Abstract
IL-12 is a major activator of tumor-killing NK cells and CTL. IFN-γ mediates most of the well-known immunological activities of IL-12. In this study, we report IFN-γ-independent activities induced by therapeutic application of rIL-12 in restricting tumor growth and metastasis in the 4T1 murine mammary carcinoma model. IFN-γ-deficient mice carrying 4T1 tumor exhibit no gross defect in the number of tumor-infiltrating lymphocytes but have exaggerated angiogenesis in the tumor. Administration of IL-12 is able to constrict blood vessels in the tumor in the absence of IFN-γ, and retains certain therapeutic efficacy even when applied late during tumor progression. IL-12 exposure in vivo does not irreversibly alter the immunogenicity of the tumor. Finally, global gene expression analysis of primary tumors reveals IL-12-induced molecular patterns and changes, implicating a number of novel genes potentially important for IFN-γ-independent immune responses against the tumor, for IL-12-mediated antiproliferation, antimetastasis, and antiangiogenesis activities.
The emergence of the concept of cancer immunosurveillance conceived by Ehrlich (1) at the beginning of 20th century and formalized by Burnet (2) in 1957, the severe challenges it faced in the 1970s and 1980s (3), and its renaissance in the last decade or so brought about by a large body of mouse and human studies in immunodeficient settings have helped solidify the belief that components of the immune system such as lymphocytes (T, NK, NKT) and cytokines such as IFN-γ, perforin, and IL-12 are critically involved in controlling primary tumor development. The refinement of this concept and its extension to immunoediting has been put forward to describe the dual host-protecting and tumor-sculpting actions of the immune system that not only impede but also mark neoplastic alterations for immune elimination (4).
Many nonimmunogenic or poorly immunogenic tumors can evade immune surveillance by down-regulation of costimulatory, adhesion, and MHC molecules, or by production of immunosuppressive factors such as TGF-β, IL-10, and PGE2, etc., which inhibit the functions of T cells and dendritic cells (5, 6). In recent years, immunotherapies have been rekindled that attempt either to mark the tumor by up-regulating the surface Ags for enhanced interaction with immune effector cells (6, 7) or to directly activate dendritic cells, T cells, and NK cells for their heightened scouting capacity and increased cytolytic potency. IL-12 is a factor belonging to the latter class (8). It is a key player in the induction of T cell-dependent and -independent activation of macrophages, NK cells, generation of Th type 1 (Th1) cells and CTL, induction of opsonic, complement-fixing Abs, and resistance to intracellular infections (9). IL-12 has powerful antitumor and antimetastatic activities against many murine tumors as well as human tumors (10). Recent encouraging developments in clinical applications of IL-12 for human T cell lymphoma (11, 12), B cell non-Hodgkin lymphoma (13), melanoma (14 –18), and renal carcinoma (19) strongly underscore the importance of understanding the cellular and molecular mechanisms of IL-12-mediated antitumor responses.
4T1 is a tumor cell line isolated from a single spontaneously arising mammary tumor from a BALB/BfC3H mouse (mouse mammary tumor virus-positive) (20). It is an excellent model system for breast cancer research, because its tumor development is well characterized both oncologically and immunologically. The 4T1 tumor closely mimics human breast cancer in its anatomical site, immunogenicity, growth characteristics, and metastatic properties (21). From the mammary gland, 4T1 tumor spontaneously metastasizes to a variety of target organs including the lung, bone, brain, and liver through primarily a hematogenous route (22). 4T1 is also poorly immunogenic in that immunization with irradiated 4T1 cells provides only slight delays in tumor growth against wild-type tumor, not sufficient to protect the animal (23). Immune suppression in the 4T1 model has been attributed in part to a STAT6-dependent inhibition of the development of tumor-specific CD8+ CTL activity (24). IL-12 administration to 4T1 tumor-bearing mice causes a substantial reduction of spontaneous metastases in the lungs and significantly prolonged their survival time (25). Tumor-draining lymph node cells obtained from 4T1 tumor-bearing mice treated with IL-12 exhibit increased NK activity and enhanced production of IFN-γ, suggesting the involvement of both NK cells and IFN-γ in this effect (25). These properties make this mammary tumor model an excellent system in which to investigate the cellular and molecular mechanisms involved in IL-12-mediated control of the malignant development.
It is well established that the primary immunological activities of IL-12 are mediated through IFN-γ. However, IFN-γ-independent antitumor and antimicrobe activities of IL-12 have been reported in IFN-γ-deficient mice (26–31). Most of these studies in various tumor models have focused on the effects of IL-12 on distant immune cells, not on the tumor cells and immune cells that infiltrate the tumor. To delineate comprehensively the cellular and molecular mechanisms in IL-12-mediated antitumor responses, we examined the genome-wide changes in gene expression induced by IL-12 in vivo directly or indirectly in tumor targets in the presence or absence of an endogenous IFN-γ-regulated response.
Materials and Methods
Mice
Female BALB/c and BALB/c.IFN-γ−/− mice (6–8 wk old) were purchased from The Jackson Laboratory (Bar Harbor, ME). All mice were housed at the Weill Medical College of Cornell University Animal Facilities in accordance with the Principles of Animal Care (National Institutes of Health publication no. 85–23). Mice bearing 4T1 tumors were all sacrificed no later than day 28 due to the morbidity caused by large tumor size and strong metastasis.
Tumor implantation, size measurement, lung metastasis assay
4T1 mammary carcinoma cells (1 × 105) were injected s.c. into the abdominal mammary gland area of recipient mice in 0.1 ml of a single-cell suspension in PBS on day 0. The dose of tumor implantation was empirically determined to give rise to tumors of ~10 mm in diameter in untreated wild-type mice in 28 days. Primary tumors were measured using electronic calipers every other day. Reported measurements are the square root of the product of two perpendicular diameters. Numbers of metastatic cells in lung were determined by the clonogenic assay (21). In brief, lungs were removed from each mouse on day 28, finely minced, and digested in 5 ml of enzyme mixture containing 1× PBS and 1 mg/ml collagenase type IV for 2 h at 37°C on a platform rocker. After incubation, samples were filtered through 70-µm nylon cell strainers and washed twice with PBS. Resulting cells were resuspended, plated, and serially diluted in 10-cm tissue culture dishes in RPMI 1640 medium containing 60 µM thioguanine for clonogenic growth. 6-Thioguanine-resistant tumor cells formed foci within 10–14 days, at which time they were fixed with methanol and stained with 0.03% methylene blue for counting.
IL-12 treatment
Recombinant murine IL-12 was provided by Genetics Institute (Cambridge, MA). IL-12 treatment was given by i.p. injection at 1 µg per mouse every other day, starting on day 7 until the end of each experiment unless otherwise described. This regimen of IL-12 was well tolerated with no signs of overt toxicity.
Histopathology
On the day of sacrifice, primary tumors were removed, fixed, sectioned, and stained with H&E, and examined by light microscopy for their architecture, evidence of lymphocyte infiltrating, and angiogenesis.
Microarray experiment
The high-density oligonucleotide microarray system of Affymetrix (Santa Clara, CA), murine Genome U74A Array, version 2, containing 12,488 genes, was used. Total RNA was isolated from freshly isolated 4T1 tumors of all four experimental groups and all surviving mice on day 28. RNA samples of each mouse within each experiment group were pooled from seven to eight mice. Ten micrograms of total RNA were used to synthesize cDNA using Superscript cDNA synthesis kit (Invitrogen, Carlsbad, CA) with a primer containing oligo(dT) and T7 RNA polymerase promoter sequences. Double-stranded cDNA was then purified by phase lock gel (Eppendorf, Westbury, NY) with phenol/chloroform extraction. The purified cDNA was used as a template to generate biotinylated cRNA using the Bioarray High Yield RNA Transcript Labeling kit (Enzo Biochem, New York, CA), and then biotinylated cRNAs were fragmented and hybridized to Affymetrix Test 3 chips. All RNA samples passed quality control (ratio of 3′ to 5′, <3). Then, the samples were hybridized to the Murine Genome Array U74Av2 array, which contains 12,488 well-substantiated mouse genes. After overnight hybridization, the arrays were washed, stained with streptavidin-PE (Molecular Probes, Eugene, OR) on the GeneChip Fluidics Station (Affymetrix), and scanned according to the standard Affymetrix protocol.
Microarray data collection and analysis
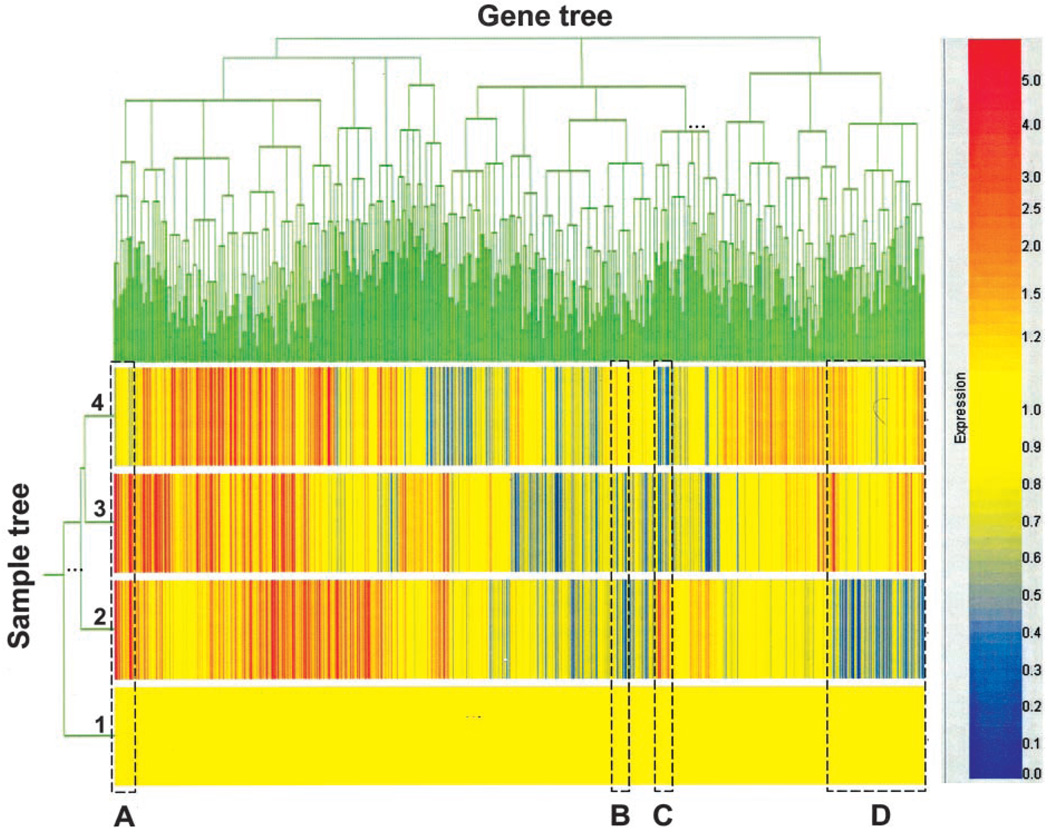
Affymetrix GeneChip 5.0 was used as the image acquisition software for the U74Av2 chips. The signal, which represents the intensity of each gene, was extracted from the image. The target intensity value from each chip was scaled to 250. Data normalization, log transformation, statistical analysis, and pattern study were performed with GeneSpring software (Silicon Genetics, Redwood City, CA). Array data were globally normalized using GeneSpring software. First, all of the measurements on each chip were divided by the 50th percentile value (per-chip normalization). Second, each gene was normalized to the baseline value of the control samples (per-gene normalization). A two-way hierarchical clustering method (32) was used to visualize the coordinated expression profiles and study coexpression patterns of all 12,000 genes over all the samples. The Standard Correlation was chosen to measure the similarity, and the minimum distance to separate genes or samples was 0.001.
