Abstract
Cellulose synthases form rosette terminal complexes in the plasma membranes of Streptophyta and various linear terminal complexes in other taxa. The sequence of a putative CESA from Griffithsia monilis (Rhodophyta, Floridiophyceae) was deduced using a cloning strategy involving degenerate primers, a cDNA library screen, and 5′ and 3′ rapid amplification of cDNA ends (RACE). RACE identified two alternative transcriptional starts and four alternative polyadenylation sites. The first translation start codon provided an open reading frame of 2610 bp encoding 870 amino acids and was PCR amplified without introns from genomic DNA. Southern hybridization indicated one strongly hybridizing gene with possible weakly related genes or pseudogenes. Amino acid sequence analysis identified a family 48 carbohydrate-binding module (CBM) upstream of the protein's first predicted transmembrane domain. There are broad similarities in predicted 3D structures of the family 48 modules from CESA, from several glycogen- and starch-binding enzymes, and from protein kinases, but there are substitutions at some residues thought to be involved in ligand binding. The module in G. monilis CESA will be on the cytoplasmic face of the plasma membrane so that it could potentially bind either low molecular weight ligands or starch which is cytosolic rather than inside membrane-bound plastids in red algae. Possible reasons why red algal CESAs have evolved family 48 modules perhaps as part of a system to regulate cellulose synthase activity in relation to cellular carbohydrate status are briefly discussed.
Keywords: Carbohydrate-binding module, cellulose synthase, CESA, family 48 CBM, Griffithsia monilis, predicted 3D structure, red algae, Rhodophyta, starch binding, sugar sensing
Introduction
Diverse prokaryotic and eukaryotic organisms synthesize cellulose microfibrils, semi-crystalline aggregates of various sizes that contain parallel β-1,4 glucan chains (Tsekos, 1999; Romling, 2002; Saxena and Brown, 2005). The glycosyltransferases involved are designated CESAs and are integral membrane proteins that show the so-called D, D, D, QxxRW signature; that is, their catalytic domain contains three spaced, probably catalytically important, glutamate residues followed by a characteristic five amino acid motif in which residues two and three are not fixed. The regions containing these four components are often referred to as U1, U2, U3, and U4. CESAs have been described from numerous embryophytes, from one streptophyte and one rhodophyte alga, from various non-plant eukaryotes (e.g. oomycetes, slime moulds), and from many prokaryotes.
In the plasma membrane, CESA proteins organize into cellulose-synthesizing complexes visible by freeze etch electron microscopy. The structure of the complexes differs between various taxa. The Streptophyta—consisting of the streptophyte algae (Mesostigmatales, Chlorokybales, Klebsormidiales, Zygnematales, Charales, and Coleochaetales) and the Embryophyta (Becker and Marin, 2009)—have rosette terminal complexes consisting of six clustered membrane particles (Mueller et al., 1976). Rosette terminal complexes are known to contain CESA proteins (Kimura et al., 1999) and disassemble in some CESA mutants (Arioli et al., 1998; Wang et al., 2008). In other taxa, cellulose synthase forms linear terminal complexes (Becker and Marin, 2009) where the particles resolved by electron microscopy occur in rows rather than forming the fixed six-member clusters characterizing Streptophyta. The length of the rows, the number of rows, and other features vary in different taxa and have been linked to variations in microfibril dimensions (Tsekos, 1999).
The red algae show linear terminal complexes with between one and four rows of particles in different species (Tsekos, 1999). The only reported rhodophyte CESA (Roberts and Roberts, 2009) is from Porphyra yezoensis (Rhodophyta, Bangiophyceae). The Rhodophyta have been reported to exist in the fossil record for >1 billion years, with representatives of Bangiophyceae and Florideophyceae recognizable in late pre-Cambrian strata (Saunders and Hommersand, 2004). Molecular clock estimates also point to very ancient origins. Cell walls of Rhodophyta vary in carbohydrate composition, and most recent cell wall studies focus on the economically important matrix polysaccharides such as the carrageenans (Cole and Sheath, 1990). The walls can contain microfibrillar xylans or mannans as well as or instead of microfibrillar cellulose. The cellulose has received little attention since early electron microscopy and X-ray diffraction studies (Myers and Preston, 1959a, b).
This study predicts the features of a CESA protein encoded by Griffithsia monilis Harvey (Rhodophyta; Florideophyceae). The protein has a family 48 carbohydrate-binding module (CBM48) in its N-terminal region which is related in sequence and predicted 3D structure to the CBMs found in some glycogen- and starch-binding proteins and to the CBMs of protein kinases involved in higher plant sugar-sensing networks. Such CBM48s are taxonomically widely distributed (Cantarel et al., 2009) but have never been reported in a CESA from any taxon.
