interfaces that bind multiple partners (putative transient interactions). This is consistent with previous studies that did not report any significant difference between these two types of hubs in terms of disorder.36,41 On the other hand, singlish or date hubs were found to be more disordered by other studies.40,42 This controversy between several recent analyses might be attributed to many factors including different definitions of multi-binding interfaces and hubs, diverse experimental datasets, and inclusion of different homologs of binding proteins in the analysis. Importantly, singlish or date hubs correspond to the full protein chains which in general are more disordered than domain footprints. There are other scenarios which might explain the mechanism of promiscuous binding. According to the “conformational selection hypothesis”, for example, proteins exist in an ensemble of conformations in dynamic equilibrium and certain conformations become energetically more favorable upon binding different partners.55–57 According to another, “dehydron hypothesis”, interaction complexity or promiscuous binding might be explained by the presence of deficiently packed backbone hydrogen bonds or dehydrons.58
Finally, we found that N- and C-termini as well as inter-domain linkers considerably contribute to the interactions by exclusively or partially forming the binding interfaces. The common practice of inferring protein–protein interactions from domain–domain interactions excludes interfaces formed by termini and linker regions. In addition, we showed that despite their high disorder content, the terminal interface regions are predicted to be less disordered than the rest of the terminal regions. Interestingly, this is not the case for inter-domain linkers which might point to their higher propensity to undergo disorder-to-order transition upon binding.
Conclusions
Our results show that analyses of disorder and protein binding should take into account all regions of the protein, as binding interfaces or disordered regions may be present on domain and extra-domain regions. While different disorder prediction methods suggest varying extent and placement of disordered residues, they agree that in general binding interfaces are more ordered and that the overall amount of disorder on a protein family diminishes with the number of interactions of its members, as may be expected as interacting proteins and protein regions are constrained in evolution. Perhaps surprisingly, reuse of a binding interface for multiple interactions across a family is not a significant indicator of disorder. A sizable but minority fraction of families have large variation in disorder content suggesting non-conserved disorder or specific interactions that utilize specific disordered regions. The diverse role of disorder in binding is further illustrated by the kinases, DNA/RNA binding, certain enzymes and regulatory proteins that exhibit putative disorder-to-order transition, in contrast to some families of enzymes with disorder outside binding interfaces pointing to the possible role of disorder in allosteric regulation and post-translational modifications.
Experimental
Assembling the dataset of physical domain interactions
In the first part of our study, we tried to decipher the role of disorder on interfaces and multibinding interfaces. In order to do this, we collected a dataset of physical protein interactions and mapped them onto a common reference frame. Physical domain–domain interactions were collected from X-ray structures in PDB with at least 3 Å resolution. Domains were assigned to protein chains from PDB using the CDD and the RPS-BLAST algorithm59 with default parameters (E-value ≤ 0.01). Among overlapping domain assignments, the domain having the longest footprint was chosen. A footprint region extends from the first to the last residue in the alignment of a CDD domain to a given sequence. Each domain family can interact with multiple domains and each domain pair can interact through multiple modes (distinct spatial orientations). To handle redundancy of similarly defined protein domains, we record interactions between superfamilies, which represent clusters of CDD families based on overlap in sequence space.60
Interacting domain pairs within each complex were identified as having 5 contacts between residues in one domain and residues in the other. A contact takes place when a non-hydrogen atom in one residue is within 6 Å of a non-hydrogen atom in the other residue. The binding interface for each domain includes all residues that make inter-domain contacts. To ensure that interactions are biological and not spurious, such as from crystal packing, we removed interactions that were not confirmed with additional instances of the same family pair interacting in the same orientation, so-called Conserved Binding Modes (CBMs).61 These CBMs are defined using structural alignments between different structural instances of the same pair of interacting domain families to confirm overlap of at least 50% of interface residue positions. All unique “interactions” described in this paper refer to interacting domains with a distinct CBM. Additionally, inter-chain interactions were confirmed to be biological using the PISA algorithm62 which is based on calculation of stability of multimeric states inferred from the crystalline state.
