Abstract
The tumor suppressor protein p53 plays a role in cellular responses to cancer-initiating events by regulating progress through the cell cycle. Several recent studies have shown that p53 transactivates expression of the members of the proapoptotic microRNA-34 family, which are underexpressed in several cancers. We demonstrate here that the latency-associated cluster of microRNAs (miRNA) encoded by an oncogenic herpesvirus, gallid herpesvirus 2 (GaHV-2), is a direct target of p53. Robust transcriptional activity was induced in three avian cell lines by a sequence mapping 600 base pairs (bp) upstream of the cluster of miRNAs. We found transcription start sites for the pri-miRNA transcript at the 3′ end of this transcription-inducing sequence. The promoter has no consensus core promoter element, but is organized into a variable number of tandem repeats of 60-bp harboring p53-responsive elements (RE). The minimal functional construct consists of two tandem repeats. Mutagenesis to change the sequence of the p53 RE abolished transcriptional activity, whereas p53 induction enhanced mature miRNA expression. The identification of a viral miRNA promoter regulated by p53 is biologically significant, because all avirulent GaHV-2 strains described to date lack the corresponding regulatory sequence, whereas all virulent, very virulent, and hypervirulent strains possess at least two tandem repeats harboring the p53 RE.
Keywords: p53, promoter, microRNA, herpesvirus
INTRODUCTION
The transcription of eukaryotic genes results from a set of combinatorial events, from chromatin recognition to the positioning of the RNA polymerase at the appropriate initiation site. This process, which includes chromatin modification and the recruitment of cofactors and basal transcription components, leads to the assembly of an elongation-competent transcription complex. Some of the mechanisms underlying the activation of transcription have been elucidated, including those involving core promoter elements, such as the TATA box and initiator element (Sandelin et al. 2007). Several core elements of the TATA box containing human promoters have been identified; these elements include the downstream core promoter element (Lee et al. 2005), the TFIIB recognition element (Deng and Roberts 2005) and the initiator element (INR). Mutagenesis of the INR sequence led to identification of the YYANWYY motif as a consensus sequence (Javahery et al. 1994; Smale and Kadonaga 2003). However, although the core promoter structure was initially thought to be invariant, it is now clear that there is extensive diversity. Thousands of different transcriptional start sites (TSSs) for mammalian class II promoters have been described, revealing the variability of potential TSSs (Frith et al. 2008). Transcription initiation may occur at various nucleotide positions within a core promoter region, but there is a preference for both a purine in position +1 and a pyrimidine in position −1 (Carninci et al. 2006; Frith et al. 2008). Otherwise, the proportion of promoters with clearly identifiable TATA boxes has tended to decrease as the number of promoters discovered increases (Suzuki et al. 2001; Sandelin et al. 2007; Yang et al. 2007). Thus, promoters lacking standard core promoter elements constitute a significant fraction of the eukaryotic genome. However, little is known about their mechanisms of transcriptional activation or the cis-activating DNA motifs involved.
MicroRNAs (miRNA) are a newly discovered class of small RNAs of ∼23 nucleotides (nt) in length that have been shown to be key mediators of RNA silencing pathways (Ambros 2004; Bartel 2004; Ason et al. 2006). By fine-tuning gene expression through the repression of target mRNA translation, miRNAs control cellular pathways governing several biological functions and the fate of the cell: cell differentiation, cell proliferation, or apoptosis (Brennecke et al. 2003; Xu et al. 2003). Both the expression and function of miRNAs are tissue-specific and temporally regulated (Pasquinelli et al. 2005; Landgraf et al. 2007). Tumorigenesis is frequently associated with disruption of the differential and dynamic expression patterns of miRNAs (Tagawa et al. 2007; Chang et al. 2008). The spatiotemporal control of miRNA expression is thought to be principally transcriptional. However, the transcriptional regulation of miRNAs is still poorly understood, because few miRNA promoters have been characterized. Some transcription factors have been identified as positive (tumor suppressor protein 53, p53, NF-κB) (Taganov et al. 2006; Chang et al. 2007) or negative (c-myc) (Chang et al. 2008) regulators of the expression of particular miRNAs. In one anti-tumoral pathway, p53 transactivates the pro-apoptotic miR-34a 30 kb downstream from the p53-responsive element (RE) (Chang et al. 2007; Raver-Shapira et al. 2007). Other roles of p53 in miRNA-dependent regulatory networks were discovered more recently, with the demonstration of p53-mediated transcriptional induction of miR-145 and miR-192 through the interaction of p53 with a potential p53 RE in their respective promoters (Song et al. 2008; Sachdeva et al. 2009). Otherwise, in a collection of 175 miRNAs for which transcription initiation regions have been described, 25 (19%) of the miRNA promoters contain a TATA element, 21% have a TFIIB recognition element, and 47% have a consensus initiator (Ozsolak et al. 2008). Thus, a significant proportion of miRNA genes lack standard core promoter elements and the transcriptional regulation of primary transcripts generating miRNAs (primiRNAs) is far from clear. Further and detailed investigation of the mechanisms governing miRNA expression is therefore required.
In this study, we investigated the transcriptional regulation of a viral microRNA cluster encoded within the latency-associated transcript (LAT) of gallid herpesvirus 2 (GaHV-2), an alphaherpesvirus model of virus-induced lymphomagenesis (Burgess et al. 2004; Osterrieder et al. 2006). Thirteen viral miRNAs are expressed by oncogenic strains of GaHV-2, which cause T-cell lymphomas in chicken (Burnside et al. 2006; Burnside and Morgan 2007; Morgan et al. 2008; Xu et al. 2008). The viral miRNA coding sequences are clustered into two separate genomic loci: cluster 1 (mdv1-miR-M1, 2, 3, 4, 5, 9, 11, 12, 31) maps upstream of and downstream from the Meq gene; cluster 2 (mdv1-miR-M6, 7, 8, 10) maps near the 5′ end of the LAT (Fig. 1). The LAT cluster encodes four pre-miRNAs that are processed to give seven mature miRNAs (miR-M6-5p and 3p; miR-M7-5p and 3p; mdv1-miR-M8-5p and 3p; and miR-M10-5p) (Fig. 1). These miRNAs have been reported to be strongly expressed in MSB-1 cells derived from a GaHV-2 lymphoma and in primary MD lymphomas (Morgan et al. 2008; Yao et al. 2008). The LAT-miRNA coding sequences are located near the 5′ end of a previously characterized large intron in LAT (Cantello et al. 1994, 1997). However, neither the promoter nor the TSS of LAT has been identified. Here, we identify and characterize a robust promoter controlling the expression of the LAT-miRNAs. This promoter was found embedded within a variable number of tandem repeats of the viral genome. No standard core promoter elements were found in this promoter, which was transactivated by p53 through a specific consensus RE.
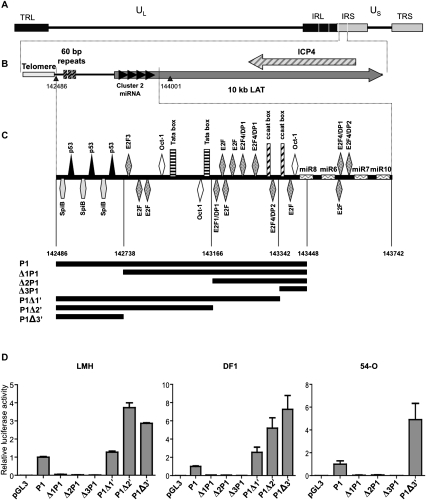
FIGURE 1.