Materials and methods
Culture of G. monilis
The source of the G. monilis isolate and the growth conditions have been described (Whitney et al., 2001). Cells in 1.0 l cultures were rinsed, blotted dry, ground to a fine powder in liquid nitrogen, and stored at –70 °C until required.
Primer design
CESA sequences from non-plant organisms with linear terminal complexes, from the green alga Mesotaenium caldariorum (Roberts et al., 2002) and the red alga P. yezoensis (Roberts and Roberts, 2009), were used for alignment by the Blockmaker program (the first step in the CODEHOP process; Supplementary Table S1 available at JXB online). The CODEHOP program (Rose et al., 1998) found primers of ≤64-fold degeneracy in regions U1–U4 when the program was weighted heavily towards the two P. yezoensis sequences, and P. yezoensis codon usage data were employed. Supplementary Table S2 shows the six degenerate primers and the other primers used.
Preparation of mRNA and cDNA
Small amounts of total RNA were prepared from ground G. monilis tissue using Total RNA Isolation Reagent (Thermo Scientific). (The manufacturers’ instructions were followed with all molecular biology reagents unless stated otherwise.) Alternatively, total RNA and genomic DNA were both extracted from 3–6 g of ground tissue (La Claire and Herrin, 1997) and separated by lithium chloride precipitation. In both cases mRNA was purified from total RNA prior to cDNA synthesis with Dynabeads Oligo dT25 (Dynal). cDNA synthesis was primed with oligo(dT18), with oligo(dT) adaptor primers (for cDNA library construction), or with primers specific to already cloned sequences [for 5′ rapid amplification of cDNA ends (RACE)]. Reverse transcriptase enzyme (MMLV) was from Promega (initial cDNA synthesis), Stratagene (cDNA library construction), or Invitrogen (Superscript III enzyme, for 5′ and 3′ RACE).
Amplifying and cloning of cDNA sequences
Oligo(dT) primer was used with MMLV reverse transcriptase (Promega) to synthesize cDNA from bead-purified mRNA in a total volume of 40 μl. PCR (Platinum Taq High Fidelity, Invitrogen) used 1.2 μl of this reaction mixture in a total volume of 15 μl with 2 mM MgCl2 and primer concentrations of 2 μM (degenerate) or 0.2 μM (pure). PCR used a ‘touchdown’ phase (six cycles each decreasing by 1 °C from 60 °C) followed by 37 cycles annealing at 58–63 °C depending on the primers. Amplified fragments were sequenced after purifying from gels (NucleoSpin Extract II columns, Machery-Nagel) and cloning into pCR2.1 (TA cloning kit, Invitrogen).
Constructing and probing a cDNA library
Bead-purified mRNA was used in a SMART cDNA library kit (Clontech/BD Biosciences), employing an initial PCR amplification step and size fractionating the cDNA on a 10–40% sucrose gradient. Library plaques were probed by standard techniques (Sambrook et al., 1989) after labelling the amplimer obtained from the degenerate U1F and U4R primers with [γ-32P]dCTP. Two hybridizing plaques obtained after plaque purification were excised into the pTriplEx2 plasmid and sequenced.
Amplification by 5′ and 3′ RACE
‘Cap-switching’ 5′ RACE (method adapted from Frohman, 2006) used cDNA synthesized from purified mRNA using a specific primer (PuGmCA1-R2, Supplementary Table S2 at JXB online) with Superscript III reverse transcriptase (Invitrogen) in a volume of 20 μl for 1 h at 55 °C. Then 1 μl of a ‘20×’ addition solution [60 mM MgCl2, 40 mM MnCl2, 20 μM CapFind_A primer (Supplementary Table S2), 2 mg ml−1 nuclease-free bovine serum albumin (BSA) and 1× first strand buffer (Invitrogen)] was added together with a further 0.25 μl of reverse transcriptase, and incubated (10 min 45 °C, 10 min 50 °C, 10 min 70 °C). After cooling, 0.8 μl of RNase H (Invitrogen) was added followed by incubation at 37 °C for 20 min, purification (Machery-Nagel NucleoSpin Extract II), and elution in 20 μl. Subsequent PCR used 1.2 μl of this in a reaction volume of 15 μl with primers CapFind_Prim and PuGmCA1-R2 or PuGmCA1-R1 (Supplementary Table S2). Gel-purified fragments were either used for further PCR with the nested primer or cloned into pCR2.1 for sequencing.
3′ RACE used PolyAfind_A (Supplementary Table S2 at JXB online) to prime cDNA synthesis as described, followed by PCR using the PolyAfind_Prim1 and GmCA1-F2 primers (Supplementary Table S2).
