Abstract
Background/Aims
Genes associated with the Helicobacter pylori (H. pylori) plasticity region may play a role in the pathogenesis of H. pylori. We compared the genes jhp0940, jhp0947, and jhp0986 in H. pylori isolates from patients with different gastroduodenal diseases and in different age groups.
Methods
The H. pylori hyperplasticity region genes jhp0940, jhp0947, and jhp0986 were studied by PCR. We also evaluated whether these genes were related to the cytotoxin-associated gene (cagA) and histology findings.
Results
Of the patient cohort, 71 (62%) were positive for jhp0940, 67 (59%) for jhp0947, 12 (10%) for jhp0986, and 69 (60%) for cagA. jhp0940 (n=18, 67%) and jhp0947 (n=23, 85%) were found more frequently in duodenal ulcer (DU) patients than in gastritis patients (n=14, 39%; p=0.029 and p<0.001, respectively). Gastric ulcer (GU) was more frequently associated with jhp0940 (17 patients, 77%; p=0.003) than with gastritis (14 patients, 39%). Gastric carcinoma (GC) was more strongly associated with both jhp0940 (22 patients, 76%; p=0.003) and jhp0947 (22 patients, 76%; p=0.003) than was gastritis (14 patients, 39%). jhp0947 was more frequently associated with chronic active inflammation (58 patients, 87%; p=0.009) than with chronic inflammation (9 patients, 13%). Multivariate analysis demonstrated that jhp0947 was associated with DU (odds ratio, 6.1; 95% confidence interval, 1.87-20).
Conclusions
The genes jhp0947 and jhp0940 were identified in H. pylori isolates from patients with GC and DU, while jhp0940 was also isolated from patients with GU. jhp0947 was independently associated with DU.
Keywords: Helicobacter pylori, Hyperplasticity region, Gastric carcinoma, Duodenal ulcer, Gastric ulcer
INTRODUCTION
Helicobacter pylori (H. pylori) is a gram-negative, spiral, microaerophilic bacterium whose infection has a worldwide distribution. Its prevalence varies from 2% in developed countries to more than 90% in developing world.1 H. pylori is the major cause of chronic gastritis and plays an important role in the pathogenesis of peptic ulcer disease, gastric carcinoma (GC), and gastric mucosa-associated lymphoid tissue lymphoma.2,3 However, why only a minority of H. pylori-positive patients develop the severe associated diseases remain unclear. Variation in clinical outcomes has been attributed to differences in environmental factors, bacterial strains, and host genetics. H. pylori virulence marker cagA, vacA, babA, and iceA influence the outcome of the associated diseases that has been evaluated in several studies.4-7 The cytotoxin-associated gene (cagA) present in H. pylori isolates is considered a marker for the presence of the cag pathogenicity island (cag PAI). This region includes a number of other genes associated with an increased virulence and severe clinical outcomes.8,9 Comparison of the genome of two H. pylori strains 26695 and J99 revealed, in addition to the cag PAI, the presence of potential PAIs regions with different G+C content so-called "plasticity region."10 Occhialini et al.11 identified in H. pylori strains isolated from patients with gastric adenocarcinoma and in patients with chronic gastritis-associated dyspepsia two new potential pathogenicity markers in the plasticity region. The jhp0947 was found frequently in strains from patients with gastric cancer (65%) than in strains from those without (35%) while the jhp0940 was detected only in some strains from patients with GC. The third gene jhp0986, which is specific to strain 26695, was detected more often in strains isolated from patients with chronic gastritis-associated dyspepsia (39%) than in strains isolated from cancer patients (12%). The aim of the study was to evaluate the association between the hyperplasticity area genes jhp0940, jhp0947 and jhp0986 genes and H. pylori infection associated gastritis, peptic ulcer disease and GC in different age groups of the patients.
