Abstract
In plants, the two-component systems (TCSs) play important roles in regulating diverse biological processes, including responses to environmental stress stimuli. Within the soybean genome, the TCSs consist of at least 21 histidine kinases, 13 authentic and pseudo-phosphotransfers and 18 type-A, 15 type-B, 3 type-C and 11 pseudo-response regulator proteins. Structural and phylogenetic analyses of soybean TCS members with their Arabidopsis and rice counterparts revealed similar architecture of their TCSs. We identified a large number of closely homologous soybean TCS genes, which likely resulted from genome duplication. Additionally, we analysed tissue-specific expression profiles of those TCS genes, whose data are available from public resources. To predict the putative regulatory functions of soybean TCS members, with special emphasis on stress-responsive functions, we performed comparative analyses from all the TCS members of soybean, Arabidopsis and rice and coupled these data with annotations of known abiotic stress-responsive cis-elements in the promoter region of each soybean TCS gene. Our study provides insights into the architecture and a solid foundation for further functional characterization of soybean TCS elements. In addition, we provide a new resource for studying the conservation and divergence among the TCSs within plant species and/or between plants and other organisms.
Keywords: soybean, two-component systems, abiotic stress
1. Introduction
Two-component systems (TCSs) are key molecular regulators that control many of the biological processes such as cell division, cell growth and proliferation, and responses to environmental stimuli and growth regulators in both eukaryotic and prokaryotic cells.1–6 The simplest form of TCS is composed of a sensory histidine kinases (His-kinases), which senses signal input, and a response regulator (RR), which mediates the output of a response. Phosphorylation of the RR modulates its ability to mediate downstream signalling.7 TCSs can also form a more complex His-to-Asp phosphorelay. In bacteria, yeast, slime moulds and plants, the so-called multiple His-to-Asp phosphorelay makes use of a ‘hybrid’ kinase that contains both a His-kinase (HK) domain and a receiver domain (Rec) in one protein. The TCSs also include a His-containing phosphotransfer (HPt) domain, which functions as a signalling module that connects to the final RRs.5 The HPts are present in a wide range of organisms from bacteria to eukaryotes.6,8 In multistep phosphorelays that involve HPts, the phosphate is transferred from HK to RR via a multistep His-to-Asp phosphorelay. It is suggested that this mechanism is advantageous because it provides multiple regulatory checkpoints for signal crosstalk or negative regulation by specific phosphatases.7,9 In some cases, the HKs may have dual functions such as possessing both HK and phosphatase activities. For instance, in Arabidopsis, the bifunctional AHK4/CRE1, a cytokinin (CK)-activated kinase exhibits a dual function depending on the presence or the absence of CK. In the presence of CK, AHK4 phosphorylates the HPt. Conversely, it removes phosphate from HPt in the absence of CK.10 The existence of multistep phosphorelay reactions in both prokaryotes and eukaryotes suggests that similar mechanisms should be more widely used in nature. However, it is interesting that the canonical His-to-Asp phosphorelay is not found in animals. Computational analyses have confirmed that two-component signalling elements are absent from the genome sequences of Homo sapiens, Drosophila melanogaster and Caenorhabditis elegans.11
Water deficit and high salinity stress limit crop productivity worldwide. In response to these stresses, plants activate a number of endogenous defense mechanisms that function to increase tolerance to adverse conditions. Phosphorylation, which is mediated by TCSs or His-to-Asp phosphorelays, is a key mechanism for stress signal transduction in cells. TCS components have been systematically identified and analysed in two completely sequenced and well-annotated model plant species: Arabidopsis thaliana and rice (Oryza sativa).2,4,12 Recently, Ishida et al.13 compiled putative TCS-associated components in Lotus japonicus, which has 67% of the genome, covering 91.3% of the gene space, sequenced.14 Increasing evidence indicates that the Arabidopsis and rice TCS pathways are involved in response to environmental stimuli. For instance, in Arabidopsis, among the identified Arabidopsis HKs (AHKs), the non-ethylene AHKs (AHK1–5) have been shown to be involved in regulation of stress and abscisic acid (ABA) signalling.15 In planta studies have demonstrated that AHK1 functions as a positive regulator, whereas AHK2, AHK3 and AHK4 as negative regulators in ABA and osmotic stress signalling in both ABA-dependent and ABA-independent pathways.15,16 AHK5/CKI2, which may also function in stress response, is the only cytoplasmic HK, which lacks transmembrane (TM) domains. Recent studies have suggested that AHK5 functions to counteract ethylene and ABA-regulated growth as well as in mediating H2O2-dependent processes in stomatal guard cells.17,18 AHK2, AHK3 and AHK4 function as CK receptors.19,20 However, at the present time, it is not known whether ABA or CK serve as the functional ligand in stress signalling. The ligands for AHK1 and AHK5 also remain to be identified. The involvement of AHK1–5 HKs in stress responses suggests that the downstream Arabidopsis HPts (AHPs) and RRs may function in relation to stress responses as well. Miyata et al.21 reported that the expression of AHP1, AHP2 and AHP3 does not change in response to cold, drought and salt stresses, but are reduced by heat stress. Currently, there are no in planta evidence that characterize the regulatory roles of AHPs in stress signalling. Among the Arabidopsis RR (ARR) genes, ARR4 and ARR5 expression is induced by low temperature, dehydration and high salinity, demonstrating their potential regulatory functions in environmental stress signalling.22 Recently, loss-of-function studies of type-A arr mutants described a complex function for the type-A ARR genes in osmotic stress regulation where ARR3 and ARR4 genes play negative roles, whereas ARR8 and ARR9 function in positive roles.16 Whether or not the type-B ARR genes are involved in stress regulation remains to be determined. As for the Arabidopsis pseudo-RRs (APRRs), recent data indicated that at least APRR5, APRR7 and APRR9 are implicated in abiotic stress responses as negative regulators. This was concluded since a prr9-11 prr7-10 prr5-10 triple mutant displayed strong tolerance against drought, salt and cold stresses.23 After the rice TCS genes have been identified,4 very little is known regarding their functional involvement in environmental stresses. However, expression studies have indicated that the transcription of several rice TCS members, including kinase, HPt and RR encoding genes, is altered by salt-stress treatments.24–26 Currently, abiotic stress-related data are not available for TCS components in L. japonicus.13
Soybean (Glycine max) is a nutritionally important crop that provides an abundant source of oil and protein for worldwide human consumption and animal feed.27–29 In addition, soybean is also viewed as an attractive crop for the production of renewable fuels such as biodiesel. The soybean genomic sequence has been recently completed, which provides an invaluable resource for functional genomics studies at a genome-wide level.30 Given the importance of TCS pathways in diverse biological and physiological processes, including our main interest area of abiotic stress responses, we performed genome-wide analyses in soybean to identify all key TCS components in this model-cropping system. We also carried out a comprehensive analysis of tissue-specific expression data generated by high-throughput microarray expression profiling experiments to discuss on the expression profile and possible functions of the soybean TCS genes. Since the Arabidopsis and rice genomes are well annotated and there is a wealth of functional information for Arabidopsis and rice TCS elements, especially abiotic stress-related functions, we performed sequence analysis and phylogenetic relationship studies of TCS elements of soybean, Arabidopsis and rice as another approach to predict the function of soybean TCS members. Since our main interest is to predict TCS genes involved in stress responses, we searched for the existence of all known abiotic stress-responsive cis-elements on the promoter regions of TCS genes to complement functional predictions using comparative analysis. By coupling knowledge gained from the presence of stress-responsive cis-elements together with data obtained from comparative analyses of Arabidopsis and/or rice TCS members for which stress-related functions are already known, we are able to effectively predict key stress-responsive TCS genes. Taken together, in this study, we performed a comprehensive and high-quality census of TCS members encoded within the soybean genome. These results provide a solid foundation for further systematic characterization of soybean TCS elements using traditional molecular approaches and/or genomic techniques at either the single-gene level or family-wide scale.
2. Materials and methods
2.1. Identification of the TCS members in soybean
As an initial step for identifying genes encoding TCS elements from the annotations of the Glyma1 model, a reciprocal similarity search method was performed between the protein sequence data set of known TCS members, including all the HKs, HPts and RRs of Arabidopsis and rice,5,31 and the modelled soybean proteome data set.30 The first search (forward search) was performed using the protein sequence data set of HKs, HPts and RRs of Arabidopsis and rice as the query against the modelled Glyma1 proteome data set with the blastp program of the NCBI BLAST with a pre-defined threshold of E < 1e−5. Hit sequences from soybean proteins were then used as the query in the second search (reverse search) against the proteome data sets of both Arabidopsis (TAIR9) and rice (TIGR/MSU Pseudomolecule ver.6) with the same threshold set for the first search. The soybean-modelled proteins showing hits in reciprocal manners and containing full open reading frames as predicted by the Glyma1 model were used for further annotation steps. If Glyma1 predicted several splicing variants for a given gene, all the alternative splice variants were carefully checked. Splice variants encoding the longest reading frames were selected, taking into account also available FL-cDNA information, as representatives for subsequent sequence alignments and phylogenetic analyses.
2.2. Annotation of the soybean genes putatively encoding TCS members
To confirm the structures of protein domains conserved in each soybean TCS member, InterProScan and the InterPro database (http://www.ebi.ac.uk/interpro/) were applied, and HMM (Hidden Markov Model) profiles corresponding to domains annotated as CCT, CHASE, HPT, HisKA, MYB, PHY, RR (or RRB) and STK were used for HMMER searches.32 To annotate the TM domain, TMHMM (ver. 2.0c) was also used for soybean putative HK proteins.33 Additionally, to search for FL-cDNA clones that are available for the TCS encoding genes, we also screened the sequence sets of ESTs and high-throughput cDNAs of RIKEN soybean FL-cDNA clones (http://rsoy.psc.riken.jp/).
2.3. Phylogenetic analysis
Sequence alignments of related proteins belonging to each class from Arabidopsis, rice and soybean proteins were performed using ClustalX with the following parameter set: gap open penalty = 10 and gap extension penalty = 0.2.34 The alignments were then visualized using GeneDoc (http://www.nrbsc.org/gfx/genedoc/) as presented in Supplementary Figs S1, S3 and S5. The sequence alignments were also used to construct the unrooted phylogenetic trees by the neighbour-joining method using MEGA4 software.35 The confidence level of monophyletic groups was estimated using a bootstrap analysis of 1000 replicates. Only the bootstrap values higher than 50% are displayed next to the branch nodes.
2.4. Gene duplications and gene clusters of soybean TCS members
Gene duplications and gene clustering formed by soybean TCS genes were estimated by analysing the amino acid sequences of TCS genes found on soybean chromosomes as described previously.32 Specifically, the presence of genes that can form pairs or clusters of closely homologous genes based on a global sequence similarity with a threshold of more than 60% amino acid sequence identity using cd-hit program of CD-HIT package was investigated.36 Gene clusters are defined as genetic loci containing three or more closely homologous genes.