Genomic DNA
Approximately 1.5 g of frozen ground alga was added to 10 ml of fresh GIT buffer [4 M guanidinium isothiocyanate, 0.1 M TRIS pH 8.0, 10 mM EDTA, 2% (w/v) sarcosyl, 5 mM dithiothreitol] and incubated at 60 °C with gentle agitation for 2 h. Caesium chloride (2.1 g) was dissolved in the supernatant after centrifugation (3500 g, 10 min, room temperature) before layering over a 3.0 ml, 5.7 M CsCl cushion in a 14×95 mm polyallomer centrifuge tube and centrifuging (SW40Ti rotor, 20 h, 24 000 rpm, 24 °C). Fractions (0.5 ml) collected from the top were spotted on an ethidium bromide plate, and DNA-containing fractions were pooled, dialysed overnight against TE buffer (10 mM TRIS pH 8.0, 1 mM EDTA), and made up to 2.0 ml with TE.
DNA was further purified by adding 1.5 ml of 2.0 M NaCl, 0.4 ml of 10% (w/v) cetyltrimethylammonium bromide (CTAB), 0.1 ml of 1.0 M TRIS pH 7.5, and 50 μl of 0.5 M EDTA. After gentle mixing at 65 °C for 30 min, 4.0 ml of chloroform was added and gently mixed for 10 min at 55 °C. After centrifugation (12 000 g, 10 min, 20 °C), the upper phase was combined with two volumes of ethanol, centrifuged immediately (12 000 g, 10 min, 25 °C), and the DNA pellet was washed twice with cold 70% ethanol. After 10 min air drying, the pellet was dissolved in 100–200 μl of TE. Successful PCR required one further CTAB treatment, and complete cutting with restriction enzymes required two further CTAB treatments.
Southern hybridization
DNA (6 μg in 50 μl) was digested (50–100 enzyme units, 37 °C, 17 h), precipitated, and dissolved in TE. DNA separated on an agarose gel was capillary transferred to a Hybond N+ membrane (GE Healthcare). The G. monilis CESA open reading frame (ORF; 2.6 kb) was amplified from genomic DNA by PCR (primers GmCA-ORF-F and GmCA-ORF-R; Supplementary Table S2 at JXB online), purified on an agarose gel, and extracted as described. A 30 ng sample with 1 pg of DNA marker ladder was labelled with 5 μl of [γ-32P]dCTP (3000 Ci mmol−1, 10 mCi ml−1, Perkin-Elmer) using a DNA labelling bead (GE Lifesciences). After overnight hybridization (60 °C in ExpressHyb, Clontech) and stringency washes at 58 °C, the membrane was exposed with two intensifying screens (20 h, room temperature).
Protein homology modelling of CBM48s
The protein modelling tools Fugue and Orchestrar provided by the Sybyl suite of programs (SYBYL 8.1, Tripos Inc., St Louis, MO, USA) were used to build 3D protein homology models for the putative glycan-binding domain of G. monilis based on the CESA1 N-terminal protein sequence. Refinement of the resulting protein structure was done using the Amber ff99 forcefield for 1000 steps of Powell optimization.
Nucleotide and protein sequences
The cDNA (and protein) sequence generated in this study has been deposited in GenBank as accession GU563823 (protein ID ADK77974).
Results
Identifying a G. monilis CesA
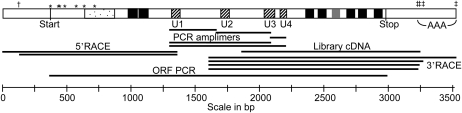
Six degenerate primers (Supplementary Table S2 at JXB online) used in PCR on G. monilis cDNA amplified five products corresponding to the CESA regions U1–U2, U2–U3, U3–U4, U1–U3, and U1–U4 (Fig. 1). A contig of the sequenced clones showed that all five originated from a single cDNA.
Fig. 1.
Summary diagram showing major features of the gene and its protein product (top) with the positions of the various cloned molecules and their origins (below). A sword symbol (†) denotes an alternative transcription initiation site; double crosses (‡) mark alternative polyadenylation sites. The first seven in-frame ATG codons are marked with asterisks (*). Regions of the sequence encoding various protein features are shown: catalytic sites (hatched), the family 48 CBM (stippled), and transmembrane helices (black). One potential transmembrane helix not reaching significance in most prediction programs is shown lightly shaded.
The cloned U1–U4 amplimer was used to probe ∼3×105 recombinant plaques in a cDNA library. Two hybridizing plaques were isolated and recombinant plasmids excised. The sequence of the insert in both plasmids ended at the same nucleotide roughly mid-way between U2 and U3 (Fig. 1) and was identical over 1370 bp until a 1 bp difference in the length of the 3′ poly(A) tract (30 bp or 31 bp). The sequence at the 5′ end matched that of the U1–U4 amplimer over 282 bp.
