Abstract
Lettuce (Lactuca sativa L.) seeds have poor shelf life and exhibit thermoinhibition (fail to germinate) above ∼25°C. Seed priming (controlled hydration followed by drying) alleviates thermoinhibition by increasing the maximum germination temperature, but reduces lettuce seed longevity. Controlled deterioration (CD) or accelerated ageing storage conditions (i.e. elevated temperature and relative humidity) are used to study seed longevity and to predict potential seed lifetimes under conventional storage conditions. Seeds produced in 2002 and 2006 of a recombinant inbred line (RIL) population derived from a cross between L. sativa cv. Salinas×L. serriola accession UC96US23 were utilized to identify quantitative trait loci (QTLs) associated with seed longevity under CD and conventional storage conditions. Multiple longevity-associated QTLs were identified under both conventional and CD storage conditions for control (non-primed) and primed seeds. However, seed longevity was poorly correlated between the two storage conditions, suggesting that deterioration processes under CD conditions are not predictive of ageing in conventional storage conditions. Additionally, the same QTLs were not identified when RIL populations were grown in different years, indicating that lettuce seed longevity is strongly affected by production environment. Nonetheless, a major QTL on chromosome 4 [Seed longevity 4.1 (Slg4.1)] was responsible for almost 23% of the phenotypic variation in viability of the conventionally stored control seeds of the 2006 RIL population, with improved longevity conferred by the Salinas allele. QTL analyses may enable identification of mechanisms responsible for the sensitivity of primed seeds to CD conditions and breeding for improved seed longevity.
Keywords: Ageing, controlled deterioration (CD), genotype×environment interactions, lettuce, quantitative trait locus (QTL), seed longevity, seed priming
Introduction
Seed deterioration is a major problem in agricultural production. It has been estimated that 25% of the annual value of seeds in inventory may be lost because of poor seed quality (McDonald and Nelson, 1986). One estimate equated this to US$500 million annually just for purchased seed, although this value would be significantly greater when reductions in yield due to poor seed quality are considered on a worldwide basis (McDonald, 1999). Seed quality is particularly important in high value crops such as lettuce (Lactuca sativa L.), as poor germination and emergence result in lack of crop uniformity and reduced yield (Cantliffe et al., 1981). In addition, lettuce produces seeds with relatively limited storage life (commercial viability lost within 5 years) (McDonald, 1999).
A number of mechanisms of seed ageing have been identified (Smith and Berjak, 1995). Lipid peroxidation, resulting in membrane damage as well as the generation of toxic byproducts, is well documented in stored seeds (Davies, 2005). Oxidative damage to DNA and proteins is also likely to be involved in seed ageing (Rao et al., 1987; Bailly et al., 2008). Formation of sugar–protein adducts (i.e. the Maillard reaction) or of isoaspartyl residues may be factors in loss of protein function during deterioration (Sun and Leopold, 1995; Oge et al., 2008; Rajjou et al., 2008). On the other hand, antioxidants, heat shock proteins (HSPs), and enzymes to repair protein damage may be involved in ameliorating the effects of ageing on seed longevity (Kibinza et al., 2006; Prieto-Dapena et al., 2006; Oge et al., 2008; Almoguera et al., 2009).
Seed priming (controlled hydration followed by drying) is a technique to improve the germination of seeds, inducing faster and more uniform germination over broader temperature ranges (Heydecker et al., 1973; Tarquis and Bradford, 1992; McDonald, 2000). This practice is commercially applied in many crops and particularly in lettuce to alleviate thermoinhibition (failure of seeds to germinate when imbibed at warm temperatures) by increasing the maximum germination temperature (Cantliffe et al., 1981; Valdes et al., 1985). However, priming often results in a reduction of seed longevity compared with non-primed seed (Tarquis and Bradford, 1992; Maude et al., 1994; Chojnowski et al., 1997), although some studies have found the opposite effect (Georghiou et al., 1987; Probert et al., 1991; Butler et al., 2009). Primed lettuce seeds are particularly prone to reduced longevity relative to non-primed seeds when stored under high moisture content conditions (Tarquis and Bradford, 1992; Schwember and Bradford, 2005; Hill et al., 2007).
Studies of seed longevity under conventional or optimal storage conditions would take years to complete, so accelerated ageing or controlled deterioration (CD) conditions are utilized to speed the loss of viability. The CD test has been used to assess the vigour of seed lots and to predict their relative longevity by ageing seeds rapidly at elevated temperature and relative humidity (RH) (Delouche and Baskin, 1973; Powell and Matthews, 2005). Proteome analysis of Arabidopsis thaliana seeds revealed common features between the CD (85% RH at 40°C for up to 7 d) and conventionally (up to 11 years at 5 °C) aged seeds (Rajjou et al., 2008). However, it remains under debate whether the mechanisms of conventional and accelerated seed deterioration are identical across broad ranges of temperature and moisture content (Walters, 1998; McDonald, 1999; Black et al., 2006).
Genetics provides a powerful approach to identify the physiological and molecular bases of phenotypic traits such as seed longevity and other quality factors. Seed longevity is a quantitative trait since it is probably controlled by multiple genes (Clerkx et al., 2004b) and is strongly affected by the environment during seed formation, harvest, and storage (Contreras et al., 2008, 2009). Genetic loci associated with seed longevity have been identified in Arabidopsis (Bentsink et al., 2000; Tesnier et al., 2002; Clerkx et al., 2004a, b) and rice (Oryza sativa) (Miura et al., 2002; Sasaki et al., 2005; Xue et al., 2008). In both species, several quantitative trait loci (QTLs) located on different chromosomes were associated with seed viability after storage. Molecular studies support the complex genetic nature of seed longevity. For example, Arabidopsis mutants affected in the tocopherol (Sattler et al., 2004) or flavonoid (Debeaujon et al., 2000) biosynthetic pathways showed reduced seed longevity, consistent with data showing that protection against reactive oxygen species production and attack are important features of Arabidopsis seed longevity (Clerkx et al., 2004a). In addition, mutations within the DOG1 (Delay of Germination) gene in Arabidopsis that shortened seed dormancy were associated with a reduced seed longevity phenotype, indicating that seed dormancy mechanisms may be involved in delaying seed deterioration (Bentsink et al., 2006).
Stress-related proteins and enzymes may also play a role in seed longevity. Prieto-Dapena et al. (2006) reported that seed-specific overexpression of the sunflower (Helianthus annuus) HaHSFA9 heat stress transcription factor in tobacco (Nicotiana tabacum) enhanced the accumulation of HSPs and significantly improved resistance of seeds to controlled deterioration, without detrimental effects on plant growth or development, seed morphology, or total seed yield. This transcription factor apparently interacts with the stress-related transcription factor HaDREB2 in a seed-specific manner to enhance tolerance to CD conditions (Almoguera et al., 2009). Conversely, activity of a membrane lipid-hydrolysing phospholipase D (PLDα1) appears to be detrimental for Arabidopsis seed quality, whereas the attenuation of PLDα1 expression has the potential to improve oil stability, seed quality, and seed longevity (Devaiah et al., 2007). Lipoxygenases (LOXs) have also been reported to be involved in seed deterioration (Suzuki et al., 1996; Li et al., 2007). Mutations in the rice aldehyde dehydrogenase 7 (OsALDH7) gene resulted in seeds that were more sensitive to CD conditions and that accumulated more malondialdehyde than the wild-type seeds, implying that this enzyme could play a role in maintaining seed viability by detoxifying the aldehydes generated by lipid peroxidation (Shin et al., 2009). The activity of protein L-isoaspartyl methyltransferase (IAMT), an enzyme repairing abnormal L-isoaspartyl residues accumulated in proteins during ageing, may be associated with the multicentenarian longevity of sacred lotus (Nelumbo nucifera) seed, since the IAMT activity of sacred lotus cotyledons persists during germination, in contrast to most seeds tested (Shen-Miller, 2002). Two Arabidopsis genes (PIMT1 and PIMT2) encode IAMT, transcripts of which increased in developing seeds in response to abscisic acid (ABA). ABA enhanced the production of one form of PIMT2 (PIMT2ω) through post-transcriptional modifications (Xu et al., 2004). Overexpressing PIMT1 in Arabidopsis enhanced seed longevity, while inactivating the gene reduced seed longevity (Oge et al., 2008), suggesting that increased ability to repair damaged proteins upon imbibition is associated with greater seed longevity.