2.5. Expression data for soybean TCS encoding genes
Gene expression data for each putative soybean TCS encoding gene were retrieved from the soybean gene expression data housed within the Genevestigator database by correspondences between soybean genes from the Glyma1 model and probe identifiers from Affymetrix GeneChip probes.37 The respective model IDs used in the Glyma1 model for the respective model IDs that were used in the Affymetrix Genechip were identified by Soybase (http://soybase.org/AffyChip/index.php).38 Gene expression data for Arabidopsis TCS encoding genes were also retrieved from Genevestigator database.
2.6. Discovery of cis-regulatory motifs in promoter regions of transcription factor genes
To discover stress-responsive cis-motifs located in the −1000-bp promoter regions of each putative soybean TCS gene, we retrieved the −1000-bp upstream sequence from either the putative transcription start site, if assigned, or the start codon, if the transcription start site was not yet assigned, for each TCS encoding gene from the Glyma1 annotation (Supplementary Tables S1–S3 and Supplementary Dataset 1). Next, we used stress-responsive cis-motifs previously reported as queries to search against the promoter sequences.39 The matched cis-sequences were counted, and presented in Supplementary Tables S1–S3.
3. Results and discussion
3.1. Identification of TCS elements in soybean
Genome analyses initially suggested that there are 16 HKs in Arabidopsis that were classified into the non-ethylene receptor AHK, the ethylene receptor and the phytochrome gene subfamilies.2 Subsequent results from kinase assays indicated that out of five ethylene receptor kinases, three members (EIN4, ERS2 and ETR2), and all five phytochrome kinases possess Ser/Thr kinase activity rather than HK activity.5,40,41 Furthermore, all five phytochrome kinases lack all five conserved signature motifs that are known to be functionally important for HKs. Collectively, these reports indicated that there are only eight HKs with HK activity in Arabidopsis. Similarly, it was initially reported that the rice genome contains 14 HKs, including all the rice kinases which showed high similarity to the non-ethylene receptor AHK, ethylene receptor and phytochrome kinases of Arabidopsis.4,12 Recently, implementation of standardized nomenclature for the rice TCS elements described eight HKs, including six non-ethylene receptor and two ethylene receptor kinases, in the rice genome.31
A bioinformatics pipeline was established to search for TCS-related genes involved in His-to-Asp phosphorelay across the soybean genome. To search for the putative HKs in soybean, all the eight HKs from both Arabidopsis and rice were used in blastp analyses against the deduced soybean proteome. A similar approach was implemented for the identification of soybean HPts and RRs. After cross-searching the soybean genome, we identified unique hits. We refined these results by performing subsequent reciprocal blastp analyses against the Arabidopsis proteome. We also performed InterProScan and HMMER searches using appropriate HMM profiles featuring the TCS proteins and a manual inspection to confirm the identified set of TCS-related genes in soybean. Our results indicate that the soybean genome contains 21 HK, 10 authentic HPt, 3 pseudo-HPt, 18 type-A RR, 15 type-B RR, 3 type-C RR and 13 pseudo-PRR encoding genes (Tables 1–3). Table 4 indicates the current numbers of TCS genes identified to date in four plant species: Arabidopsis, rice, L. japonicus and soybean. A summary including gene ID defined by the Glyma1 model for each of the predicted genes, corresponding available full-length cDNA (FL-cDNA) accession numbers (RIKEN) and the chromosomal location of all the TCS elements identified in soybean are presented in Supplementary Tables S1–S3. In addition to these aforementioned data, we also listed hyperlinks to available expression data. Additionally, the cDNAs, protein sequences and promoter regions (−500, −1000 and −1500 bp) annotated by the Glyma1 model are provided as additional data that can be easily downloaded (Supplementary Dataset 1). For those genes that have alternate splice variants as putatively predicted by Glyma1, we only selected the splice variants which encode full and longest reading frames as representatives for using in sequence alignments and phylogenetic analyses.
Table 1.
Predicted HK proteins in soybean
| Gene name | Chromosome locusa | Featuresb | Familyc | Length (amino acids) | Identity (%)d |
|---|---|---|---|---|---|
| GmHK01 | Glyma03g37760 | HK, Rec | CKI1 like | 955 | 34.6 |
| GmHK02 | Glyma04g06190 | HK, Rec | CKI2/AHK5 like | 903 | 59.7 |
| GmHK03 | Glyma14g12330 | HK, Rec | CKI2/AHK5 like | 936 | 55.7 |
| GmHK04 | Glyma17g33670 | HK, Rec | CKI2/AHK5 like | 998 | 55.6 |
| GmHK05 | Glyma06g06240 | HK, Rec | CKI2/AHK5 like | 788 | 52.6 |
| GmHK06 | Glyma06g06180 | HK, Rec | CKI2/AHK5 like | 730 | 45.6 |
| GmHK07 | Glyma01g36950 | HK, Rec | AHK1 like | 1174 | 63.5 |
| GmHK08 | Glyma11g08310 | HK, Rec | AHK1 like | 1196 | 62.4 |
| GmHK09 | Glyma02g05220 | HK, Rec | AHK1 like | 1226 | 60.5 |
| GmHK10 | Glyma02g47610 | CHASE, HK, Rec | AHK2 like | 1077 | 56.6 |
| GmHK11 | Glyma14g01040 | CHASE, HK, Rec | AHK2 like | 1011 | 56 |
| GmHK12 | Glyma08g11060 | CHASE, HK, Rec | AHK3 like | 1030 | 67.9 |
| GmHK13 | Glyma05g28070 | CHASE, HK, Rec | AHK3 like | 1030 | 67.7 |
| GmHK14 | Glyma07g27540 | CHASE, HK, Rec | AHK4 like | 983 | 65.6 |
| GmHK15 | Glyma02g09550 | CHASE, HK, Rec | AHK4 like | 984 | 65.4 |
| GmHK16 | Glyma05g34310 | CHASE, HK, Rec | AHK4 like | 997 | 63.4 |
| GmHK17 | Glyma08g05370 | CHASE, HK, Rec | AHK4 like | 1010 | 63.2 |
| GmERS1 | Glyma19g40090 | C2H4, HK | ERS1 like | 636 | 72.9 |
| GmERS2 | Glyma03g37470 | C2H4, HK | ERS1 like | 636 | 72.6 |
| GmETR1 | Glyma09g00490 | C2H4, HK, Rec | ETR1 like | 740 | 81.6 |
| GmETR2 | Glyma12g37050 | C2H4, HK, Rec | ETR1 like | 739 | 81.6 |
aChromosome locus predicted in the Glyma1 model.
bFeatures indicate conserved HK domain, Rec domain, CHASE domain for CK binding (CHASE) and ethylene-binding domain (C2H4).
cFamily indicates classification based on the highest amino acid sequence identity with the Arabidopsis HK counterpart (Supplementary Fig. S2).
dIdentity to the closest Arabidopsis orthologue.
Table 2.
HPt proteins in soybean
| Gene name | Chromosome locusa | Featuresb | Familyc | Length (amino acid) | Identity (%)d |
|---|---|---|---|---|---|
| GmHP01 | Glyma15g10660 | HPt | AHP1 like | 155 | 73.7 |
| GmHP02 | Glyma13g28390 | HPt | AHP1 like | 155 | 73 |
| GmHP03 | Glyma07g38310 | HPt | AHP1 like | 155 | 68.3 |
| GmHP04 | Glyma10g29890 | HPt | AHP1 like | 146 | 63.2 |
| GmHP05 | Glyma20g37450 | HPt | AHP1 like | 150 | 62.5 |
| GmHP06 | Glyma19g33420 | HPt | AHP3 like | 131 | 37.5 |
| GmHP07 | Glyma07g01840 | HPt | AHP4 like | 143 | 60.8 |
| GmHP08 | Glyma08g21510 | HPt | AHP4 like | 140 | 61.4 |
| GmHP09 | Glyma02g16980 | HPt | AHP5 like | 152 | 52.8 |
| GmHP10 | Glyma10g02820 | HPt | AHP5 like | 152 | 53.5 |
| GmPHP01 | Glyma07g03390 | Pseudo-HPt | AHP6 like | 156 | 79.4 |
| GmPHP02 | Glyma08g22720 | Pseudo-HPt | AHP6 like | 156 | 78.8 |
| GmPHP03 | Glyma15g01010 | Pseudo-HPt | AHP6 like | 154 | 72.6 |
aChromosome locus predicted in the Glyma1 model.
bFeatures indicate whether the proteins possess a conserved His-containing phosphotransfer domain (HPt) or a pseudo-HPt lacking the His phosphorylation site.
cFamily indicates classification based on the highest amino acid sequence identity with the AHP counterpart (Supplementary Fig. S4).
dIdentity to the closest Arabidopsis orthologue.
Table 3.