When further library screening failed to detect longer cDNA clones, two nested reverse primers (PuGmCA1-R2 and PuGmCA1-R1; Supplementary Table S2) were designed from the U1–U2 sequence, and used in 5′ RACE. Two bands of ∼1.3 kb resulted when primers PuGmCA1-R1 and CapFind_Prim were used to PCR amplify from cDNA generated using primers PuGmCA1-R2 and CapFind_A (Supplementary Fig. S1A at JXB online). Ten of 13 clones examined from the combined bands had inserts of 1341 bp and three had 1210 bp inserts. The sequences were identical except for additional upstream sequence in the larger clone. This indicated that two alternative transcriptional initiation sites existed (Fig. 1). The 3′ ends of these RACE fragments had 63 bp that was identical to the U1–U2 sequence.
The 3′ RACE technique was performed on G. monilis cDNA made using the 3′ RACE primer PolyAfind_A, amplifying with the internal forward primer GmCA1-F2 and the primer PolyAfind_Prim1 (Supplementary Table S2). This generated two amplimer bands (Supplementary Fig. S1B), which were cloned together. Sequencing four clones revealed four different polyadenylation sites at nucleotides 3239, 3256, 3311, and 3539 of the cDNA sequence (Fig. 1).
Predicted sequence of a full-length cDNA
The sequence of a full-length cDNA of 3539 bp that was the consensus of all clones identified in Fig. 1 was predicted. It contained a potential ORF of 2610 bp beginning at the first ATG codon. Upstream of this are three in-frame stop codons in the 366 bp 5′ untranslated region (UTR), two of them in the 131 bp region unique to the longer 5′ RACE product. A BLASTN search with this sequence returned many CESAs, the most similar being the P. yezoensis sequence, followed by CESA sequences from species of the oomycete Phytophthora. Similarities between the two red algal nucleotide sequences extended into the 5′ UTRs where the P. yezoensis sequence terminated 286 bp downstream of the G. monilis terminus (Supplementary Fig. S2 at JXB online).
Copy number and lack of introns in the CESA gene
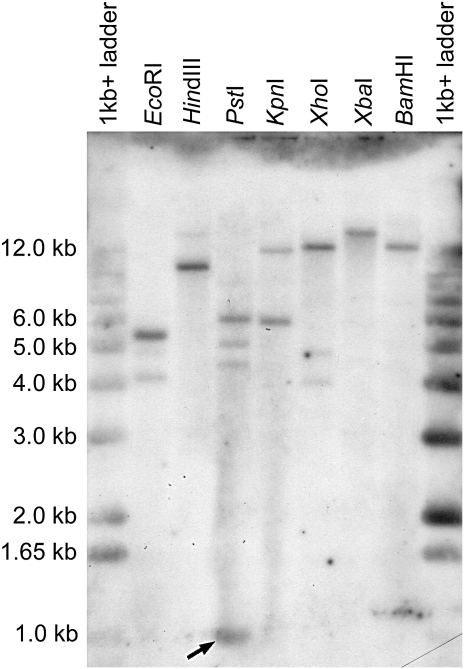
The lack of introns in the coding region of the gene was shown by PCR amplifying from genomic DNA a 2.6 kb sequence containing the entire ORF with no insertions (primers GmCA-ORF-F and GmCA0ORF-R in Supplementary Table S2; ORF PCR in Fig. 1). Southern analysis with this as probe revealed bands consistent with the predicted number and position of cutting sites including an internal 981 bp band after digestion with PstI (Fig. 2). Additional, fainter bands that occurred in some lanes from enzymes having no internal site (HindIII, XhoI, and BamHI) may indicate weakly related genes or pseudogenes.
Fig. 2.
Southern blot analysis of G. monilis genomic DNA digested with the labelled restriction enzymes and separated on a 0.7% agarose gel. The intense band at ∼1 kb in the PstI digest (arrowed) is predicted by the cDNA sequence. Numbers of sites in the cDNA sequence are: EcoRI, 1; HindIII, 0; PstI, 3; KpnI, 1; XhoI, 0; XbaI, 0; BamHI, 0.