RT-PCR
Primers used for PCR were as follows: 1) monokine induced by IFN-γ (MIG),3 sense, AGAACTCAGCTCTGCCATGAAGTC, and antisense, CTAGGCAGGTTTGATCTCCGTTC T; 2) IFN-γ-inducible protein 10 (IP-10), sense, GGATGGCTGTCCTAGCTCTG, and antisense, ATAAC CCCTTGGGAAGATGG; and 3) HPRT, sense, GTTGGATACAGGCCA GACTTTGTTG, and antisense, GAGGGTAGGCTGGCCTATGGCT. The annealing temperature was 60°C for all genes. All cDNAs were amplified for 28 cycles.
Statistical tests
Tumor growth and metastasis data to be compared were first subjected to normality test. Where the samples studied were normally distributed, statistical comparisons were performed using Student’s t test. Where the samples deviated from normality, a nonparametric, Mann-Whitney rank sum test was used for comparisons. Statistical analyses were performed using SigmaStat software (SPSS, Chicago, IL). For all experiments, the mean and the SD are depicted.
Results
IL-12 has IFN-γ-independent antitumor activities
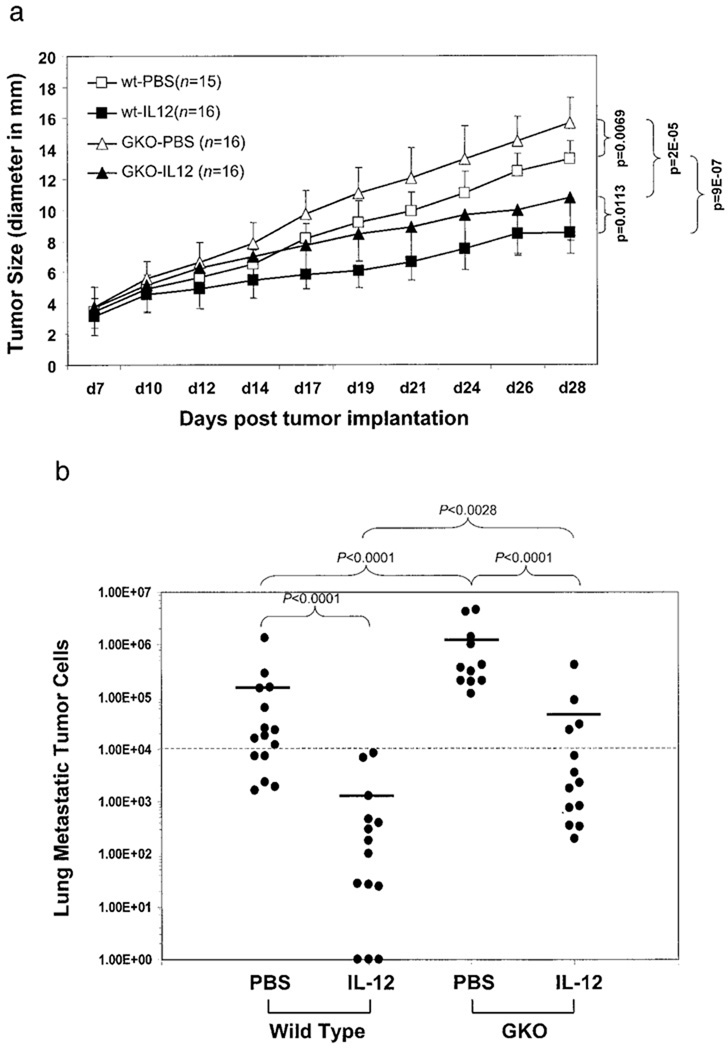
To assess the role of IFN-γ in tumor regression and metastasis induced by IL-12, 4T1 mammary carcinoma was initiated by s.c. injection of 4T1 cells into two types of mice on the syngeneic BALB/c background: wild-type (WT) and IFN-γ knockout (GKO). rIL-12 was given i.p. every 2 days starting on day 7 post-tumor injection to both types of mice when the primary tumor had grown to ~3–5 mm in diameter. The timing of IL-12 administration was based on potential therapeutic considerations to mimic clinical situations in which breast cancer patients do not get therapy until the presence of malignant growth in the breast has been identified by mammogram or other means. As shown in Fig. 1a, by day 28, there were significant differences in tumor growth between wild-type and GKO mice with or without IL-12 treatment. As expected, GKO mice were more permissive for tumor growth than wild-type mice. However, IL-12 was still able to restrict tumor growth almost to the same extent (31% reduction in tumor size) as in the wild type (36% tumor reduction). The pattern of lung metastasis in these mice in response to IL-12 treatment was similar to that of the primary tumor growth (Fig. 1b). IL-12 treatment prevented 4T1 lung metastasis in wild-type mice from crossing the lethal threshold of 104 metastatic tumor cells per lung (33). IL-12 retained partial efficacy in GKO mice and reduced 4T1 lung metastasis. These results clearly demonstrate both IFN-γ-dependent and -independent antitumor activities of IL-12.
FIGURE 1.
IFN-γ-dependent and -independent antitumor activities induced by IL-12. 4T1 tumor cells were injected into the recipient mice on day 1. Recombinant murine IL-12 was applied every other day starting on day 7. Tumor size (a) was measured every 2–3 days. The number of mice in each group is indicated by their respective n, which was compiled from two independent experiments. The p values of various statistical comparisons for the end of point (day 28) are given. Lung metastatic tumor cell number (b) was determined following the termination of the experiment (day 28). Each filled circle (●) represents an individual mouse. The horizontal bars represent the mean values of metastasis in each group. Values of p are given respectively for their bracketed comparisons. The dash lines in a and b mark the lethal threshold of lung metastasis (104).
IL-12 treatment increases lymphocyte infiltration and inhibits neovascularization in the tumor
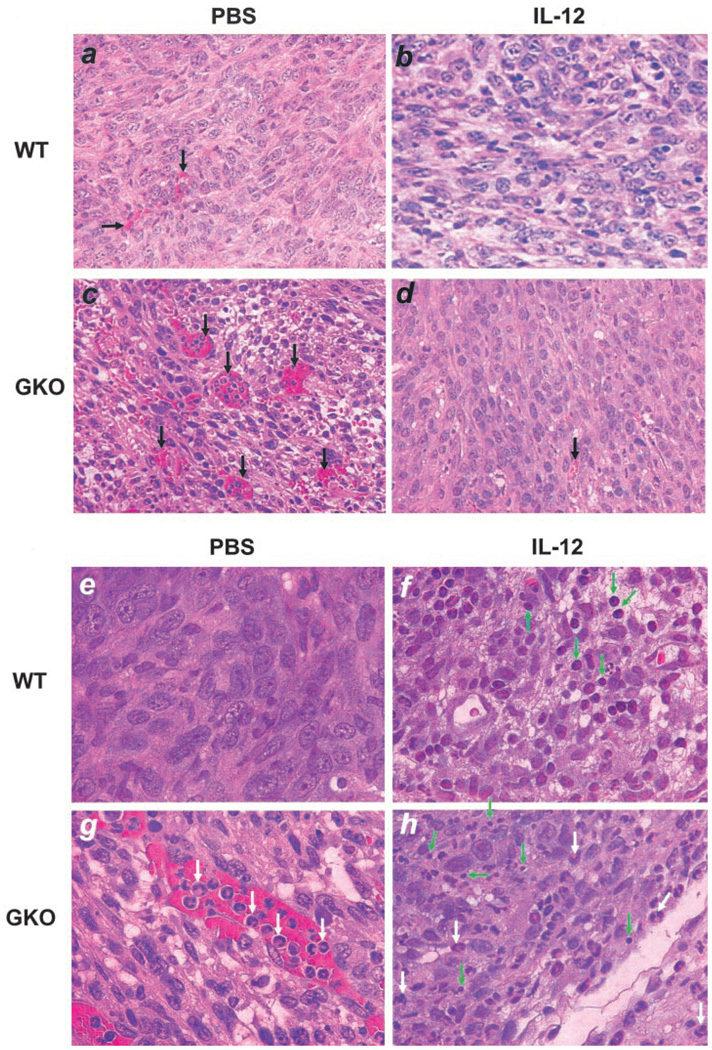
We examined microscopically tumor sections of the four groups of mice, i.e., wild type with or without IL-12 treatment and GKO mice with or without IL-12 treatment (Fig. 2). One striking feature of these tumor sections is the amount of blood vessels rampant in nontreated GKO mice (Fig. 2c, black arrows) compared with other groups, confirming the well-established role of IFN-γ in inhibition of angiogenesis. Surprisingly, IL-12 treatment largely blocked the formation of these vessels in GKO mice (Fig. 2d). Tumors not treated with IL-12 had few infiltrating T lymphocytes (Fig. 2e). IL-12 treatment in the wild-type mice significantly increased the number of tumor-infiltrating lymphocytes (TILs) (Fig. 2f, green arrows). Tumors in GKO mice had few TILs other than neutrophils present in the blood vessels (Fig. 2g, white arrows). These polymorphonuclear cells may have come in response to necrosis that likely occurred because of the rapid tumor growth outpacing blood supply. However, in the GKO mice treated with IL-12, there were considerably greater numbers of TILs (Fig. 2h, green arrow) than those in nontreated GKO mice, although not as frequent as in wild-type mice treated with IL-12. There were also many polymorphonuclear cells present in necrotic areas in the tumor (white arrows). These results are not only confirmatory for the antiangiogenic activity of IFN-γ and TIL-inducing activity of IL-12, but also reveal an interesting IFN-γ-independent antiangiogenic effect induced by IL-12.
FIGURE 2.
Lymphocyte infiltration and angiogenesis in primary tumors. Paraffin-embedded tumor sections were stained with H&E and microscopically examined for blood vessels (a–d) (×200 magnification) and TILs (e–h) (×400 magnification). Shown for each group is one representative slide of three tumor samples randomly picked from the four groups. Black arrows, Blood vessels; white arrows, neutrophils; green arrows, TILs.