Response regulator proteins in soybean
| Gene name | Chromosome locusa | Featuresb | Familyc | Length (amino acids) | Identity (%)d |
|---|---|---|---|---|---|
| Type-A | |||||
| GmRR01 | Glyma02g03140 | Rec | ARR4 like | 240 | 48.1 |
| GmRR02 | Glyma17g10170 | Rec | ARR6 like | 207 | 55.7 |
| GmRR03 | Glyma05g01730 | Rec | ARR6 like | 211 | 55.1 |
| GmRR04 | Glyma08g40330 | Rec | ARR6 like | 223 | 49.1 |
| GmRR05 | Glyma18g17330 | Rec | ARR6 like | 222 | 49.1 |
| GmRR06 | Glyma15g37770 | Rec | ARR8 like | 179 | 48.8 |
| GmRR07 | Glyma04g42680 | Rec | ARR8 like | 235 | 40.9 |
| GmRR08 | Glyma06g12100 | Rec | ARR8 like | 232 | 40.3 |
| GmRR09 | Glyma03g28570 | Rec | ARR9 like | 248 | 60 |
| GmRR10 | Glyma19g31320 | Rec | ARR9 like | 246 | 59.9 |
| GmRR11 | Glyma11g21650 | Rec | ARR9 like | 187 | 50.8 |
| GmRR12 | Glyma04g29250 | Rec | ARR9 like | 172 | 48.7 |
| GmRR13 | Glyma13g03560 | Rec | ARR9 like | 211 | 35.3 |
| GmRR14 | Glyma06g19870 | Rec | ARR15 like | 204 | 55.6 |
| GmRR15 | Glyma04g34820 | Rec | ARR15 like | 204 | 56.6 |
| GmRR16 | Glyma06g14750 | Rec | ARR17 like | 146 | 65.5 |
| GmRR17 | Glyma04g40100 | Rec | ARR17 like | 146 | 64.9 |
| GmRR18 | Glyma13g26770 | Rec | ARR17 like | 179 | 39.6 |
| Type-B | |||||
| GmRR19 | Glyma17g03380 | Rec, Myb | ARR1 like | 677 | 46.1 |
| GmRR20 | Glyma09g04470 | Rec, Myb | ARR1 like | 673 | 45.4 |
| GmRR21 | Glyma07g37220 | Rec, Myb | ARR2 like | 679 | 49.7 |
| GmRR22 | Glyma15g15520 | Rec, Myb | ARR2 like | 672 | 46.4 |
| GmRR23 | Glyma11g37480 | Rec, Myb | ARR11 like | 497 | 42.3 |
| GmRR24 | Glyma05g27670 | Rec, Myb | ARR11 like | 584 | 39.3 |
| GmRR25 | Glyma08g10650 | Rec, Myb | ARR11 like | 543 | 35.4 |
| GmRR26 | Glyma04g06650 | Rec, Myb | ARR12 like | 630 | 36.7 |
| GmRR27 | Glyma17g33230 | Rec, Myb | ARR12 like | 667 | 36.5 |
| GmRR28 | Glyma14g13320 | Rec, Myb | ARR12 like | 642 | 36.2 |
| GmRR29 | Glyma15g24770 | Rec, Myb | ARR12 like | 697 | 35.8 |
| GmRR30 | Glyma09g14650 | Rec, Myb | ARR12 like | 698 | 35 |
| GmRR31 | Glyma06g06730 | Rec, Myb | ARR12 like | 690 | 34.3 |
| GmRR32 | Glyma13g22320 | Rec, Myb | ARR12 like | 619 | 31.2 |
| GmRR33 | Glyma07g26890 | Rec, Myb | ARR14 like | 633 | 28.2 |
| Type-C | |||||
| GmRR34 | Glyma03g32720 | Rec | ARR24 like | 132 | 55 |
| GmRR35 | Glyma19g35470 | Rec | ARR24 like | 116 | 46.4 |
| GmRR36 | Glyma19g35480 | Rec | ARR24 like | 108 | 43.1 |
| Pseudo | |||||
| GmPRR37 | Glyma04g33110 | Pseudo-Rec, CCT | APRR1 like | 575 | 45.8 |
| GmPRR38 | Glyma05g00880 | Pseudo-Rec, CCT | APRR1 like | 455 | 36.3 |
| GmPRR39 | Glyma06g14150 | Pseudo-Rec, CCT | APRR5 like | 731 | 31.8 |
| GmPRR40 | Glyma19g44970 | Pseudo-Rec, CCT | APRR5 like | 735 | 24 |
| GmPRR41 | Glyma13g19870 | Pseudo-Rec | APRR7 like | 549 | 36.4 |
| GmPRR42 | Glyma12g07860 | Pseudo-Rec, CCT | APRR7 like | 549 | 11.5 |
| GmPRR43 | Glyma04g40640 | Pseudo-Rec, CCT | APRR9 like | 691 | 30.6 |
| GmPRR44 | Glyma16g02050 | Pseudo-Rec, CCT | APRR9 like | 709 | 23.8 |
| GmPRR45 | Glyma07g05530 | Pseudo-Rec, CCT | APRR9 like | 722 | 23.6 |
| GmPRR46 | Glyma05g06070 | Pseudo-Rec, Myb | APRR2 like | 524 | 46.1 |
| GmPRR47 | Glyma17g16360 | Pseudo-Rec, Myb | APRR2 like | 553 | 45.5 |
| GmPRR48 | Glyma01g40900 | Pseudo-Rec, Myb | APRR2 like | 532 | 44.4 |
| GmPRR49 | Glyma11g04440 | Pseudo-Rec, Myb | APRR2 like | 389 | 36.3 |
aChromosome locus predicted in the Glyma1 model.
bFeatures indicate receiver domain (Rec), pseudo-receiver domain (Pseudo-Rec) lacking the conserved D, Myb-like domain (Myb), CCT motif found in clock proteins.
cFamily indicates classification based on the highest amino acid sequence identity with the ARR counterpart (Supplementary Fig. S6).
dIdentity to the closest Arabidopsis orthologue.
Table 4.
TCS members identified in A. thaliana, O. sativa, L. japonicus and G. max
| Species | HKs | HPt | Type A RR | Type B RR | Type C RR | Pseudo RR | Total | Reference |
|---|---|---|---|---|---|---|---|---|
| Arabidopsisthaliana | 8 | 6a | 10 | 11 | 2 | 9 | 46 | Schaller et al.5 |
| Oryza sativa | 8 | 5b | 13 | 13 | 2 | 8 | 49 | Schaller et al.31 |
| Lotus japonicus | 14 | 7 | 7 | 11 | 1 | 5c | 40 | Ishida et al.13 |
| Glycine max | 21 | 13 | 18 | 15 | 3 | 13 | 83 | This work |
aFive authentic and one pseudo-HPts.
bTwo authentic and three pseudo-HPts.
cOnly clock-associated.
3.2. The HK protein family in soybean
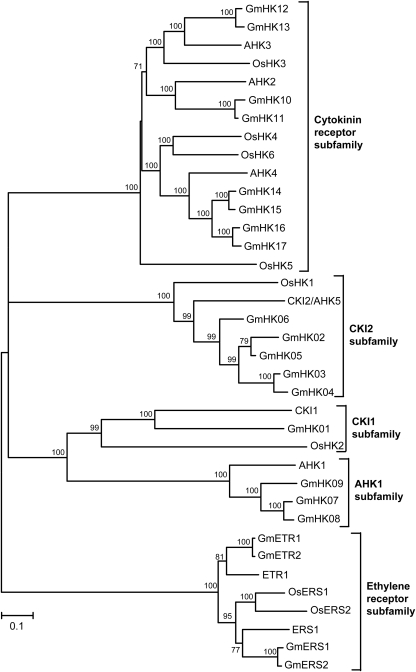
Genome-wide analysis supports the existence of 21 putative hybrid HKs in soybean. This number is considerably larger than the 8-member family in both Arabidopsis and rice and the 14 genes from L. japonicus (Tables 1 and 4). The size of the predicted proteins range from 636 to 1226 amino acids with overall 8.1–98.5% identity and 16.82–99.6% similarity in amino acid sequence (Table 1; Supplementary Figs S1 and S2). Among these soybean HKs, similarity searches with Arabidopsis and rice HKs have identified 17 as non-ethylene receptor (GmHK) and four as ethylene receptor-like soybean HKs. The non-ethylene GmHKs were further classified into eight CK receptor-like GmHKs, three AHK1-like GmHKs, five AHK5-like and one CKI1-like GmHKs (Table 1). Domain analysis of these GmHKs confirmed that all the 17 non-ethylene GmHKs have a typical hybrid HK-type structure with a conserved HK domain which contains the conserved His phosphorylation site. In addition, the non-ethylene GmHKs also contain a complete Rec domain, which contains a highly conserved Asp as the phospho-acceptor, although the number of the TM domains is variable (Table 1; Supplementary Table S1 and Fig. S1). Additionally, all the eight CK receptor-like GmHKs contain the conserved cyclases/HK-associated sensory extracellular (CHASE) domain. In contrast, none of the other non-ethylene GmHKs and ethylene receptor-like soybean HKs contains CHASE domains (Table 1).
In planta studies in Arabidopsis have provided strong evidence that the non-ethylene receptor AHK1 is involved with osmotic stress responses by functioning as a positive regulator of drought and salt-stress responses in ABA signalling.15,16 On the basis of sequence alignments and phylogenetic analyses, we identified three GmHKs which share over 60% high identity with AHK1 (Fig. 1; Table 1). In addition, recent genetic and molecular studies also demonstrated that among the non-ethylene receptor HKs of Arabidopsis, the CK receptors AHK2, AHK3 and AHK4 can act as negative regulators of osmotic stress signalling in both ABA-dependent and ABA-independent pathways.15,42 Our phylogenetic analysis supported the existence of eight CK receptor-like GmHKs in soybean which share 42.3–67.9% amino acid sequence identity with the established sequences for AHK2, AHK3 and AHK4 proteins (Fig. 1, Table 1 and Supplementary Table S2). In this subfamily of soybean CK receptor-like kinases, two proteins show more than 56% to AHK2, two display >67.7% to AHK3, and the remaining four show >63.2% sequence identity to AHK4, respectively (Table 1). In the two annotated AHK2-like GmHKs, we found two putative TM domains instead of the three domains that are reported for AHK2. The number of putative TM domains in several predicted rice CK receptor HKs is also different compared with that of their Arabidopsis counterparts. The OsHK5, which shows the highest amino acid identity to AHK2, has only one putative TM domain (Supplementary Fig. S2).4 Each of the AHK3-like and AHK4-like GmHKs contains three and two TMs, respectively, in their deduced amino acid sequences as their AHK3 and AHK4 orthologues (Supplementary Table S1). In addition to the AHK1–AHK4 proteins, AHK5 has been recently shown to be involved in stress responses.18 We found that five GmHKs show significant homology with AHK5 with overall 45.6–59.7% sequence identity (Table 1). Similar to what has been reported for AHK5, no putative TM domain could be predicted in the five AHK5-like GmHKs (Supplementary Table S1).
Figure 1.
Phylogenetic relationship of HKs from Arabidopsis, rice and soybean. The bar indicates the relative divergence of the sequences examined. Bootstrap values higher than 50% are displayed next to the branch.
According to our knowledge, CKI, which is the remaining non-ethylene receptor HK in Arabidopsis, does not function in stress responses. Loss-of-function studies have indicated that CKI1 function is required for megagametophyte development.43 It has also been reported that CKI1 can regulate His-to-Asp phosphorelay independently of CK. This CK-independent activity of CKI1 and the CK-induced functions of AHK2 and AHK3 are important for vascular bundle formation in Arabidopsis shoots.44 One protein (GmHK01) was detected within Glyma1 which displays significant homology with CKI1 (34.6% sequence identity). The GmHK01 protein and the predicted rice CKI1-like kinase do not contain the same number of TMs as Arabidopsis CKI1.4 The GmHK01 and the rice CKI1-like kinase contain one and two TM domains, respectively, whereas the Arabidopsis CKI1 possesses three TMs (Fig. 1; Table 1 and Supplementary Table S1).