MATERIALS AND METHODS
1. H. pylori strains
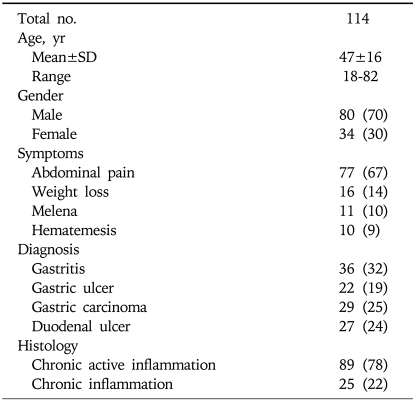
H. pylori strains were isolated from the gastric biopsy of 114 patients who underwent endoscopy for evaluation of symptoms related to the upper gastrointestinal tract with gastritis associated with non-ulcer dyspepsia in 36 (32%), gastric ulcer (GU) in 22 (19%), GC in 29 (25%) and duodenal ulcer (DU) in 27 (24%) (Table 1). They attended the gastroenterology outpatient and endoscopy suite from June 2004 to December 2008. The study was approved by the Ethics Review Committee of the Aga Khan University. All patients gave an informed consent for endoscopy and participation in the study. None of the patients had received antibiotics, acid reducing drugs such as H2-receptor antagonists, acid pump inhibitors, non-steroidal anti-inflammatory drugs or bismuth compounds in the last 4 weeks.
Table 1.
Details of Patients from Which Helicobacter pylori Isolates Were Obtained
Values are presented as number (%).
2. Culture and identification of H. pylori
The specimens were transported immediately in sterile normal saline to isolate H. pylori. Thus, within three hours of collection each specimen was homogenized in sterile eppendorf tubes with electric homogenizer. The resulting suspension was inoculated onto Columbia Blood Agar (Oxoid) medium and Dents supplement (containing vancomycin, trimethoprim, and polymyxin) and incubated at 37℃ under microaerophilic conditions for 4-6 days. Plates were then examined for bacterial growth and typical colonies were selected for identification. The identity of H. pylori was confirmed by Gram stain and by the production of urease and catalase. One half of the homogenate was used for culture, and the other half was kept at -80℃. H. pylori isolates were defined as gram-negative spiral-shaped bacilli that were catalase positive and rapidly (less than 1 hour) urease positive. H. pylori NCTC 11637 (type strain) was used as a positive control for the culture conditions and identification tests.
3. Histological analysis
Formalin-fixed and paraffin-embedded gastric biopsy specimens were routinely processed. All biopsy specimens for histological examination were fixed in 10% formalin, embedded in paraffin wax on the oriented edge, and cut into 5µm thick sequential sections. All tissue sections were stained with hematoxylin and eosin for histological examination. The degree of acute and chronic inflammation, as well as the H. pylori density was scored according to the updated Sydney system.12 The bacterial density was graded from 0 to 3 (0, absent; 1 to 3, from few and isolated bacteria to colonies). The infiltration of gastric mucosa by mononuclear cells and polymorphonuclear leucocytes, atrophy, and intestinal metaplasia were graded as follows: 0, none; 1, mild; 2, moderate; 3, marked. Chronic inflammation was defined according to an increase in lymphocytes and plasma cells in the lamina propria graded into mild, moderate or marked increase in density. Chronic active gastritis indicated chronic inflammation with neutrophilic polymorph infiltration of the lamina propria, pits or surface epithelium graded as 0, nil; mild, <1/3 of pits and surface infiltrated; moderate, 1/3-2/3; and marked, >2/3. Antrum and corpus gastritis were scored by total sum of grade of gastritis (mild, 1; moderate, 2; marked, 3 infiltration with lymphocytes and plasma cells) and activity of gastritis (mild, 1; moderate, 2; marked, 3 infiltration with neutrophilic granulocytes) either in the antrum or in the corpus, a maximum of a sum of 6 points for each individual patient. Atrophy was defined as the loss of inherent glandular tissue, with or without replacement by intestinal-type epithelium. For optimal histological evaluation, all gastric biopsy specimens included surface epithelium and muscularis mucosae. Lymphoid aggregates were defined as accumulations of lymphocytes and plasma cells without a germinal centre.
4. Extraction of genomic DNA
The bacterial cells on chocolate agar plate was washed twice with phosphate buffer saline (PBS, pH 8.0) then centrifuged at 3,000 rpm for 20 minutes. H. pylori DNA was extracted by a phenol/chloroform method similar to the method previously described.13 Briefly, bacterial pellet was resuspended in Tris-Cl buffer containing ethylenediaminetetraacetate (T.E pH 8.0) and lysozyme and was then incubated at 37℃ for 30 minutes. The suspension was treated with sodium dodecyl sulphate (SDS), proteinase K and RNase A. DNA was extracted with phenol/chloroform/isoamyl alcohol, precipitated by sodium acetate and ice-cold absolute alcohol, and washed with ice cold alcohol (70%). The pellet of DNA was finally resuspended in TE buffer. DNA content and purity was determined by measuring the absorbance at 260 nm and 280 nm using a spectrophotometer (Beckman DU-600; Beckman Instruments, Fullerton, CA, USA).