IL-12 is able to inhibit the growth of the metastatic tumor cells in the lung
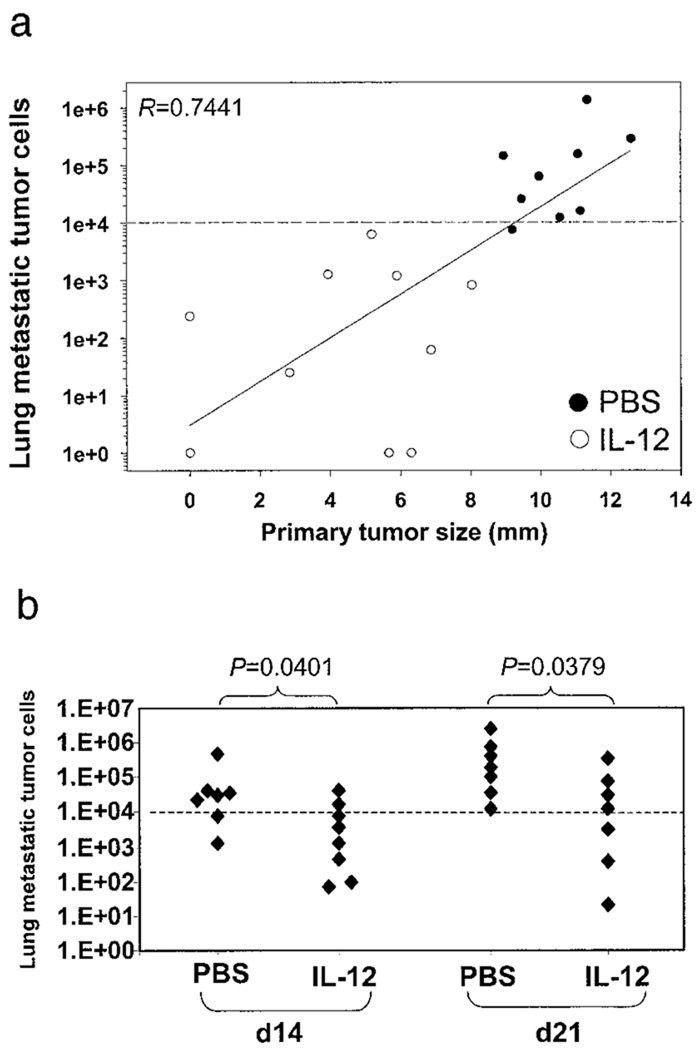
We observed a positive correlation between primary tumor size and the number of metastatic cells in the lung, i.e., the larger the tumor, the greater the lung metastases (Fig. 3a). This prompted us to ask whether the restrictive effect of IL-12 was on the growth of the primary tumor only or on the expansion of metastatic cells in the lung also. To address this question, mice were implanted with 4T1 tumor cells on day 0. On days 7, 14, or 21, the primary tumors were removed surgically, and mice were then given the IL-12 treatment for 21, 14, and 7 days, respectively. Lung metastasis was measured on day 28 post-primary tumor injection (Fig. 3b). There was little lung metastasis for the group of mice that had the primary tumor (~4 mm in diameter) removed on day 7 and that were given the IL-12 treatment for 21 days (<104 metastatic cells per lung). The other two groups whose primary tumors were removed later (on days 14 and 21 with tumor sizes being ~7 and 10 mm, respectively) showed progressively greater lung metastasis in the absence of therapeutic intervention. However, IL-12 treatment even at these later stages for shorter periods of time (14 and 7 days, respectively) was still able to reduce significantly the number of 4T1 tumor cells in the lung. Because lung metastasis had already occurred at the time of surgical removal of the primary tumors on days 14 and 21, and there was no more source of fresh metastatic cells at the primary tumor site, it is likely, thus, that the inhibitory effect of IL-12 seen in this study was on the tumor cells already residing in the lung, not on tumor cells on their way to the lung.
FIGURE 3.
Effects of IL-12 on metastatic cells in the lung in the absence of the primary tumor. 4T1 tumor cells were implanted in BALB/c mice with or without the IL-12 treatment. Lung metastasis was measured at the end of the experiment. a, Correlation between primary tumor size and lung metastatic cell number. Analysis of linear regression was performed with the indicated R value (0.7441). Each circle represents an individual mouse. ●, Mice that received PBS. ○, Mice that received IL-12. b, Effects of IL-12 treatment at different times post-tumor implantation in the absence of the primary tumor. The primary tumor was implanted without IL-12 treatment and removed surgically from recipient mice on days 14 and 21. These mice were then given PBS or IL-12 treatment for 14 and 7 days, respectively, until day 28. Lung metastasis was measured. Each filled diamond (♦) represents an individual mouse. Values of p are given for the bracketed comparisons. The dash line marks the lethal threshold of lung metastasis. Data are representative of two separate experiments.
IL-12-induced immunogenicity on the primary tumor is not irreversible
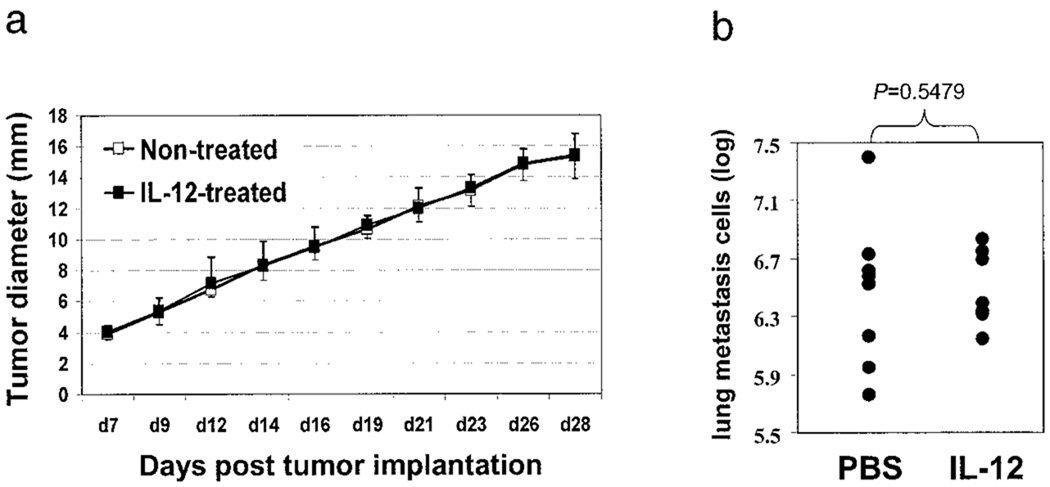
We were interested in whether the IL-12 treatment could induce immunogenicity in the tumor in one host that can be transferred to another. This was addressed by treating 4T1 tumor-bearing mice with or without IL-12, removing the primary tumors (on day 28) from the two groups of mice, and reimplanting purified tumor cells (by removing other types of cells such as TILs) into naive mice. As shown in Fig. 4a, growth of tumors derived from nontreated or IL-12-treated mice was almost identical. Lung metastasis of the two types of tumors was very similar as well (Fig. 4b). Of note is the observation that these freshly isolated 4T1 tumor cells were much more aggressive in vivo than the in vitro-passaged tumor cell line, evidenced by the larger sizes of the primary tumors and greater lung metastasis. These results suggest that the effect of IL-12 on the primary tumor is not permanent and is maintained in an appropriate physiological setting that is absent in animals not treated with rIL-12, or with tumor cells depleted of TILs.
FIGURE 4.
Effects of previous exposure of 4T1 tumor to IL-12 on its progression in naive mice. 4T1 tumor cells were implanted in BALB/c mice with or without IL-12 treatment. On day 28, primary tumors were removed surgically and digested with collagenase to make single-cell suspension and to remove nontumor cells. These purified tumor cells were then injected into naive mice and monitored for their growth (a) without any treatment. On day 28, mice were sacrificed, and lung metastasis was measured (b). The p value of the bracketed statistical comparison is indicated.
Genome-wide analysis of gene expression in primary tumors
To obtain a detailed molecular appreciation of what was going on in 4T1 tumors exposed in vivo to IL-12 in the presence or absence of a functional endogenous IFN-γ gene, we performed global gene expression analysis of the four groups of tumors on day 28 using the Affymetrix oligonucleotide microarray system (Murine Genome U74A Array, version 2, containing 12,488 genes). To reduce variations between individual mice, RNA samples were pooled from all mice within each group for the microarray analysis. Microarray data were normalized and subjected to hierarchical clustering analysis of the entire 12,488 genes to reveal groups of genes whose expression changes specifically in response to IL-12 treatment with or without IFN-γ-mediated activities (Fig. 5). The group of untreated wild-type mice was set as the baseline (with an expression value of 1.0) to which the other three groups were compared. On the sample tree view, the two groups of GKO mice treated with IL-12 or with PBS were placed together (groups 3 and 4), indicating that they were more similar to each other than to the two groups of wild-type mice. The IL-12-treated wild-type mice (group 2) were more similar to the two GKO mouse groups than to PBS-treated wild type (group 1). Four clusters of genes are of interest (labeled A, B, C, and D, respectively). Genes in cluster A were induced by IL-12 in both groups 2 (WT treated with IL-12) and 3 (GKO treated with IL-12), but not in group 4 (GKO treated with PBS), suggesting that these genes are independent of IFN-γ for their induction. In contrast, cluster C was only induced by IL-12 in wild-type mice (group 2), but not in group 3. In fact, expression of this cluster of genes was suppressed in GKO mice, suggesting that these genes depend on IFN-γ not only for their IL-12-induced expression but also for the maintenance of their basal level of expression during tumor progression. Cluster B was suppressed in IL-12-treated tumors regardless of the genotype (groups 2 and 3), indicating that these genes are independent of IFN-γ for their silencing. However, cluster D was repressed only in wild-type mice (group 2), but not in GKO mice (group 3), suggesting that this cluster of genes is dependent on IFN-γ to mediate IL-12’s suppressive effects on their expression.
FIGURE 5.
Hierarchical clustering analyses of the microarray data. The microarray data were analyzed by the hierarchical clustering method with two views of the clusters: sample view and gene tree view. The sample tree consists of four experimental groups: 1) wild type treated with PBS; 2) wild type treated with IL-12; 3) IFN-γ−/− mice treated with IL-12; and 4) IFN-γ−/− mice treated with PBS. Four clusters of genes labeled A, B, C, and D marked by dashed rectangular boxes are described in detail in Results. The color bar on the right provides quantitative visual references for the magnitude, in log of 2, of induction (red) or suppression (blue) from the baseline (yellow), which is the level of expression of group 1 genes (wild type treated with PBS).
IFN-γ-dependent and -independent genes induced by IL-12
To further examine the details of the genes in clusters A and C and other similar clusters in the dendrogram shown in Fig. 5 in an effort to delineate IFN-γ-dependent and -independent genes induced by IL-12 in its antitumor activities, we categorized these genes using additional stringencies, i.e., by choosing only those genes that were expressed 2-fold or higher in IL-12-treated tumors with statistically significant detection p values (p < 0.05) and present calls. By these criteria, 147 genes are defined as IL-12-induced genes of which 30 are expressed sequence tags. The remaining 117 genes can be divided into 67 IFN-γ-dependent (Table I) and 50 IFN-γ-independent genes (Table II).
Table I.