Localization of the latency-associated transcript promoter. (A) The organization of the GaHV-2 genome is similar to that of an E alpha herpes virus. UL and US indicate unique long and unique short sequences; TRL/TRS indicate terminal repeats, long and short, respectively; and IRL/IRS indicate internal repeats, long and short, respectively. (B) Localization of miRNA cluster 2 in the TRS and IRS. The gray arrow indicates a noncoding transcript and black arrows indicate cluster 2 miRNAs. (C) Schematic diagram of the various cis elements identified in the promoter by in silico analysis (Genomatix). Coordinates of the pre-mdv1-miRNAs are as follows: pre-mdv1-miR-M8 (143403-143482), pre-mdv1-miR-M6 (143524-143594), pre-mdv1-miR-M7 (143695-143772), and pre-mdv-miR-M10 (143815-143895). Potential promoter sequences are segmented and each potential promoter is represented in black under the schematic DNA sequence. (D) Relative luciferase activity for each promoter-luciferase construct, normalized with respect to the luciferase activity obtained with the P1 pGL3 vector is shown. These values reflect the transcriptional strength of the various promoters in LMH, DF1, and 54-O cells. Each value corresponds to the mean of three independent assays performed simultaneously. Error bars indicate the SEM for three replicates.
RESULTS
Robust transcriptional activity is induced by the tandem repeat sequences upstream of the latency-associated transcript of GaHV-2 encoding viral miRNAs
We investigated the regulation of miRNA expression from the LAT, by studying putative promoter DNA sequences between the telomeric repeats and the locus encoding LAT-derived miRNAs (Fig. 1A,B). Genomatix software (www.genomatix.de/cgi) was used for in silico predictions of the cis response elements (RE) in this DNA sequence (Fig. 1C). The 5′ region of this sequence amplified from RB-1B Bacmide DNA consists of three repeats of 60 base pairs (bp) containing only two types of RE (p53 and SpiB), with a number of potential REs predicted downstream (Oct-1, E2F, CAAT, and TATA boxes) (Fig. 1C). We defined the largest potential promoter, P1, between the first nucleotide downstream from the telomeric repeats of the IRS of GaHV-2 and the mdv1-miR-M8-5p sequence (extending from nucleotides 142486 to 143448 in the GaHV-2 reference genome, RB-1B). The promoter sequence was amplified by PCR from RB-1B Bacmide, and inserted into a promoter-less luciferase reporter plasmid (pGL3basic), to generate the P1 construct. The transcriptional activity associated with P1 was tested in three avian cell lines (LMH, DF-1, and 54-O) (Fig. 1D): P1 yielded robust reporter activity in all three cell lines (Fig. 1D), similar to that of the CMV promoter in the positive control (pcDNA luciferase; data not shown). We identified the sequence involved in the transcriptional activity of P1, by amplifying nested-set genomic fragments resulting from three deletions each at the 5′ and 3′ends of P1 and testing them in luciferase reporter assays, as described above for P1 (Fig. 1C,D). Constructs Δ1P1, Δ2P1, and Δ3P1 correspond to nested deletions from the 5′ end of P1 deleting the 60-bp repeats. They had much lower levels of transcriptional activity than P1 (by factors of 20, 10 and 50 in LMH, DF1, and 54-O cells, respectively). The P1Δ1′, P1Δ2′, and P1Δ3′ constructs, corresponding to nested deletions from the 3′ end of P1 and retaining all the 60-bp repeats had promoter activities similar to that of unmodified P1. Thus, the 60-bp repeats at the 5′ end of P1 appear to be required for promoter activity, whereas the RE located downstream do not appear to be essential.
To confirm that the DNA promoter sequence was functional, we used rapid amplification of cDNA ends (RACE) to determine the 5′ end of the LAT-miRNA transcript from the 54-0 cell line, a GaHV-2 lymphoma cell line expressing high levels of LAT-encoded miRNAs (B. Muylkens, pers. comm.). All of the TSSs mapped to the last block of the 60-bp repeats or the proximal downstream region (Fig. 2A). We found that 16 of the 17 identified TSSs were spread over a 13-nt sequence that most of these TSSs mapped to the proximal downstream sequence (nucleotide +6, genomic coordinate 142734 in the RB-1B genome). Similarly, all of the TSSs corresponding to the RNA isolated from the LMH cell line transfected with the luciferase reporter vector P1Δ3′ (chimeric LAT-Luc) mapped to this region (Fig. 2B). Thus, all the transcripts analyzed in two independent RACE experiments were found to have been initiated in the vicinity of the last block of 60-bp repeats, suggesting that the promoter of the LAT-encoded miRNAs is essentially located in the sequence corresponding to P1Δ3′, from the telomeric sequences to the downstream proximal region of the last 60-bp repeat.
FIGURE 2.
Identification of transcription start sites by 5′ RACE-PCR. (A) Localization and frequencies of various TSSs identified by 5′ RACE-PCR in 54-O cells, a cell line isolated from a lymphoma induced by GaHV-2. (B) Localization and frequencies of various TSSs identified by 5′ RACE-PCR in LMH cells transfected with the P1Δ3′ pGL3 vector (luciferase gene under the control of the P1Δ3′ promoter). Transcription start sites are shown in bold for RNA from 54-O cells and are underlined for LMH cells transfected with the P1Δ3′ pGL3 vector. Each TSS is labeled with the number of initiations found. The DNA sequence represents GaHV-2 genome, with the last 60-bp repeat highlighted in gray.
The promoter consisting of 60-bp repeats is responsible for the production of mature miRNAs from the LAT-miRNA-cluster2
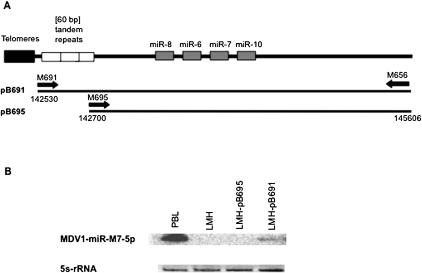
The miRNA cluster is located within the LAT gene. We therefore investigated whether the promoter of this gene mediated the production of mature miRNAs. Two constructs, pB691 and pB695, encompassing the 5′ end of the LAT mRNA defined above and the 4 miRNAs, with and without, respectively, the 60-bp repeats, were generated (Fig. 3A). The sequences corresponding to pB691 and pB695 were amplified from the bacmide p-RB-1B, with primers binding 18 nt upstream of the first 60-bp repeat (M691) or in the terminal part of the last 60-bp repeat (M695), respectively, and 1719 nt downstream from miR10 (M656). These sequences were then inserted into the pBS-SK vector (Fig. 3A). The resulting constructs were then used to transfect LMH cells and the expression of MDV-miR-7-5p was estimated from by Northern blots of extracted total RNA with a radiolabeled probe specific for MDV-miR-7-5p, which is classically considered to be representative of miRNA cluster transcription (Yao et al. 2008). RNA from peripheral blood leukocytes (PBL) of GaHV-2-infected chickens overexpressing mdv1-miR-M7-5p during natural infection (B. Muylkens, pers. comm.) was used as a positive control, and RNA from nontransfected LMH cells was used as a negative control. A specific signal of the expected size for MDV-miR-7-5p was detected in LMH cells transfected with pB691, but not in LMH cells transfected with pB695 (Fig. 3B). Mature miRNA production therefore is strongly associated with the presence of the three 60-bp tandem repeats.
FIGURE 3.
Northern blot analysis of the role of the core promoter in mature miRNA expression. (A) Schematic diagram of the LAT-miRNA promoter and the miRNA cluster. Each black line under the representation corresponds to a DNA fragment amplified from the bacmide p-RB-1B genome and inserted into the pBS-SK vector. (B) Northern blot analysis. Each lane was loaded with 15 μg of total RNA extracted with Trizol. Peripheral blood leukocytes were recovered 21 d after infection, from highly susceptible B13 chickens infected intramuscularly with RB-1B (1000 PFU of RB-1B). RNA was extracted from infected PBL, untransfected LMH cells and LMH cells transfected with pB691 or pB695. A 32P-5′ end-labeled DNA oligonucleotide probe complementary to the miRNA was used to detect the miR-7-specific signal. Ribosomal 5s RNA (ethidium bromide staining, EB) levels were used to check for equal loading.