5. Polymerase chain reaction
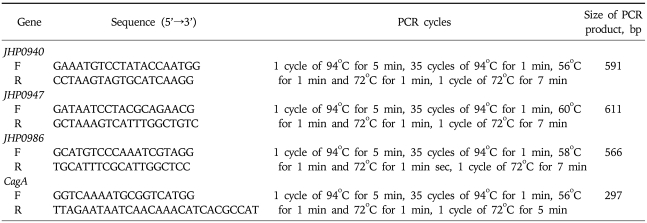
Amplification of jhp0940, jhp0947, jhp0986 and cagA genes by PCR was performed in a volume of 50 µL containing 10 mmol/L Tris-HCl (pH 8.3), 50 mmol KCl, 1.5-2.5 mmol/L MgCl2, 200µmol/L deoxynucleoside triphosphates, 2 units Taq DNA polymerase (Promega) and 25 pmol of both forward and reverse primers (Table 2) used before (synthesized by MWG Automatic synthesizer).11,14 PCR was performed in a Perkin Elmer 9700 thermal cycler. The amplification cycles for different genes are given in Table 1. Positive and negative reagent control reactions were performed with each batch of amplifications. DNA from H. pylori strains ATCC 43504 (cagA positive), ATCC 51932 (cagA negative) was used to define the accuracy of the cagA. After PCR, the amplified PCR products were electrophoresed in 2% agarose gels containing 0.5% x Tris/acetate/ethylenediaminetetraacetic acid, stained with ethidium bromide, and visualized under a short wavelength ultraviolet light source.
Table 2.
Oligonucleotide Primers Used for PCR
6. Statistical assessment
The statistical package for social science SPSS (Release 16, standard version; SPSS Inc., Chicago, IL, USA) was used for data analysis. The descriptive analysis was done for demographic and clinical features. Results were presented as mean±standard deviation for quantitative variables and number (percentage) for qualitative variables. Differences in proportion were assessed by using Pearson chi square, Fisher exact or likelihood ratio test where appropriate. Variables such as gender, age, jhp0947, jhp0940, jhp0986 and cagA status (negative or positive), and the intensity and activity of gastritis (scored as defined above) were evaluated. Data were examined by logistic regression analysis for association of jhp0947, jhp0940 and jhp0986 gene status (the dependent variable) with disease, e.g., DU, GU, etc with adjustments made for potential confounding factors such as age group, cagA, chronic gastritis. A p value of ≤0.20 in the univariate analysis was used as a significant level for inclusion in the full model. In the multivariate analysis, a p value of ≤0.05 was considered significant. The odds ratios (OR) of significant covariates, as well as their 95% confidence intervals (CI), were determined.
RESULTS
1. Distribution of the hyperplasticity region genes
Seventy-one (62%) of the H. pylori isolates were positive for jhp0940, 67 (59%) for jhp0947 and 12 (10%) for jhp0986. The jhp0986 was identified in 12 (100%) (p=0.017) of the H. pylori strains isolated from only male patients.