Protection of proteins and membranes during desiccation and storage probably depends on multiple mechanisms. During dehydration, water molecules are replaced by sugars at hydrogen-bonding sites to preserve the native structure of proteins and the spacing between membrane phospholipids (Hoekstra et al., 2001). At low moisture contents, glass formation increases viscosity and slows deteriorative chemical reactions (Walters, 1998; Hoekstra et al., 2001). Specific anti-oxidative mechanisms may also play a role in the protection of seeds against ageing (Bailly et al., 2008). While these processes and deterioration itself occur in dry seeds where metabolism is prevented, the genes involved during seed development in establishing the pre-conditions for these mechanisms might be revealed by genetic analysis.
Storage experiments and QTL analyses were conducted to study potential relationships between CD and conventional ageing of control and primed seeds and to investigate the genetic basis of seed longevity in lettuce. A recombinant inbred line (RIL) population derived from a cross between L. sativa cv. Salinas×L. serriola accession UC96US23 was utilized to test whether QTLs associated with seed longevity under CD and conventional storage conditions could be identified.
Materials and methods
Recombinant inbred lines and seed production
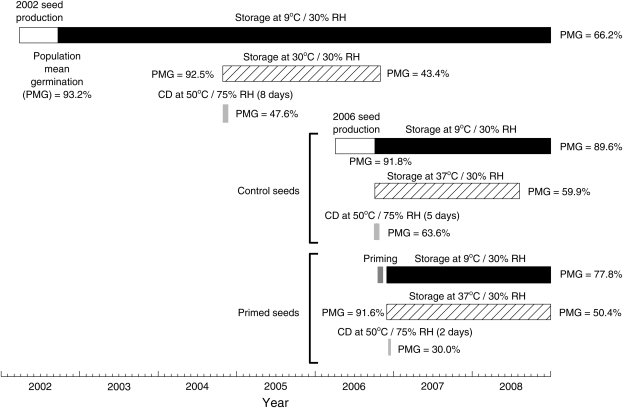
Seeds from 89 F8 RILs in a population derived by single-seed descent from an interspecific cross between L. sativa cv. Salinas and L. serriola accession UC96US23 were utilized. This population has been analysed previously for seed germination and quality traits (Argyris et al., 2005, 2008a, b) and responses to seed priming (Schwember and Bradford, 2010). This lettuce mapping population was developed by Dr Richard Michelmore's group at the University of California, Davis and analysed as part of the Compositae Genome Project (Truco et al., 2007; http://compgenomics.ucdavis.edu). Seeds of this RIL population were produced in 2002 and 2006 in the field in Davis, California and were stored subsequently at 9 °C and 30% RH [∼5–6% seed moisture content (MC), dry weight basis] (Fig. 1). These are hereafter termed the 2002 and 2006 populations.
Fig. 1.
Timeline of seed production and storage experiments using seeds of a lettuce RIL population produced in 2002 or in 2006. Population mean germination (PMG) is the average of the germination percentages of all RILs tested at the times indicated. Specific storage experiments and conditions are described in the text.
Seed priming protocol
Seeds of the RILs produced in 2006 were osmoprimed using a standard protocol (Heydecker et al., 1973; Tarquis and Bradford, 1992) as follows: 1.2 g of seeds of each line were primed in an 8.8 cm Petri dish over one blotter saturated with 10 ml of –1.25 MPa solution of polyethylene glycol 8000 for 48 h at 9 °C under continuous fluorescent light. Seeds were then rinsed briefly with water and surface water was removed by suction in a Buchner funnel. Subsequently, seeds were rapidly dried for 2 h at 32 °C and 25–30% RH in a ventilated oven before transfer to a chamber at 9 °C and 30% RH and equilibration for at least 2 d to constant MC. Seeds were stored thereafter in the same condition (Fig. 1).
Conventional storage
Seeds of the 2002 and 2006 populations were stored at 30% RH and different temperatures for various durations to result in ∼50% mean viability across the RILs (Fig. 1). These low RH (or MC) and moderate temperature conditions are termed here as ‘conventional storage’. Subsamples of control (not primed) seeds of the 2002 RIL population were stored for 2 years at 30 °C and 30% RH for after-ripening and deterioration studies beginning in 2004. The overall mean germination percentages had fallen to 43.4% by November 2006. The control seeds of the 2006 RIL population were stored at 37 °C for 21 months, with overall mean germination of 59.9% in July 2008. The primed seeds of the 2006 RIL population were stored at 37 °C for 24 months, when overall mean germination had fallen to 50.4% (December 2008). The germination percentages of the RILs at these times were used in the QTL analyses. Control seeds of the parental lines produced in Davis in 2002 were stored at 9 °C and 30% RH for almost 7 years, and germination was scored in 2002, 2006, and 2009.
Controlled deterioration
For CD, control seeds of the 2002 RIL population and control and primed seeds of the 2006 RIL population were adjusted to higher MC by incubation over saturated NaCl solution (75% RH) for 4 d at 25 °C in a hermetic chamber (Zhang and McDonald, 1997). The seeds were then sealed in vials and stored for different durations in an oven at 50 °C, followed by standard germination tests to assess longevity. The overall mean germination percentage of the control seeds of the 2002 RIL population was 47.6% after 8 d at 50 °C (Fig. 1). The control and the primed seeds of the 2006 RIL population exhibited mean germination percentages of 63.6% and 30.0% after incubation at 50 °C for 5 d and 2 d, respectively.
Germination tests for phenotypic analyses
All the seeds exposed to either CD or conventional storage conditions were equilibrated for 24 h at 9 °C/25–30% RH before conducting the germination tests. For the control seeds, three replicates of 50 seeds each were placed in 4 cm diameter Petri dishes over two blotters wetted with distilled water (4 ml). For the primed seeds, three replicates of 25 seeds were utilized using the same conditions. In all the tests, the seeds were incubated under continuous fluorescent light at 20 °C, and germination (radicle emergence) was scored after 72 h. Variation in germination percentages among replicates of individual RILs was small relative to variation among RILs. The mean germination percentages of RILs were transformed to probits to normalize variances in data distributions (0.1% and 99.9% were used for 0% and 100% of germination, respectively). Viability was measured by scoring radicle emergence, as the viability curves for normal seedlings are consistently offset from those for radicle emergence (Ellis and Roberts, 1981; Bradford et al., 1993). Using radicle emergence to assess seed longevity removes additional experimental error due to analyst interpretation of normal seedling characteristics. Primed seeds often exhibited damage to the radicle tips after ageing before radicle emergence itself was affected. However, radicle emergence was scored in this case also, so storage periods for primed seeds are overestimated relative to the periods for obtaining normal or usable seedlings.