Our genome-wide analysis detected four HKs in soybean that can be classified as ERS1 and ETR1 ethylene receptor HK orthologues based on high sequence identity with their Arabidopsis counterparts (Fig. 1). These soy proteins share 56.6–81.6% sequence identity to Arabidopsis ERS1 and ETR1 proteins (Supplementary Fig. S2). Similar to their Arabidopsis counterparts, all the four putative soybean ethylene receptor HKs possess three TM domains, a GAF (cyclic GMP, adenylyl cyclase, FhlA) domain and a HK domain. However, only the two ETR1-like GmETR1 and GmETR2 (both have 81.6% identity to ETR1) carry the Rec domain, but the two ERS1-like GmERS1 (72.6% identity to ERS1) and GmERS2 (72.9% identity to ERS1) do not (Table 1, Supplementary Fig. S1). Although the Arabidopsis ERS1- and ETR1-type HKs have been shown to display HK activity,41,45 their major role in ethylene signalling remains to be elucidated.46–48 Additionally, ethylene has long been regarded as a stress hormone, and ethylene receptor HKs have been shown to function in stress signalings.49–51 It is known that ethylene is required for plant salt tolerance, and alteration of ethylene signalling affects plant salt-stress responses. ETR1 has been characterized as a negative regulator in salt signalling because a gain-of-function etr1-1 mutant exhibits a salt-sensitive phenotype.49 It is also possible that ERS1-like kinases play a role in stress responses. Expression of gene encoding a wheat ERS1-like kinase, which shares over 70% similarity to Arabidopsis ERS1, is up-regulated in treatments known to induce the senescence of detached leaves including jasmonate, ABA and wounding.52
3.3. The HPt protein family in soybean
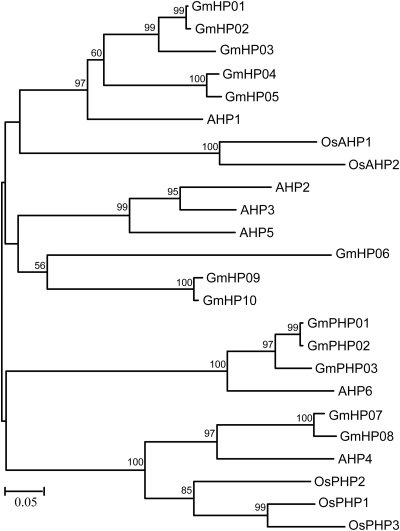
The current Glyma1 model allowed us to identify 10 authentic (GmHP1-10) and 3 pseudo (GmHP11–13) HPt proteins in soybean (Table 2). The numbers of authentic and pseudo-HPt proteins identified in Arabidopsis, rice and L. japonicus are summarized in Table 4. The percentage identity and similarity in amino acid sequences among the GmHPs and GmPHPs are between 27% and 98.1% and 39.6% and 99.4%, respectively (Supplementary Figs S3 and S4). The 10 putative GmHP genes encode GmHP proteins that all contain a typical phosphotransfer intermediate sequence with the conserved His phosphorylation site. Moreover, similar to Arabidopsis and rice counterparts, these 10 GmHPs share the HQXKGSSXS(I/V)G consensus sequence that contains the conserved His residue (underlined). The three pseudo-GmPHP genes encode pseudo-proteins which lack the invariant and phospho-accepting His residue. These occurrences are similar to those observed in the Arabidopsis pseudo-protein (AHP6/APHP1) and the three from rice (OsPHP1-3). The GmPHPs contain an N-residue instead of H at the conserved phosphorylation site (Supplementary Fig. S3).
In Arabidopsis, AHP proteins have been shown to interact with both hybrid HKs and RRs, consistent with an ability to function in a multistep phosphorelay.53–56 Analyses of ahp knockout mutants have suggested their redundant function in CK signalling and plant development.57 The phylogentic tree constructed from all the authentic and pseudo-HPts of Arabidopsis, rice and soybean indicates the presence of subfamilies (Fig. 2). The GmHP01–06 and GmHP09 and 10 have a closed relationship with the established CK positive regulators AHP1, AHP2, AHP3 and AHP5, which are functionally redundant positive regulators in CK signalling.57 The GmHP07 and 08 proteins are close to AHP4, which is evolutionarily distinct from the other AHPs and may play a negative role in CK signalling (Fig. 2).57 Microarray analysis suggested that AHP4 is the only AHP protein that is down-regulated in response to osmotic stresses.58 The three atypical GmPHP11–13 proteins showed 72.6–79.4% identity to Arabidopsis AHP6, which functions as a competitor of other AHPs and plays a negative role in CK responses by interfering with phosphorelay.59 Interestingly, our phylogenetic analysis indicated that unlike the pseudo-GmPHPs, the three pseudo-OsPHPs (OsPHP1–3) of rice show higher sequence homology to the authentic AHP4 than the pseudo-AHP6 (Fig. 2).
Figure 2.
Phylogenetic relationship of phosphotransfer (HPt) proteins from Arabidopsis, rice and soybean. The bar indicates the relative divergence of the sequences examined. Bootstrap values higher than 50% are displayed next to the branch.
3.4. The RR proteins family in soybean
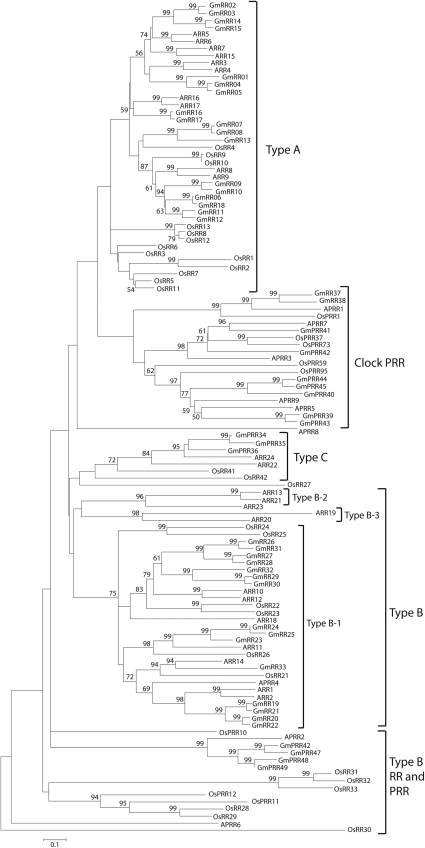
The completion of the soybean genome sequence has allowed us to predict a total of 49 RRs in soybean, including both the authentic and the pseudo-RRs. Previous reports have indicated that there are 32 genes in Arabidopsis, 36 genes in rice and 26 genes in L. japonicus encoding authentic and pseudo-RRs (Tables 3 and 4). In the TCS signalling pathway, the RRs function as terminal components by acting as phosphorylation-activated switches that catalyse the transfer of the phosphoryl group to a conserved Asp in its own regulatory domain. Structural analysis of the soybean RR proteins has enabled us to classify the soybean RRs into type-A GmRR, type-B GmRR, type-C GmRR and pseudo-GmPRR categories similar to those in Arabidopsis and rice based on their structural features (Fig. 3 and Supplementary Fig. S5).
Figure 3.
Phylogenetic relationship of response regulator proteins from Arabidopsis, rice and soybean. The bar indicates the relative divergence of the sequences examined. Bootstrap values higher than 50% are displayed next to the branch.
Analysis of the genomes of several plant species, ranging from unicellular algae, moss and lycophytes to higher plants, including Arabidopsis and rice, revealed that the type-A RRs first appeared in the land plant species.6 Among the 49 soybean RRs, we identified 18 type-A GmRRs (GmRR01–18), each of which contains a receiver domain along with a divergent C-terminal extension. The overall percentage identity of the type-A GmRRs ranges from 27.6% to 97.7%, and similarity from 39.5% to 99.3% (Supplementary Fig. S6). The phylogenetic tree developed from the RRs collected from Arabidopsis, rice and soybean indicates closed relationship among the type-A RRs of the three species, which might suggest similar functions for the soybean type-A GmRR (Table 3, Fig. 3). In Arabidopsis, the type-A ARR3–9 and 15 were reported to function as negative regulators of CK signalling.60–63 Some of the type-A ARRs have been shown to be involved in the regulation of light response (ARR4), circadian period (ARR3 and ARR4) and meristem size (ARR5, ARR6, ARR7 and ARR15).61,64–66 The GmRR01, GmRR06–08 and GmRR09–13 showed significant homology to ARR4, ARR8 and ARR9 proteins, respectively, which have been reported to function in stress signalling (Table 3, Fig. 3).16,22 Additionally, the expression of the six type-A OsRR genes (OsRR1, 3, 4, 6, 9 and 13) in rice has been shown to be stress inducible. When these functional data are taken into consideration with the information provided by our phylogenetic relationship study, we would get a hint about the potential stress-responsive type-A GmRR genes in soybean.24–26 Further studies of the type-A GmRR orthologues either in Arabidopsis or in soybean are warranted to shed light on their in planta function roles.
The type-B GmRR subfamily consists of 15 members compared with 11 members in Arabidopsis and 13 in rice. Similar to the type-B RRs identified in other plant species, the type- B GmRRs are transcription factors (TFs). In addition, each of these type-B RRs contains an N-terminal receiver domain and a long C-terminal extension with an Myb-like DNA binding domain (GARP domain). The amino acid sequences of the type-B GmRRs share from 18.5% to 95.4% sequence identity and 27.37% to 98.8% similarityamong themselves (Supplementary Fig. S6). Interestingly, although the Arabidopsis type-B ARRs could be classified into three subfamilies (type B1, B2 and B3) based on a phylogenetic study,67 an analysis of phylogenetic relationship of soybean and Arabidopsis type-B RRs has revealed that without exception all the predicted type-B GmRR proteins were grouped into type-B1 subfamily (Fig. 3). The absence of the type-B2 and type-B3 GmRRs may be due to the following reasons: (i) the sequenced soybean genome comprised of only 950 Mb, representing ∼85% of the predicted 1115-Mb genome; and/or (ii) approximately one-third of 66 400 putative protein-coding loci annotated in the Glyma1 model were predicted with low confidence.30 Therefore, we may expect that future update of soybean genomic sequence and/or fine-tuning of genome annotation might enable us to identify the type-B2 and type-B3 GmRRs.
With their signature combination of RR and Myb domains, the type-B RRs are plant specific.68 Unlike the type-A RRs, the type-B RRs are present not only in land plant species but also in the unicellular algae, moss and lycophytes, suggesting that they might have originally performed a function in the regulation of photosynthesis and later been recruited for other functions such as CK signal transduction.6,69 The soybean type-B GmRRs may be capable of controlling various biological processes in similar manner. The Arabidopsis ARR1, ARR10 and ARR12 play key roles in CK signalling, and ARR2 may modulate ethylene signalling.69–73 Overexpression of activated type-B ARR20 results in plants with small flowers, abnormal siliques and reduced fertility, whereas that of activated type-B ARR21 created seedlings in which cell proliferation is activated to form callus-like structures.74 The rice OsRR30/Ehd1 is hypothesized to regulate developmental processes and environmental signalling mediated by CK, ethylene and light.75 With the exception of the stress-inducible profile of three type-B OsRR genes (OsRR21, 23 and 24), no information about the function of type-B RRs of any plant species in stress response is currently available.26 Thus, our phylogenetic analysis might serve as a comparative genomics approach for the selection of GmRR orthologues of stress-inducible type-B OsRR genes for further genetic studies in response to stresses (Fig. 3).
Similar to the type-A RRs, both Arabidopsis and rice contain two type-C RRs which only have the receiver domain without the long C-terminal extension. However, the type-C RRs are not closely related to the type-A RRs as indicated by phylogenetic studies. The L. japonicus genome possesses at least one type-C RR (Table 4). In soybean, we identified three type-C GmRRs which grouped into the same group as Arabidopsis and rice type-C RRs (Fig. 3). Expression of the type-C ARRs of Arabidopsis, which is predominantly observed in flowers and siliques, is not regulated by CK, like that of Arabidopsis type-A ARRs. Overexpression of ARR22 showed reduced shoot growth, poor root development, reduced CK-responsive gene induction and insensitivity to CK under conditions for callus production. Collectively, these observations suggest that ARR22 possess an inhibitory function in CK signalling.76–78 The type-C RRs of soybean might have similar functions as their corresponding Arabidopsis orthologues.