2. Hyperplasticity region genes and diseases
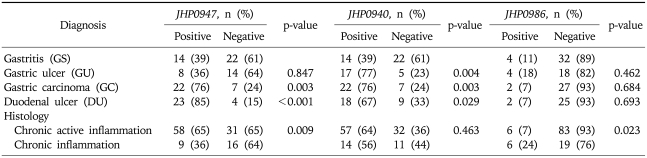
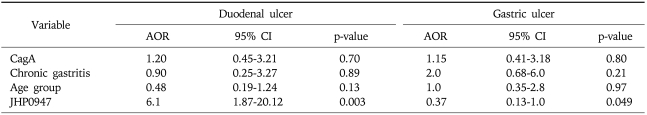
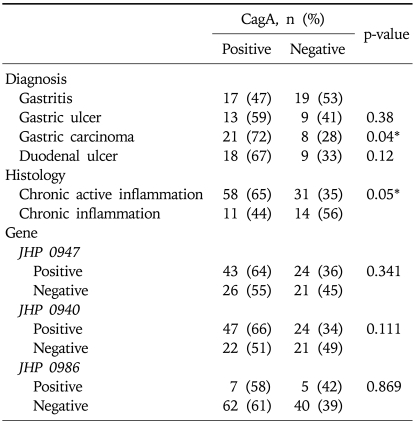
Jhp0947 gene was found in 23 (85%) (p<0.001) of the H. pylori strains isolated from patients with DU and 22 (76%) (p=0.003) with GC compared to 14 (39%) with NUD gastritis (Table 3). Jhp0940 was present in 18 (67%) (p=0.029) of the H. pylori strains isolated from DU, 17 (77%) (p=0.004) with GU and 22 (76%) (p=0.003) with GC compared to 14 (39%) with NUD gastritis (Table 3). There was a low frequency of jhp0986 gene in H. pylori isolates. It was positive in 4 (11%) isolates from NUD gastritis, 4 (18%) with GU, 2 (7%) with GC and 2 (7%) with DU (Table 3). Multivariate analysis demonstrated jhp0947 was associated with DU (OR, 6.1; 95% CI, 1.87-20.12) while jhp0940 was associated with GU (OR, 2.8; 95% CI, 0.92-9) (Table 4).
Table 3.
Association of Hyperplasticity Region Genes with Diseases
GU, GC and DU were compared against GS. Differences in proportion were assessed by using Pearson chi square, Fisher exact or likelihood ratio test where appropriate. A p-value <0.05 was considered significant.
Table 4.
Factors Associated with Duodenal and Gastric Ulcer in the Multivariate Analysis
A p value of ≤0.20 in the univariate analysis was used as a significant level for inclusion in the full model. In the multivariate analysis, a p value of ≤0.05 was considered significant. The adjusted odds ratios (AOR) of significant covariates, as well as their 95% confidence intervals (95% CI), were determined.
3. Association of cagA and hyperplasticity region genes with diseases
CagA was positive in 69 (60%) isolates. It was significantly associated with GC in 21 (72%) (p=0.04) compared to 17 (47%) from gastritis (Table 5). In cagA positive strains, jhp0940 gene was associated with GU in 11 (85%) (p=0.010) and GC in 18 (86%) (p=0.002) compared to NUD gastritis 6 (35%) while jhp0947 was associated with GC in 17 (81%) (p=0.011) and DU 16 (89%) (p=0.003) compared to 7 (41%) in NUD gastritis.
Table 5.
Association between cagA and Hyperplasticity Region Genes and Diseases
*p-value <0.05 was considered significant.
4. Association of cagA and hyperplasticity area genes with histology
H. pylori isolates were associated with chronic active inflammation in 89 (78%) and with chronic inflammation in 25 (22%) patients (Table 1). Jhp0947 was significantly associated with chronic active inflammation in 58 (65%) (p=0.009) compared to chronic inflammation in 9 (36%) (Table 3). Jhp0986 was significantly associated with chronic inflammation in 6 (24%) (p=0.023) compared to chronic active inflammation in 6 (7%) (Table 3). Jhp0940 was associated with both chronic active inflammation 57 (64%) and chronic inflammation in 14 (56%) (p=0.463) (Table 3). However, in the presence of cagA, jhp0947 was not significantly associated with chronic active inflammation in 39 (67%) (p=0.087) and neither jhp0986 3 (27%) (p=0.075) with chronic inflammation.
DISCUSSION
The study showed that the hyperplasticity region genes jhp0947 and jhp0940 were present in 60% of H. pylori isolates while jhp0986 was infrequently demonstrated. The jhp0940 was demonstrated in strains isolated from patients in comparatively older age group though they did not show any gender distribution. The H. pylori strains with jhp0947 and jhp0940 were significantly associated with GC and DU while jhp0940 was also associated with GU (Table 3). In these H. pylori strains, cagA was only significantly associated with GC but not with DU and GU (Table 5). However, when these cagA positive strains were positive for jhp0947 it was significantly associated with both GC and DU while jhp0940 with GC and GU. H. pylori strains with jhp0947 were significantly associated with chronic active inflammation in 65% (Table 3). However, histological inflammation was not augmented when they were concurrently cagA status positive.