QTL analyses
For low-resolution mapping, a genetic linkage map based on the F7 RIL population comprising >1700 amplified fragment length polymorphism (AFLP), simple sequence repeat (SSR), and expressed sequence tag (EST) markers consisting of nine chromosomal linkage groups and spanning 1254 cM was constructed using JOINMAP v2.0. A subset of 486 robust markers ∼2–3 cM apart was chosen as the framework map for QTL analysis (Argyris et al., 2005). For QTLs associated with seed longevity, the probit-transformed germination percentage values for control and primed seeds of each RIL were analysed separately using Windows QTL Cartographer V. 2.5. QTL analyses were performed using composite interval mapping, and QTLs were verified by 1000 permutations of phenotypic data and declared significant (P <0.05) above the permutated log likelihood ratio (LOD score) threshold of 3.2 (control seeds) and 2.7 (primed seeds). QTLs having LOD scores close to or below these thresholds were subsequently tested for significance using multilocus mixed model analysis of variance (ANOVA; PROC MIXED) (Argyris et al., 2005). For high-resolution mapping, single-position polymorphisms (SPPs) were obtained for unigenes by analysis of 113 RILs hybridized in duplicate to a lettuce Affymetrix GeneChip® genotyping microarray (van Leeuwen et al., 2007). SPPs were ordered using MadMapper (http://cgpdb.ucdavis.edu/XLinkage/MadMapper). Markers from the high density mapping array (van Leeuwen et al., 2009) were combined with the framework markers comprising linkage groups 1 and 4 from the Lactuca integrated map (Truco et al., 2007) to obtain a more densely populated genetic map for the refinement of the QTL intervals on those chromosomes.
The naming of the Seed longevity (Slg) QTLs was based upon the seed treatment, storage condition, and linkage group (LG). QTLs identified for conventionally stored control seeds were named Slg[LG].[number on LG], such as Slg4.1, Slg4.2, and so on in the order that they occurred within the LG. QTLs identified in conventionally stored primed seeds were termed SlgP, in CD-stored control seeds were termed SlgCD, and in CD-stored primed seeds were termed SlgPCD, followed by the LG and number on the LG, as above. For LGs 1 and 4 that were analysed using the high-resolution map, markers from that map were integrated with the markers of the low-resolution map, but map distances are not strictly comparable between the low- and high-resolution maps. Nonetheless, some markers were on both maps within the QTL intervals, and while they were not exact, QTLs that largely overlapped based on common markers were assigned the same numbers to reduce confusion.
Mean comparisons
RILs homozygous for Salinas or UC96US23 alleles at Slg4.1 and Slg4.2 were sorted by parental allele indicated by the molecular markers CLS_S3_Contig5097 and CLSM10124.b1_H11.ab1, and significant differences in their mean viabilities were calculated based upon the t distribution test (Argyris et al., 2005).
Results
Seed longevity of parental lines
Control seeds of the parent lines Salinas and UC96US23 produced in Davis in 2002 were conventionally stored at 9 °C and 30% RH (Table 1; Fig. 1). Germination percentages remained high throughout 2006, when Salinas and UC96US23 seeds exhibited 96% and 100% germination, respectively. However, by July 2009, germination of the UC96US23 seeds had fallen to 32% while germination of Salinas seeds was still 98.7%. UC96US23 seeds also aged more rapidly under CD conditions, with the Salinas and UC96US23 seeds germinating 81.3% and 0%, respectively, after 90 d at 75% RH and 50 °C (Table 1).
Table 1.
Germination of control seeds of the parent lines Lactuca sativa cv. Salinas and L. serriola accession UC96US23 stored under conventional or controlled deterioration conditions
| Type of ageing | Test date/days at 75% RH–50 °C | Germination (%) ±SE |
|
| Salinas | UC96US23 | ||
| Conventional ageing | October 2002 | 99.3±0.7 | 96.7±1.8 |
| October 2006 | 96.0±2.3 | 100.0±0.0 | |
| January 2009 | 98.7±1.3 | 44.0±3.1 | |
| July 2009 | 98.7±0.7 | 32.0±2.3 | |
| Controlled deterioration | 0 d | 93.3±1.3 | 90.7±1.3 |
| 4 d | 89.3±1.3 | 90.7±4.8 | |
| 7 d | 88.0±2.3 | 84.0±4.6 | |
| 10 d | 89.3±2.7 | 82.7±1.3 | |
| 14 d | 88.0±2.3 | 81.3±7.1 | |
| 21 d | 86.7±1.3 | 80.0±0.0 | |
| 30 d | 88.0±0.0 | 72.0±6.7 | |
| 40 d | 86.7±5.8 | 48.0±7.1 | |
| 60 d | 84.0±2.3 | 26.7±6.7 | |
| 90 d | 81.3±6.7 | 0.0±0.0 | |
Seeds of both parental lines were produced in 2002 and stored under conventional conditions (9 °C and 30% RH). For the CD tests, seeds of both lines produced in 2008 were equilibrated at 75% RH for 4 d, and then transferred to an oven at 50 °C for different times. Three replicates of 50 and 25 seeds each were germinated at 20 °C in light for the conventional and CD tests, respectively. Radicle emergence was scored 72 h after planting. Mean germination percentages and standard errors (SE) are shown.
No correlation between storage conditions and production years
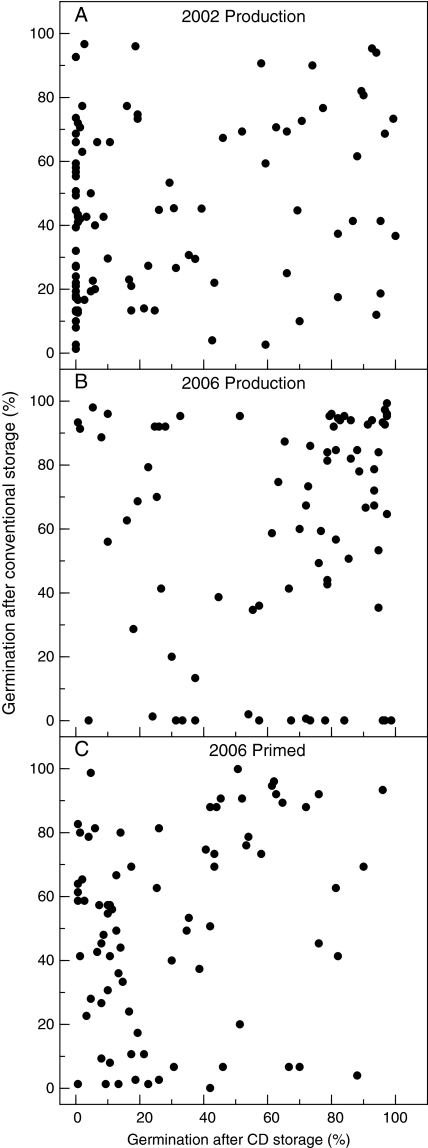
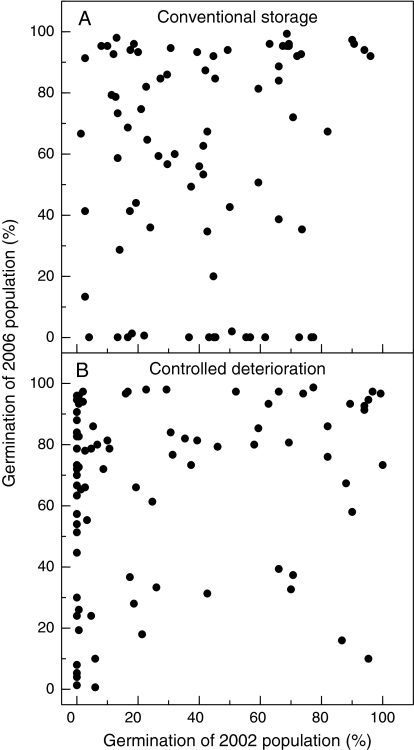
The longevity of seeds from individual RILs was assessed under both conventional and CD storage conditions (Fig. 1). Seed viabilities of the individual RILs after conventional and CD storage were not significantly correlated for either control or primed seeds (Fig. 2). In the case of the control seeds, both the 2002 and the 2006 RIL populations exhibited very low correlations between the two types of storage conditions (Fig. 2A, B). The primed seeds of the 2006 RIL population also showed a very low correlation between viabilities after conventional and CD storage (Fig. 2C). In addition, the viabilities of seeds of individual RILs produced in 2002 and in 2006 were poorly correlated after storage under either condition (Fig. 3).