Previous works reported that there are nine pseudo-RRs (APRRs) in Arabidopsis and eight such genes (OsPRRs) in rice, with five clock-associated PRR genes in each.5,31 The total number of PRR genes in L. japonicus is unknown, but there may be at least five clock-associated PRR genes in L. japonicus (Table 4). We identified 13 pseudo-RR proteins (GmPRRs) in soybean which share from 5.23% to 87.1% identity and from 11.51% to 93% similarity among themselves (Supplementary Fig. S6). These soy GmPRRs contain receiver-like domains at their N-terminal end but lack the conserved Asp residue required for phosphorylation (Supplementary Fig. S5). With the exception of GmPRR38, 41 and 42 proteins, which completely lack the Asp, the remaining GmPRRs have the Glu in the place of the Asp residue. Similar to the structure of pseudo-RRs in Arabidopsis and rice, the C-terminal domains of GmPRRs contain either a CCT motif (GmPRR37–40 and 42–45) or a Myb-like motif (GmPRR46–49). An exception occurred with the GmPRR41 which lacks both the CCT and the Myb motifs (Table 3 and Supplementary Table S3). Expression of the CCT-motif PRR genes varies in a circadian manner and loss-of-function mutants have altered circadian periods, suggesting that these types of PRRs participate in circadian rhythms.79–82 Our phylogenetic analysis suggest that the GmPRR37–40 and 42–45 proteins, which contain the CCT motif, may function in circadian rhythm as they show close relationship with the known clock-associated pseudo members of Arabidopsis (Fig. 3). No functional evidence has been provided to suggest the role for Myb-motif PRRs in any plant species. Since we are interested to identify stress-related GmPRR genes, we carefully examined the phylogenetic tree to identify those GmPRRs that show the highest homology to the clock-associated APPR5, APRR7 and APRR9 of Arabidopsis, which have been reported previously to implicate in abiotic stress regulation.23 Such GmPRR genes can be listed as GmPRR39–41 and 42–45 proteins (Fig. 3).
3.5. Chromosomal distribution and gene duplications of TCS members
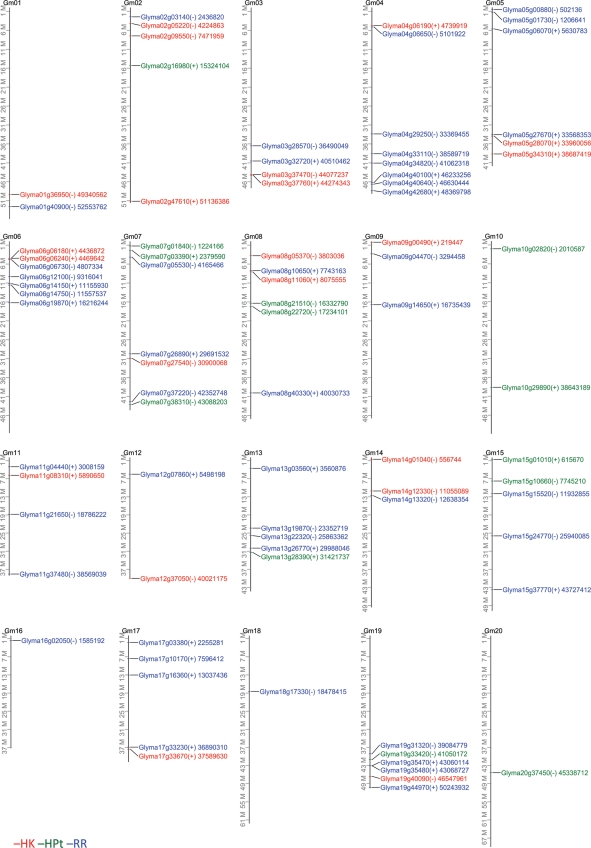
We are also interested to characterize the local distribution and homology of TCS genes in each family relative to each other. Our analysis indicated that the soybean TCS members are distributed on every chromosome in soybean. For instance, the 21 identified soybean HKs are distributed among almost all 20 chromosomes with the exception for chromosomes 10, 13, 15, 16, 18 and 20 (Fig. 4). The GmRR genes are scattered throughout the soybean genome, with the exception of chromosomes 10 and 20. The members of GmHP family, which contains the smallest number of genes relative to kinase and RR families, are located on chromosomes 2, 7, 8, 10, 13, 15, 19 and 20 (Fig. 4). We also observed that a large number of TCS genes encode amino acid sequences that show high identity to each other (Supplementary Figs S2, S4 and S6). Previous studies have described duplications and clusters of highly homologous genes in Arabidopsis and soybean. Closely related or closely homologous genes, which are defined by >60% amino acid sequence identity,36 account for ∼77.75% of the total number in the TF families of soybean.32 In Arabidopsis, gene duplications on either same chromosomes or different chromosomes may account for >60% of the genome.36 Two types of duplications and clusters can be distinguished based on the evolutionary history of the genes that they contain. The first type of duplications and clusters consists of a series of paralogous genes, suggesting that they arose through repeated tandem duplications which originated from a founding locus. In contrast, the second type of duplications and clusters contains genes that arose independently from each other at diverse locations within the genome. Over time, it is likely that they relocated to form these duplications and clusters.32
Figure 4.
Graphical representation of locations for putative TCS genes on soybean chromosomes. The (+) and (−) indicate the sense and antisense strands, respectively. Number displayed next to each gene ID indicates the position of the annotation start for each TCS gene in bases.
By analysing alignments of the TCS proteins, we found that a large number of TCS genes form pairs or clusters of duplicated genes (Table 5). Pairs of duplicated genes on different chromosomes are most common and gene clusters of three or more highly related genes are also widely found. On the basis of the distance of their occurrence, a few of the duplicated genes could be classified arbitrarily as genes that were duplicated on the same chromosome. However, none of the duplicated genes could be categorized as tandemly duplicated. This categorization is based on a classification of tandemly duplicated genes as those which are duplicated in same chromosome but reside <50 kb apart from each other (Table 5).32,36 Within the soybean TCS members, 20 genes, accounted for 95.24%, encoding soybean HKs, may form pairs and clusters of closely homologous genes that have more than 60% amino acid sequence identity (Table 5). For instance, the CKI2-like GmHK02–06 proteins form eight pairs of closely related genes. These highly homologous genes are located on either the same (such as GmHK05 and 06 are located on the same chromosome 6, and share 77.2% identity) or different chromosomes (such as GmHK02 and 03 are located on chromosome 4 and 14, respectively, and share 70.7% identity; Fig. 4 and Supplementary Fig. S2). These distributions suggest that they were the resultants of duplications on either the same or different chromosomes, respectively. Among the soybean HPts, 9 of 10 authentic GmHP proteins form three very highly homologous pairs (GmHP04 and GmHP05 share 94%, GmHP07 and GmHP08 94.4%, and GmHP09 and GmHP10 97.3% identity, respectively) and one highly homologous cluster of three proteins (GmHP01, GmHP02 and GmHP03 have 82.8–98% amino acid identity to each other). The three pseudo GmPHP01–03 make a highly homologous cluster with 89.8–99.3% identity as well. If the 60% amino acid sequence identity criterion is applied for defining the closely related genes, we can define a cluster consisting of five genes of GmHP01–05 which encode authentic HPts sharing overall 66.6–98% identity. The HPt genes encoding highly homologous proteins are perhaps the products of genome duplications on different chromosomes as they are distributed on different chromosomes (Fig. 4). We were also able to discover several pairs and clusters formed by genes encoding RR proteins (Table 5). For example, we identified three pairs (GmRR04 and 05, GmRR07 and 08, and GmRR09 and 10) and two clusters of closely related genes (cluster of GmRR02, 03, 14 and 15, and cluster of GmRR06, 11, 12, 18), encoding proteins that share more than 60% amino acid sequence identity.
Table 5.
Classification of holomologous soybean TCS genes
| Protein family | Number of genes with close homologuea | Percentage of genes with close homologue | Number of homologous genes on different chromosomesb | Number of homologous genes on same chromosomesc | Number of genes involved in tandem duplicationsd | Number of gene clusters/Number of genes in cluster (Number of chromosmes)e |
|---|---|---|---|---|---|---|
| GmHK | 20 | 95.24 | 18 | 2 | 0 | 4/14 (12) |
| GmHP | 12 | 92.31 | 12 | 0 | 0 | 2/8 (6) |
| Type A GmRR | 18 | 100.00 | 18 | 0 | 0 | 4/14 (10) |
| Type B GmRR | 11 | 73.33 | 11 | 0 | 0 | 2/7 (6) |
| Type C GmRR | 3 | 100.00 | 1 | 2 | 0 | 1/3 (2) |
| Pseudo GmRR | 6 | 46.15 | 6 | 0 | 0 | 0/0 (0) |
aGenes were considered closely homologues if they showed >60% amino acid sequence identity as defined in Riechmann et al.36
bNumber of closely homologous genes which are involved in duplications on different chromosomes.
cNumber of closely homologous genes which are involved in duplications on same chromosome but resided >50 kb apart from each other.
dNumber of closely homologous genes which are involved in duplications on same chromosome and resided <50 kb apart from each other.
eClusters of three or more closely homologous genes.
It is not surprising that the multigene families of soybean TCS contain highly related genes. Evolutionary studies have suggested that the paleopolyploid soybean genome experienced two whole-genome duplication events at ∼59 and 13 million years ago, resulting in a highly duplicated genome with nearly 75% of the genes present in multiple copies. Since then, the soybean genome has gone through extensive gene rearrangements and deletions to become diploidized.30,83
3.6. Analysis of expression patterns of TCS genes using Genevestigator database
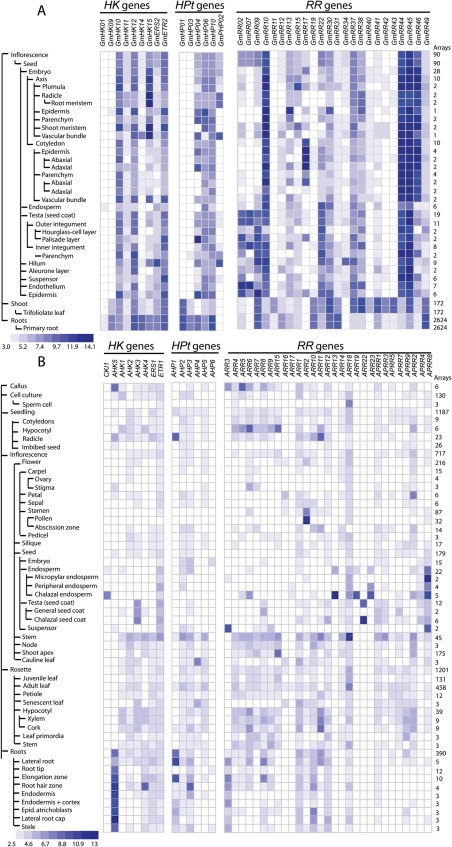
Knowledge about specific expression of TCS genes is interesting because it tells us about the genes which are involved in defining the precise nature of individual tissues. The specific expression of TCS genes was examined using the data set that is publicly available on the Genevestigator database.37 This bioinformatics resource contains measurements of transcript levels for 35 different organs and tissues. These data were obtained by microarray experiments using the Affymetrix Soybean Array GeneChip, which was designed specifically to analyse ∼37 500 soybean, 15 800 Phytophthora sojae as well as 7500 Heterodera glycines transcripts. First, we identified the respective gene ID used in the Glyma1 model for the gene ID which was used in the Affymetrix GeneChip using Soybase (http://soybase.org/AffyChip/index.php). We confirmed that probes exist for a total of 9 GmHK (out of 21), 6 GmHP (out of 13) and 24 GmRR (out of 49) genes on the soybean GeneChip. The information on the expression patterns of these TCS genes, including those in 35 different organs and tissues, can be accessed through hyperlinks which link the genes directly to their respective pages on Genevestigator (Supplementary Tables S1–S3). The heat maps shown in Fig. 5 display the patterns of expression of these TCS genes across 35 major organs and tissues that were examined. The data supplied here are useful to assess the extent of TCS gene expression as they provide the first line of temporal and spatial evidence which links them to putative in planta functions.