Features of the CESA protein
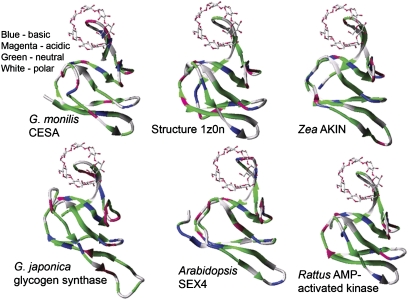
There are seven potential in-frame start codons upstream of the first predicted transmembrane domain in the G. monilis CESA (Fig. 1, and Supplementary Fig. S2 at JXB online) giving a potential protein of up to 870 amino acids. In choosing between possible translational start sites, BLASTP searches were made with conceptual translations beginning from the first potential start codon at the 5′ end. These identified a conserved domain (residues 94–166) belonging to the category exemplified by the glycogen-binding region of the β-subunit of the AMP-activated protein kinase of rats and also seen in some higher plant protein kinases, starch-binding proteins, and in enzymes synthesizing or metabolizing α-1,4 glucans. The latter include a putative glycogen synthase encoded by a G. japonica expressed sequence tag (Fig. 3). Such CBMs show wide taxonomic distribution in prokaryotes and eukaryotes and are assigned to family 48 in the classification of CBMs at CAZy (www.cazy.org; Cantarel et al., 2008; Christiansen et al., 2009). The overall sequence conservation shown by the full set of CBM48s is not high, but the G. monilis sequence shows many of the conserved residues highlighted by Christiansen et al. (2009; their Fig. 1). Sequences of CBM48s are quite distinct from those of known cellulose-binding modules, making it very unlikely that the CESA CBM48 binds a nascent cellulose chain (a β-1,4 glucan) before it exits the cytoplasm. Such a CBM48 is also present in the P. yezoensis CESA if it is translated from one of the potential start codons upstream of the one chosen by Roberts and Roberts (2009) that lies within the CBM48 (Fig. 4).
Fig. 3.
Alignment (ClustalW) of residues 94–166 of the G. monilis CESA sequence with four of the CBM48 sequences listed at CAZy and in Table 1 of Christiansen et al. (2009). Asterisks above the sequences denote residues identified by Polekhina et al. (2005) as important for binding the model substrate cyclodextrin. The sequences are: AMP-activated protein kinase of Rattus norvegicus, GenBank AAH62008; AKIN-β-γ-1 protein of Zea mays, AF276085; starch excess 4 protein (SEX4) of Arabidopsis thaliana, AAN28817; and glycogen synthase of G. japonica, AAM93999. Amino acids are classified as acidic, basic, neutral, and polar, and conservative substitutions are denoted by lighter shading.
Fig. 4.
Alignment of the G. monilis CESA1 and P. yezoensis CESA1 N-terminal protein sequences when their respective nucleotide sequences (GenBank 1312137 and EU279853.1) are translated from their first in-frame start codons. Identical residues are darkly shaded, and similar residues are lightly shaded. Residues in the family 48 carbohydrate-binding module are boxed; the early residues in the first transmembrane helix are enclosed with a dashed line. Methionine residues are in bold, and an asterisk marks the methionine at the start of the P. yezoensis CESA1 protein as annotated by Roberts and Roberts (2009).
Features of the protein homology model for the putative CBM48
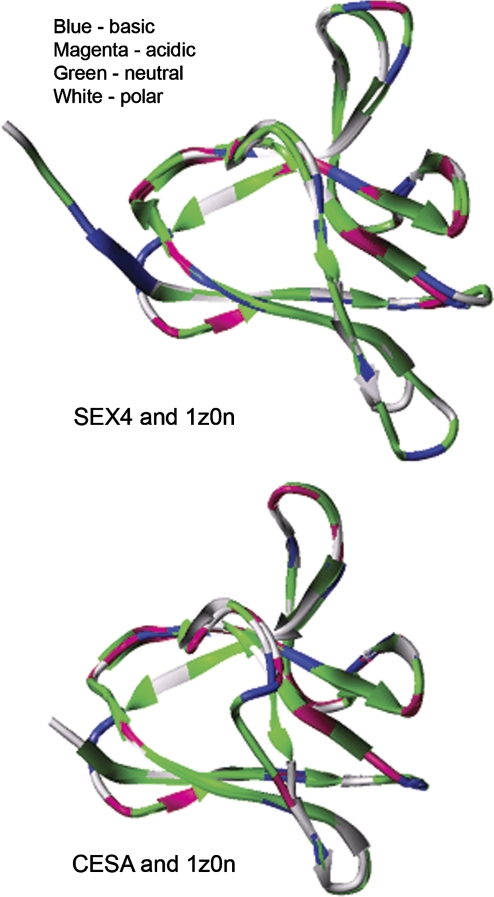
A homology model of the G. monilis CESA CBM48 could be constructed based on the X-ray structure of the glycogen-binding domain of the AMP-activated protein kinase from rat (pdb code: 1Z0N, normalized Z-score: 19.32) (Fig. 5). Some of the other protein sequences classified as CBM48s by Christiansen et al. (2009) (Zea AKIN1, G. japonica putative glycogen synthase, Arabidopsis SEX4) also gave similar predicted structures (Fig. 5). Some structural similarity (Z-score: 8.89) to the CBM20 family that contains other starch-binding domains (Christiansen et al., 2009) did not lead to satisfactory models. Superposition of the overall 3D structures, the cyclodextrin-binding motif of the model, and the X-ray structure showed an excellent steric fit (Figs 5, 6). However, despite the good overall similarity between protein and homology models, the physicochemical properties of some potentially critical amino acids in the G. monilis CESA homology model and, to a lesser degree, in the SEX4 starch-binding protein, differed markedly (Figs 3, 6) from those present in the rat AMP-activated kinase. They included some suggested to be directly involved in binding (Polekhina et al., 2005).