Fig. 2.
Relationships between germination after conventional and CD storage of the control lettuce seeds of RILs produced in 2002 (A) and of control (B) and primed (C) seeds of RILs produced in 2006. The control 2002 RIL population was conventionally stored at 30 °C during 2 years and the 2006 RIL population was stored at 30% RH and 37 °C for 21 (control seeds) or 24 months (primed seeds). For CD conditions, the seeds were incubated in 75% RH at 25 °C for 4 d before storage at 50 °C for 8 (2002 population), 5 (2006 population), or 2 d (2006 primed). Each point represents the average percentage of radicle emergence of three replicates for each test. Correlations between the two types of ageing tests were not significant: 2002 population, R2=0.054; 2006 population, R2=0.0099; and 2006 primed, R2=0.068.
Fig. 3.
Relationships between germination of control conventional (A) and CD (B) stored seeds of the RILs produced in 2002 and 2006. The 2002 RIL population was conventionally stored at 30 °C during 2 years while the 2006 RIL population was stored at 30% RH and 37 °C for 21 months. For CD conditions, the lettuce seeds were incubated at 75% RH for 4 d before storage at 50 °C for 8 d (2002 population) or 5 d (2006 population). Each point represents the average percentage of radicle emergence of three replicates for each test. The correlations between production years of both conventionally and CD aged seeds were not significant (R2=0.0093 and 0.0307, respectively).
Longevity-associated QTLs of control conventionally stored seeds
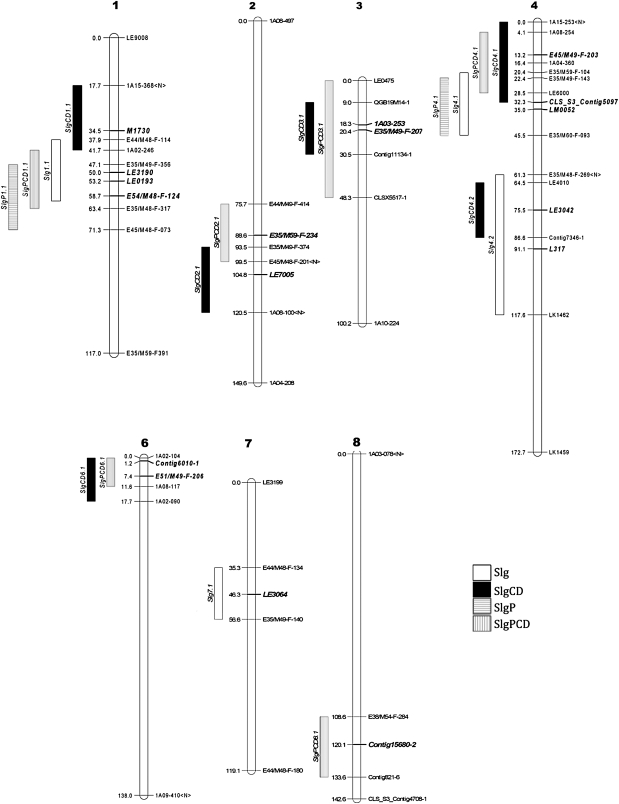
For QTL analyses, the probit-transformed viability percentage of each RIL after storage was used as the phenotypic character, and an overall germination percentage of the RIL population of ∼50% was targeted in order to have diversity in viability among RILs (Fig. 1). Using low-resolution mapping, four significant Slg QTLs were identified for conventionally aged seeds (Table 2; Fig. 4). Among these, two QTLs on LG4 (Slg4.1 and Slg4.2) together were responsible for 30.4% of the variance in viability in the 2006 control RIL population (Table 2). High-resolution mapping conducted on chromosome 4 using a more densely populated genetic map to refine the QTL interval confirmed two QTLs on LG4 (Fig. 4A; Table 3). (Note that cM locations are not directly comparable between the two maps.)
Table 2.
Seed longevity-associated QTLs of seeds stored under conventional and CD conditions—low-resolution mapping
| Type of seeds | Year | Type of ageing | LG | QTL | Closest marker | 1–LOD interval (cM) | LOD score | R2 | Additive effect | Multilocus analysis (Pr >F) |
| Control | 2002 | Conv | 7 | Slg7.1 | LE3064 | 35–57 | 3.07 | 0.11 | B | 0.016 |
| 2006 | Conv | 1 | Slg1.1 | LE3190 | 38–61 | 2.62 | 0.076 | B | 0.021 | |
| 4 | Slg4.1 | 1A01-129 | 20–46 | 3.79 | 0.112 | A | ||||
| 4 | Slg4.2 | L317 | 61–118 | 5.95 | 0.192 | B | ||||
| 2002 | CD | 2 | SlgCD2.1 | LE7005 | 94–120 | 3.76 | 0.148 | A | ||
| 2006 | CD | 1 | SlgCD1.1 | M1730 | 18–42 | 3.02 | 0.082 | A | <0.0001 | |
| 3 | SlgCD3.1 | E35/M49-F-207 | 9–30 | 3.34 | 0.091 | A | <0.0001 | |||
| 4 | SlgCD4.1 | E45/M49-F-203 | 0–32 | 2.61 | 0.072 | A | 0.004 | |||
| 4 | SlgCD4.2 | LE3042 | 64–87 | 3.90 | 0.11 | B | ||||
| 6 | SlgCD6.1 | E51/M49-F-206 | 0–18 | 3.60 | 0.103 | A | ||||
| Primed | 2006 | Conv | 1 | SlgP1.1 | E54/M48-F-124 | 47–71 | 2.88 | 0.126 | A | 0.001 |
| 4 | SlgP4.1 | LM0052 | 22–45 | 2.72 | 0.123 | A | 0.012 | |||
| 2006 | CD | 1 | SlgPCD1.1 | LE0193 | 42–63 | 2.99 | 0.093 | A | <0.0001 | |
| 2 | SlgPCD2.1 | E35/M59-F-234 | 76–100 | 3.95 | 0.136 | A | ||||
| 3 | SlgPCD3.1 | 1A03-253 | 0–48 | 3.17 | 0.076 | A | ||||
| 6 | SlgPCD6.1 | Contig6010-1 | 0–18 | 4.11 | 0.138 | A | ||||
| 8 | SlgPCD8.1 | Contig15680-2 | 109–134 | 4.29 | 0.127 | B |
Low-resolution mapping was performed across all linkage groups. Seeds of the RIL populations were produced in 2002 (control) and 2006 (control and primed). Multilocus ANOVA results are shown for loci with marginally significant LOD (log of odds) scores. Additive effects were conferred by alleles from A, L. sativa; and B, L. serriola.
CD, controlled deterioration; conv, conventional; LG, linkage group; P, primed; Slg, Seed longevity.
Fig. 4.
Summary of seed longevity-associated QTLs identified by low-resolution mapping across the lettuce genome. Markers associated with the peak and the 1–LOD confidence interval of each QTL are shown, as well as the initial and final marker of each linkage group. Slg, control seeds–conventional storage; SlgCD, control seeds–CD storage; SlgP, primed seeds–conventional storage; SlgPCD, primed seeds–CD storage. Flanking and peak (bold) markers are shown for each QTL.
Table 3.