Figure 5.
Heat map representation for tissue-specific expression of soybean and Arabidopsis TCS genes. Heat map representation for the tissue-specific expression of 9 HK, 6 HPt and 24 RR encoding soybean TCS genes (A). Heat map representation for the tissue-specific expression of Arabidopsis TCS genes (B). Elevated expression levels are indicated by increasing intensities of blue colour expressed in log2 scale.
Data analysis indicates high variability in transcript abundance of the soybean TCS genes. For instance, GmHK01 show weak expression in root and endosperm and insignificant in other tissues, whereas GmHK10, GmHK12 and GmETR2 are more abundant in all the tissues examined (Fig. 5A). The spatial expression patterns of the soybean TCS genes shown in the heat maps appear to be tissue-specific, suggesting that the functions of the soybean TCS members are diversified. Several TCS genes, such as GmHP01, GmHP03 and GmRR11, are root- and shoot-specific, whereas GmHP02, GmRR17 and GmRR34 show expression only in inflorescence and some of the seed tissues (Fig. 5A). None of the predicted type-B GmRR genes, for which expression data available, displayed predominant expression in endosperm (Fig. 5A), which was found as a unique feature for the type-B2 and type-B3 Arabidopsis ARR genes (Fig. 5B).84 This observation strengthens the result of phylogenetic analysis which grouped all the predicted type-B GmRR proteins into only the type-B1 subfamily (Fig. 3). On the other hand, we cannot rule out that the expression of some of other TCS genes, for which data are currently not available, may be ubiquitous. Ubiquitous TCS genes alone, if they exist, in isolation or in combination with each other, may control the general cellular machinery. It is possible that combinations of specific TCS members might be involved in the regulation of tissue-specific genes. It is noteworthy that the type-B RRs are TFs. It is therefore important to note that TF activity often depends on the post-translational events and that the levels of gene expression are not necessarily directly correlated with their regulatory activity. Moreover, since co-operativity between TFs has been shown to involve extensive protein–protein interactions, both within families of homomeric and heteromeric TFs and between structurally unrelated TFs, analysis of such interactions may help us to elucidate the patterns of combinatorial regulation and ultimately decipher the regulatory functions of the type-B RRs.85–87
Comparison between the tissue-specific expression profiles of soybean and Arabidopsis TCS genes might shed the light into the functions of the soybean TCS genes (Fig. 5A and B). For example, the expression of both Arabidopsis CKI1 and its soybean orthologous GmHK01 was detected in endosperm of seed, suggesting that GmHK01 may play an important role in megagametogenesis as CKI1. GmHK09, which was predicted to encode an AHK1 orthologue, expresses mainly in shoot and root tissues as AHK1, whereas both GmHK14 and its orthologous AHK4 showed highest transcript abundance in root organs (Table 1; Figs 1 and 5).19,43,88 Collectively, the similar expression profiles may suggest similar function for Arabidopsis orthologous soybean TCS members. Taken together, our analyses of expression data and phylogenetic relationship of Arabidopsis and soybean TCS members indicate a correlation between gene expression, sequence conservation and gene function, which is consistent with the results reported previously.32,89,90
One of our main interests in the expression analysis of soybean TCS elements is to predict abiotic stress-responsive TCS genes that will be taken in priority for further in planta functional studies. However, at the present time, the Genevestigator and other public resources do not contain data of high-throughput microarray transcript profiling experiments for tissues subjected to either drought or high salinity or cold or flooding which are the major abiotic stresses affecting soybean productivity. A larger search for other chemical stress inducers found a microarray analysis of soybean leaves, which were treated with polyethylene glycol (PEG), using an array containing 5760 soybean cDNAs (<10% of the soybean predicted genes). PEG is widely known as an osmotic stress inducer. Analysis of the down- and up-regulated data set identified only one down-regulated gene (AW567584, down-regulated 2.14-fold), which is identical to the GmPRR41, suggesting that GmPRR41 may play a role in abiotic stress response.91 Previously, our sequence analysis indicated that GmPRR41 shares significant identity with the Arabidopsis APRR7, which was shown to function as a negative regulator in drought, cold and salt stresses (Fig. 3, Table 3).23 This observation also indicates that expression correlates with sequence conservation. Thus, sequence-based comparative genomics might be used as an approach to predict the function of a protein. It is hoped that with the availability of the complete soybean genomic sequence, genome-wide transcriptome analyses will be accelerated, which would make abiotic stress-related large-scale expression data available in the near future so that the scientific research community can be supported in a more comprehensive manner.
3.7. Cis-element- and comparative sequence analysis-based approach for prediction of abiotic stress-responsive TCS genes in soybean
Plants respond to environmental changes by altering large-scale transcriptional responses. The exquisite sensitivity and specificity of these responses are controlled in large part by cis-regulatory elements. The molecular mechanisms regulating gene expression in response to abiotic stresses have been studied by analysing the cis- and the trans-acting elements, i.e. the sequence-specific binding TFs.85,86 Over the years, extensive promoter analyses have identified a large number of cis-elements, which are important molecular switches that are functionally involved in the transcriptional regulation of a dynamic network of gene activities controlling various biological processes, including abiotic stress responses, hormone responses and developmental processes.39,92 Among these cis-elements, several members have been reported for their essential roles in determining the stress-induced expression patterns of genes.39
Increasing evidence indicates that the cis-motifs are highly conserved among orthologous or paralogous genes and co-regulated genes. In addition, defined cis-elements can effectively aid in the genome-wide screening of ABA and abiotic stress-responsive genes.93,94 Therefore, a careful inspection of the existence of cis-regulatory elements in the promoter regions of the TCS genes can enable the prediction of functions for the respective TCS members in response to various stress stimuli. To facilitate the prediction and functional characterization of stress-responsive soybean TCS elements, we retrieved the −1000 promoter regions for all the TCS genes from soybean genomic sequence database and subjected them to an extensive in silico analyses to search for the existence of all known stress-responsive cis-regulatory motifs. The identified cis-sequences were then counted. Results of the search for cis-elements located in the −1000 promoter region of each TCS gene are provided on Supplementary Tables S1–S3. Table 6 indicates the TCS genes which contain stress-responsive cis-motif(s) in their −1000 bp promoter region. The occurrences of these motifs suggest that these TCS genes may have a potential functional role in stress signalling. Out of 12 stress-responsive cis-motifs used in the search, we found the occurrence of four cis-motifs. Among the TCS genes, we discovered that out of 21 soybean genes encoding HKs, 8 genes have stress-responsive cis-motifs in their promoter regions. It is worth mentioning that all these eight promoters contain cis-motifs for MYB/MYC-binding TFs. We could also detect stress-responsive cis-motifs (ABRE, MYB or MYC) in the promoter regions of three GmHP genes, seven type-A, four type-B, all three type-C and seven pseudo-GmRR genes, suggesting that these genes may be under the control of ABRE-binding TFs, such as the bZIP-type TFs, and/or MYB/MYC-binding TFs, such as bHLH- and MYB-related TFs (Table 6).32 The MYB-like type-B RRs have been shown to regulate the expression of some type-A RR genes.95 It would be interesting to see whether the MYB-like type-B RRs could also control the expression of the kinase and/or HPt encoding genes in a feedback regulation of stress signalling. The majority of the TCS genes predicted to function in stress signalling by cis-annotation were also predicted by comparative genomics with Arabidopsis counterparts (Table 6). Although the Arabidopsis counterpart of the AHK1-like soy HKs are stress inducible and function in stress signalling,15,16,88 it is important to note that none of the 12 major stress-responsive cis-motifs was detected in the −1000 bp promoter regions of the AHK1-like soy HKs (GmHK07–09; Tables 1 and 6). However, when we screened the −1000 bp promoter region of the AHK1 gene, we could not discover the existence of any of the 12 major stress-responsive cis-motifs in the −1000 bp promoter of AHK1 gene either (data not shown). We hypothesize that more stress-responsive TCS genes could be predicted if more stress-responsive cis-motifs and/or longer promoter regions were used in the predictive searches. Although we have included all the currently known major stress-responsive (drought, high salinity and cold stresses-responsive) cis-motifs in our study, we cannot rule out the possibility that additional stress-responsive cis-motifs exist in the plant genome. Furthermore, it should also be noted that all the cis-motifs used in this study were stress-inducible motifs, which can be used to predict the stress-inducible genes. However, there are also stress-repressive genes among the TCS genes,21 thus stress-repressive cis-motifs should be included into the study as well. Unfortunately, the identity of these types of cis-motifs is currently not known.
Table 6.