IFN-γ-dependent genes induced by IL-12 (67 genes)a
| Common Name | GenBank | Map | WT—IL-12- Treated |
GKO—Not Treated |
GKO—IL-12- Treated |
Function |
|---|---|---|---|---|---|---|
| Monokine induced by γ-IFN (MIG), CXCL9 | M34815 | 5 | 9.16 | 0.01 | 0.01 | Chemoattractant for activated T cells |
| Hemopexin | U89889 | 7.37 | 0.37 | 0.33 | Protective antioxidant | |
| Suppressor of cytokine signaling-1 (SOCS-1) | U88325 | 16 | 5.07 | 0.44 | 0.91 | IFN signaling |
| Sex-limited protein Slp(w7)αγ chain | X06454 | 17 | 4.95 | 0.56 | 1.54 | Complement activation |
| GTPase IGTP | U53219 | 11 | 4.63 | 0.23 | 0.25 | IFN-induced GTP binding protein |
| Histocompatibility 2, class II Ag A, β1 | M21932 | 17 | 4.27 | 0.29 | 0.26 | Ag presentation to CD4 T |
| Predicted GTP binding protein (IRG-47) | M63630 | 4.20 | 0.28 | 0.27 | GTP binding and metabolism | |
| Histocompatibility 2, class II, locus Mb1 | U35330 | 17 | 3.98 | 0.24 | 0.19 | Ag presentation to CD4 T |
| Xanthine dehydrogenase, XD | X75129 | 17 | 3.83 | 0.84 | 0.99 | Nucleotide metabolism |
| STAT1 | U06924 | 1 | 3.82 | 0.08 | 0.11 | IFN-γ signaling |
| Histocompatibility 2, class II Ag E-β | X00958 | 17 | 3.80 | 0.24 | 0.26 | Ag presentation |
| Guanylate nucleotide binding protein 1 | M55544 | 3 | 3.63 | 0.26 | 0.18 | G protein signaling |
| MPS1 (macrophage-specific), Mpq-1 | L20315 | 3.59 | 0.97 | 0.85 | Regulates centrosome duplication | |
| Histocompatibility 2, class II Ag E-α | V00833 | 17 | 3.51 | 0.19 | 0.21 | Ag presentation to CD4 + T |
| IFN-inducible GTPase | AJ007971 | 3.50 | 0.08 | 0.06 | IFN-induced GTP binding protein | |
| Similar to X64449 proteasome subunit MC13 | AV355612 | 3.45 | 0.57 | 0.71 | Ag processing | |
| Class II transactivator (CIITA) form III | AF042158 | 3.42 | 0.71 | 0.84 | Transcription factor for MHC class II genes | |
| Serine/cysteine proteinase inhibitor, clade G (C1 inhibitor) | AF010254 | 2 | 3.41 | 0.53 | 1.27 | Inhibitor of serine/cysteine proteases |
| NKG2-D | AF054819 | 3.40 | 0.64 | 1.39 | NK cell-activating immunoreceptor | |
| Complement component 2 (within H-2S) | M57891 | 3.37 | 0.92 | 0.80 | Complement activation | |
| Fatty acid CoA ligase, long chain 2 | U15977 | 3.19 | 1.30 | 1.96 | Lipid metabolism | |
| Tissue inhibitor of metalloproteinases-3 (TIMP-3) | U26437 | 3.13 | 1.00 | 0.82 | Apoptosis inducer/angiogenesis blocker | |
| H-2Ma, H2-Ma | U35323 | 17 | 3.11 | 0.55 | 0.44 | Ag presentation to CD4 T |
| IFN-g induced GTPase | AJ007972 | 3.10 | 0.29 | 0.26 | IFN-γ induced GTP binding protein | |
| Histocompatibility 2, class II Ag A, α | X52643 | 17 | 3.08 | 0.25 | 0.26 | Ag presentation |
| Guanylate nucleotide binding protein 2 | AJ007970 | 3 | 3.06 | 0.32 | 0.25 | G protein signaling |
| IL-12Rβ component, IL-12Rβ1 | U23922 | 8 | 3.03 | 0.55 | 0.27 | IL-12 signaling in T/NK cells |
| κ Casein precursor, CSN10 | M10114 | 5 | 2.92 | 0.94 | 1.36 | Stabilizes the micelle structure |
| Glutamyl aminopeptidase; BP-1/6C3 Ag (Enpep) | M29961 | 2.89 | 1.68 | 1.70 | Cleaves angiotensin II, etc. | |
| IFN-γ induced (Mg11) | U15635 | 2.88 | 0.69 | 0.82 | Unknown | |
| TRAIL, Tnfsf10 | U37522 | 2.82 | 0.69 | 1.06 | Cell death | |
| Macrophage IFN inducible protein 10 (IP-10), CXCL10 | M33266 | 5 | 2.79 | 0.61 | 0.41 | Chemoattractant for activated Th1 cells |
| IL-18 binding protein, MC54L, IL-18BP | AB019505 | 2.78 | 0.46 | 0.42 | Inhibitor of IL-18 | |
| Fibrinogen-like protein, flp | M16238 | 5 | 2.74 | 0.35 | 0.57 | Thrombosis, cytotoxic T-specific protein |
| Secreted frizzied-related sequence protein 1 | U88566 | 2.71 | 1.38 | 1.62 | Inhibitor of Wnt, induces apoptosis | |
| Ag processing-associated transporter TAP1-g7 | U60020 | 17 | 2.66 | 0.37 | 0.36 | Ag processing |
| Eukaryotic translation initiation factor 1A | U28419 | 2.63 | 0.91 | 0.62 | Initiation of protein synthesis | |
| Cd1d1, CD1.1 | M63695 | 2.60 | 0.72 | 0.76 | NKT cell marker | |
| uridine phosphorylase, UPase, UdRPase | D44464 | 2.60 | 0.48 | 0.74 | catalyzes reversible phosphorolysis of uridine to uracil | |
| Follistatin | Z29532 | 13 | 2.55 | 1.04 | 1.33 | Inhibitor of activin, role in embryogenesis |
| Low-molecular-mass polypeptide complex subunit 2 | D44456 | 17 | 2.51 | 0.48 | 0.38 | IFN-γ-induced, Ag processing |
| WRS, tryptophan-tRNA ligase | X69656 | 2.47 | 0.70 | 0.66 | Protein synthesis, antiangiogenic | |
| IDO, indoleamine 2,3-dioxygenase | M69109 | 2.43 | 0.35 | 0.19 | Enzyme in the catabolism of tryptophan | |
| Mg21, with GTP binding motif; T-cell-specific protein | L38444 | 2.41 | 0.13 | 0.06 | IFN-γ-induced, possible G protein | |
| Ig germline D-J-C regionα gene and secreted tail, Iga | J00475 | 12 | 2.32 | 0.38 | 0.53 | Ab component (IgA) |
| IFN regulatory factor 1, IRF-1 | M21065 | 11 | 2.31 | 0.65 | 0.58 | IFN signaling |
| Class II, T region locus 18, thymus-leukemla Ag Tla(a)-3 | X03052 | 17 | 2.30 | 1.07 | 1.27 | Ag presentation to CD4 T |
| Proteasome (prosome, macropain) subunit, βtype 10 | Y10875 | 8 | 2.30 | 0.82 | 0.66 | Ag processing |
| His, Hsd, histidase, histidine ammonia-lyase | L07645 | 2.23 | 1.37 | 0.94 | Histidine metabolism | |
| Castaneus TAP2 (TAP2-cas) | U60091 | 17 | 2.21 | 0.74 | 0.56 | Ag transporter |
| MHC class I H2-TL-T10 (b haplotype) | M35244 | 17 | 2.20 | 0.52 | 0.50 | Ag presentation to CD8 + T |
| IL-15 Ra | U22339 | 2 | 2.20 | 1.62 | 1.41 | IL-15 signaling (NK and CTL) |
| Cellular repressor of E1A-stimulated genes (CREG) | AF084524 | 2.18 | 0.96 | 0.87 | Inhibits cell growth and/or promotes differentiation | |
| Protein tyrosine phosphatase, nonreceptor type 18 | U49853 | 1 | 2.18 | 0.89 | 1.05 | Expressed in hemopoletic progenitors |
| IFN-γ-inducible protein 1, G-protein-like LRG-47 | U19119 | 2.17 | 0.41 | 0.22 | Resistance to intracellular infection | |
| IFN consensus binding protein (ICSBP) | M32489 | 8 | 2.15 | 0.75 | 0.75 | IFN-γ induced transcription |
| CD1d2 Ag, CD1.2 | M63697 | 3 | 2.10 | 1.02 | 0.84 | NKT cell marker |
| CD157, BST-1 | D31788 | 2.09 | 1.15 | 1.94 | ADP ribosyl cyclase | |
| NO synthase 2, inducible NO synthase | U43428 | 11 | 2.08 | 0.83 | 0.92 | Cytotoxic killing |
| Ly69, integrinβ7 subunit | M68903 | 2.08 | 0.69 | 1.09 | Lymphocyte homing and retention | |
| Ifit2, glucocorticoid-attenuated response gene 39 (GARG-39) | U43085 | 19 | 2.08 | 0.18 | 0.09 | LPS and IFN-inducible gene |
| GTP binding protein, γ8 transducing activity polypeptide 2 | AI882325 | 11 | 2.07 | 0.59 | 0.57 | G protein signaling |
| Histocompatibility 2, class II, locus Mb2 | X90807 | 17 | 2.06 | 0.94 | 1.01 | Ag presentation to CD4 T |
| Hephaestin | AF082567 | 2.03 | 0.60 | 0.96 | Membrane multi-copper ferroxidase | |
| pK30, Ly-6C variant | D86232 | 15 | 2.03 | 0.82 | 0.91 | Regulates endothelial adhesion and homing of CD8+ T cells via integrin |
| Mer, c-mer tyrosine kinase receptor | U21301 | 2 | 2.02 | 1.20 | 1.07 | Clearance of apoptotic cells |
| IFN-stimulated protein (15 kDa) | X56602 | 2.01 | 0.87 | 0.64 | Unknown |
Genes are selected based on the following criteria: 1) they must be expressed more than two times than the respective control samples (untreated wild type or GKO) to qualify as IL-12-induced genes; 2) their expression detection must be statistically valid as present with detection p < 0.05; 3) IFN-γ-dependent genes are defined as those whose expression must be induced by IL-12 in GKO mice less than two times compared with untreated GKO mice; and 4) IFN-γ-independent genes are defined as those whose expression must be induced by IL-12 in GKO mice greater than two times compared with untreated GKO mice. GenBank accession numbers are given. Chromosomal locations (map) are given when they are known. The numbers in columns 4–6 are relative fold of expression over the baseline (control untreated, wild-type group), which is arbitrarily set as 1.
Table II.