Seed longevity-associated QTLs of seeds stored under conventional and CD conditions—high-resolution mapping
| Type of seeds | Year | Type of ageing | LG | QTL | Closest marker | 1–LOD interval (cM) | LOD score | R2 | Additive effect | Multilocus analysis (Pr >F) |
| Control | 2006 | Conv | 4 | Slg4.1 | CLS_S3_Contig5097-1 | 20–49 | 6.27 | 0.228 | A | |
| 4 | Slg4.2 | CLSM10124.b1_H11.ab1 | 96–118 | 3.57 | 0.135 | B | ||||
| 2006 | CD | 1 | SlgCD1.2 | QGG9J14.yg.ab1 | 84–116 | 3.76 | 0.174 | A | ||
| 4 | SlgCD4.1 | 1A04-360 | 0–21 | 2.52 | 0.099 | A | 0.039 | |||
| 4 | SlgCD4.2 | CLSS8112.b1_011.ab1 | 74–100 | 2.50 | 0.094 | B | 0.045 | |||
| Primed | 2006 | CD | 1 | SlgPCD1.2 | CLR_S1_Contig138 | 75–88 | 3.10 | 0.131 | A | |
| 4 | SlgPCD4.1 | 1A04-360 | 4–29 | 3.10 | 0.139 | A |
High resolution mapping was conducted on linkage groups 1 and 4. Seeds of the RIL populations were produced in 2002 (control) and 2006 (control and primed). Multilocus analysis results are shown for loci with marginally significant LOD (log of odds) scores. Additive effects were conferred by alleles from A, L. sativa; and B, L. serriola
CD, controlled deterioration; conv, conventional; LG, linkage group; P, primed; Slg, Seed longevity.
The first QTL (Slg4.1) spanned 20–49 cM in the high-resolution map with a LOD score of 6.3 and explained 22.8% of the variance, with the improved viability being conferred by the Salinas parent line (Table 3). The second QTL (Slg4.2) spanned 96–118 cM in the high-resolution map with a LOD score of 3.6 and explained 13.5% of the variance, with the enhanced viability attributed to the UC96US23 allele (Table 3). Together, the two QTLs on LG4 explained 36.3% of the variance of the conventionally aged control seeds of the 2006 RIL population. All of the identified polymorphic loci mapping near these QTLs are shown in Supplementary Table S1 available at JXB online.
For each RIL of the 2006 population, molecular markers positioned close to the peaks of Slg4.1 (CLS_S3_Contig5097) and Slg4.2 (CLSM10124.b1_H11.ab1) were categorized by genotypic class (i.e. either Salinas or UC96US23) and their germination percentages after 21 months of conventional storage were averaged. RILs containing the Salinas and the UC96US23 alleles for the marker associated with Slg4.1 showed mean viabilities of 71.1% and 49.2%, respectively, while for the marker associated with Slg4.2, the mean viabilities were 51.3% and 67.8% (differences significant at P <0.05). These inverse relationships with viability were consistent with the predicted effects of each allele on longevity.
Longevity-associated QTLs of control seeds stored under CD conditions
QTL analyses were also conducted using the control seeds of the 2002 and 2006 RIL populations after storage under CD conditions (i.e. 75% RH and 50 °C for 8 d and 5 d, respectively) (Fig. 1). Different viability-associated QTLs were detected depending upon the year when the RIL population was grown, indicating strong genotype by environment interactions (Table 2). The 2002 RIL population exhibited only one significant QTL (SlgCD2.1) above the 3.2 LOD score threshold. SlgCD2.1 extended between 94 cM and 120 cM with a LOD score of 3.8 and explained 14.8% of the variation in viability. The RIL population produced in 2006 showed five significant viability-related QTLs on chromosomes 1, 3, 4, and 6 that collectively explained 45.8% of the phenotypic variation in viability (Table 2; Fig. 4). Using high-resolution mapping, a distinct QTL was identified on chromosome 1 (84–116 cM), SlgCD1.2, which was responsible for 17.4% of the phenotypic variation in viability, but QTLs on LG3 and LG6 were not confirmed (Table 3). Mapped genes identified within this QTL interval are listed in Suppplementary Table S2 at JXB online. Although viability-associated QTLs were identified by high-resolution mapping on LG4 after both conventional and CD ageing of the 2006 RIL population, overlap of QTLs between the two conditions was minimal (Fig. 5).
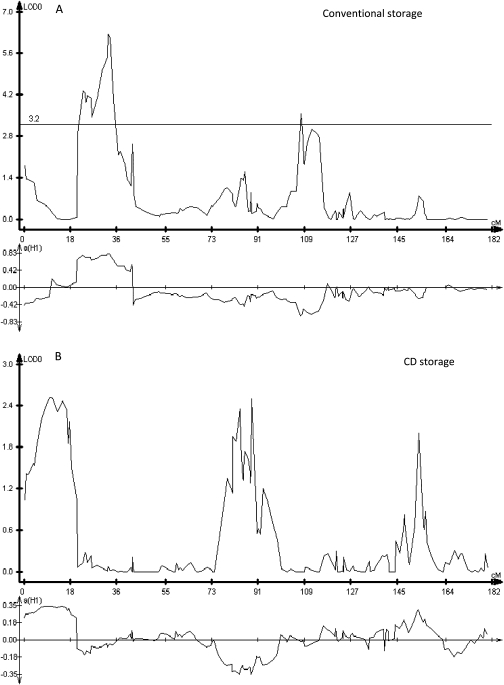
Fig. 5.
High-resolution mapping on linkage group (chromosome) 4 of QTLs associated with increased germination during conventional (A) and CD storage (B) using seeds from a RIL population derived from L. sativa cv. Salinas and L. serriola UC96US23 and produced in 2006. The lower graph in each panel indicates which parent contributed to the trait, with positive values indicating Salinas and negative values indicating UC96US23.
Longevity-associated QTLs of primed seeds
Two viability-associated QTLs were detected for primed seeds conventionally stored for 2 years (Table 2; Fig. 4). SlgP1.1 showed a LOD score of 2.9 and explained 12.6% of the variance in viability. The improved viability was attributed to the Salinas allele. A second viability-associated QTL (SlgP4.1) extended between 22 cM and –45 cM on LG4 with a LOD score of 2.7 and explained 12.3% of the variation in viability. However, these two QTLs were not confirmed using high-resolution mapping (Table 3).
When primed seeds of the 2006 RIL population were stored under CD conditions, five viability-associated QTLs were identified by low-resolution mapping (Table 2; Fig. 4). Two of these QTLs on LG1 and LG4 were significant using high-resolution mapping (Table 3). SlgPCD1.2 overlapped slightly with one detected in CD-stored control seeds (SlgCD1.2). Genes mapping within this chromosomal region are listed in Supplementary Table S2 at JXB online.
Discussion
Genetic markers for improved seed longevity would be useful to allow breeders to incorporate this trait into seeds, as conventional ageing tests are generally too slow to be useful in selection. Early indicators of post-harvest deterioration would also be valuable to enable seed companies to harvest, process, store, ship, market, and replenish seed stock costs effectively. To date, there are no reliable indirect genetic markers for seed longevity, though a number of candidate genes have been reported to influence longevity (Debeaujon et al., 2000; Clerkx et al., 2004b; Sattler et al., 2004; Xu et al., 2004; Bentsink et al., 2006; Prieto-Dapena et al., 2006; Devaiah et al., 2007; Oge et al., 2008; Rajjou et al., 2008; Almoguera et al., 2009). All research on the mechanisms of seed deterioration and longevity is hampered by the lack of convenient assays to assess potential seed longevity. A particular problem is that many seed lots exhibit a lag period of variable length during which viability remains essentially constant, followed by a relatively rapid decline in viability (e.g. Table 1; Tarquis and Bradford, 1992; Walters, 1998). Thus, seeds may appear to have high viability but may be susceptible to rapid deterioration in the near future or when moved to less optimal storage conditions. A recent report of a method to distinguish aged seeds prior to loss of viability therefore represents a potentially valuable development (Kranner et al., 2010). Modelling approaches have also been proposed that can predict potential seed longevity based upon changes in germination rates prior to loss of viability (Bradford et al., 1993).