Soybean TCS genes having stress-responsive cis-motif(s) in their promoter region
| Gene name | Familya | Function in stressb | ABRE1 | ABRE2 | MYBR | MYCR |
|---|---|---|---|---|---|---|
| GmHK02 | CKI2/AHK5 like | Yes | 0 | 0 | 0 | 1 |
| GmHK05 | CKI2/AHK5 like | Yes | 0 | 0 | 0 | 1 |
| GmHK10 | AHK2 like | Yes | 0 | 0 | 1 | 0 |
| GmHK11 | AHK2 like | Yes | 0 | 0 | 1 | 0 |
| GmHK14 | AHK4 like | Yes | 0 | 0 | 1 | 0 |
| GmHK15 | AHK4 like | Yes | 0 | 0 | 1 | 0 |
| GmERS1 | ERS1 like | Yes | 0 | 0 | 0 | 1 |
| GmETR1 | ETR1 like | Yes | 0 | 0 | 1 | 1 |
| HPt proteins | ||||||
| GmHP01 | AHP1 like | Yes | 0 | 0 | 0 | 2 |
| GmHP04 | AHP1 like | Yes | 1 | 1 | 0 | 0 |
| GmHP05 | AHP1 like | Yes | 1 | 1 | 0 | 0 |
| RR proteins | ||||||
| Type A | ||||||
| GmRR01 | ARR4 like | Yes | 0 | 1 | 0 | 0 |
| GmRR02 | ARR6 like | Unknown | 0 | 0 | 0 | 1 |
| GmRR03 | ARR6 like | 0 | 0 | 0 | 1 | |
| GmRR06 | ARR8 like | Yes | 0 | 0 | 0 | 1 |
| GmRR09 | ARR9 like | Yes | 0 | 0 | 0 | 2 |
| GmRR10 | ARR9 like | Yes | 0 | 0 | 1 | 1 |
| GmRR18 | ARR17 like | Unknown | 0 | 0 | 0 | 1 |
| Type B | ||||||
| GmRR25 | ARR11 like | Unknown | 0 | 0 | 0 | 1 |
| GmRR26 | ARR12 like | Unknown | 0 | 0 | 0 | 1 |
| GmRR29 | ARR12 like | Unknown | 0 | 0 | 1 | 1 |
| Type C | ||||||
| GmRR34 | ARR24 like | Unknown | 0 | 0 | 0 | 1 |
| GmRR35 | ARR24 like | Unknown | 0 | 1 | 0 | 1 |
| GmRR36 | ARR24 like | Unknown | 0 | 0 | 0 | 1 |
| Pseudo | ||||||
| GmPRR37 | APRR1 like | Unknown | 0 | 1 | 0 | 0 |
| GmPRR38 | APRR1 like | Unknown | 0 | 1 | 0 | 0 |
| GmPRR39 | APRR5 like | Yes | 0 | 1 | 0 | 1 |
| GmPRR40 | APRR5 like | Yes | 0 | 1 | 0 | 0 |
| GmPRR43 | APRR9 like | Yes | 0 | 0 | 0 | 1 |
| GmPRR45 | APRR9 like | Yes | 0 | 0 | 0 | 2 |
aFamily indicates classification based on sequence identity with the ARR proteins.
bStress-related function of the Arabidopsis TCS members listed in ‘Family’ has been determined (Yes) or is still unknown (Unknown) based on literature analysis.
At the present time, stress-related expression data are not yet available for soybean TCS genes. Our study aims to aid in selection of stress-related TCS members for further functional studies and genetic engineering through a systematic identification of stress-related TCS genes. Specifically, the integration of stress-responsive cis-motif annotation and comparative sequence analyses, which were carried out and described previously by sequence alignments and phylogenetic relationship studies, has been shown to effectively aid in the prediction of stress-related TCS members.
3.8. Conclusions
Recent progress has provided compelling evidence that multistep component systems play important roles in signal transduction in response to environmental stimuli and plant growth regulators.15,16,23 In this regard, we have identified and characterized the complete set of TCS elements in soybean. Our analysis of the TCS machinery of soybean and comparative analysis with those of Arabidopsis and rice has revealed that the overall structure TCS machinery is very similar between these three species despite their disjunctive evolutionary histories. Individual TCS members from these three species also show significant sequence conservation. Since the assemblies of the soybean genome analysed in this study (the Glyma1 model) still need to be improved, part of the soybean TCS machinery identified in this study may be affected by future fine-tuning of their annotations.
A number of studies have substantiated that sequence similarity-based clustering of the members of several gene families correlate with their function.32,89,90 Experimental characterizations of the TCS members of Arabidopsis, rice and maize in the CK signalling also provide evidence for the correlation between phylogenetic relationship and functional conservation.19,96,97 Moreover, a great deal of evidence demonstrates that defined cis-elements can effectively aid in prediction of stress-responsive genes.93,94 Therefore, the results of phylogenetic relationship studies of soybean TCS members with their Arabidopsis and rice TCS counterparts, among which stress-related functions of some members have been identified, can be combined with the results of cis-motif annotations for systematic functional predictions of the stress-related soybean TCS genes.
In this study, we have provided a resource for (i) further elucidation of regulatory mechanisms of the soybean TCS machinery underlying different developmental and physiological processes, including environmental stress responses, and for (ii) comparative genomics and interspecific analysis of evolutionary relationship of TCSs within plants and comparisons to other organisms.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
Research in Tran's Lab is supported by Grants-in-Aid (Start-up) for Scientific Research (21870046) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a Start-up Support grant (M36-57000) from the RIKEN Yokohama Institute Director Discretionary Funds. This work was also supported by Grant-in-Aid for Young Scientists (B) (21780011) to KM from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Material
Footnotes
Edited by Mikio Nishimura
References
- 1.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. doi:10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Hwang I., Chen H.C., Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–15. doi: 10.1104/pp.005504. doi:10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuno T. Two-component phosphorelay signal transduction systems in plants: from hormone responses to circadian rhythms. Biosci. Biotechnol. Biochem. 2005;69:2263–76. doi: 10.1271/bbb.69.2263. doi:10.1271/bbb.69.2263. [DOI] [PubMed] [Google Scholar]
- 4.Pareek A., Singh A., Kumar M., Kushwaha H.R., Lynn A.M., Singla-Pareek S.L. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–97. doi: 10.1104/pp.106.086371. doi:10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaller G.E., Kieber J.J., Shiu H. Two-component signaling elements and histidyl-aspartyl phosphorelays. In: Somerville C., Meyerowitz E., editors. The Arabidopsis Book, The American Society of Plant Biologists. 2008. Rockville, MD, USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pils B., Heyl A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009;151:782–91. doi: 10.1104/pp.109.139188. doi:10.1104/pp.109.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urao T., Yamaguchi-Shinozaki K., Shinozaki K. Two-component systems in plant signal transduction. Trends Plant Sci. 2000;5:67–74. doi: 10.1016/s1360-1385(99)01542-3. doi:10.1016/S1360-1385(99)01542-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Shi L. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology. 2005;151:2159–73. doi: 10.1099/mic.0.27987-0. doi:10.1099/mic.0.27987-0. [DOI] [PubMed] [Google Scholar]
- 9.Urao T., Yamaguchi-Shinozaki K., Shinozaki K. Plant histidine kinases: an emerging picture of two-component signal transduction in hormone and environmental responses. Sci. STKE. 2001;2001:re18. doi: 10.1126/stke.2001.109.re18. [DOI] [PubMed] [Google Scholar]
- 10.Mahonen A.P., Higuchi M., Tormakangas K., et al. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 2006;16:1116–22. doi: 10.1016/j.cub.2006.04.030. doi:10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 11.Schultz J., Copley R.R., Doerks T., Ponting C.P., Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–4. doi: 10.1093/nar/28.1.231. doi:10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito Y., Kurata N. Identification and characterization of cytokinin-signalling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. doi:10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Ishida K., Niwa Y., Yamashino T., Mizuno T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009;16:237–47. doi: 10.1093/dnares/dsp012. doi:10.1093/dnares/dsp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. doi:10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran L.S., Urao T., Qin F., et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:20623–8. doi: 10.1073/pnas.0706547105. doi:10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wohlbach D.J., Quirino B.F., Sussman M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20:1101–17. doi: 10.1105/tpc.107.055871. doi:10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwama A., Yamashino T., Tanaka Y., et al. AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:375–80. doi: 10.1093/pcp/pcl065. doi:10.1093/pcp/pcl065. [DOI] [PubMed] [Google Scholar]
- 18.Desikan R., Horak J., Chaban C., et al. The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS One. 2008;3:e2491. doi: 10.1371/journal.pone.0002491. doi:10.1371/journal.pone.0002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi M., Pischke M.S., Mahonen A.P., et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl Acad. Sci. USA. 2004;101:8821–6. doi: 10.1073/pnas.0402887101. doi:10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner T., Schmulling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–38. doi: 10.1016/j.pbi.2009.07.002. doi:10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Miyata S., Urao T., Yamaguchi-Shinozaki K., Shinozaki K. Characterization of genes for two-component phosphorelay mediators with a single HPt domain in Arabidopsis thaliana. FEBS Lett. 1998;437:11–4. doi: 10.1016/s0014-5793(98)01188-0. doi:10.1016/S0014-5793(98)01188-0. [DOI] [PubMed] [Google Scholar]
- 22.Urao T., Yakubov B., Yamaguchi-Shinozaki K., Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427:175–8. doi: 10.1016/s0014-5793(98)00418-9. doi:10.1016/S0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]
- 23.Nakamichi N., Kusano M., Fukushima A., et al. Transcript profiling of an Arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–62. doi: 10.1093/pcp/pcp004. doi:10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 24.Jain M., Tyagi A.K., Khurana J.P. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa) BMC Plant Biol. 2006;6:1. doi: 10.1186/1471-2229-6-1. doi:10.1186/1471-2229-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain M., Tyagi A.K., Khurana J.P. Differential gene expression of rice two-component signaling elements during reproductive development and regulation by abiotic stress. Funct. Integr. Genomics. 2008;8:175–80. doi: 10.1007/s10142-007-0063-6. doi:10.1007/s10142-007-0063-6. [DOI] [PubMed] [Google Scholar]
- 26.Karan R., Singla-Pareek S.L., Pareek A. Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct. Integr. Genomics. 2009;9:411–7. doi: 10.1007/s10142-009-0119-x. doi:10.1007/s10142-009-0119-x. [DOI] [PubMed] [Google Scholar]
- 27.Ososki A.L., Kennelly E.J. Phytoestrogens: a review of the present state of research. Phytother. Res. 2003;17:845–69. doi: 10.1002/ptr.1364. doi:10.1002/ptr.1364. [DOI] [PubMed] [Google Scholar]
- 28.Shinozaki K. Acceleration of soybean genomics using large collections of DNA markers for gene discovery. DNA Res. 2007;14:235. doi: 10.1093/dnares/dsm031. doi:10.1093/dnares/dsm031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai T., Kogiso M. Soy isoflavones and immunity. J. Med. Invest. 2008;55:167–73. doi: 10.2152/jmi.55.167. doi:10.2152/jmi.55.167. [DOI] [PubMed] [Google Scholar]
- 30.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 31.Schaller G.E., Doi K., Hwang I., et al. Nomenclature for two-component signaling elements of rice. Plant Physiol. 2007;143:555–7. doi: 10.1104/pp.106.093666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009;16:353–69. doi: 10.1093/dnares/dsp023. doi:10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. doi:10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. doi:10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 36.Riechmann J.L., Heard J., Martin G., et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. doi: 10.1126/science.290.5499.2105. doi:10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 37.Hruz T., Laule O., Szabo G., et al. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant D., Nelson R.T., Cannon S.B., Shoemaker R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010;38:D843–6. doi: 10.1093/nar/gkp798. doi:10.1093/nar/gkp798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi-Shinozaki K., Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Yeh K.C., Lagarias J.C. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl Acad. Sci. USA. 1998;95:13976–81. doi: 10.1073/pnas.95.23.13976. doi:10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moussatche P., Klee H.J. Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J. Biol. Chem. 2004;279:48734–41. doi: 10.1074/jbc.M403100200. doi:10.1074/jbc.M403100200. [DOI] [PubMed] [Google Scholar]