IFN-γ-independent genes induced by IL-12 (50 genes)a
| Common Name | GenBank | Map | WT—IL-12- Treated |
GKO—Not Treated |
GKO—IL-12- Treated |
Function |
|---|---|---|---|---|---|---|
| RANTES, CCL5 | AF065947 | 11 | 38.79 | 0.34 | 4.44 | Chemoattractant for Mϕ/DC, T/NK/basophil/eosinophil |
| CD6 Ag | U37543 | 19 | 14.97 | 0.70 | 19.14 | Binds CD166, activated leukocyte-cell adhesion molecule |
| CD8 Ag β-chain | AV316162 | 6 | 13.15 | 0.52 | 10.25 | Cytotoxic T and DC markers |
| Ig κ-chain variable 28 (V28) | M18237 | 6 | 11.51 | 0.61 | 17.59 | Ab component |
| Preproinsulin-like growth factor IA, Igf1 | X04480 | 10 | 9.86 | 1.05 | 5.61 | Growth hormone mediator |
| CXCL13, BLC, BCA-1 | AF030636 | 9.37 | 0.30 | 12.44 | Chemoattractant for naive B and activated CD4+ T cells | |
| Similar to ζ-chain (TCR)-associated protein kinase ZAP-70 | AI386093 | 1 | 8.56 | 2.30 | 9.61 | T cell activation signaling |
| Putative castaneus IgK chain gene, C-region, IgM | M80423 | 8.48 | 0.08 | 17.41 | Natural Ab | |
| CD3 Ag, δ polypeptide | X02339 | 9 | 7.27 | 0.96 | 6.12 | T lymphocyte marker |
| T cell receptor α chain | M16118 | 14 | 6.82 | 1.00 | 9.47 | T lymphocyte marker |
| Adipsin | X04673 | 10 | 5.83 | 0.33 | 4.46 | Complement D activity, activates alternative pathway |
| IL-18R1 | U43673 | 5.81 | 0.66 | 9.16 | Receptor for an inflammatory cytokine | |
| CD3-γ | M18228 | 4.84 | 0.74 | 7.52 | T lymphocyte marker | |
| Adipocyte-specific Fsp27 | M61737 | 4.27 | 0.10 | 3.23 | Cell death-inducing factor-like | |
| ζ-chain (TCR)-associated protein kinase ZAP-70 | U04379 | 4.25 | 0.95 | 4.80 | T cell activation signaling | |
| α-1 acid glycoprotein (Agp-1B) | M27008 | 4 | 4.21 | 1.39 | 2.61 | Serum acute phase protein |
| Apolipoprotein D | X82648 | 16 | 4.08 | 1.48 | 4.77 | Androgen-regulated, plasma lipid transport |
| Apolipoprotein C1 (Apoc1) | Z22661 | 7 | 3.69 | 0.75 | 2.01 | Mediator of lipid binding in plasma |
| Similar to human lipoprotein lipase | AA726364 | 8 | 3.69 | 0.71 | 2.38 | Hydrolyzes triglycerides in lipid metabolism |
| Stromal cell-derived factor, SDF-1, Sdf1a | L12029 | 3.41 | 0.33 | 2.83 | Chemoattractant for T/B/DC and CD34+ progenitor cells | |
| spi2/eb4 protease inhibitor | M64086 | 3.37 | 1.15 | 2.89 | Inhibitor of serine protease | |
| Activator of S phase kinase, DBF-4-related protein | AJ003132 | 3.33 | 0.35 | 2.26 | G1/S phase transition in cell cycle | |
| Monocyte chemoattractant protein 2 (MCP2), CCL8 | AB023418 | 11 | 3.30 | 0.16 | 1.46 | Chemoattractant for T/monocyte/basophil/eosinophil |
| ICAM-1 | M90551 | 9 | 3.00 | 0.44 | 1.94 | Cell adhesion/migration |
| c-myb | M12848 | 10 | 3.00 | 0.70 | 1.52 | Proto-oncogene |
| Lipoprotein lipase | M63335 | 8 | 2.80 | 0.53 | 2.36 | Hydrolyzes triglycerides in lipid metabolism |
| Stearoyl-CoA desaturase, ab, SCD, Scd-1 | M21285 | 19 | 2.79 | 0.52 | 2.65 | Synthesis of monounsaturated fatty acids |
| PGE receptor subtype EP2 | AB007696 | 14 | 2.73 | 0.23 | 2.16 | PGE2 signaling |
| Voltage-dependent Na+ channel β 1 subunit | L48687 | 2.65 | 1.31 | 2.67 | Regulates Na+ channels | |
| C57BL/6J ob/ob haptoglobin, Hp | M96827 | 8 | 2.68 | 0.42 | 1.91 | Serum acute phase protein |
| TCR germline β-chain gene constant region (CT) | M26056 | 2.59 | 1.00 | 3.61 | T cell marker and signaling | |
| Killer cell lectin-like receptor, subfamily D, member 1, CD94 | AF030311 | 2.59 | 0.59 | 3.24 | Cytotoxic killer cell inhibitory receptor | |
| Lck, protein tyrosine kinase | M12056 | 2.58 | 0.89 | 3.87 | Lymphocyte-specific signaling | |
| Pentraxin related gene, PTX3 | X83601 | 3 | 2.54 | 1.24 | 2.23 | Secreted pattern-recognition receptor |
| Nuclear matrix attachment DNA-binding protein SATB1 | U05252 | 17 | 2.49 | 0.66 | 4.06 | Regulator of chromatin remodeling |
| Killer cell lectin-like receptor subfamily C, member 1, NKG2A | AF095447 | 2.42 | 1.11 | 2.45 | Inhibitory receptor of cytotoxic cells | |
| Granzyme B | M12302 | 2.41 | 0.58 | 2.52 | Cytotoxic killing | |
| b allele; Ig active J chain | M90766 | 5 | 2.38 | 1.00 | 4.16 | Ab component |
| Nuclear orphan receptor LXR-α | AF085745 | 2.34 | 0.70 | 2.42 | cAMP-responsive transcriptional regulator | |
| Ig-like receptor PIRA3 (7M1) | U96684 | 2.34 | 0.93 | 1.91 | Mast cell activating | |
| Germ line gene fragment for µ-Ig C terminus (secreted form), Igh-6 | V00817 | 12 | 2.31 | 0.48 | 4.00 | Ab component |
| Plasma phospholipid transfer protein | U28960 | 2 | 2.31 | 0.73 | 1.58 | Suppressor of cancer cell |
| Lymphotoxin B | U16985 | 17 | 2.27 | 0.61 | 3.44 | Cytotoxicity |
| Serum amyloid A (SAA) 3 protein | X03505 | 7 | 2.25 | 0.80 | 1.95 | Serum acute phase protein |
| Stroma cell-derived factor, SDF-1 β | L12030 | 2.21 | 1.10 | 2.84 | Chemoattractant for T/B/DC and CD34+ progenitor cells | |
| Gpcr25, putative G protein-coupled receptor TDAG8 | U39827 | 12 | 2.11 | 0.66 | 1.40 | T cell death-associated gene |
| GM-CSF 2 receptor, β1, low affinity | M34397 | 15 | 2.10 | 0.60 | 1.96 | Growth and differentiation of granulocytes and Mϕ |
| Close homolog of L1, CHS-1-like protein | X94310 | 2.09 | 0.37 | 3.28 | Novel neural recognition molecule | |
| Adipocyte complement-related protein of 30 kDa,AdipoQ | U49915 | 2.06 | 0.34 | 2.07 | Regulates insulin sensitivity | |
| XCL1, lymphotactin | U15607 | 2.01 | 1.34 | 2.71 | Chemoattractant for NK and T lymphocytes |
Genes are selected based on the following criteria: 1) they must be expressed more than two times than the respective control samples (untreated wild type or GKO) to qualify as IL-12-induced genes; 2) their expression detection must be statistically valid as present with detection p< 0.05; 3) IFN-γ-dependent genes are defined as those whose expression must be induced by IL-12 in GKO mice less than two times compared with untreated GKO mice; and 4) IFN-γ-independent genes are defined as those whose expression must be induced by IL-12 in GKO mice greater than two times compared with untreated GKO mice. GenBank accession numbers are given. Chromosomal locations (map) are given when they are known. The numbers in columns 4–6 are relative fold of expression over the baseline (control untreated, wild-type group), which is arbitrarily set as 1.
Many of the genes listed in Table I are well-known IFN-γ-induced targets: chemokines and antiangiogenic factors such as MIG and IP-10; signaling molecules such as Stat1 and several IFN-γ-inducible G proteins; IL-12Rβ1 and IL-15Rα; complement components; numerous MHC I and II molecules, and Ag-processing and transport proteins; apoptosis- and cell death-inducing factors, such as TRAIL, secreted frizzled-related sequence protein 1, phospholipid transfer protein, G protein-coupled receptor 25, and inducible NO synthase (reviewed in Ref. 34); NK cell-activating immunoreceptor NKG2-D; and NKT cell markers such as CD1.1 and CD1.2. Indoleamine dioxygenase, an enzyme responsible for the degradation of tryptophan, could function to starve the tumor of this essential amino acid (35). These confirmatory results instill great confidence in the validity of the microarray data.
The list of IL-12-induced, IFN-γ-independent genes (Table II) reveals many interesting molecules that could potentially be involved in the inhibition of tumor progression by IL-12 in the absence of IFN-γ. Many T cell markers such as CD6, CD8, CD3, and TCRs fall into this group, supporting the histopathological evidence of substantial T cell infiltration in GKO mice. This is also consistent with the fact that several immune cell-recruiting chemokines such as RANTES, CXC ligand (CXCL)13 (B lymphocyte chemoattractant), monocyte chemoattractant protein 2, stromal cell-derived factor-1α, stromal cell-derived factor-1β, and lymphotactin (XCL1) were induced normally or substantially in GKO mice treated with IL-12, so were some of the TCR-associated protein kinases such as ζ-associated protein 70 and lck that are important signaling molecules in T cell activation. In the absence of a functional IFN-γ gene, two well-known cytotoxic proteins were induced by IL-12: granzyme B and lymphotoxin β, which at least in part explains the ability of the IFN-γ-deficient host to restrict 4T1 tumor growth in response to IL-12 treatment. Several other potential apoptotic effector molecules are present in this list. The serine protease inhibitor Spi2 has been shown to mediate apoptosis of olfactory neurons in response to neurodegenerative insults (36). The human homolog of mouse adipocyte-specific FSP27 gene (37), cell-death-inducing DNA fragmentation factor (DFF45)-like effector-3 (CIDE-3), has very recently been reported to be expressed not only in adipocytes, but also in small intestines, heart, colon, and stomach, and has death-inducing activity by nuclear DNA fragmentation assay in CIDE-3-transfected 293T cells (38). Another gene on this list, adipsin, has complement D activity in the alternative pathway of complement activation (39).
IFN-γ-dependent and -independent genes inhibited by IL-12
We were also interested in the genes whose expression was inhibited by the IL-12 treatment in the presence or absence of the IFN-γ gene, because they could reveal potential targets involved in the major promotional mechanisms by which a tumor progresses. We summarized these genes using the same criteria as discussed above except that the expression of the genes must be at least 2-fold less in IL-12-treated tumor than the nontreated tumor. An added criterion for IFN-γ-independent genes is that their expression must be at least 2-fold less in IL-12-treated GKO tumor than in nontreated GKO tumor. These criteria resulted in 65 genes as IFN-γ-dependent, IL-12-inhibited targets (Table III) and 2 genes as IFN-γ-independent targets (Table IV).
Table III.