Although the parent lines used in the present genetic studies were not selected in advance for differences in potential seed longevity, they did differ for this trait (Table 1). The control Salinas seeds that had been conventionally stored since their production in 2002 retained high viability up to July 2009, while UC96US23 seeds produced at the same time and stored in the same conditions were only 32% viable (Table 1). The same trend was observed between recently produced seeds of the parent lines in the CD tests (Table 1), although it is unclear why these seeds exhibited much greater resistance to deterioration, requiring a few months to lose viability while seeds from the RILs lost viability after only a few days using the same CD conditions. In the conventional storage tests, the genetic differences on chromosome 4 associated with longevity were attributed to both Salinas and UC96US23 alleles in the RIL population studied, although overall relatively more Salinas alleles seem to be involved in enhancing lettuce seed longevity in this population (Tables 2, 3).
The low correlations between the results of the conventional and CD storage conditions (Fig. 2) were rather unexpected, since the 75% RH/50 °C CD test is commonly used for assessing vigour and longevity of lettuce seeds (Zhang and McDonald, 1997; Schwember and Bradford, 2005). In the present case, this CD test did not predict the deterioration that occurs under moderate RH and temperature storage conditions. This suggests that different deterioration mechanisms may be involved in CD and conventional storage (Walters, 1998; McDonald, 1999). For example, studies with soybeans found little increase in free radical levels of conventionally aged seeds but a doubling of free radicals in accelerated aged seeds (Priestley and Leopold, 1983; Priestley et al., 1985). Powell and Harman (1985) questioned whether the physiological events occurring under CD conditions reflected those found during conventional storage of pea (Pisum sativum) seeds. Conversely, Likhatchev et al. (1984) concluded that physiological changes in seeds exposed to accelerated ageing conditions were the same as those under conventional storage conditions, with the exception of the rate at which they occur. Longevities of Arabidopsis seeds were also well correlated when seeds from various ecotypes, mutants, and RILs were aged at temperatures ranging between ambient conditions up to 85% RH at 50 °C (Clerkx et al., 2004a, b). In keeping with this, proteome analyses in Arabidopsis showed that the extent of protein carbonylation was strongly increased by both storage conditions but the protein targets of carbonylation were nearly the same (Rajjou et al., 2008). Thus, species-specific processes that are involved in the loss of viability may be independent of (i.e. Arabidopsis) or dependent on (i.e. lettuce, soybean) the environment in which the seeds are stored. While CD tests of lettuce seeds may be useful for rapid comparisons among seed lots, an alternative rapid ageing test is needed for studying the mechanisms responsible for seed deterioration under conditions more closely resembling conventional or commercial storage conditions.
Another striking result was the complete lack of correlation among seeds of the same RIL population produced in different years following either conventional or CD storage (Fig. 3). This absence of correlation between the two types of ageing tests and the general failure to identify the same QTLs when the 2002 and 2006 RIL populations were compared, regardless of the conditions in which the seeds were stored (Tables 2, 3), indicate that lettuce seed longevity exhibits strong genotype by environment interactions. This is consistent with reports that lettuce seed longevity in high MC storage was enhanced by exposure to far-red light during seed development (Contreras et al., 2008, 2009). Thus, seed longevity is influenced by the environment during seed formation, harvest, and storage (Ellis et al., 1993), contributing to the strong genotype by environment interactions. However, multiple longevity-associated QTLs identified in specific genetic×environment combinations in this study can be the basis for more targeted genetic and physiological studies in the future.
A major QTL on chromosome 4 termed Slg4.1 was responsible for almost 23% of the phenotypic variation in viability of the conventionally stored control seeds of the 2006 RIL population, and the improved longevity was conferred by the Salinas allele (Table 3; Fig. 5A). The maximum LOD score at 32.5 cM coincided with CLS_S3_Contig5097, which has high sequence homology with an Arabidopsis gene AtMAT3 (accession no. NM_129243) encoding a protein with methionine adenosyltransferase activity (Supplementary Table S1 at JXB online). A nearby sequence mapping at 36.4 cM has sequence homology to eukaryotic initiation factor 5 (eIF-5) (Supplementary Table S1), variants of which in Arabidopsis may be involved in regulating a switch between cell division and cell death (Thompson et al., 2004). Another sequence (CLS_S3_Contig10071) that mapped within this QTL interval has strong homology with an Arabidopsis gene encoding a heat shock transcription factor (AtHSF4, accession no. NM_119862) (Supplementary Table S1). Small HSPs contribute to processes that have been associated with seed longevity, such as tolerance of embryo desiccation, membrane stabilization, and oxidative stress resistance (Wehmeyer and Vierling, 2000; Scharf et al., 2001). Post-priming treatments using heat shock enhanced the expression of an HSP that encodes a chaperone protein (i.e. 78 kDa binding protein or BiP), which may have contributed to improved longevity of primed tomato seeds following post-priming treatments (Gurusinghe et al., 2002). Overexpression of a specific heat stress transcription factor (HaHSFA9) in transgenic tobacco plants increased the accumulation of HSPs and significantly improved resistance of seeds to deterioration (Prieto-Dapena et al., 2006; Almoguera et al., 2009). Interestingly, Slg4.1 and SlgPCD4.1 overlapped in the region containing the putative heat shock transcription factor (23.9 cM) (Table 3; Supplementary Table S1). While it is not possible at this time to link Slg4.1 directly with HSPs, these results suggest the potential value of testing the association of HSPs with seed longevity in lettuce seeds.
A sequence encoding a DNA replication factor complex-related protein (Contig7346-1) mapped at the centre of the QTL peak for Slg4.2 (108.4 cM) (Supplementary Table S1 at JXB online). A gene mapping nearby at 105.5 cM (QGD6M21.yg.ab1) has sequence homology to an ABC transporter family protein (Supplementary Table S1); a member of this family in Arabidopsis (COMATOSE) is involved in the transition from dormancy to germination (Footitt et al., 2002, 2006). A sequence encoding a LOX (QGD7P09.yg.ab1) also mapped within the Slg4.2 confidence interval at 114.4 cM (Supplementary Table S1). Li et al. (2007) found that cultivars of maize (Zea mays) with low or no LOX activity had longer storage life when the seeds were stored under CD conditions. However, this was not the case in soybean (Glycine max), where longevity under CD conditions was unaffected in LOX mutants, and the loss of one or two of the three LOX isozymes had no effect on seed deterioration (Trawatha et al., 1995). Further work would be required to determine whether ABC transporters or lipoxygenases contribute to lettuce seed ageing characteristics.
Primed lettuce seeds are more susceptible to ageing under CD conditions than are the control seeds based upon the overall population viability percentages of the 2006 RIL population. The control and the primed seeds of this population showed mean viability percentages of 63.6% and 30.0% after exposure at 75% RH and 50º C for 5 d and 2 d, respectively. However, this large discrepancy in viability is not as evident when conventionally stored control and primed seeds of the 2006 RIL population are compared, as their overall germination percentages were 59.9% (21 months at 30% RH/37 °C) and 50.4% (24 months at 30% RH/37 °C), respectively. These results confirm that primed lettuce seeds are more sensitive to the adverse effects of higher seed MC than are control seeds during storage at elevated temperatures, as was reported previously (Tarquis and Bradford, 1992; Schwember and Bradford, 2005; Hill et al., 2007). QTL analyses revealed potential genetic loci that may have influenced the loss of viability of the primed seeds of the 2006 RIL population (Tables 2, 3). The majority of these were associated with CD storage conditions. It is possible that RILs responded differently to the standard priming treatment with respect to its influence on longevity, contributing to differences in the QTL identified in control and primed seeds. It should also be noted that the longevity of primed seeds was overestimated by using radicle protrusion as the indicator of viability, as primed seeds often exhibited damaged root tips that would not be scored as normal seedlings, but were counted as germinated here as long as radicle protrusion occurred. Thus, the differences between longevity of primed and control seeds are larger from a practical viewpoint than is indicated here. Since loss of normal seedlings and loss of radicle protrusion are completely parallel during storage of both primed and control lettuce seeds (Bradford et al., 1993), genetic analyses will not be affected by the use of radicle emergence rather than normal seedlings as the phenotype.