The core promoter of the LAT-encoded miRNAs consists of at least two 60-bp repeats
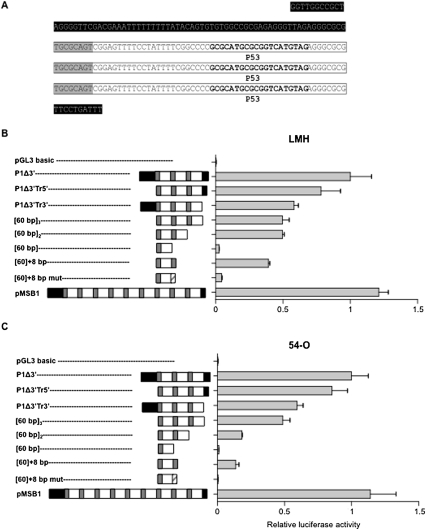
We found that the DNA sequence from the telomeres to the last 60-bp repeat upstream of the LAT locus displayed robust promoter activity, despite the absence of TATA and CAAT sequences. This region of tandem repeats in GaHV-2 isolates is hypervariable (Spatz and Silva 2007a): there are between 0 and 13 copies of the 60-bp direct repeat, depending on the viral strain. The P1Δ3′ construct obtained from RB-1B Bacmide DNA contains three copies, with 63 nt of upstream and 10 nt of downstream sequences. We investigated the effects of both the sequences flanking the tandem repeats and the copy number of tandem repeats on promoter activity in the LMH and 54-O cell lines, by studying eight additional reporter constructs (Fig. 4). The reporter activities generated by these constructs were compared with that for the promoter construct P1Δ3′ (arbitrarily set at 100%) (Fig. 4B,C). Removal of the sequence upstream of the tandem repeats did not significantly affect promoter activity (construct P1Δ3′Tr5′) (Fig. 4B,C). By contrast, removal of the short 3′ sequence flanking the tandem repeats (construct P1Δ3′Tr3′) reduced reporter activity to ∼50% that of the P1Δ3′ construct (Fig. 4B,C). This is consistent with the removal of one of the major TSSs identified within this sequence (Fig. 2). Deletion of both the 5′ and 3′ sequences surrounding the tandem repeats (construct [60 bp]3) had no further effect on the activity of the promoter (Fig. 4B,C). Therefore, these surrounding sequences, although involved in positive regulation of the promoter, are not essential for core promoter function, because three direct repeats devoid of the surrounding sequences displayed >50% of the reporter activity of the highly active control construct.
FIGURE 4.
Determination of the core promoter of the LAT-miRNA coding sequence. (A) DNA sequence of the P1Δ3′ promoter. White boxes correspond to the 60-bp repeats and sequences shaded in black correspond to nonrepeated sequences. Sequences shaded in gray correspond to the first eight nucleotides of a 60-bp block; p53 responsive elements are indicated in bold. (B,C) Relative luciferase activity obtained with various promoter constructs: [60 bp]3, [60 bp]2, or [60 bp]1 carrying of three, two, or one 60-bp repeats, respectively, and [60 bp] + 8 bp and [60 bp] + 8 bp mut, contain one block of 60 bp with the first eight bases of a second repeat or eight mutated bases (polyG), respectively. The luciferase activities obtained with these constructs were normalized with respect to that of the P1Δ3′luciferase construct. Tests were performed with LMH (B) and 54-O (C) cells. Each value corresponds to the mean of three independent assays performed simultaneously. Error bars indicate the SEM of three replicates. Schematic diagrams of the various promoters are shown to the left of each histogram: sequences shaded in black correspond to nonrepeated sequences; white boxes correspond to the 60-bp repeats; gray boxes correspond to the first eight base pairs of the 60-bp repeat; dashed boxes correspond to the mutation of the first eight base pairs to eight guanines.
We next investigated whether the number of copies of the repeats affected promoter activity (Fig. 4B,C). We used the MSB-1 cell line currently used as a reference, derived from a lymphoma induced by the virulent GaHV-2 strain BC-1, to obtain a construct with nine copies. The transcriptional activity of this construct was not markedly different from that of the reference construct P1Δ3′, which has three copies of the tandem repeat. Constructs with two and three copies of the 60-bp repeat had 50% of the activity of the reference P1Δ3′ construct in the LMH cell line, and no significant differences were observed between these two constructs (Fig. 4B). By contrast, the construct harboring a single copy of the 60-bp repeat displayed significantly lower levels of activity in LMH cells (3% that of P1Δ3′; p < 0.0001) (Fig. 4B). The effect of the number of copies of the 60-bp repeat on reporter activity was even more pronounced in the 54-O cell line (Fig. 4C): The construct with two copies displayed only 18% of the activity of P1Δ3′. The transcriptional activity of the construct with one copy did not exceed the background activity of a promoter-less, negative control plasmid (Fig. 4C). The addition of 8 bp at the 3′ end of a single 60-bp repeat (construct [60 bp] + 8 bp) restored activity to 80% that for two full-length 60-bp repeats, in both LMH and 54-O cells (Fig. 4B,C). Invalidation of the 8-bp sequence at the end of construct ([60 bp] + 8 bp) by mutation severely impaired its activity (by 90% and 95% in LMH and 54-O cells, respectively) (Fig. 4B,C).
These findings indicate that the tandem repeat region upstream of the TSS of LAT requires a minimum of two copies of the 60-bp repeat for strong promoter activity. Nevertheless, a single 60-bp sequence with an 8-bp sequence from a second copy of the 60-bp repeat at the 3′end constituted a minimal functional construct.
The transcriptional activity of the LAT-miRNA promoter is dependent on the p53 responsive element
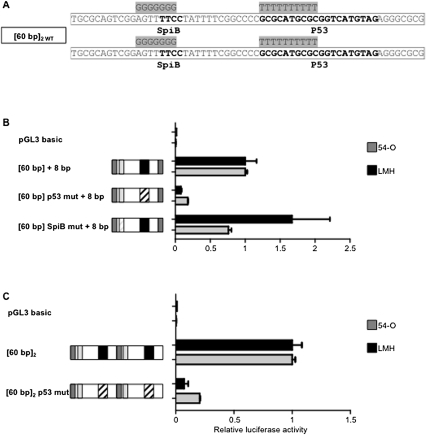
The sequence of the region of tandem repeats contains DNA stretches with similarities to transcription factor response elements (RE) and to potential transcription initiator signals (Fig. 5): the 60-bp sequence contains one potential M22 transcription initiator site (TGCGCANK-TGCGCAGT), one possible RE specific for SpiB (TTCC in reverse orientation), one canonical INR transcription initiator site (YYANWYY-CTATTTT), and one highly conserved consensus p53-binding site (GCGCATGCGCGGTCATGTAG). The consensus p53-binding sequence is a 20-bp DNA sequence consisting of two repeats of the RRRCWWGYYY motif separated by 0–13 nt. We investigated the possible involvement of these REs in the transcriptional activity of the 60-bp repeats, by evaluating their function with the shortest functional construct. This construct included a single copy of the 60-bp fragment followed by the first eight base pairs of a second copy ([60 bp] + 8 bp). We used site-directed mutagenesis to modify the two candidate REs and studied reporter activity in cells (Fig. 5B). The mutation of the p53 RE abolished 82% and 92% of the transcriptional activity in 54-O and LMH cells, respectively (Fig. 5B). By contrast, mutation of the SpiB RE did not significantly impair transcriptional activity in either cell line (Fig. 5B). We confirmed the effect of the p53 RE mutation in a construct containing two full-length copies of the 60-bp repeats, in which both p53 REs were invalidated (construct [60 bp p53mut]2) (Fig. 5C). The transcriptional activity of construct [60 bp]2 p53 mut was 7% that of the control in 54-O cells and 20% that for the control in LMH cells. These findings show that the p53 RE is directly involved in the transcriptional activity of the promoter.
FIGURE 5.