IFN-γ-dependent genes inhibited by IL-12 (65 genes)a
| Common Name | GenBank | Map | WT—IL-12- Treated |
GKO—Not Treated |
GKO—IL-12- Treated |
Function |
|---|---|---|---|---|---|---|
| Proliferin (PLF) | AV290437 | 13 | 15.84 | 2.89 | 1.79 | Angiogenic placental hormone |
| Macrophage-inflammatory protein-2 (MIP2) | X53798 | 4q21 | 6.16 | 2.24 | 1.78 | Chemokine |
| Platelet-derived growth factor-inducible KC protein (GRO1) | J04596 | 5 E–F | 4.79 | 4.74 | 1.75 | A secretory protein related to platelet α-granule proteins |
| Adrenomedullin | U77630 | D7Wsu1 3De | 4.79 | 2.06 | 1.51 | Mitogenic, antiapoptosis factor |
| Matrix metalloproteinase 10, stromelysin-2 | Y13185 | 9 | 4.61 | 1.40 | 1.92 | Degradation of extracellular matrix |
| ADAMTS-1, a disintegrin and metalloprotease with thrombospondin motifs | D67076 | 16 C3–C5 | 4.27 | 1.27 | 1.85 | Cleaves versican in the expanded cumulus oocyte complex |
| Carboxypeptidase X2 | AF017639 | 4.17 | 1.29 | 1.77 | Peptidase | |
| IL-1α | M14639 | 2 F | 4.13 | 1.38 | 1.63 | Inflammatory cytokine |
| Amphiregulin | L41352 | 5 | 4.09 | 0.79 | 1.58 | Ligand for EGFR, growth factor |
| Lymphocyte Ag 57, BLNK | AF068182 | d1 | 4.09 | 4.05 | 3.85 | B lymphopoiesis |
| Intracellular calcium-binding protein (MRP14) | M83219 | 3 | 4.02 | 1.27 | 1.52 | Cell adhesion |
| Placental growth factor, Plgf | X80171 | 12 D | 3.36 | 0.80 | 1.51 | Placental growth factor |
| Urate oxidase, Uox | M27695 | 3 | 3.34 | 1.25 | 1.46 | Converts uric acid to allantoin |
| Ornithine decarboxylase antizyme | AV212241 | 10 | 3.26 | 2.67 | 2.36 | Cell growth inhibition |
| Immediate early response 3, gly96 | X67644 | 3.25 | 1.54 | 1.45 | Unknown | |
| Basonuclin | U88064 | Unknown | 3.14 | 0.86 | 1.41 | Cellular proliferation |
| HMGCR | X07888 | 3.04 | 1.35 | 2.17 | Key enzyme for cholesterol synthesis | |
| Cellular retinoic acid binding protein (CRABP) | X15789 | 9 | 3.04 | 1.45 | 1.39 | Regulation of retinoid acid availability for nuclear receptors |
| c-Fos | V00727 | 12 | 2.99 | 0.82 | 1.31 | Oncogene for cellular proliferation |
| IL-1R, type II | X59769 | 1 | 2.91 | 1.14 | 1.28 | Type II IL-1R |
| Growth factor-inducible immediate early gene (3CH134), Ptpn 16 | X61940 | 17 A2-C | 2.87 | 1.16 | 1.26 | Cell adhesion, migration, and proliferation |
| Parathyroid hormone-related peptide gene, PTHRP | M60057 | 6 F–G | 2.86 | 1.81 | 1.22 | Regulates endochondral ossification during skeletal development |
| Wnt-5a | M89798 | 14 | 2.83 | 1.39 | 1.22 | Mammary cell adhesion |
| mts1 | M36579 | 3 | 2.78 | 1.56 | 1.16 | Associated with tumor metastasis |
| Endothelial-specific receptor tyrosine kinase, Tie 2 | X71426 | 4 | 2.74 | 1.31 | 1.15 | Endothelial cell-specific receptor tyrosine kinase involved in angiogenesis |
| Intracellular calcium-binding protein (MRP8) | M83218 | 3 | 2.72 | 0.66 | 1.14 | Cell adhesion |
| Mature T cell proliferation 1 (MTCP1) | U32332 | X | 2.70 | 0.93 | 1.14 | Oncogene |
| Matrix metalloproteinase 13 | X66473 | 11q22.3 | 2.66 | 1.62 | 2.17 | Degradation of extracellular matrix |
| Short stature homeobox 2, OG-12a homeodomian protein | U66918 | 3 E3-F1 | 2.64 | 0.62 | 1.12 | Involved in growth |
| Chemokine orphan receptor 1, RDC1 | AF000236 | 1 | 2.52 | 1.51 | 1.12 | A coreceptor for HIV and SIV |
| Mglap | D00613 | 6 | 2.50 | 1.76 | 1.11 | Regulation of extracellular matrix calcification |
| Endostatin, α1 (XVIII) collagen | L22545 | 10 | 2.46 | 1.31 | 1.10 | Angiogenic factor |
| Invasion-inducing protein (Tiam1) | U05245 | 16 | 2.44 | 1.89 | 1.06 | Invasion-inducing protein |
| Melanoma Ag H52 | D10049 | 2.37 | 1.16 | 1.04 | Encodes the env gene and LTR of endogenous ecotropic murine leukemia provirus of AKV-type | |
| NS1-associated protein 1-like, RRM RNA binding protein GRY-RBP | AF093821 | Unknown | 2.33 | 1.24 | 1.02 | A component of apobec-1 editosome |
| Eps15 | L21768 | 2.33 | 1.00 | 0.99 | EGF receptor pathway substrate involved in mitogenic activity | |
| Heat shock protein, 70 kDa, 3 | M12571 | 17 | 2.32 | 1.81 | 0.97 | Chaperone of proteins exposed to stress |
| IL-1β | M15131 | 2 F | 2.32 | 0.91 | 0.97 | Inflammatory cytokine |
| Granzyme E | M36901 | 14 | 2.31 | 1.11 | 0.94 | Serine protease |
| Bone sialoprotein (IBSP) | L20232 | 5 | 2.27 | 0.61 | 0.92 | Involved in nucleation of hydroxyapatiteduring de novo bone formation |
| Cyclin-dependent kinase 4 | AA791962 | 2.26 | 0.61 | 0.92 | Regulates cell cycle progression through the G1 phase | |
| Connexin (Cx26) | M81445 | 14 D1-E1 | 2.25 | 1.18 | 0.92 | Gap junctional communication |
| Slfn4 | AF099977 | Unknown | 2.23 | 1.63 | 0.89 | Growth regulatory gene that affects thymocyte development |
| Matrix metalloproteinase 3, stromelysin 1 | X66402 | 9 | 2.22 | 0.59 | 0.89 | Degradation of extracellular matrix |
| RIKEN cDNA 1200008D14 gene | AW208938 | 2.17 | 1.22 | 0.88 | Unknown | |
| Keretin 6a, MK6a, mK6a, Krt2–6c, 2310016L08Rik | AB012033 | 15 | 2.16 | 0.85 | 0.86 | Structural protein of epithelial cells |
| Retinoblastoma susceptibility protein (pp105 Rb) | M26391 | 13q14.2 | 2.15 | 0.64 | 0.83 | Antiapoptotic |
| Cyclin 1 | AF005886 | 5 E3.3-F1.3 | 2.15 | 1.52 | 2.16 | Cell cycle progression |
| Plasminogen activator inhibitor 2 | X16490 | 1 | 2.14 | 0.74 | 0.82 | Cell invasion |
| Adenylate kinase 3 | AB020239 | 4 | 2.10 | 0.81 | 0.81 | Metabolism of ATP |
| Transcription factor AP-2 | U17291 | 13 A5-B1 | 2.10 | 0.94 | 0.81 | Oncogenic transcription factor |
| Early growth response 2 | AV232606 | 10 | 2.10 | 0.75 | 0.81 | Candidate gene for the regulation of pattern formation in the hindbrain |
| Matrilin 2 | U69262 | 2.09 | 1.28 | 0.78 | Component of extracellular filamentous networks | |
| Histone H1 | X72805 | 13 | 2.07 | 1.11 | 0.78 | Nuclear protein |
| Connexin 43 (α1 gap junction) | M63801 | 10 | 2.07 | 1.97 | 0.73 | Gap junctional communication |
| Smooth muscle γ actin gene | U20365 | 6 | 2.06 | 0.76 | 0.71 | Cell structural protein |
| Matrix metalloproteinase 9 | AV239570 | 2 | 2.05 | 0.78 | 0.69 | Degradation of extracellular matrix |
| Hyaluronan synthase 2 | U52524 | c-myc/Dhfr-rs1 | 2.04 | 1.17 | 0.63 | Membrane-bound enzyme for the polymerization of hyaluronan |
| Mast cell protease 8 | X78545 | Unknown | 2.04 | 0.84 | 0.61 | Granule constituent protease |
| Similar to latent TGF-β binding protein-4. | AA838868 | 2.03 | 0.72 | 0.59 | Unknown | |
| Ephrin-A1; ligand for Eph-related receptor tyrosine kinases | U90662 | 3 | 2.02 | 0.65 | 0.57 | Involved in cell migration and adhesion |
| Forkhead box C1 | AF045017 | D13Mit3/D13Bir9 | 2.02 | 1.45 | 2.04 | A TGFβ1-responsive gene |
| ATP-binding cassette, subfamily B (MDR/TAP), member 4 | M24417 | 5 | 2.01 | 1.55 | 0.53 | Compensatory transporter for vincristine and etoposide |
| Tm4sf, Tspan6, TSPAN-6, transmembrane 4 superfamily | AF053454 | 2.01 | 0.35 | 0.51 | Cell motility and metastasis | |
| Viral envelope like protein (G7e) | U69488 | 18.96–18.98 | 2.01 | 0.70 | 0.49 | Unknown (absent in human) |
Genes are selected based on the following criteria: 1) they must be expressed two times less than the controls (untreated wild type or GKO) to qualify as IL-12-inhibited genes; 2) their expression detection must be statistically valid as present with detection p < 0.05; 3) IFN-γ-dependent genes are defined as those whose expression must be inhibited by IL-12 in GKO mice less than two times compared with untreated GKO mice; and 4) IFN-γ-independent genes are defined as those whose expression must be inhibited by IL-12 in GKO mice greater than two times compared with untreated GKO mice. GenBank accession numbers are given. Chromosomal locations (map) are given when they are known. The numbers in columns 4–6 are relative fold of expression over the baseline (control untreated, wild-type group), which is arbitrarily set as 1. These numbers are negative, meaning that they represent the degree of inhibition of the expression of these genes by the IL-12 treatment, compared with the baseline.
Table IV.
IFN-γ-independent genes inhibited by IL-12 (two genes)a
| Common Name | Genbank | Map | WT—IL12- Treated |
GKO—Not Treated |
GKO—IL12- Treated |
Function |
|---|---|---|---|---|---|---|
| Decysin | AJ242912 | Unknown | 5.93 | 2.64 | 6.67 | Metalloprotease |
| CCP5 | X56990 | 14 | 3.07 | 0.92 | 2.87 | Cytotoxic cell protease |
Genes are selected based on the following criteria: 1) they must be expressed two times less than the controls (untreated wild type or GKO) to qualify as IL-12-inhibited genes; 2) their expression detection must be statistically valid as present with detection p < 0.05; 3) IFN-γ-dependent genes are defined as those whose expression must be inhibited by IL-12 in GKO mice less than two times compared with untreated GKO mice; and 4) IFN-γ-independent genes are defined as those whose expression must be inhibited by IL-12 in GKO mice greater than two times compared with untreated GKO mice. GenBank accession numbers are given. Chromosomal locations (map) are given when they are known. The numbers in columns 4–6 are relative fold of expression over the baseline (control untreated, wild-type group), which is arbitrarily set as 1. These numbers are negative, meaning that they represent the degree of inhibition of the expression of these genes by the IL-12 treatment, compared with the baseline.
IFN-γ dependency and inducibility by IL-12 of some important chemokine genes
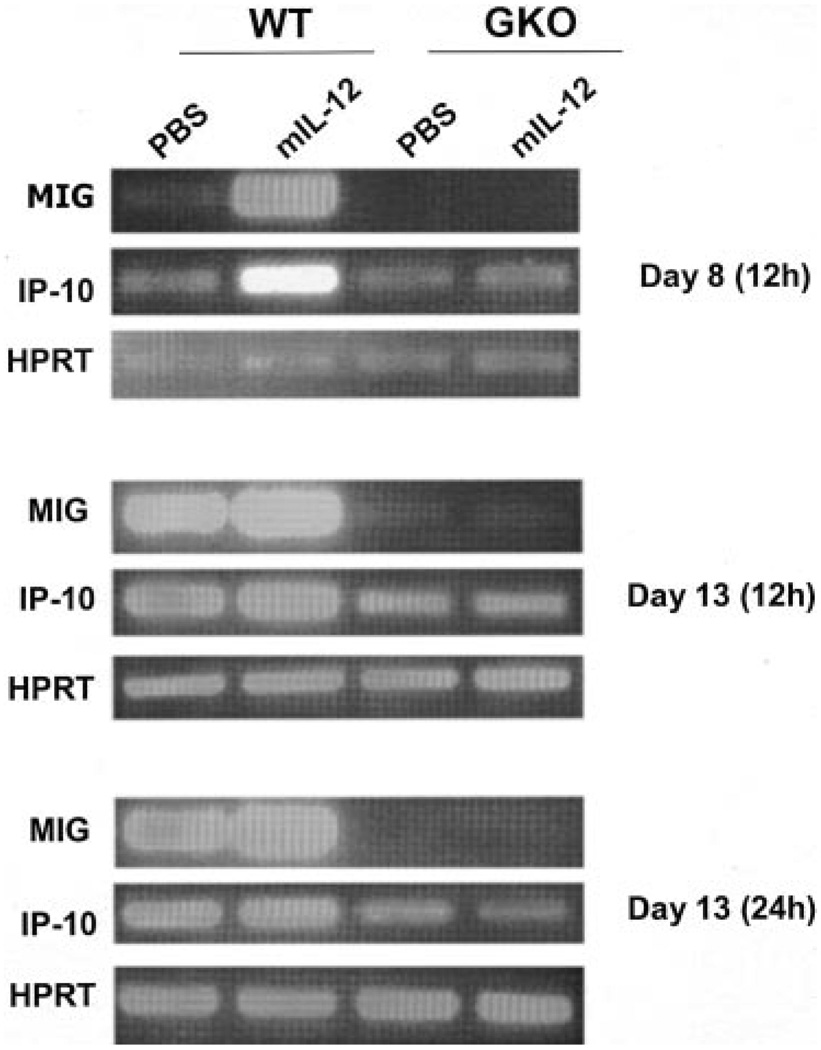
An interesting question to address is how IL-12 was able to induce intratumoral T cell infiltration and reduce blood vessel formation in GKO mice. MIG and IP-10 are IFN-γ-inducible chemoattractants for activated T cells and are purported to be inhibitors of angiogenic activity. Their expression was up-regulated within the tumor in wild-type mice during the course of IL-12 therapy (Table I). Thus, it is likely that, in wild-type animals, these chemokines contribute to the antitumor mechanisms. However, whereas MIG expression is completely IFN-γ dependent, IP-10 expression is not strictly so (40). We decided to analyze the expression of MIG and IP-10 by RT-PCR. To assess whether these chemokines could be induced transiently following IL-12 treatment in vivo in the presence or absence of a functional IFN-γ gene, we analyzed mRNA expression of wild-type and GKO mice bearing 4T1 tumor 12 h following IL-12 treatment on days 8 (first injection) and 13 (third injection), respectively (Fig. 6). On day 8 of tumor progression, MIG expression was greatly induced by the IL-12 treatment in wild-type mice, whereas no MIG expression was detectable in GKO mice either treated or not with IL-12. On day 13, there was a strong spontaneous expression of MIG mRNA, which was further enhanced by IL-12 in wild-type mice but not in GKO mice. IP-10 mRNA expression was very similar to that of MIG in wild-type mice on days 8 and 13. However, in contrast to MIG, expression of IP-10 mRNA was readily detectable in GKO mice on both days, albeit at a lower level than in wild-type mice. Interestingly, IL-12 did not enhance IP-10 expression 12 or 24 h (data not shown) after the treatment. These results are highly consistent with the microarray data obtained at a much later time frame, and indicate that, although IP-10 expression is not tightly dependent on IFN-γ, IL-12 cannot induce IP-10 in the absence of endogenous IFN-γ.