To characterize disorder on multibinding regions for each domain family, interfaces from each family were mapped on a common reference frame following the procedure described previously.5 A template or representative structure was chosen for each domain family. Other members of the family, their interfaces, and predicted disordered regions were mapped to the template using VAST63 structural alignments. The resulting dataset contains 60 296 interactions of 57 055 domains from 1364 domain families. Those interface positions of a given domain family that participated in interactions with at least two different domain families or binding modes comprise the so-called multibinding interface.
Identifying disordered regions
Disordered regions were predicted for full chain sequences using the Disopred2,44 FoldUnfold,45 and VSL246,47 algorithms. VSL2, the top-performing method for disorder prediction at CASP7,64 combines specialized predictors to balance accuracy on long and short disordered regions using features from sequence profiles and secondary structures. Disopred2, another of the best-scoring methods at CASP7, employs a support vector machine classifier to identify disordered regions from sequence profiles. FoldUnfold is a very rapid method that assigns disorder directly from sequence based on low packing density, using pre-determined average packing density values for each amino acid. The default prediction thresholds were used for all of the above-mentioned programs. Because VSL2 only accepts standard amino acids as input, we deleted masked residues (X’s) from sequences for prediction with VSL2. Short stretches of masked residues (1–2 residues) located within a disordered region were assigned as disordered, and the remaining were considered to be ordered. Mapping the disordered regions on the common reference frame (template representative structure) allowed us to analyze the tendency of disordered regions to be located on multibinding interfaces for each individual domain family. Residues on the template representative structures were labeled as disordered if disordered residues from at least two non-redundant sequences were mapped to the template position. Redundant sequences were defined as having more than 90% sequence identity and less than 90% difference in sequence lengths and were clustered using the CD-HIT program.65
Assembling the dataset of physical chain–chain interactions
In the second part of our study, we tried to understand the effect of disorder outside domain footprints, in inter-domain linker regions and terminal protein regions. Therefore we considered interactions between full protein chains. For all protein chains from the previous section, that is, the chains containing a domain mapped to its family representative, their biological interactions with other chains in the respective complexes were identified, and interfaces, disordered regions, and CDD domains were mapped following the procedures described previously. Domain footprints (for all domains on those proteins, not all of which are included in the domain interaction dataset) were used to partition each chain into domain, inter-domain (linker), and N- and C-terminal regions. Redundant sequences were clustered as described in the previous section, and region sizes and disorder counts were averaged over each group of non-redundant sequences. We gathered interactions for 35 812 chains in 4615 non-redundant chain clusters.
Supplementary Material
Acknowledgments
We thank Vladimir Uversky for insightful discussions. This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
Electronic supplementary information (ESI) available: Supplementary figures and tables. See DOI: 10.1039/c005144f
Contributor Information
Jessica H. Fong, Email: fongj@ncbi.nlm.nih.gov.
Anna R. Panchenko, Email: panch@ncbi.nlm.nih.gov.