Conclusions
A lettuce RIL population was utilized to search for QTLs associated with seed longevity under different storage conditions. The longevities of seeds stored in conventional and CD conditions were not correlated for both control and primed seeds. Thus, for lettuce seeds, CD tests at elevated temperature and MC may not be predictive of how seeds would store under conditions more closely resembling commercial storage. It may therefore be necessary to test longevity under more moderate storage conditions, making the development of methods to predict potential storage life well before the loss of viability even more important (Bradford et al., 1993; Kranner et al., 2010). Seed longevity exhibits strong genotype by environment interactions, as in general the same QTLs were not identified when seeds of the RIL population produced in 2002 and 2006 were compared, regardless of the conditions in which the seeds were stored. This supports the general concept that seed physiological properties (e.g. dormancy, vigour, and longevity) are strongly influenced by the environment during seed development (Donohue, 2009). Despite this complexity, regions on chromosomes 1 and 4 containing overlapping QTLs may highlight genomic regions containing loci affecting seed deterioration (Fig. 4). Further work identifying the genes associated with these QTLs and developing molecular markers for them may enable breeding for improved seed longevity or assist in elucidating mechanisms responsible for seed deterioration during storage. It will also be of interest to identify loci associated with the environmental sensitivity of potential seed longevity.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Candidate genes mapped near two longevity-associated QTLs on chromosome 4.
Table S2. Candidate genes mapped near two longevity-associated QTLs on chromosome 1.
Supplementary Material
Acknowledgments
This work was supported by the Western Regional Seed Physiology Research Group and the US National Science Foundation (grants 0421630 and 0820451) through the Compositae Genome Project (http://compgenomics.ucdavis.edu). Richard Michelmore provided the RIL population, Allen Van Deynze and Richard Michelmore provided the high-density genetic map, Hamid Ashrafi and Maria Truco assisted with the QTL analyses, Jason Argyris produced the seeds of the RIL population in 2002, and Anthony Joudi assisted with the germination tests.
Glossary
Abbreviations
- ABA
abscisic acid
- AFLP
amplified fragment length polymorphism
- CD
controlled deterioration
- EST
expressed sequence tag
- HSP
heat shock protein
- LOX
lipoxygenase
- MC
seed moisture content (dry weight basis)
- LOD
log of the odds or log likelihood ratio
- QTL(s)
quantitative trait locus (loci)
- RH
relative humidity
- RIL
recombinant inbred line(s)
- SPP
single-position polymorphism
- SSR
simple sequence repeat
References
- Almoguera C, Prieto-Dapena P, Diaz-Martin J, Espinosa J, Carranco R, Jordano J. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biology. 2009;9:75. doi: 10.1186/1471-2229-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Hayashi E, Still DW, Bradford KJ. Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiology. 2008a;148:926–947. doi: 10.1104/pp.108.125807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J, Dahal P, Truco MJ, Ochoa O, Still DW, Michelmore RW, Bradford KJ. Genetic analysis of lettuce seed thermoinhibition. Acta Horticulturae. 2008b;782:23–33. [Google Scholar]
- Argyris J, Truco MJ, Ochoa O, Knapp SJ, Still DW, Lenssen GM, Schut JW, Michelmore RW, Bradford KJ. Quantitative trait loci associated with seed and seedling traits in Lactuca. Theoretical and Applied Genetics. 2005;111:1365–1376. doi: 10.1007/s00122-005-0066-4. [DOI] [PubMed] [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F. From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. Comptes Rendus Biologies. 2008;331:806–814. doi: 10.1016/j.crvi.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiology. 2000;124:1595–1604. doi: 10.1104/pp.124.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M, Bewley JD, Halmer P. The encyclopedia of seeds: science, technology and uses. Wallingford, UK: CAB International; 2006. [Google Scholar]
- Bradford KJ, Tarquis AM, Duran JM. A population-based threshold model describing the relationship between germination rates and seed deterioration. Journal of Experimental Botany. 1993;44:1225–1234. [Google Scholar]
- Butler LH, Hay FR, Ellis RH, Smith RD, Murray TB. Priming and re-drying improve the survival of mature seeds of Digitalis purpurea during storage. Annals of Botany. 2009;103:785–794. doi: 10.1093/aob/mcp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantliffe DJ, Shuler KD, Guedes AC. Overcoming seed thermodormancy in a heat-sensitive romaine lettuce by seed priming. Hortscience. 1981;16:196–198. [Google Scholar]
- Chojnowski M, Corbineau F, Come D. Physiological and biochemical changes induced in sunflower seeds by osmopriming and subsequent drying, storage and aging. Seed Science Research. 1997;7:323–331. [Google Scholar]
- Clerkx EJM, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M. Genetic differences in seed longevity of various Arabidopsis mutants. Physiologia Plantarum. 2004a;121:448–461. [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijin-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiology. 2004b;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras S, Bennett MA, Metzger JD, Tay D. Maternal light environment during seed development affects lettuce seed weight, germinability, and storability. HortScience. 2008;43:845–852. [Google Scholar]
- Contreras S, Bennett MA, Metzger JD, Tay D, Nerson H. Red to far-red ratio during seed development affects lettuce seed germinability and longevity. HortScience. 2009;44:130–134. [Google Scholar]
- Davies MJ. The oxidative environment and protein damage. Biochimica et Biophysica Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delouche JC, Baskin CC. Accelerated aging techniques for predicting the relative storability of seed lots. Seed Science and Technology. 1973;1:427–452. [Google Scholar]
- Devaiah SP, Pan XQ, Hong YY, Roth M, Welti R, Wang XM. Enhancing seed quality and viability by suppressing phospholipase D in Arabidopsis. The Plant Journal. 2007;50:950–957. doi: 10.1111/j.1365-313X.2007.03103.x. [DOI] [PubMed] [Google Scholar]
- Donohue K. Completing the cycle: maternal effects as the missing link in plant life histories. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:1059–1074. doi: 10.1098/rstb.2008.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RH, Hong TD, Jackson MT. Seed production environment, time of harvest, and the potential longevity of seeds of 3 cultivars of rice (Oryza sativa L.) Annals of Botany. 1993;72:583–590. [Google Scholar]
- Ellis RH, Roberts EH. The quantification of aging and survival in orthodox seeds. Seed Science and Technology. 1981;9:373–409. [Google Scholar]
- Footitt S, Marquez J, Schmuths H, Baker A, Theodoulou FL, Holdsworth M. Analysis of the role of COMATOSE and peroxisomal beta-oxidation in the determination of germination potential in Arabidopsis. Journal of Experimental Botany. 2006;57:2805–2814. doi: 10.1093/jxb/erl045. [DOI] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu YS, Larson T, Graham I, Baker A, Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO Journal. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georghiou K, Thanos CA, Passam HC. Osmoconditioning as a means of counteracting the aging of pepper seeds during high-temperature storage. Annals of Botany. 1987;60:279–285. [Google Scholar]
- Gurusinghe S, Powell ALT, Bradford KJ. Enhanced expression of BiP is associated with treatments that extend storage longevity of primed tomato seeds. Journal of the American Society for Horticultural Science. 2002;127:528–534. [Google Scholar]
- Heydecker W, Higgins J, Gulliver RL. Accelerated germination by osmotic seed treatment. Nature. 1973;246:42–44. [Google Scholar]