FIGURE 6.
IFN-γ dependency and inducibility by IL-12 of chemokines. Total RNA was isolated from 4T1 tumor cells on days 8 and 13 from wild-type and GKO mice treated or not with murine IL-12 for 12 h (12 and 24 h for the D13 samples). The RNA samples were subjected to RT-PCR analysis for the detection of MIG and IP-10. As an internal control, HPRT mRNA was also analyzed.
Discussion
IFN-γ is a primary effector cytokine mediating a wide range of antitumor activities of the host through its ability to activate several classes of genes that control Ag processing and presentation, leukocyte trafficking, induction of Th1 and CTL, antiangiogenesis, direct and indirect tumor cytotoxicity (34, 41). IFN-γ -independent antitumor mechanisms mediated by IL-12 have been reported in which cytokines such as GM-CSF (27) and IL-15 (29) were shown to be of importance in the absence of IFN-γ. However, to date, there have been no studies in which IL-12-mediated, IFN-γ-independent control of malignant development has been examined systematically and comprehensively, from the perspective of either the immune system or the tumor. Our study will help lead to a greater understanding of the cellular and molecular pathways whereby IL-12 induces tumor regression and protective immunity.
The approach we took to examine the global gene expression profile in the whole tumor with its stroma and lymphocytic infiltrates, as opposed to looking at certain purified populations of cells within the tumor, provides us with the ability to view all of the transcriptional changes (limited by the number of genes on the microarray) effected by IL-12, thus permitting a more comprehensive analysis of the interactions between malignant cells and their microenvironment.
Our data demonstrate that, in the absence of IFN-γ, IL-12 retains the ability, at least partially, to recruit T lymphocytes to and inhibit neoangiogenesis in the tumor. RANTES, the most induced gene in IL-12-treated GKO mice, has been shown to elicit strong antitumor and recall responses as well as tumor-specific cytotoxic T cell activity when delivered intratumorally using an HSV amplicon vector in a murine model of pre-established lymphoma (42). The microarray analysis of the primary tumors indicates that T lymphocytes bearing CD6, a pan-T cell structure that binds to the activated leukocyte cell adhesion molecule (ALCAM/CD166) (43), were recruited to the tumor site by IL-12 in an IFN-γ-independent manner. mAbs to CD6 exclusively stimulate T cell clones with γδ rather than αβ Ag receptors (44). Furthermore, simultaneously cross-linking the accessory and costimulatory molecules CD6 and CD28 can induce T cell activation through an Ag-independent pathway involving signaling steps distinct between CD6 and CD28 (45).
The inhibitory program effected by IL-12 includes predominantly genes in two major functions: cell proliferation and migration. Top on the list is Proliferin (PLF). PLF is angiogenic (46) and important for the migration of uterine vascular endothelial cells toward the giant cells (47), the outer layer of the extraembryonic compartment at the implantation site. Secreted PLF protein accumulates to high levels in both the maternal circulation and in the fetal compartment (48), and PLF binding to developing structures in the fetus suggests roles on both endothelial cells and on other cell types (49). Toft et al. (50) showed that PLF gene expression can also occur in a progressive fibrosarcoma mouse tumor cell model. PLF expression is greatly augmented in cell lines from the aggressively invasive stage of development, a stage at which the tumor becomes highly angiogenic, and PLF expression remains high in cell lines from the end stage of fibrosarcoma. Secreted PLF protein is functionally important in tumor cell angiogenic activity. These results suggest that an extraembryonic genetic program, which has evolved to support fetal growth, may be reactivated in certain tumors and contribute to tumor growth.
Structural changes in the extracellular matrix are necessary for cell migration during tissue remodeling and tumor invasion. The matrix metalloproteinases and their inhibitors have been shown to be critical modulators of extracellular matrix composition and are thus crucial in neoplastic cell invasion and metastasis (51). Thus, the inhibition of several of metalloproteases by IL-12 in IFN-γ-dependent (Table III) or -independent (Table IV) manners very likely represents one of the major mechanisms whereby IL-12 could impede tumor progression.
A number of genes are also implicated as IFN-γ-independent antitumor factors in the microarray-based analysis, such as some of the lymphocyte chemoattractants and cytotoxic molecules that had not been previously appreciated in this context. One enigma is that there appear to be no obvious candidates of the 50 genes induced by IL-12 in the absence of IFN-γ that have a reported role in restricting angiogenesis in the tumor. However, the effect of IL-12 on blood vessels was strikingly apparent (Fig. 2h). There could be a number of explanations for this. First, the responsible gene(s) may not be represented in the U74A chip that covers about one-third of the mouse genome. Second, the responsible factor(s) may not be regulated at the level of mRNA expression. Third, the responsible gene(s) may be a known gene(s) with an undescribed or underdescribed function in regulating angiogenesis. For example, B lymphocyte chemoattractant (CXCL13) has been reported to have an inhibitory effect in vitro on endothelial cell proliferation induced by fibroblast growth factor (FGF)-2 through competitive displacement of FGF-2 binding to endothelial cells, suggesting that CXCL13 may modulate angiogenesis by interfering with FGF-2 activity (52). Fourth, there could be angiogenic factors that are suppressed by IL-12. However, the two genes that were inhibited by IL-12 in the tumors, decysin and cytotoxic cell proteinase 5, have no reported function related to neovascularization. It would be of great interest to further explore this aspect of IL-12-mediated, IFN-γ-independent antiangiogenic activities.
Footnotes
This research was supported by National Institutes of Health Grant CA79772 (to X.M.). S.C. was supported by a fellowship from the Susan G. Komen Breast Cancer Foundation.
Abbreviations used in this paper: MIG, monokine induced by IFN-γ; IP-10, IFN-γ-inducible protein 10; GKO, IFN-γ knockout; WT, wild type; TIL, tumor-infiltrating lymphocyte; CXCL, CXC ligand; PLF, Proliferin; FGF, fibroblast growth factor.