Fig. 5.
Predicted structures for the five sequences presented in Fig. 3 and the structure of the rat AMP-activated kinase as determined experimentally in a complex with the model substrate cyclodextrin (structure 1z0n). Their structures show many similarities, but the limited conservation in amino acid sequence seen in Fig. 3 is shown here spatially by using colour coding to classify the amino acids as acidic, basic, neutral, and polar.
Fig. 6.
Superposition of the rat kinase structure (1z0n) with the Arabidopsis SEX4 model and with the G. monilis CESA model emphasizes the high structural similarity and various non-conservative amino acid substitutions.
The remaining 11 sequences listed as CBM48s by Christiansen et al. (2009; see their Fig. 1) had significant amino acid insertions compared with those shown here to give a good overall fit with the structure of the rat protein kinase. Their predicted structures did not closely resemble structure 1Z0N but resembled other experimentally determined structures—the N-terminal domain of Escherichia coli branching enzyme (structure 1M7X); Sulfolobus solfataricus glycosyltrehalose trehalohydrolase (1EH9); Klebsiella pneumoniae pullulanase (2FHF); Pseudomonas isoamylase (1BF2); and Bacillus acidopullulyticus pullulanase (2WAN) (data not shown).
Discussion
The CESA gene in G. monilis encodes a family 48 CBM in the N-terminal domain of the predicted protein. Such modules have been found only in enzymes that metabolize polysaccharides containing α-1,4 glucans (starch, glycogen, or pullulan) or in protein kinases that are part of sugar-sensing networks (see listing at the CAZy database). Specifically, they have not been found in an enzyme such as cellulose synthase that makes β-1,4 glucans, raising many questions about evolution and function.
General features of the G. monilis CESA gene and protein
The use of 5′ and 3′ RACE generated extensive UTR sequence and demonstrated two alternative transcription start sites and at least four alternative polyadenylation sites. The three in-frame stop codons in the 5′ UTR showed that the upstream limit of translation was reached. The nucleotide sequence shows high similarity to the CESA gene from P. yezoensis (Rhodophyta, Bangiophyceae; Roberts and Roberts, 2009; Supplementary Fig. S2 at JXB online). Both CESA-coding regions lacked introns, as did almost all genes in the Cyanidioschyzon merolae genome (Rhodophyta, Bangiophyceae) (Matsuzaki et al., 2004). Southern analysis of G. monilis suggests a single copy with perhaps some pseudogenes or weakly related genes, although no evidence for other genes emerged when using the degenerate primers or library screening. Roberts and Roberts (2009) also reported a single CESA and documented two pseudogenes in P. yezoensis, a contrast with the multiple CESAs invariably found in higher plants. A requirement for several different CESAs to make a functional cellulose synthase may be a feature of organisms with rosette terminal complexes (Somerville, 2006) that many, perhaps all, of the diverse organisms having linear terminal complexes lack. Since higher plants also employ different CESAs to make primary and secondary wall celluloses (Somerville, 2006), the absence of secondary walls in most Rhodophyta (Martone et al., 2009) removes a further driver for genomes to encode multiple CESAs.
Translation initiation
BLASTP searches with conceptual translations beginning from the first of seven potential start codons in G. monilis showed a CBM48 between residues 94 and 166 that would be part of the protein if translation began from any of the first six ATGs. The improbability of a nucleotide sequence capable of encoding this CBM being conserved in evolution if it were part of the 5′ UTR allowed the seventh ATG to be excluded as the translation start site (Fig. 1). Further support for the importance of the CBM comes from re-examining the P. yezoensis nucleotide sequence (EU279853.1) since the CESA protein it encodes would also contain a complete family 48 CBM if translation began at one of three start codons upstream of the one that Roberts and Roberts (2009) chose to start the ORF (Fig. 4). Their choice was based on nucleotide context around the fourth ATG, although a recent genomic analysis of C. merolae (Rhodophyta, Bangiophyceae) found comparatively little bias in the identities of nucleotides immediately up- and downstream of start codons (Nakagawa et al., 2008). Using the highest contextual biases observed in C. merolae (at the minus 1 and minus 3 positions; Nakagawa et al., 2008), the most favoured ATG codons in the G. monilis sequence are numbers one, four and five, and number one in the P. yezoensis sequence. All would produce a protein with the complete CBM. The weaker similarity between the potential amino acid sequences on the N-terminal side of the CBM in the two red algae than in the CBM (Fig. 4) and in the catalytic domain (Supplementary Fig. S3 at JXB online), and the lack of resemblance to N-terminal sequences in CESAs from other taxa provide no further clues to choose between the other ATG codons. For reasons of simplicity, the first ATG in both proteins has been favoured here, but it is recognized that protein sequencing will be required to settle the matter definitively.