- Hill HJ, Cunningham JD, Bradford KJ, Taylor AG. Primed lettuce seeds exhibit increased sensitivity to moisture content during controlled deterioration. HortScience. 2007;42:1436–1439. [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends in Plant Science. 2001;6:431–438. doi: 10.1016/s1360-1385(01)02052-0. [DOI] [PubMed] [Google Scholar]
- Kibinza S, Vinel D, Côme D, Bailly C, Corbineau F. Sunflower seed deterioration as related to moisture content during ageing, energy metabolism and active oxygen species scavenging. Physiologia Plantarum. 2006;128:496–506. [Google Scholar]
- Kranner I, Kastberger G, Hartbauer M, Pritchard HW. Noninvasive diagnosis of seed viability using infrared thermography. Proceedings of the National Academy of Sciences, USA. 2010;107:3912–3917. doi: 10.1073/pnas.0914197107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JK, Zhang Y, Yu ZL, Wang YJ, Yang Y, Liu Z, Jiang JY, Song M, Wu YJ. Superior storage stability in low lipoxygenase maize varieties. Journal of Stored Products Research. 2007;43:530–534. [Google Scholar]
- Likhatchev BS, Zelensky GV, Kiashko YG, Shevchenko ZN. Modeling of seed ageing. Seed Science and Technology. 1984;12:385–393. [Google Scholar]
- Maude RB, Drew RLK, Gray D, Bujalski W, Nienow AW. The effect of storage on the germination and seedling abnormalities of leek seeds primed and dried by different methods. Seed Science and Technology. 1994;22:299–311. [Google Scholar]
- McDonald MB. Seed deterioration: physiology, repair and assessment. Seed Science and Technology. 1999;27:177–237. [Google Scholar]
- McDonald MB, Nelson CJ. Physiology of seed deterioration. CSSA Special Publication No. 11. Madison, WI: Crop Science Society of America; 1986. [Google Scholar]
- McDonald MB. Seed priming. In: Black M, Bewley JD, editors. Seed technology and its biological basis. Sheffield, UK: Sheffield Academic Press; 2000. pp. 287–325. [Google Scholar]
- Miura K, Lin SY, Yano M, Nagamine T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2002;104:981–986. doi: 10.1007/s00122-002-0872-x. [DOI] [PubMed] [Google Scholar]
- Oge L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin J-P, Job D, Jullien M, Grappin P. Protein repair l-isoaspartyl methyltransferase1 is involved in both seed longevity and germination vigor in Arabidopsis. The Plant Cell. 2008;20:3022–3037. doi: 10.1105/tpc.108.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AA, Harman GE. Absence of a consistent association of changes in membranal lipids with the aging of pea seeds. Seed Science and Technology. 1985;13:659–667. [Google Scholar]
- Powell AA, Matthews S. Towards the validation of the controlled deterioration vigour test for small seeded vegetables. Seed Testing International. 2005;129:21–24. [Google Scholar]
- Priestley DA, Leopold AC. Lipid changes during natural aging of soybean seeds. Physiologia Plantarum. 1983;59:467–470. [Google Scholar]
- Priestley DA, Werner BG, Leopold AC. The susceptibility of soybean seed lipids to artificially-enhanced atmospheric oxidation. Journal of Experimental Botany. 1985;36:1653–1659. [Google Scholar]
- Prieto-Dapena P, Castano R, Almoguera C, Jordano J. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiology. 2006;142:1102–1112. [Google Scholar]
- Probert RJ, Bogh SV, Smith AJ, Wechsberg GE. The effects of priming on seed longevity in Ranunculus sceleratus L. Seed Science Research. 1991;1:243–249. [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghaz M, Job C, Job D. Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiology. 2008;148:620–641. doi: 10.1104/pp.108.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao NK, Roberts EH, Ellis RH. Loss of viability in lettuce seeds and the accumulation of chromosome damage under different storage conditions. Annals of Botany. 1987;60:85–96. [Google Scholar]
- Sasaki K, Fukuta Y, Sato T. Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breeding. 2005;124:361–366. [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity, and for preventing lipid peroxidation during germination. The Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress and Chaperones. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwember AR, Bradford KJ. Drying rates following priming affect temperature sensitivity of germination and longevity of lettuce seeds. HortScience. 2005;40:778–781. [Google Scholar]
- Schwember AR, Bradford KJ. A genetic locus and gene expression patterns associated with the priming effect on lettuce seed germination at elevated temperatures. Plant Molecular Biology. 2010;73:105–118. doi: 10.1007/s11103-009-9591-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Miller J. Sacred lotus, the long-living fruits of China Antique. Seed Science Research. 2002;12:131–143. [Google Scholar]
- Shin JH, Kim SR, An G. Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiology. 2009;149:905–915. doi: 10.1104/pp.108.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MT, Berjak P. Deteriorative changes associated with the loss of viability of stored desiccation tolerant and -sensitive seeds. In: Kigel J, Galili G, editors. Seed development and germination. New York: Marcel Dekker, Inc; 1995. pp. 701–746. [Google Scholar]
- Sun WQ, Leopold AC. The Maillard reaction and oxidative stress during aging of soybean seeds. Physiologia Plantarum. 1995;94:94–104. [Google Scholar]
- Suzuki Y, Yasui T, Matsukura U, Terao J. Oxidative stability of bran lipids from rice variety [ Oryza sativa (L)] lacking lipoxygenase-3 in seeds. Journal of Agricultural and Food Chemistry. 1996;44:3479–3483. [Google Scholar]
- Tarquis AM, Bradford KJ. Prehydration and priming treatments that advance germination also increase the rate of deterioration of lettuce seeds. Journal of Experimental Botany. 1992;43:307–317. [Google Scholar]
- Tesnier K, Strookman-Donkers HM, Van Pijlen JG, Van der Geest AHM, Bino RJ, Groot SPC. A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Science and Technology. 2002;30:149–165. [Google Scholar]
- Thompson JE, Hopkins MT, Taylor C, Wang T-W. Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends in Plant Science. 2004;9:174–179. doi: 10.1016/j.tplants.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Trawatha SE, Tekrony DM, Hildebrand DF. Soybean lipoxygenase mutants and seed longevity. Crop Science. 1995;35:862–868. [Google Scholar]
- Truco MJ, Antonise R, Lavelle D, et al. A high-density, integrated genetic linkage map of lettuce (Lactuca spp.) Theoretical and Applied Genetics. 2007;115:735–746. doi: 10.1007/s00122-007-0599-9. [DOI] [PubMed] [Google Scholar]
- Valdes VM, Bradford KJ, Mayberry KS. Alleviation of thermodormancy in coated lettuce seeds by seed priming. Hortscience. 1985;20:1112–1114. [Google Scholar]
- van Leeuwen H, Caldwell DG, Stoffel K, Chen F, Kozik A, Truco MJ, Michelmore RW, Van Deynze AE. Plant and animal genome XV. San Diego, CA: Scherago International; 2007. GeneChip for massively parallel genotyping and gene expression analysis. W452. [Google Scholar]
- van Leeuwen H, Stoffel K, Kozik A, et al. Plant and animal genome XVII. San Diego, CA; 2009. High-density mapping of the lettuce genome with SFP markers in over 15,000 unigenes. [Google Scholar]
- Walters C. Understanding the mechanisms and kinetics of seed aging. Seed Science Research. 1998;8:223–244. [Google Scholar]
- Wehmeyer N, Vierling E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiology. 2000;122:1099–1108. doi: 10.1104/pp.122.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu QL, Belcastro MP, Villa ST, Dinkins RD, Clarke SG, Downie AB. A second protein l-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiology. 2004;136:2652–2664. doi: 10.1104/pp.104.046094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhang S, Yao Q, Peng R, Xiong A, Li X, Zhu W, Zhu Y, Zha D. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.) Euphytica. 2008;164:739–744. [Google Scholar]
- Zhang JH, McDonald MB. The saturated salt accelerated aging test for small-seeded crops. Seed Science and Technology. 1997;25:123–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.