Identification of transcription factors involved in activation of the promoter. (A) Sequence of the core promoter [60 bp]2, with various REs probably involved in transcriptional activity shown in bold. The various mutations of the [60 bp] repeats are shaded in gray. (B) Identification, by mutagenesis, of transcription factors involved in the function of the promoter. The [60 bp] + 8 bp promoter was used for mutagenesis. Each reported luciferase activity value is normalized with respect to that of the [60 bp] + 8 bp promoter construct. (C) Identification, by mutagenesis, of transcription factors involved in the function of the promoter. The [60 bp]2 promoter was used for mutagenesis. Each reported luciferase activity value is normalized with respect to that of the [60 bp]2 promoter construct. White boxes indicate 60-bp repeats containing the various RE sites and gray boxes correspond to the first eight base pairs of the 60-bp repeat. Black boxes correspond to the p53 RE, and light gray boxes correspond to the SpiB RE. Dashed boxes indicate mutated sites.
p53 transactivates the promoter of the LAT-miRNA-cluster2
The minimal promoter construction, [60 bp]2, has robust promoter activity, strongly associated with the presence of at least one p53 RE. We investigated the essential role of p53 in the transactivation of this core promoter, by three complementary approaches: (1) ChIP assays were performed from MSB-1 cells derived from a GaHV-2 lymphoma, (2) the [60 bp]2 construct was used to transfect p53-deficient human osteosarcoma SAOS-2 cells complemented with human or chicken p53, and (3) the levels of three miRNAs from the cluster were measured in MSB-1 cells treated with adriamycin, a genotoxic drug that induces p53 accumulation, through phosphorylation of the Ser15 residue close to the mdm2-binding domain (Vassilev 2005).
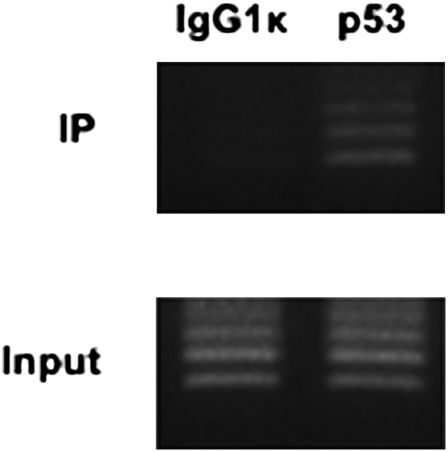
For the first approach, we assessed the direct association between endogenous p53 and the LAT-miRNA promoter, by performing ChIP experiments with MSB-1 cells. Immunoprecipitated DNA was analyzed by PCR using specific primers, surrounding the p53 RE. We found that p53 was recruited to the LAT-miRNA promoter (Fig. 6). As expected, ladders resulting from primers hybridization to different consecutive repeats were observed after PCR amplification from immunoprecipitated DNA (Fig. 6).
FIGURE 6.
Direct association of p53 with the LAT-miRNA promoter The PCR amplification is carried out using primers pair M645 and M735 (Table 1). Electrophoretic analyses of the PCR amplification products, using immunoprecipitated DNA (IP) or total input DNA (Input), are shown. Mouse IgG1 antibodies (left column) were used in ChIP assays as a negative control. The right column shows the PCR amplification products from DNA immunoprecipitated with p53 antibodies HP64. LAT-miRNA-promoter specific primers amplified products of various sizes from 154 bp to more with an increment of 60 bp corresponding to one 60-bp repeat.
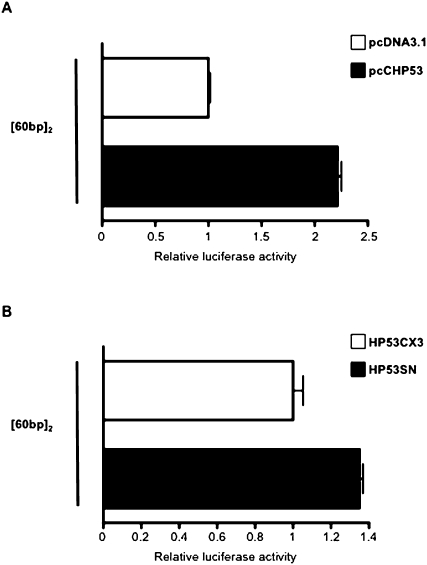
For the second approach, two related sets of complementation experiments were carried out. SAOS-2 cells were cotransfected with the minimal construct [60 bp]2 and an expression plasmid for chicken p53 (pcCHP53) or with the empty expression vector, pcDNA3.1(+), as control (Fig. 7A). The pcCHP53 construct was generated by inserting the chicken p53 cDNA downstream from the CMV promoter of the expression vector, pcDNA3.1(+). The ectopic expression of chicken p53 led to promoter activity of the minimal construct [60 bp]2 twice as strong as that of the empty pcDNA3.1(+) vector control (Fig. 7A). The other complementation experiment used an expression plasmid for wild-type human p53 (pHP53SN) or for human p53 with a mutation in codon 143 (pHP53CX3) (Fig. 7B). Similar results were obtained: The promoter activity of the minimal construct [60 bp]2 was 1.35 times higher when SAOS-2 cells were cotransfected with the expression plasmid for wild-type human p53 (pHP53SN) than when transfected with the construct encoding the mutated protein HP53CX3 (Fig. 7B). Residual promoter activity was observed in SAOS-2 cells transfected with the [60 bp]2 construct alone. Other proteins of the p53 family have similar activities and the residual activity in SAOS-2 cells may be due to the effects of p73 or p63, which are not knocked out (Irwin and Kaelin 2001).
FIGURE 7.
Tumor protein p53 transactivates the [60 bp]2 minimal promoter SAOS-2 cells were cotransfected with the minimal promoter [60 bp]2 and an empty pcDNA3.1(+) expression plasmid and an expression plasmid for chicken p53 (pcCHP53) (A) or plasmids encoding wild-type human p53 (pHP53SN) and mutated human p53 (pHP53CX3) (B). Luciferase activities following cotransfection with pcCHP53 and pHP53SN were standardized with respect to those of pcDNA3.1 (A) and pHP53CX3 (B), respectively. PRL-TK (A) and pRL-CMV (B) vectors were used to normalize the transfection rate. Each value corresponds to the mean of at least three independent assays performed simultaneously. Error bars indicate the SEM for three replicates.
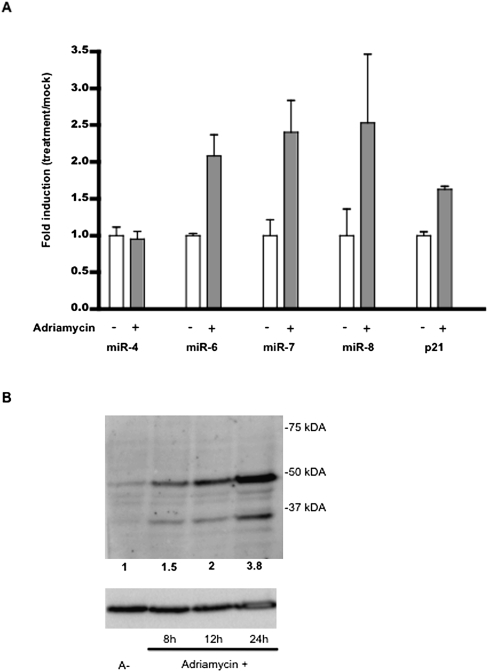
We evaluated the expression of three mature miRNAs of the LAT-miRNA-cluster2 (mdv1-miR-M6, mdv1-miR-M7, and mdv1-miR-M8) in MSB-1 cells treated with the genotoxic drug adriamycin, by quantitative RT-PCR, to confirm the p53-dependent activity of the promoter in natural conditions. The level of expression of mdv1-miR-M4, which belongs to the second cluster of MDV miRNAs, was determined as a negative control. Conversely, p21 mRNA levels were determined as a positive control of the transactivation effect dependent on the stabilization of p53 after adriamycin treatment. Western blot analysis of MSB-1 p53 expression after adriamycin treatment was carried out to prove the involvement of adriamycin in p53 stabilization. After 8, 12, and 24 h adriamycin treatment, the p53 expression level increases by a factor equal to 1.5, 2, and 3.8 respectively (Fig. 8B).
FIGURE 8.