Family 48 CBMs occur in diverse taxa but in a limited range of enzymes
The CAZy database lists >2000 examples of family 48 CBMs drawn from Archaea (38), Bacteria (1921), and Eukaryota (307). The majority occur in enzymes that metabolize polysaccharides with α-1,4-linked glucose residues such as starch, glycogen, or pullulan, but CBM48s also occur in many eukaryotic protein kinases. Rhodophyte CESAs seem to be the first example of an enzyme that contains such a module that is not a protein kinase or involved in metabolizing α-1,4 glucans.
Comparing the sequence and predicted structures of family 48 CBMs in different proteins
The extent to which the CBM48 that occurs in rhodophyte CESAs resembles other CBM48s in amino acid sequence and predicted 3D structure was explored. Table 1 of Christiansen et al. (2009) compares the sequences of members of the evolutionarily related CBM families 20, 21, 48, and 53. There is obvious sequence heterogeneity (most obviously seen in some substantial inserts) within the 16 aligned CBM48s that comprise regulatory subunits of protein kinases and various enzymes metabolizing α-1,4 glucans. According to the present homology models, the CBM48 of the G. monilis CESA closely resembles the first five members of the CBM48 family listed by Christiansen et al. (2009). These comprise three protein kinases (the β-subunit of the AMP-activated protein kinase of Rattus norvegicus, AKIN-β-γ-1 of Zea mays, and the β1 regulatory subunit of the SNF-related kinase from Oryza sativa) and two glucan-metabolizing enzymes (SEX4, the starch excess 4 protein of Arabidopsis thaliana, and the putative glycogen synthase from G. japonica). All have the glycogen-binding domain of the β-subunit of the AMP-activated protein kinase of rat as their closest 3D homologue (structure 1Z0N).
Although the structural models support the view that the CESA CBM48 is similar to several other CBM48s, there are non-conservative substitutions in several of the residues that Polekhina et al. (2005) suggest are involved in carbohydrate binding in the glycogen-binding domain of the rat AMP-activated protein kinase (pdb code: 1Z0N) (see their Fig. 6). Some of the mainly hydrophobic and slightly polar residues of the cyclodextrin-binding motif in 1Z0N are mutated to acidic and basic amino acids in the G. monilis CESA as judged by superposition of models (Fig. 6) or by sequence alignments (Fig. 3). Less extensive substitutions of those key residues occurs in other CBM48s. For example, both tryptophan residues (W100 and W133 in the rat kinase) are fully conserved in the other sequences, whereas CESA has a conservative valine substitution at one but replaces the other with an acidic glutamate. Such substitutions raise doubts about the ability of the CESA CBM48 to bind glycans but, partially counteracting these doubts, the Arabidopsis SEX4 protein binds starch (Kerk et al., 2006) in spite of some non-conservative substitutions (Figs 3, 6). An experimental demonstration that the CESA CBM48 shows glycan-binding activity is required to settle the issue of binding. Interestingly, in the context of a CBM48, Rhodophyta have cytosolic rather than plastidic starch (Viola et al., 2001; Deschamps et al., 2008), making it potentially possible for α-1,4-glucan chains to bind to the CESA CBM48 that is predicted to lie on the cytoplasmic face of the plasma membrane.
A unique feature of CESAs in the Rhodophyta
The presence of a CBM48 in CESAs from members of both the Bangiophyceae and Florideophyceae points to strong conservation over an extended period of evolution since representatives of the two families have been distinguished in the fossil record of late pre-Cambrian strata (Saunders and Hommersand, 2004). The sequenced genome (Matsuzaki et al., 2004) of Cyanidioschyzon merolae (Rhodophyta, Bangiophyceae) does not shed further light on whether CESAs and perhaps other wall polysaccharide synthases contain CBM48s in red algae because it is wall-less. Interestingly, however, none of its limited set of protein kinases contains a CBM48, leaving it without some major components of the sugar-sensing pathways that regulate carbohydrate metabolism in many other eukaryotes. If the absence of protein kinases with CBM48s is widespread in the Rhodophyta, sugar sensing through CBM48s may occur on some individual carbohydrate-synthesizing enzymes including CESAs, and perhaps modulate their activity in response to cellular carbohydrate status.
In conclusion, the CESA of G. monilis has been described and it was shown that it (and probably the P. yezoensis CESA) contains a potential CBM48. Although showing a predicted 3D structure similar to that of other CBM48s, substitutions at key residues leave doubts about its glycan-binding ability and thus a potential role in regulating cellulose synthase.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. DNA fragments amplified from G. monilis cDNA by rapid amplification of cDNA ends (RACE) procedures.