In a previous study, Occhialini et al.11 found that the jhp0947 gene was present more frequently in strains isolated from patients with GC (64.7%; 11 of 17) than in those isolated from patients with gastritis alone (34.6%; 9 of 26). Also, Santos et al.15 demonstrated that jhp0947 gene was present in Brazilian strains from patients with GC and DU and appeared to be implicated in the development of both DU and GC. In our study, there was an independent association between the presence of jhp0947-positive H. pylori strains and DU. This is consistent with the previous studies.11,15 However, in contrast we also noted that there was an association not only between the jhp0940 gene and GC and DU but also between jhp940 and GU in our H. pylori strains.11,15 The jhp0986 gene was demonstrated in only few of our H. pylori isolates. As, we did not use different modality such as hybridization to confirm this finding, failure with PCR does not necessarily mean that jhp0986 gene was absent in our H. pylori isolates. In this study, we have demonstrated an association between the presence of the jhp0940 gene and GU. The jhp0947 and jhp0940 genes, as well as the cagA gene, which is a marker of the cag PAI, may be implicated in the development of not only DU and GC but also GU in patients with H. pylori infection. This is also the first study in Pakistan that has suggested a relation between positive cagA and jhp0947 or jhp0940 with GC, DU and GU. We noted that the hyperplasticity genes were not associated with the cagA. In the presence of cagA positivity, no increase in severity of histological gastritis was demonstrated. In conclusion, this study demonstrated that the new hyperplasticity region genes jhp0947 and jhp0940 as virulence marker of H. pylori that are associated with GC and peptic ulcer disease. The functional analysis of these genes in a larger number of H. pylori isolates will provide further insight into its role in the virulence of H. pylori and the evolution of the infection.
ACKNOWLEDGEMENTS
This work was done by the research grant to J.Y. from the Aga Khan University. The authors are grateful to the staff of the Juma Research Laboratory for the help provided during the course of this work.
References
- 1.Ahmad MM, Ahmed DS, Rowshon AH, et al. Long-term re-infection rate after Helicobacter pylori eradication in Bangladeshi adults. Digestion. 2007;75:173–176. doi: 10.1159/000107046. [DOI] [PubMed] [Google Scholar]
- 2.Ernst PB, Gold BD. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol. 2000;54:615–640. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 3.Kume K, Hirakoba M, Murata I, Yoshikawa I, Otsuki M. Disappearance of both MALT lymphoma and hyperplastic polyps in the stomach after eradication of Helicobacter pylori. Am J Gastroenterol. 2001;96:2796–2797. doi: 10.1111/j.1572-0241.2001.04143.x. [DOI] [PubMed] [Google Scholar]
- 4.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40:297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SPrinz C, Schöniger M, Rad R, et al. Key importance of the Helicobacter pylori adherence factor blood group antigen binding adhesin during chronic gastric inflammation. Cancer Res. 2001;61:1903–1909. [PubMed] [Google Scholar]
- 6.Chomvarin C, Namwat W, Chaicumpar K, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30–36. doi: 10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 7.van Doorn LJ, Figueiredo C, Sanna R, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 8.Censini S, Lange C, Xiang Z, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Occhialini A, Marais A, Urdaci M, et al. Composition and gene expression of the cag pathogenicity island in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2001;69:1902–1908. doi: 10.1128/IAI.69.3.1902-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 11.Occhialini A, Marais A, Alm R, Garcia F, Sierra R, Mégraud F. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2000;68:6240–6249. doi: 10.1128/iai.68.11.6240-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis The updated Sydney System International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Yakoob J, Hu GL, Fan XG, et al. Diversity of Helicobacter pylori among Chinese persons with H. pylori infection. APMIS. 2000;108:482–486. doi: 10.1034/j.1600-0463.2000.d01-86.x. [DOI] [PubMed] [Google Scholar]
- 14.Covacci A, Rappuoli R. PCR amplification of gene sequences from Helicobacter pylori strains. In: Lee A, Meégraud F, editors. Helicobacter pylori: techniques for clinical diagnosis and basic research. Philadelphia: W.B. Saunders; 1996. pp. 94–109. [Google Scholar]
- 15.Santos A, Queiroz DM, Ménard A, et al. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J Clin Microbiol. 2003;41:1651–1655. doi: 10.1128/JCM.41.4.1651-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]