References
- 1.Ehrlich P. Ueber den jetzigen stand der Karzinomforschung. Ned. Tijdschr. Geneeskd. 1909;5:273. [Google Scholar]
- 2.Burnet F. Cancer—a biological approach. Brit. Med. J. 1957;1:841. doi: 10.1136/bmj.1.5023.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stutman O. Tumor development after 3-methylcholanthrene in immunologically deficient athymic-nude mice. Science. 1974;183:534. doi: 10.1126/science.183.4124.534. [DOI] [PubMed] [Google Scholar]
- 4.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 2002;3:991. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 5.Browning MJ, Bodmer WF. MHC antigens and cancer: implications for T-cell surveillance. Curr. Opin. Immunol. 1992;4:613. doi: 10.1016/0952-7915(92)90036-e. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Ashe S, Brady WA, Hellstrom I, Hellstrom KE, Ledbetter JA, McGowan P, Linsley PS. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell. 1992;71:1093. doi: 10.1016/s0092-8674(05)80059-5. [DOI] [PubMed] [Google Scholar]
- 7.Townsend SE, Allison JP. Tumor rejection after direct costimulation of CD8+ T cells by B7-transfected melanoma cells. Science. 1993;259:368. doi: 10.1126/science.7678351. [DOI] [PubMed] [Google Scholar]
- 8.Coughlin CM, Salhany KE, Gee MS, LaTemple DC, Kotenko S, Ma X, Gri G, Wysocka M, Kim JE, Liu L, et al. Tumor cell responses to IFN-γ affect tumorigenicity and response to IL-12 therapy and antiangiogenesis. Immunity. 1998;9:25. doi: 10.1016/s1074-7613(00)80585-3. [DOI] [PubMed] [Google Scholar]
- 9.Trinchieri G. Interleukin-12: a cytokine at the interface of inflammation and immunity. Adv. Immunol. 1998;70:83. doi: 10.1016/s0065-2776(08)60387-9. [DOI] [PubMed] [Google Scholar]
- 10.Trinchieri G, Scott P. Interleukin-12: basic principles and clinical applications. Curr. Top. Microbiol. Immunol. 1999;238:57. doi: 10.1007/978-3-662-09709-0_4. [DOI] [PubMed] [Google Scholar]
- 11.Rook AH, Wood GS, Yoo EK, Elenitsas R, Kao DM, Sherman ML, Witmer WK, Rockwell KA, Shane RB, Lessin SR, Vonderheid EC. Interleukin-12 therapy of cutaneous T-cell lymphoma induces lesion regression and cytotoxic T-cell responses. Blood. 1999;94:902. [PubMed] [Google Scholar]
- 12.Rook AH, Zaki MH, Wysocka M, Wood GS, Duvic M, Showe LC, Foss F, Shapiro M, Kuzel TM, Olsen EA, et al. The role for interleukin-12 therapy of cutaneous T cell lymphoma. Ann. NY Acad. Sci. 2001;941:177. doi: 10.1111/j.1749-6632.2001.tb03721.x. [DOI] [PubMed] [Google Scholar]
- 13.Ansell SM, Witzig TE, Kurtin PJ, Sloan JA, Jelinek DF, Howell KG, Markovic SN, Habermann TM, Klee GG, Atherton PJ, Erlichman C. Phase 1 study of interleukin-12 in combination with rituximab in patients with B-cell non-Hodgkin lymphoma. Blood. 2002;99:67. doi: 10.1182/blood.v99.1.67. [DOI] [PubMed] [Google Scholar]
- 14.Mortarini R, Borri A, Tragni G, Bersani I, Vegetti C, Bajetta E, Pilotti S, Cerundolo V, Anichini A. Peripheral burst of tumor-specific cytotoxic T lymphocytes and infiltration of metastatic lesions by memory CD8+ T cells in melanoma patients receiving interleukin 12. Cancer Res. 2000;60:3559. [PubMed] [Google Scholar]
- 15.Gollob JA, Mier JW, Veenstra K, McDermott DF, Clancy D, Clancy M, Atkins MB. Phase I trial of twice-weekly intravenous interleukin 12 in patients with metastatic renal cell cancer or malignant melanoma: ability to maintain IFN-γ induction is associated with clinical response. Clin Cancer Res. 2000;6:1678. [PubMed] [Google Scholar]
- 16.Lee P, Wang F, Kuniyoshi J, Rubio V, Stuges T, Groshen S, Gee C, Lau R, Jeffery G, Margolin K, et al. Effects of interleukin-12 on the immune response to a multipeptide vaccine for resected metastatic melanoma. J. Clin. Oncol. 2001;19:3836. doi: 10.1200/JCO.2001.19.18.3836. [DOI] [PubMed] [Google Scholar]
- 17.Kang WK, Park C, Yoon HL, Kim WS, Yoon SS, Lee MH, Park K, Kim K, Jeong HS, Kim JA, et al. Interleukin 12 gene therapy of cancer by peritumoral injection of transduced autologous fibroblasts: outcome of a phase I study. Hum. Gene Ther. 2001;12:671. doi: 10.1089/104303401300057388. [DOI] [PubMed] [Google Scholar]
- 18.Gajewski TF, Fallarino F, Ashikari A, Sherman M. Immunization of HLA-A2+ melanoma patients with MAGE-3 or MelanA peptide-pulsed autologous peripheral blood mononuclear cells plus recombinant human interleukin 12. Clin. Cancer Res. 2001;7:895s. [PubMed] [Google Scholar]
- 19.Portielje JE, Kruit WH, Schuler M, Beck J, Lamers CH, Stoter G, Huber C, de Boer-Dennert M, Rakhit A, Bolhuis RL, Aulitzky WE. Phase I study of subcutaneously administered recombinant human interleukin 12 in patients with advanced renal cell cancer. Clin. Cancer Res. 1999;5:3983. [PubMed] [Google Scholar]
- 20.Miller FR, Miller BE, Heppner GH. Characterization of metastatic heterogeneity among subpopulations of a single mouse mammary tumor: heterogeneity in phenotypic stability. Invasion Metastasis. 1983;33:22. [PubMed] [Google Scholar]
- 21.Pulaski BA, Ostrand-Rosenberg S. Reduction of established spontaneous mammary carcinoma metastases following immunotherapy with major histocompatibility complex class II and B7.1 cell-based tumor vaccines. Cancer Res. 1998;58:1486. [PubMed] [Google Scholar]
- 22.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399. [PubMed] [Google Scholar]
- 23.Morecki S, Yacovlev L, Slavin S. Effect of indomethacin on tumorigenicity and immunity induction in a murine model of mammary carcinoma. Int. J. Cancer. 1998;75:894. doi: 10.1002/(sici)1097-0215(19980316)75:6<894::aid-ijc12>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Ostrand-Rosenberg S, Grusby MJ, Clements VK. Cutting edge: STAT6-deficient mice have enhanced tumor immunity to primary and metastatic mammary carcinoma. J. Immunol. 2000;165:6015. doi: 10.4049/jimmunol.165.11.6015. [DOI] [PubMed] [Google Scholar]
- 25.Rakhmilevich AL, Janssen K, Hao Z, Sondel PM, Yang NS. Interleukin-12 gene therapy of a weakly immunogenic mouse mammary carcinoma results in reduction of spontaneous lung metastases via a T-cell-independent mechanism. Cancer Gene Ther. 2000;7:826. doi: 10.1038/sj.cgt.7700176. [DOI] [PubMed] [Google Scholar]
- 26.Wang ZE, Zheng S, Corry DB, Dalton DK, Seder RA, Reiner SL, Locksley RM. Interferon-γ-independent effects of interleukin-12 administered during acute or established infection due to Leishmania major. Proc. Natl. Acad. Sci. USA. 1994;91:12932. doi: 10.1073/pnas.91.26.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zilocchi C, Stoppacciaro A, Chiodoni C, Parenza M, Terrazzini N, Colombo MP. Interferon-γ-independent rejection of interleukin 12-transduced carcinoma cells requires CD4+ T cells and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 1998;188:133. doi: 10.1084/jem.188.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hafner M, Falk W, Echtenacher B, Mannel DN. Interleukin-12 activates NK cells for IFN-γ-dependent and NKT cells for IFN-γ-independent antimetastatic activity. Eur. Cytokine Netw. 1999;10:541. [PubMed] [Google Scholar]
- 29.Gri G, Chiodoni C, Gallo E, Stoppacciaro A, Liew FY, Colombo MP. Antitumor effect of interleukin (IL)-12 in the absence of endogenous IFN-γ: a role for intrinsic tumor immunogenicity and IL-15. Cancer Res. 2002;62:4390. [PubMed] [Google Scholar]
- 30.Comes A, Di Carlo E, Musiani P, Rosso O, Meazza R, Chiodoni C, Colombo MP, Ferrini S. IFN-γ-independent synergistic effects of IL-12 and IL-15 induce anti-tumor immune responses in syngeneic mice. Eur. J. Immunol. 2002;32:1914. doi: 10.1002/1521-4141(200207)32:7<1914::AID-IMMU1914>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 31.Pulaski BA, Smyth MJ, Ostrand-Rosenberg S. Interferon-γ-dependent phagocytic cells are a critical component of innate immunity against metastatic mammary carcinoma. Cancer Res. 2002;62:4406. [PubMed] [Google Scholar]
- 32.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulaski BA, Clements VK, Pipeling MR, Ostrand-Rosenberg S. Immunotherapy with vaccines combining MHC class II/CD80+ tumor cells with interleukin-12 reduces established metastatic disease and stimulates immune effectors and monokine induced by interferon-γ. Cancer Immunol. Immunother. 2000;49:34. doi: 10.1007/s002620050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tannenbaum CS, Hamilton TA. Immune-inflammatory mechanisms in IFN-γ-mediated anti-tumor activity. Semin. Cancer Biol. 2000;10:113. doi: 10.1006/scbi.2000.0314. [DOI] [PubMed] [Google Scholar]
- 35.Burke F, Knowles RG, East N, Balkwill FR. The role of indoleamine 2,3-dioxygenase in the anti-tumour activity of human interferon-γ in vivo. Int. J. Cancer. 1995;60:115. doi: 10.1002/ijc.2910600117. [DOI] [PubMed] [Google Scholar]
- 36.Thiemmara V, Pays L, Danty E, Jourdan F, Moyse E, Mehlen P. Serine protease inhibitor Spi2 mediated apoptosis of olfactory neurons. Cell Death Differ. 2002;9:1343. doi: 10.1038/sj.cdd.4401098. [DOI] [PubMed] [Google Scholar]
- 37.Danesch U, Hoeck W, Ringold GM. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27: CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J. Biol. Chem. 1992;267:7185. [PubMed] [Google Scholar]
- 38.Liang L, Zhao M, Xu Z, Yokoyama KK, Li T. Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem. J. 2003;370:195. doi: 10.1042/BJ20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen BS, Cook KS, Yaglom J, Groves DL, Volanakis JE, Damm D, White T, Spiegelman BM. Adipsin and complement factor D activity: an immune-related defect in obesity. Science. 1989;244:1483. doi: 10.1126/science.2734615. [DOI] [PubMed] [Google Scholar]
- 40.Asensio VC, Maier J, Milner R, Boztug K, Kincaid C, Moulard M, Phillipson C, Lindsley K, Krucker T, Fox HS, Campbell IL. Interferon-independent, human immunodeficiency virus type 1 gp120-mediated induction of CXCL10/IP-10 gene expression by astrocytes in vivo and in vitro. J. Virol. 2001;75:7067. doi: 10.1128/JVI.75.15.7067-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colombo MP, Trinchieri G. Interleukin-12 in anti-tumor immunity and immunotherapy. Cytokine Growth Factor Rev. 2002;13:155. doi: 10.1016/s1359-6101(01)00032-6. [DOI] [PubMed] [Google Scholar]
- 42.Kutubuddin M, Federoff HJ, Challita-Eid PM, Halterman M, Day B, Atkinson M, Planelles V, Rosenblatt JD. Eradication of pre-established lymphoma using herpes simplex virus amplicon vectors. Blood. 1999;93:643. [PubMed] [Google Scholar]
- 43.Bowen MA, Bajorath J, D’Egidio M, Whitney GS, Palmer D, Kobarg J, Starling GC, Siadak AW, Aruffo A. Characterization of mouse ALCAM (CD166): the CD6-binding domain is conserved in different homologs and mediates cross-species binding. Eur. J. Immunol. 1997;27:1469. doi: 10.1002/eji.1830270625. [DOI] [PubMed] [Google Scholar]
- 44.Pawelec G, Buhring HJ. Monoclonal antibodies to CD6 preferentially stimulate T-cell clones with γ/δ rather than α/β antigen receptors. Hum. Immunol. 1991;31:165. doi: 10.1016/0198-8859(91)90022-2. [DOI] [PubMed] [Google Scholar]
- 45.Osorio LM, Rottenberg M, Jondal M, Chow SC. Simultaneous cross-linking of CD6 and CD28 induces cell proliferation in resting T cells. Immunology. 1998;93:358. doi: 10.1046/j.1365-2567.1998.00442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jackson D, Volpert OV, Bouck N, Linzer DI. Stimulation and inhibition of angiogenesis by placental proliferin and proliferin-related protein. Science. 1994;266:1581. doi: 10.1126/science.7527157. [DOI] [PubMed] [Google Scholar]
- 47.Ma GT, Roth ME, Groskopf JC, Tsai FY, Orkin SH, Grosveld F, Engel JD, Linzer DI. GATA-2 and GATA-3 regulate trophoblast-specific gene expression in vivo. Development. 1997;124:907. doi: 10.1242/dev.124.4.907. [DOI] [PubMed] [Google Scholar]
- 48.Lee S, Talamantes F, Wilder E, Linzer D, Nathans D. Trophoblastic giant cells of the mouse placenta as the site of proliferin synthesis. Endocrinology. 1988;122:1761. doi: 10.1210/endo-122-5-1761. [DOI] [PubMed] [Google Scholar]
- 49.Jackson D, Linzer DIH. Proliferin transport and binding in the mouse fetus. Endocrinology. 1997;138:149. doi: 10.1210/endo.138.1.4828. [DOI] [PubMed] [Google Scholar]
- 50.Toft DJ, Rosenberg SB, Bergers G, Volpert O, Linzer DIH. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc. Natl. Acad. Sci. USA. 2001;98:13055. doi: 10.1073/pnas.231364798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bodey B, Bodey B, Jr, Groger AM, Siegel SE, Kaiser HE. Invasion and metastasis: the expression and significance of matrix metalloproteinases in carcinomas of the lung. In Vivo. 2001;15:175. [PubMed] [Google Scholar]
- 52.Spinetti G, Camarda G, Bernardini G, Romano Di Peppe S, Capogrossi MC, Napolitano M. The chemokine CXCL13 (BCA-1) inhibits FGF-2 effects on endothelial cells. Biochem. Biophys. Res. Commun. 2001;289:19. doi: 10.1006/bbrc.2001.5924. [DOI] [PubMed] [Google Scholar]