References
- 1.Cortese MS, Uversky VN, Dunker AK. Prog Biophys Mol Biol. 2008;98:85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turoverov KK, Kuznetsova IM, Uversky VN. Prog Biophys Mol Biol. 2010 doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobeli I, Favia AD, Thornton JM. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- 5.Tyagi M, Shoemaker BA, Bryant SH, Panchenko AR. Protein Sci. 2009;18:1674–1683. doi: 10.1002/pro.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauling L. J Am Chem Soc. 1940;62:2643–2657. [Google Scholar]
- 7.Meador WE, Means AR, Quiocho FA. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 8.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uversky VN. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 10.Wright PE, Dyson HJ. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 11.Fink AL. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 13.Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Trends Biochem Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 14.Tompa P. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 15.Hegyi H, Schad E, Tompa P. BMC Struct Biol. 2007;7:65. doi: 10.1186/1472-6807-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong J, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. PLoS Comput Biol. 2009;5(3):e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mészáros B, Tompa P, Simon I, Dosztanyi Z. J Mol Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunasekaran K, Tsai CJ, Nussinov R. J Mol Biol. 2004;341:1327–1341. doi: 10.1016/j.jmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Levy Y, Wolynes PG, Onuchic JN. Proc Natl Acad Sci U S A. 2004;101:511–516. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordelon T, Montegudo SK, Pakhomova S, Oldham ML, Newcomer ME. J Biol Chem. 2004;279:43085–43091. doi: 10.1074/jbc.M406139200. [DOI] [PubMed] [Google Scholar]
- 23.Sugase K, Dyson HJ, Wright PE. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 24.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 25.Lobanov MY, Shoemaker BA, Garbuzynskiy SO, Fong JH, Panchenko AR, Galzitskaya OV. Nucleic Acids Res. 2010;38:D283–D287. doi: 10.1093/nar/gkp963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 27.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. BMC Genomics. 2008;9(suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhalla J, Storchan GB, MacCarthy CM, Uversky VN, Tcherkasskaya O. Mol Cell Proteomics. 2006;5:1212–1223. doi: 10.1074/mcp.M500315-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Mészáros B, Simon I, Dosztanyi Z. PLoS Comput Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dosztanyi Z, Meszaros B, Simon I. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Tan H, Rost B. J Mol Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 33.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 34.Patil A, Nakamura H. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnell S, Fortunato S, Roy S. Proteomics. 2007;7:961–964. doi: 10.1002/pmic.200600455. [DOI] [PubMed] [Google Scholar]
- 37.Manna B, Bhattacharya T, Kahali B, Ghosh TC. Gene. 2009;434:50–55. doi: 10.1016/j.gene.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 39.Kim PM, Lu LJ, Xia Y, Gerstein MB. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 40.Singh GP, Ganapathi M, Dash D. Proteins: Struct, Funct, Genet. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 41.Higurashi M, Ishida T, Kinoshita K. Protein Sci. 2008;17:72–78. doi: 10.1110/ps.073196308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim PM, Sboner A, Xia Y, Gerstein M. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Galzitskaia OV, Garbuzinskii SA, Lobanov M. Mol Biol. 2006;40:341–348. [PubMed] [Google Scholar]
- 46.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Proteins: Struct, Funct, Genet. 2005;61(Suppl 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 47.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. J Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basu MK, Carmel L, Rogozin IB, Koonin EV. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 51.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher RA. The genetical theory of natural selection. The Clarendon Press; Oxford: 1930. [Google Scholar]
- 53.Worth CL, Gong S, Blundell TL. Nat Rev Mol Cell Biol. 2009;10:709–720. doi: 10.1038/nrm2762. [DOI] [PubMed] [Google Scholar]
- 54.Abeln S, Frenkel D. PLoS Comput Biol. 2008;4:e1000241. doi: 10.1371/journal.pcbi.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma B, Kumar S, Tsai CJ, Nussinov R. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 56.Boehr DD, Wright PE. Science. 2008;320:1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- 57.Humphris EL, Kortemme T. PLoS Comput Biol. 2007;3:e164. doi: 10.1371/journal.pcbi.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez A, Berry RS. Proc Natl Acad Sci U S A. 2004;101:13460–13465. doi: 10.1073/pnas.0405585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchler-Bauer A, Bryant SH. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geer LY, Domrachev M, Lipman DJ, Bryant SH. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shoemaker BA, Panchenko AR, Bryant SH. Protein Sci. 2006;15:352–361. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krissinel E, Henrick K. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Gibrat JF, Madej T, Bryant SH. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 64.Bordoli L, Kiefer F, Schwede T. Proteins: Struct, Funct, Genet. 2007;69(Suppl 8):129–136. doi: 10.1002/prot.21671. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Godzik A. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.