Induction of mature LAT-miRNA by adriamycin. (A) Adriamycin treatment induces mature LAT-miRNA expression. MSB-1 cells derived from a Gallid herpes virus-2 (GaHV-2)-induced lymphoma were treated with adriamycin for 8 h. RNA was extracted with Trizol and analyzed by quantitative primer-extension PCR assay, with the Biorad analysis kit. Expression profiles for cluster 2 miRNAs (mdv1-miR-M7, -M8 and -M6) were recorded. The expressions of non-p53-specific mdv1-miR-M4 and p53-specific p21 were evaluated as negative and positive controls, respectively. Fold induction after adriamycin treatment is shown for each RNA. Each value corresponds to the mean of three independent assays performed simultaneously. Errors bars indicate the SEM of three replicates. (B) Western blot analysis of p53 and the GAPDH level after adriamycin treatment. MSB-1 cells were treated with adriamycin (0.2 μg/mL) for 8, 12, and 24 h. Each blot was probed with mouse anti-p53 (HP64) and mouse anti-GaPDH primary antibodies, followed by rabbit anti-mouse horseradish-conjugated secondary antibody. GaPDH was used to check for equal protein loading. Relative signal intensities for p53 were calculated with respect to the GaPDH loading control, with Adobe Photoshop element 6 software. The values reported under the blots correspond to the respective level of p53 detected after adriamycin treatment, relative to the signal for p53 without adriamycin treatment.
As expected, p21 mRNA transcription levels were 1.8 times higher in GaHV-2-infected MSB-1 cells treated with adriamycin for 8 h than in untreated MSB-1 cells. By contrast, the expression of mdv1-miR-M4, which does not belong to the LAT-miR-cluster2, was not affected by adriamycin treatment. Adriamycin increased the level of expression of the three miRNAs (mdv1-miR-M6, mdv1-miR-M7, and mdv1-miR-M8) of LAT-miRNA-cluster2 by factors of 1.8, 2.4, and 2, respectively (Fig. 8A). Same experiments performed in homolog cell line 54-O have shown similar results (data not shown). Collectively, these data provided strong evidence for the involvement of p53 in the production of mature LAT-miRNAs.
DISCUSSION
Herpesviruses have a large double-stranded DNA genome and have several conserved mechanisms for regulating gene expression, including complex alternative transcription and post-transcriptional regulation mechanisms triggered by miRNAs. They also produce noncoding RNAs (ncRNAs), which are involved in the establishment and maintenance of latency (Jones 2003). These latency-associated ncRNAs may be variants of long intranuclear noncoding RNAs antisense to various mRNAs described in mammals (Birney et al. 2007; Carninci 2008). In the avian oncogenic herpesvirus GaHV-2, LAT belongs to a family of RNAs spliced from a 10-kb ncRNA antisense to the mRNA for the major lytic transactivator protein ICP4 (Cantello et al. 1994, 1997). Recent studies have shown that LAT contains a cluster of miRNAs (Burnside et al. 2006; Burnside and Morgan 2007; Morgan et al. 2008; Xu et al. 2008). GaHV-2 lymphomagenesis occurs during the latency phase. The miRNA-LAT gene is overexpressed in GaHV-2-transformed and latently infected cells, and it has therefore been suggested that the miRNA cluster is involved in latency and/or lymphomagenesis.
In this study, we successfully mapped the core promoter of this LAT-miRNA transcript 600 nt upstream of miRNA cluster 2 and showed that this promoter led to the transcription of mature miRNAs (Fig. 3). The LAT-miRNA minimal promoter consists of at least two identical blocks of 60 bp. Each block contains one potential RE for p53 and one for SpiB. Only the p53 RE was found to be functional in the avian cell lines tested (Fig. 5). Its sequence (GCGCATGCG CGGTCATGTAG) is very similar to the consensus p53 RE sequence reported for humans and several other species. It is consistent with the 10-nt consensus RRRCWWGYYY (half-site) tandem repeat sequences separated by 0–13 nt (el-Deiry et al. 1992). Systematic analysis of p53 REs and a recent in vivo study have shown that the most efficient promoters of the murine Mdm2 gene have no spacer between the two half-sites of the p53 RE (Jordan et al. 2008). In light of these reports, the perfect palindrome for the 5′ half-site and the lack of spacer bases between the two half-sites in the GaHV-2 miRNA-LAT promoter may explain its considerable transcriptional strength, almost as great as that of the strong CMV promoter.
A common feature identified in several p53 target genes is the presence of multiple p53-binding sites (el-Deiry et al. 1992; Resnick-Silverman et al. 1998; Hwang et al. 1999; Thornborrow and Manfredi 1999; Espinosa and Emerson 2001; Nakano and Vousden 2001). The distances between these p53 REs are variable. Synergies have been described between these multiple p53 REs (Thornborrow and Manfredi 1999; Jordan et al. 2008). However, the space between the p53 REs of the murine mdm2 promoter cannot be significantly modified without a loss of function (Jordan et al. 2008). In the miRNA-LAT promoter described here, the p53 REs are 60 bp apart. The relative rates of transcription from the [60 bp] + 8 bp, [60 bp], and [60 bp]2 constructs highlight the essential role of a transcription initiator sequence downstream from the p53 RE (Fig. 4), but we cannot exclude the possibility of synergy potentiating the p53 RE in neighboring 60-bp blocks. In lymphoid cells (54-O), derived from GaHV-2 lymphomas, the constructs harboring three full p53-binding sites displayed transcriptional activities twice as high as that of the construct possessing only two p53-binding sites. Furthermore, the promoter structure found to be functional in transcriptional analysis (Fig. 4) is biologically relevant, because genotyping data for a large panel of GaHV-2 strains showed that oncogenic strains possess at least two blocks of 60-bp harboring p53 REs (Spatz and Silva 2007b). However, the presence of more than three tandem repeats did not result in significantly greater transcriptional activity (Fig. 4).
The use of a reporter gene to analyze promoter activity showed that, in addition to the p53-binding site, one downstream functional INR played a significant role in the promoter activity of the LAT gene. The tandem repeats sequence contains a consensus initiator element (INR; CTATTTT) starting at nucleotide +19 of the 60-bp repeats. However, this INR appears to be a minor TSS, because only one (of 16) and three (of 17) transcripts started within this sequence in 5′ RACE assays. By contrast, both 5′-end analyses identified a major TSS of LAT in a sequence very similar to the INR consensus sequence YYANWYY, immediately downstream from the last 60-bp repeat (CTGATTT; position 142734 in the RB-1B genome). Several observations have implicated this INR-like sequence in promoter function. A construct based on three 60-bp repeats, but with the deletion of this downstream INR, displayed two-thirds the activity of an equivalent construct containing the INR motif in LMH cells and about one-third that of the same construct in 54-O cells. These findings were consistent with the numbers of TSSs found in these cell lines, as one-third of the TSSs in LMH cells and two-thirds of those in 54-O cells corresponded to this INR-like site. All the experiments indicated that a p53 RE was required upstream of any potential start site signal, for optimal transcription. Activity was retained if the INR-like sequence was replaced by the first eight nucleotides of the 5′ end of the 60-bp repeat. These eight nucleotides correspond to the M22 site (TGCGCANK), which has been predicted to have a high prevalence in TATA-less core promoters (Birney et al. 2007).