- 42.Tran L.S., Shinozaki K., Yamaguchi-Shinozaki K. Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav. 2010;5:148–50. doi: 10.4161/psb.5.2.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pischke M.S., Jones L.G., Otsuga D., Fernandez D.E., Drews G.N., Sussman M.R. An Arabidopsis histidine kinase is essential for megagametogenesis. Proc. Natl Acad. Sci. USA. 2002;99:15800–5. doi: 10.1073/pnas.232580499. doi:10.1073/pnas.232580499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hejatko J., Ryu H., Kim G.T., et al. The histidine kinases cytokinin-independent1 and Arabidopsis histidine kinase2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell. 2009;21:2008–21. doi: 10.1105/tpc.109.066696. doi:10.1105/tpc.109.066696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z.G., Zhou H.L., Chen T., et al. Evidence for serine/threonine and histidine kinase activity in the tobacco ethylene receptor protein NTHK2. Plant Physiol. 2004;136:2971–81. doi: 10.1104/pp.103.034686. doi:10.1104/pp.103.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Binder B.M., O'Malley R.C., Wang W., et al. Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 2004;136:2913–20. doi: 10.1104/pp.104.050369. doi:10.1104/pp.104.050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qu X., Schaller G.E. Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 2004;136:2961–70. doi: 10.1104/pp.104.047126. doi:10.1104/pp.104.047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho Y.H., Yoo S.D. Ethylene response 1 histidine kinase activity of Arabidopsis promotes plant growth. Plant Physiol. 2007;143:612–6. doi: 10.1104/pp.106.091504. doi:10.1104/pp.106.091504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao W.H., Liu J., He X.J., et al. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007;143:707–19. doi: 10.1104/pp.106.094292. doi:10.1104/pp.106.094292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierik R., Tholen D., Poorter H., Visser E.J., Voesenek L.A. The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 2006;11:176–83. doi: 10.1016/j.tplants.2006.02.006. doi:10.1016/j.tplants.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 51.Yoo S.D., Cho Y., Sheen J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009;14:270–9. doi: 10.1016/j.tplants.2009.02.007. doi:10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Q.H., Wang X.M. Characterization of an ethylene receptor homologue from wheat and its expression during leaf senescence. J. Exp. Bot. 2003;54:1489–90. doi: 10.1093/jxb/erg142. doi:10.1093/jxb/erg142. [DOI] [PubMed] [Google Scholar]
- 53.Imamura A., Hanaki N., Umeda H., et al. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl Acad. Sci. USA. 1998;95:2691–6. doi: 10.1073/pnas.95.5.2691. doi:10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urao T., Miyata S., Yamaguchi-Shinozaki K., Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000;478:227–32. doi: 10.1016/s0014-5793(00)01860-3. doi:10.1016/S0014-5793(00)01860-3. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka Y., Suzuki T., Yamashino T., Mizuno T. Comparative studies of the AHP histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2004;68:462–5. doi: 10.1271/bbb.68.462. doi:10.1271/bbb.68.462. [DOI] [PubMed] [Google Scholar]
- 56.Dortay H., Gruhn N., Pfeifer A., Schwerdtner M., Schmulling T., Heyl A. Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J. Proteome Res. 2008;7:3649–60. doi: 10.1021/pr0703831. doi:10.1021/pr0703831. [DOI] [PubMed] [Google Scholar]
- 57.Hutchison C.E., Li J., Argueso C., et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18:3073–87. doi: 10.1105/tpc.106.045674. doi:10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Argueso C.T., Ferreira F.J., Kieber J.J. Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 2009;32:1147–60. doi: 10.1111/j.1365-3040.2009.01940.x. doi:10.1111/j.1365-3040.2009.01940.x. [DOI] [PubMed] [Google Scholar]
- 59.Mahonen A.P., Bishopp A., Higuchi M., et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–8. doi: 10.1126/science.1118875. doi:10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 60.To J.P., Haberer G., Ferreira F.J., et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–71. doi: 10.1105/tpc.018978. doi:10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leibfried A., To J.P., Busch W., et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–5. doi: 10.1038/nature04270. doi:10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 62.Lee D.J., Park J.Y., Ku S.J., et al. Genome-wide expression profiling of Arabidopsis response regulator 7(ARR7) overexpression in cytokinin response. Mol. Genet. Genomics. 2007;277:115–37. doi: 10.1007/s00438-006-0177-x. doi:10.1007/s00438-006-0177-x. [DOI] [PubMed] [Google Scholar]
- 63.To J.P., Deruere J., Maxwell B.B., et al. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–14. doi: 10.1105/tpc.107.052662. doi:10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sweere U., Eichenberg K., Lohrmann J., et al. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science. 2001;294:1108–11. doi: 10.1126/science.1065022. doi:10.1126/science.1065022. [DOI] [PubMed] [Google Scholar]
- 65.Salome P.A., To J.P., Kieber J.J., McClung C.R. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell. 2006;18:55–69. doi: 10.1105/tpc.105.037994. doi:10.1105/tpc.105.037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mira-Rodado V., Sweere U., Grefen C., et al. Functional cross-talk between two-component and phytochrome B signal transduction in Arabidopsis. J. Exp. Bot. 2007;58:2595–607. doi: 10.1093/jxb/erm087. doi:10.1093/jxb/erm087. [DOI] [PubMed] [Google Scholar]
- 67.Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2004;135:927–37. doi: 10.1104/pp.103.038109. doi:10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riano-Pachon D.M., Correa L.G., Trejos-Espinosa R., Mueller-Roeber B. Green transcription factors: a chlamydomonas overview. Genetics. 2008;179:31–9. doi: 10.1534/genetics.107.086090. doi:10.1534/genetics.107.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mason M.G., Mathews D.E., Argyros D.A., et al. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell. 2005;17:3007–18. doi: 10.1105/tpc.105.035451. doi:10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hass C., Lohrmann J., Albrecht V., et al. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 2004;23:3290–302. doi: 10.1038/sj.emboj.7600337. doi:10.1038/sj.emboj.7600337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rashotte A.M., Mason M.G., Hutchison C.E., Ferreira F.J., Schaller G.E., Kieber J.J. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl Acad. Sci. USA. 2006;103:11081–5. doi: 10.1073/pnas.0602038103. doi:10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama A., Yamashino T., Amano Y., et al. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007;48:84–96. doi: 10.1093/pcp/pcl040. doi:10.1093/pcp/pcl040. [DOI] [PubMed] [Google Scholar]
- 73.Ishida K., Yamashino T., Yokoyama A., Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. doi:10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- 74.Tajima Y., Imamura A., Kiba T., Amano Y., Yamashino T., Mizuno T. Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol. 2004;45:28–39. doi: 10.1093/pcp/pcg154. doi:10.1093/pcp/pcg154. [DOI] [PubMed] [Google Scholar]
- 75.Doi K., Izawa T., Fuse T., et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–36. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kiba T., Aoki K., Sakakibara H., Mizuno T. Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 2004;45:1063–77. doi: 10.1093/pcp/pch128. doi:10.1093/pcp/pch128. [DOI] [PubMed] [Google Scholar]
- 77.Kiba T., Naitou T., Koizumi N., Yamashino T., Sakakibara H., Mizuno T. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His->Asp Phosphorelay circuitry. Plant Cell Physiol. 2005;46:339–55. doi: 10.1093/pcp/pci033. doi:10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- 78.Gattolin S., Alandete-Saez M., Elliott K., et al. Spatial and temporal expression of the response regulators ARR22 and ARR24 in Arabidopsis thaliana. J. Exp. Bot. 2006;57:4225–33. doi: 10.1093/jxb/erl205. doi:10.1093/jxb/erl205. [DOI] [PubMed] [Google Scholar]
- 79.Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T. Pseudo-response regulators, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–98. doi: 10.1093/pcp/pci086. doi:10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 80.Nakamichi N., Kita M., Ito S., Sato E., Yamashino T., Mizuno T. The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 2005;46:609–19. doi: 10.1093/pcp/pci061. doi:10.1093/pcp/pci061. [DOI] [PubMed] [Google Scholar]
- 81.Salome P.A., McClung C.R. Pseudo-response regulator 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. doi:10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ito S., Niwa Y., Nakamichi N., Kawamura H., Yamashino T., Mizuno T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008;49:201–13. doi: 10.1093/pcp/pcm178. doi:10.1093/pcp/pcm178. [DOI] [PubMed] [Google Scholar]
- 83.Schlueter J.A., Lin J.Y., Schlueter S.D., et al. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics. 2007;8:330. doi: 10.1186/1471-2164-8-330. doi:10.1186/1471-2164-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Day R.C., Herridge R.P., Ambrose B.A., Macknight R.C. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 2008;148:1964–84. doi: 10.1104/pp.108.128108. doi:10.1104/pp.108.128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. doi:10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 86.Tran L.S., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 2007;428:109–28. doi: 10.1016/S0076-6879(07)28006-1. doi:10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 87.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–63. doi: 10.1038/nrg2538. doi:10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 88.Urao T., Yakubov B., Satoh R., et al. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–54. doi: 10.1105/tpc.11.9.1743. doi:10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fang Y., You J., Xie K., Xie W., Xiong L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics. 2008;280:547–63. doi: 10.1007/s00438-008-0386-6. doi:10.1007/s00438-008-0386-6. [DOI] [PubMed] [Google Scholar]
- 90.Tran L.S., Quach T.N., Guttikonda S.K., et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics. 2009;281:647–64. doi: 10.1007/s00438-009-0436-8. doi:10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 91.Irsigler A.S., Costa M.D., Zhang P., et al. Expression profiling on soybean leaves reveals integration of ER- and osmotic-stress pathways. BMC Genomics. 2007;8:431. doi: 10.1186/1471-2164-8-431. doi:10.1186/1471-2164-8-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150:535–46. doi: 10.1104/pp.109.136028. doi:10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang W., Ruan J., Ho T.H., You Y., Yu T., Quatrano R.S. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21:3074–81. doi: 10.1093/bioinformatics/bti490. doi:10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- 94.Won S.K., Lee Y.J., Lee H.Y., Heo Y.K., Cho M., Cho H.T. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–73. doi: 10.1104/pp.109.140905. doi:10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taniguchi M., Sasaki N., Tsuge T., Aoyama T., Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–77. doi: 10.1093/pcp/pcl063. doi:10.1093/pcp/pcl063. [DOI] [PubMed] [Google Scholar]
- 96.Yonekura-Sakakibara K., Kojima M., Yamaya T., Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004;134:1654–61. doi: 10.1104/pp.103.037176. doi:10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Du L., Jiao F., Chu J., Jin G., Chen M., Wu P. The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707. doi: 10.1016/j.ygeno.2007.02.001. doi:10.1016/j.ygeno.2007.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.