Figure S2. Comparison of nucleotide sequences encoding CESAs from G. monilis (GmCESA1) and P. yezoensis (PyCESA1).
Figure S3. Comparison of protein sequences for CESAs from G. monilis and P. yezoensis.
Table S1. CesA proteins used to design degenerate primers for PCR with G. monilis cDNA.
Table S2. Sequences of the six degenerate primers and of other primers used.
Supplementary Material
Acknowledgments
We thank the Australian Research Council for support through the Linkage Program (LP0669276) to REW and TA, and Dr Spencer Whitney for supplying the G. monilis culture and advice on purifying genomic DNA from it.
Glossary
Abbreviations
- CBM
carbohydrate-binding module
- CESA
cellulose synthase A
- ORF
open reading frame
- RACE
rapid amplification of cDNA ends
- UTR
untranslated region
References
- Arioli T, Peng L, Betzner AS, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Annals of Botany. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Research. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen C, Hachem MA, Janecek S, Vikso-Nielsen A, Blennow A, Svensson B. The carbohydrate-binding module family 20—diversity, structure and function. FEBS Journal. 2009;276:5006–5029. doi: 10.1111/j.1742-4658.2009.07221.x. [DOI] [PubMed] [Google Scholar]
- Cole KM, Sheath RG. Biology of the red algae. New York, NY: Cambridge University Press; 1990. [Google Scholar]
- Deschamps P, Haferkamp I, d'Hulst C, Neuhaus HE, Ball SG. The relocation of starch metabolism to chloroplasts: when, why and how. Trends in Plant Science. 2008;13:574–582. doi: 10.1016/j.tplants.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Frohman MA. Cold Spring Harbor protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. Cap-switching RACE. [Google Scholar]
- Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB. A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. The Plant Journal. 2006;46:400–413. doi: 10.1111/j.1365-313X.2006.02704.x. [DOI] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown RM., Jr Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. The Plant Cell. 1999;11:2075–2086. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Claire J, Herrin D. Co-isolation of high quality DNA and RNA from coenocytic green algae. Plant Molecular Biology Reporter. 1997;15:263–272. [Google Scholar]
- Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somervill C, Ralph J. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Current Biology. 2009;19:169–175. doi: 10.1016/j.cub.2008.12.031. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin IT, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Mueller SC, Brown RM, Jr, Scott TK. Cellulosic microfibrils: nascent stages of synthesis in a higher plant cell. Science. 1976;194:949–951. doi: 10.1126/science.194.4268.949. [DOI] [PubMed] [Google Scholar]
- Myers A, Preston RD. Fine structure in the red algae. II. The structure of the cell wall of Rhodymenia palmata. Proceedings of the Royal Society B: Biological Sciences. 1959a;150:447–455. doi: 10.1098/rspb.1959.0033. [DOI] [PubMed] [Google Scholar]
- Myers A, Preston RD. Fine structure in the red algae. III. A general survey of cell-wall structure in the red algae. Proceedings of the Royal Society B: Biological Sciences. 1959b;150:456–459. doi: 10.1098/rspb.1959.0034. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Research. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure. 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Roberts EM, Delmer DP. Cellulose synthase (CesA) genes in the green alga Mesotaenium caldariorum. Eukaryotic Cell. 2002;1:847–855. doi: 10.1128/EC.1.6.847-855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Roberts AW. A cellulose synthase (Cesa) gene from the red alga Porphyra yezoensis (Rhodophyta) Journal of Phycology. 2009;45:203–212. doi: 10.1111/j.1529-8817.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- Romling U. Molecular biology of cellulose production in bacteria. Research in Microbiology. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Research. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Saunders GW, Hommersand MH. Assessing red algal supraordinal diversity and taxonomy in the context of contemporary systematic data. American Journal of Botany. 2004;91:1494–1507. doi: 10.3732/ajb.91.10.1494. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown RM., Jr Cellulose biosynthesis: current views and evolving concepts. Annals of Botany. 2005;96:9–21. doi: 10.1093/aob/mci155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annual Review of Cell and Developmental Biology. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Tsekos I. The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. Journal of Phycology. 1999;35:635–655. [Google Scholar]
- Viola R, Nyval P, Pedersen M. The unique features of starch metabolism in red algae. Proceedings of the Royal Society B: Biological Sciences. 2001;268:1417–1422. doi: 10.1098/rspb.2001.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Elliott JE, Williamson RE. Features of the primary wall CESA complex in wild type and cellulose-deficient mutants of Arabidopsis thaliana. Journal of Experimental Botany. 2008;59:2627–2637. doi: 10.1093/jxb/ern125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. The Plant Journal. 2001;26:535–547. doi: 10.1046/j.1365-313x.2001.01056.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.