Several examples of TATA-less promoters with various transcription start sites have been described (Carninci et al. 2006; Birney et al. 2007). Furthermore, studies on class II mammalian promoters have revealed that TSSs are closely clustered within the core promoter region (Carninci et al. 2006; Frith et al. 2008). We identified several TSSs in the last 58 bp of the promoter (Fig. 2). The TSSs of the chimeric LAT-Luc transcript are distributed throughout this 60-bp DNA motif, although most (94%) were found immediately downstream from the last 60-bp block near the INR-like CTGATTT in analyses of the LAT transcripts from the homologous system (Fig. 2); 64% of the TSSs were found at the classical position (third base) in this INR-like sequence (coordinate 142734 in the RB-1B genome). The LAT RNA is processed into spliced small RNAs by complex alternative splicing (Cantello et al. 1997), so the differences in the TSS distributions between the two systems may arise from this cotranscriptional event. Interestingly, detailed analysis of the LAT TSSs revealed that transcription started preferentially at sites with a purine in position +1 and a pyrimidine at position −1, as previously reported for the human and murine transcriptomes (Carninci et al. 2006). Indeed, 79% of TSSs (76% and 82% of the LAT-Luc and LAT transcripts, respectively) display a YR configuration at positions −1, +1 (Fig. 2). In addition, the composition of the surrounding sequence of most of the TSSs was consistent with recent observations based on exhaustive CAGE analyses (Frith et al. 2008)
Tumor suppressor protein p53 is a major transcription factor specifically activated in response to numerous stress signals, including those associated with cancers (Oren 2003; Harris and Levine 2005; Vousden and Prives 2005; Toledo and Wahl 2006). Alterations to this protein are among the most frequent causes of cancer. To date, three miRNA genes have been described as direct transcriptional targets of p53. One, miR-34a, an miRNA from the miR-34 family involved in senescence, cell-cycle arrest, and apoptosis, maps 30 kb downstream from a p53-binding site. This miRNA displays p53-dependent up-regulation (Chang et al. 2007; Raver-Shapira et al. 2007). Several groups have reported that the miR-34 family, including miR-34a, miR-34b, and miR-34c, which target the Bcl-2, Notch, and HMGA2 mRNAs, respectively, are induced by DNA damage and oncogenic stress, in a p53-dependent manner (Tazawa et al. 2007; Ji et al. 2008). In 2008, miR-192 was also identified as a direct target of p53, suggesting that this miRNA is involved in cell-cycle regulation through the p53-miRNA pathways (Song et al. 2008). The recent identification of miR-145 as a direct p53 target has expanded the repertoire of p53-regulated genes. The induction of miR-145 by p53 leads to the down-regulation of c-Myc through miR-145-mediated silencing (Sachdeva et al. 2009). In our study, p53 seemed to transactivate the miRNA-LAT promoter directly, through a specific p53 RE (Figs. 5–8). Previous studies revealed that LAT and miRNA cluster 2 in GaHV-2 are abundantly expressed during the latency and lymphomagenesis stages of viral infection (Burnside et al. 2006, 2008; Xu et al. 2008). Although p53 is considered to be a major tumor repressor, it has been shown that nonmutated p53 is present in GaHV-2-induced lymphomas (Gimeno et al. 2005). Further experiments are required to investigate the apparent contradiction between the production of native p53 and the development of lymphomas. The p53 cellular response to GaHV-2 infection may lead to overtranscription of the viral LAT-miRNA gene, potentially interfering with the pro-apoptotic pathways induced by p53.
Epidemiological observations support an association between the p53 target sequences driving the expression of the LAT locus and the oncogenic properties of GaHV-2. Several strains of GaHV-2 with different oncogenic capacities, correlated with their level of pathogenicity, have been isolated and partially sequenced. Recent published alignments of the sequences from a panel of 14 strains, including the promoter and part of the LAT gene, have shown that all the virulent, very virulent, and hypervirulent oncogenic GaHV-2 strains harbor two or more 60-bp tandem repeats (Spatz and Silva 2007a). By contrast, avirulent, mildly virulent, and vaccine strains have no copies of the tandem repeat. In addition, the minor effect on promoter activity of the short upstream sequence separating the telomeres from the 60-bp tandem repeats (Fig. 4) is reflected in vivo: this sequence has been lost from the genome of one of the very virulent strains (vv GaHV-2 strain 595).
In conclusion, we found sequences robustly initiating and inducing transcription of the pri-miRNA LAT transcript embedded within variable number tandem repeats harboring functional p53 binding sites in the GaHV-2 genome. The minimal functional construct consisted of two tandem repeats, consistent with the pathobiological characteristic of oncogenic and nononcogenic GaHV-2 strains. Transcriptional analysis indicated that a transcription initiator sequence downstream from a p53 binding site is required for the function of this TATA-less promoter. This p53-dependent promoter controlling viral miRNAs expression during viral latency and lymphomagenesis, allows the viruses to exploit the cellular p53 regulatory network.
MATERIALS AND METHODS
Bioinformatic analysis
Genomatix software (www.genomatix.de/cgi) was used for the in silico prediction of promoter sequences. This program identifies potential transcription factor response elements within a DNA sequence.
Cell culture, virus, and treatments
Five cell lines were used. The LMH line (ATCC no. CRL-217), established from chicken hepatic carcinoma epithelial cells, was cultured in DMEM:F12 medium (Cambrex) supplemented with 10% fetal bovine serum and 5% chicken serum. The avian fibroblast cell line DF1 (ATCC no. CRL-12203) was cultured in a DMEM medium similarly supplemented with sera. The MSB-1 cell line is derived from a spleen lymphoma induced by a virulent strain of MDV-1 (Akiyama et al. 1973). It was previously shown to be coinfected with both MDV-1 (BC-1) and MDV-2 (HPRS24) (Hirai et al. 1990). The 54-O cell line was isolated in the TLVI laboratory (INRA, Nouzilly) from an ovary lymphoma induced by recombinant RB1-B virus reconstituted from an infectious genome cloned as a bacterial artificial chromosome (kindly provided by Venugopal Nair). These cell lines were cultured in RPMI-1640 medium (Cambrex) supplemented with 1 mM sodium pyruvate, 10% fetal bovine serum, and 5% chicken serum. Both latently infected MDV cell lines display comparable characteristics and express high levels of MDV microRNAs and the MSB-1 cell line is currently used as a reference. The p53-deficient osteosarcoma SAOS-2 cells (ATCC n°HTB-85) were cultured in a DMEM medium supplemented with 10% fetal bovine serum.
We treated cells with 0.2 μg/mL adriamycin for 8 h to induce p53 before the recovery of total RNA.
Plasmid constructs
The entire sequence of the putative promoter of the GaHV-2 LAT transcript was amplified from Bacmide DNA of the RB-1B strain (Petherbridge et al. 2003). The initial construct extended from nucleotide 142486 to nucleotide 143448 of the reference RB-1B genome (GenBank accession number EF523390), covering the region between the telomeric repeat and the LAT-miRNA cluster. Various primers were used to obtain different fragments of potential promoters (Fig 1; Table 1). PCR was performed as follows: 94°C for 4 min, followed by 25 cycles of denaturation (94°C 30 sec), annealing (55°C 30 sec), and extension (72°C 1 min), in a final volume of 50 μL containing 2.5 units of Taq DNA polymerase (NEB, New England Biolabs), 0.1 μM of each primer (Eurogentec), 0.2 mM of each deoxyribonucleotide, 10 mM Tris-HCl pH 9, 50 mM KCl, 0.5 mM MgCl2, 0.1% TritonX-100, and 50 ng DNA.
TABLE 1.
Primers used to construct different sequences of promoters and to perform RACE-PCR
A wild-type monomer of the 60-bp repeat was generated by PCR amplification of a 60-nt single-stranded oligonucleotide (Eurogentec). Multimers ([60 bp]2 and [60 bp]3) were constructed by annealing two partially complementary oligonucleotides of 60 nt. The resulting double-stranded fragments harbored overlapping ends that were used for concatemerization and ligation. The mutated promoters were generated in the same way, from 60-nt single-stranded oligonucleotides carrying mutations at the target sites. These sequences then amplified by PCR, with specific primers, in the conditions described above.
All the putative promoter sequences were amplified with primer pairs (Table 1) including KpnI and HindIII sites in their tails. The KpnI/HindIII-digested PCR products were inserted in the correct orientation between the KpnI and HindIII sites of the pGL3 basic vector (Promega).
The p53 expression vectors were provided by Thierry Soussi (UPMC/Karolinska Institutet). pHP53SN and pHP53CX3 encode the wild-type human p53 and a mutated form of human p53, respectively, under control of the CMV promoter. The chicken p53 cDNA was also provided by T. Soussi, flanked by EcoRI restriction sites in the pBS-SK plasmid. The p53 fragment was digested and inserted into the vector pcDNA3.1(+) linearized by digestion with EcoRI.
The two constructs, pB691 and pB695, encompassing the 5′ end of the LAT mRNA and the four miRNAs, with and without, respectively, the 60-bp repeats, were amplified from the bacmide p-RB-1B, with primers binding 18 nt upstream of the first 60-bp repeat (M691) or in the terminal part of the last 60-bp repeat (M695), respectively, and 1719 nt downstream from miR10 (M656) (Table 1). Each primer used contains a BamHI restriction site in their tail. The BamHI-digested PCR products were inserted into the pBS-SK vector (Stratagene).
Plasmid DNA was purified with the Nucleobond Xtra midi kit (Macherey Nagel), and the inserts of all constructs were completely sequenced.
Luciferase assays
The pGL3basic (Promega) expression vector was used to test the strength of promoters inserted upstream of the firefly luciferase (F-Luc) gene. The pRL-TK (Promega) vector includes an expression cassette for the Renilla luciferase (R-Luc) under the control of the thymidine kinase promoter of human herpes virus 1 (HHV-1). Cotransfection with these two vectors (pGL3 carrying the test promoter and pRL-TK) made it possible to normalize measurements as a function of the rate of transfection. The pRL-CMV vector contains an expression cassette for the Renilla luciferase (R-Luc) under the control of the CMV promoter. The use of this vector also makes it possible to normalize the transfection rate.
Lipofectamine reagent (Invitrogen) was used for the transfection of adherent cells. Transfection assays were performed with 2 × 104 cells per well, in 96-well plates, with a mixture of 300 ng of firefly luciferase reporter construct and 5 ng of Renilla luciferase control vector.
We transfected 54-O lymphoblastoid cells with plasmids by electroporation. We washed 54-O cells twice with RPMI medium and adjusted the cell density to 8 × 106 cells/ 800 μL of serum-free medium. For each electroporation, 40 μg of the firefly luciferase reporter construct and 600 ng of Renilla luciferase control vector were mixed with cells. The cells were then electroporated in a 4 mm cuvette subjected to 100 V/mm for 5 ms. The contents were then dispensed into 2.5 mL of complete medium.
Luciferase assays were performed 24 h after transfection, with the dual luciferase reporter assay system (Promega) and a Centro LB960 luminometer (Berthold Technologies). Student's t-test was used for statistical analysis.
We transfected SAOS-2 cells in 24-well plates seeded with 1 × 105 cells per well, using Lipofectamine reagent (Invitrogen). Each well was transfected with a mixture of 1 μl Lipofectamine, 400 ng promoter construct, 100 ng inductive expression plasmid (p53chicken pcCHP53, human wild-type HP53SN or human mutated HP53CX3), and 10 ng of pRL-TK or pRL-CMV (for cotransfection with chicken and human p53 constructs, respectively). For SAOS-2 cells, we carried out the dual luciferase reporter assay 12 h after transfection.
5′ RACE analysis
5′ Rapid amplification of cDNA ends (5′ RACE) was carried out on total RNA isolated from two cell cultures. The first RNA sample was isolated from 54-O cells and the second was isolated from LMH cells transfected with pGL3-P1Δ3′ (containing the P1Δ3′ promoter). Total RNA was extracted with the Trizol extraction kit. cDNAs were obtained with the GeneRacer kit (Invitrogen). This technique is based on RNA ligase-mediated and oligo-capping RACE methods, and results in the selective ligation of an RNA oligonucleotide to the 5′ ends of decapped RNA. The resulting cDNA was amplified by PCR, using forward primers annealing to the ligated RNA oligonucleotide and reverse primers annealing to the target gene (Table 1).
The 5′ end of the ligated cDNA obtained from 54-O cells was determined with reverse primers binding to a published exon of LAT. The 5′ end of the ligated cDNA obtained from transfected LMH cells was determined with reverse primers binding to the firefly luciferase ORF. PCR products of various sizes were obtained and inserted into the cloning vector PCR4-TOPO TA. We sequenced 20 and 42 positive clones for LMH and 54-O cells, respectively, to determine the 5′ end in each case.
Northern blot analysis
RNA was extracted in Trizol and 15 μg of total RNA per lane was resolved by electrophoresis in a 15% acrylamide/bisacrylamide (19:1) gel containing 7 M urea, in Tris-borate-EDTA (TBE) buffer. Gels were briefly stained with ethidium bromide, and RNA was transferred, by electroblotting, onto a Nylon Plus membrane (Amersham) for 1.5 h at 350 mA in 0.50X TBE. The membrane was cross-linked by exposure to UV (Stratalinker, Stratagene). The membrane was subjected to prehybridization for 1 h and was then hybridized overnight in Perfect Hyb TM Plus hybridization buffer (Sigma) at 50°C, with a 20 nM 32P-5′ end-labeled DNA oligonucleotide probe complementary to the miRNA. It was then washed in low-stringency wash buffer. Blots were analyzed by phosphorimaging with a FLA7000 scanner from Fuji.
ChIP assays
ChIP assays were carried out as previously described (Shkreli et al. 2007). Briefly, chromatin from 107 MSB-1 cells was cross-linked, washed, resuspended in lysis buffer and sonicated 12 times 6 sec with an 18J-pulse (VibraCell Bioblock Scientific). A fraction of the total chromatin was taken as the total input DNA control. After preclearing with protein G agarose/salmon sperm DNA beads, protein/DNA complexes were immunoprecipitated overnight at 4°C with anti-p53 (70 μL of hybridomas supernatant of HP64 antibody kindly provided by T. Soussi) or with mouse immunoglobulin G1κ (6 μg of IgG1κ, Sigma Aldrich) as a negative control. Immunoprecipitated complexes were collected with Protein G beads. Immunoprecipitation products were washed with low salinity buffer, high-salinity buffer, lithium chloride washing buffer, and finally with TE (Tris-EDTA) buffer. Immunocomplexes were extracted and cross-linking was reversed by incubation at 65°C overnight. DNA fragments were then purified using Nucleospin Extract II (Macherey-Nagel) and eluted in 30 μL of ultrapure water. PCR were performed in 50 μL volume with Promega Taq DNA polymerase and with primers pair M645/M735 (Table 1). PCR was initiated with 3 min at 94°C followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, and finally 7 min at 72°C.
Western blot analysis
MSB-1 cell lines were treated with 0.2 μg/mL of adriamycin during 8, 12, and 24 h, respectively. Five hundred thousand cells were treated with lysis buffer (15 mM Tris, 15 mM NaCl, 20 mM EDTA, 1% SDS, 5% beta-mercaptoethanol, 20% glycerol, bromophenol blue, [pH 6.8]). Proteins were separated by electrophoresis in 10% polyacrylamide gels containing SDS and passively blotted onto nitrocellulose membranes. The membranes were blocked by incubation with 3% skim milk powder in PBST (PBS, 0.5% Tween 20) and probed with monoclonal mouse anti-p53 (HP64 antibody kindly provided by T. Soussi) and monoclonal mouse anti-GaPDH (Millipore) primary antibodies, followed by polyclonal rabbit anti-mouse horseradish peroxidase-conjugated secondary antibodies. The immunoreactions of interest were detected with the Femto Super Signal Western detection kit (Pierce Chemical Co.). GaPDH was used as a control to demonstrate equal protein loading. Adobe Photoshop element 6 software was used to calculate the signal intensities for p53 level after different timing induction relative to the GaPDH loading control.
miRNA quantification by q-RT-PCR
Mdv1-miR-M4-5p, -M6-5p, -M7-5p, and -M8-5p RNA levels were quantified as described by Raymond et al. (2005). Briefly, total RNA was isolated by guanidinium thiocyanate-phenol-chloroform extraction (TRIzol, Invitrogen). After total RNA quantification by Nanodrop methods, the cDNAs were synthesized in primer extension assays containing 1 μg of total RNA (Table 1). Following reverse transcription, triplicate measurements were made on 2 μL of cDNA in a final reaction volume of 20 μL, by qPCR, in a 96-well optical PCR plate, with an iCycler qPCR device (Biorad). SYBR green PCR mixture, containing 10 μL iQ SYBR green PCR master mix (Biorad), 5 μL water, 1 μL of 10 mM universal primer, 1 μL of 10 mM LNA-R primer, and 3 μL of sample. The primer sequences are reported in Table 1. qPCR was carried out in the conditions recommended by the manufacturer, and dissociation curves were generated post-run for the analysis of amplicon species. The temperature at the dissociation peak is reported in Table 1. For the detection and quantification of chicken p21 mRNA, we used the SYBR green method with p21 sense and antisense oligomers as primers (Table 1), as described elsewhere (Sato et al. 2006).
ACKNOWLEDGMENTS
We thank Dr. Nair Venugopal (IAH, Compton) for providing us with the Bacmide p-RB1B and Sascha Trapp for the 54-O cell line. We also thank Dr. Thierry Soussi (UPMC; Karolinska), who kindly provided us with the antibody anti-p53 (HP64), the expression vectors for wild-type human p53 (pHP53SN) and mutated human p53 (pHP53CX3), and the cDNA for chicken p53. This work was supported by the “Ligue Nationale contre le cancer, Comité du Cher, Comité de l'Indre” and the “Agence Nationale de la Recherche” (ANR-07-MIME-012-01). B.M. is a postdoctoral researcher supported by the Fonds National de la Recherche Scientifique (FNRS).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2121210.
REFERENCES
- Akiyama Y, Kato S, Iwa N 1973. Continuous cell culture from lymphoma of Marek's disease. Biken J 16: 177–179 [PubMed] [Google Scholar]
- Ambros V 2004. The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH 2006. Differences in vertebrate microRNA expression. Proc Natl Acad Sci 103: 14385–14389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Burgess SC, Young JR, Baaten BJ, Hunt L, Ross LN, Parcells MS, Kumar PM, Tregaskes CA, Lee LF, Davison TF 2004. Marek's disease is a natural model for lymphomas overexpressing Hodgkin's disease antigen (CD30). Proc Natl Acad Sci 101: 13879–13884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J, Morgan RW 2007. Genomics and Marek's disease virus. Cytogenet Genome Res 117: 376–387 [DOI] [PubMed] [Google Scholar]
- Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J Virol 80: 8778–8786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, Meyers BC, Green PJ, Markis M, Isaacs G, Huang E, et al. 2008. Deep sequencing of chicken microRNAs. BMC Genomics 9: 185 doi: 10.1186/1471-2164-9-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantello JL, Anderson AS, Morgan RW 1994. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek's disease virus. J Virol 68: 6280–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantello JL, Parcells MS, Anderson AS, Morgan RW 1997. Marek's disease virus latency-associated transcripts belong to a family of spliced RNAs that are antisense to the ICP4 homolog gene. J Virol 71: 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P 2008. Non-coding RNA transcription: Turning on neighbours. Nat Cell Biol 10: 1023–1024 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Roberts SG 2005. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev 19: 2418–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B 1992. Definition of a consensus binding site for p53. Nat Genet 1: 45–49 [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol Cell 8: 57–69 [DOI] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A 2008. A code for transcription initiation in mammalian genomes. Genome Res 18: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno IM, Witter RL, Fadly AM, Silva RF 2005. Novel criteria for the diagnosis of Marek's disease virus-induced lymphomas. Avian Pathol 34: 332–340 [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ 2005. The p53 pathway: Positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Hirai K, Yamada M, Arao Y, Kato S, Nii S 1990. Replicating Marek's disease virus (MDV) serotype 2 DNA with inserted MDV serotype 1 DNA sequences in a Marek's disease lymphoblastoid cell line MSB1-41C. Arch Virol 114: 153–165 [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Ford JM, Hanawalt PC, Chu G 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci 96: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin MS, Kaelin WG 2001. p53 family update: p73 and p63 develop their own identities. Cell Growth Differ 12: 337–349 [PubMed] [Google Scholar]
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol 14: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L 2008. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 8: 266 doi: 10.1186/1471-2407-8-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin Microbiol Rev 16: 79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JJ, Menendez D, Inga A, Nourredine M, Bell D, Resnick MA 2008. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet 4: e1000104.doi: 10.1371/journal.pgen.1000104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA 2005. Functional characterization of core promoter elements: The downstream core element is recognized by TAF1. Mol Cell Biol 25: 9674–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, et al. 2008. Sequence conservation and differential expression of Marek's disease virus microRNAs. J Virol 82: 12213–12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- Oren M 2003. Decision making by p53: Life, death and cancer. Cell Death Differ 10: 431–442 [DOI] [PubMed] [Google Scholar]
- Osterrieder N, Kamil JP, Schumacher D, Tischer BK, Trapp S 2006. Marek's disease virus: From miasma to model. Nat Rev Microbiol 4: 283–294 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev 22: 3172–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Hunter S, Bracht J 2005. MicroRNAs: A developing story. Curr Opin Genet Dev 15: 200–205 [DOI] [PubMed] [Google Scholar]
- Petherbridge L, Howes K, Baigent SJ, Sacco MA, Evans S, Osterrieder N, Nair V 2003. Replication-competent bacterial artificial chromosomes of Marek's disease virus: Novel tools for generation of molecularly defined herpesvirus vaccines. J Virol 77: 8712–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM 2005. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick-Silverman L, St Clair S, Maurer M, Zhao K, Manfredi JJ 1998. Identification of a novel class of genomic DNA-binding sites suggests a mechanism for selectivity in target gene activation by the tumor suppressor protein p53. Genes Dev 12: 2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY 2009. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 106: 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA 2007. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat Rev Genet 8: 424–436 [DOI] [PubMed] [Google Scholar]
- Sato F, Kurokawa M, Yamauchi N, Hattori MA 2006. Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. Am J Physiol Cell Physiol 291: C538–C545 [DOI] [PubMed] [Google Scholar]
- Shkreli M, Dambrine G, Soubieux D, Kut E, Rasschaert D 2007. Involvement of the oncoprotein c-Myc in viral telomerase RNA gene regulation during Marek's disease virus-induced lymphomagenesis. J Virol 81: 4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT 2003. The RNA polymerase II core promoter. Annu Rev Biochem 72: 449–479 [DOI] [PubMed] [Google Scholar]
- Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J 2008. miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res 14: 8080–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz SJ, Silva RF 2007a. Polymorphisms in the repeat long regions of oncogenic and attenuated pathotypes of Marek's disease virus 1. Virus Genes 35: 41–53 [DOI] [PubMed] [Google Scholar]
- Spatz SJ, Silva RF 2007b. Sequence determination of variable regions within the genomes of gallid herpesvirus-2 pathotypes. Arch Virol 152: 1665–1678 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, et al. 2001. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res 11: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang KJ, Baltimore D 2006. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M 2007. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci 98: 1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H 2007. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci 104: 15472–15477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornborrow EC, Manfredi JJ 1999. One mechanism for cell type-specific regulation of the bax promoter by the tumor suppressor p53 is dictated by the p53 response element. J Biol Chem 274: 33747–33756 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM 2006. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Vassilev LT 2005. p53 Activation by small molecules: Application in oncology. J Med Chem 48: 4491–4499 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2005. p53 And prognosis: new insights and further complexity. Cell 120: 7–10 [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13: 790–795 [DOI] [PubMed] [Google Scholar]
- Xu H, Yao Y, Zhao Y, Smith LP, Baigent SJ, Nair V 2008. Analysis of the expression profiles of Marek's disease virus-encoded microRNAs by real-time quantitative PCR. J Virol Methods 149: 201–208 [DOI] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E 2007. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene 389: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: Predominance of virus-encoded microRNAs. J Virol 82: 4007–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]