Abstract
As RNAs fold to functional structures, they traverse complex energy landscapes that include many partially-folded and misfolded intermediates. For structured RNAs that possess catalytic activity, this activity can provide a powerful means of monitoring folding that is complementary to other biophysical approaches. RNA catalysis can be used to track accumulation of the native RNA specifically and quantitatively, readily distinguishing the native structure from intermediates that resemble it and may not be differentiated by other approaches. Here we outline how to design and interpret experiments using catalytic activity to monitor RNA folding, and we summarize adaptations of the method that have been used to probe aspects of folding well beyond determination of the folding rates.
1. INTRODUCTION
The discovery in 1982 by Cech and co-workers that a group I intron from Tetrahymena thermophila folds into a specific tertiary structure and catalyzes the chemical reactions necessary to excise itself from a precursor rRNA (Kruger et al., 1982) ushered in a new age of interest in understanding how RNAs fold to functional structures. Since then, interest in RNA folding has continued to grow as structured RNAs have been implicated in a dizzying array of cellular functions. In addition to performing self-splicing and cleavage reactions in a wide variety of contexts (Fedor, 2009), RNAs process and translate messages as core components of the spliceosome and ribosome (Moore et al., 2002; Noller, 2005; Wahl et al., 2009), process tRNA precursors (Evans et al., 2006), protect chromosomal ends (Collins, 2006; Theimer and Feigon, 2006), mediate protein trafficking events (Egea et al., 2005), and regulate gene expression by a variety of mechanisms (Bartel, 2004; Carthew and Sontheimer, 2009).
In parallel, the known catalytic RNAs and the corresponding catalytic repertoire have increased dramatically. The natural catalytic RNAs include group I and group II introns, RNase P, and self-cleaving RNA elements including the hairpin, hammerhead, Varkud satellite, and hepatitis delta virus (HDV) ribozymes, and most recently the GlmS riboswitch (Cech, 2002; Fedor, 2009). Self-cleaving elements have been found in the genomes of humans and other mammals (Teixeira et al., 2004; Salehi-Ashtiani et al., 2006; Martick et al., 2008), and it seems likely that many more remain undiscovered (Katayama et al., 2005; Carninci et al., 2005; Salehi-Ashtiani et al., 2006). The catalytic capabilities of RNA have even been extended by in vitro selection methods beyond the range identified in nature to include phosphorylation and ligation reactions (Lorsch and Szostak, 1994; Lincoln and Joyce, 2009), and even more diverse reactions such as alkylations and condensations (Chen et al., 2007).
As described throughout this volume, a powerful arsenal of biophysical approaches has been developed and applied to study the formation of specific structures by RNA. These approaches have been used to probe questions such as what provides the energetic driving force for formation of compact RNA structures? What features of the RNAs provide specificity to stabilize the functional structures relative to others? How fast do RNAs form specific structures, and what physical steps limit the folding rates? More generally, what do the energy landscapes of RNA folding look like?
Beginning in the early 1990s, catalytic RNAs became prominent in studies of RNA folding, and work using these RNAs has fundamentally advanced our understanding of folding processes and landscapes. Two powerful time-resolved footprinting methods were developed and first applied to the Tetrahymena group I ribozyme. One of them followed accessibility of the RNA to complementary oligonucleotide probes (Zarrinkar and Williamson, 1994; Zarrinkar and Williamson, 1996), and the other used hydroxyl radicals to monitor backbone exposure of RNA within milliseconds after initiation of folding by addition of Mg2+ ions (Sclavi et al., 1997; Sclavi et al., 1998).
The most general conclusion from these studies was that RNA folding involves multiple steps, with highly structured intermediates being populated along the way. Since then, the presence of intermediates has been shown to be a general feature of RNA folding (Pan and Sosnick, 1997; Treiber et al., 1998; Pan and Woodson, 1998; Russell and Herschlag, 1999; Chadalavada et al., 2002; Chauhan and Woodson, 2008). The extensive formation of intermediates creates a significant challenge for studying RNA folding. It is critical to know whether the RNA is completely folded to the native, functional structure or instead is present as a folding intermediate, but this can be exceedingly difficult for folding intermediates that are extensively structured and bear a strong resemblance to the native structure.
Catalytic RNAs provide a unique window to view the folding process, because the onset of catalytic activity can provide a powerful and specific readout for formation of the native structure. This approach allows the native state to be distinguished from alternative long-lived intermediates, regardless of their physical similarities, as only the native state is capable of performing the catalytic reaction. In the most favorable cases, this method can provide an unambiguous measure of the amount of native ribozyme as a function of time, which can then be integrated with data from other approaches. It was used in early studies of ribosome assembly (Traub and Nomura, 1969; Held and Nomura, 1973), and since then has been applied to a broad range of RNAs [e.g. (Woodson and Cech, 1991; Emerick et al., 1996; Pan and Sosnick, 1997; Russell and Herschlag, 1999; Chadalavada et al., 2002; Su et al., 2003; Xiao et al., 2005; Brooks and Hampel, 2009)]. Catalytic activity has also been used to follow protein folding (Kiefhaber et al., 1990; Kiefhaber, 1995), but it is uniquely well-suited to RNA because the propensity of RNA to form kinetically-trapped intermediates causes folding of many RNAs to be slower than their catalytic reactions (Treiber and Williamson, 1999).
Nevertheless, there are also limitations to the activity approach. Of course, it is limited to RNAs that possess catalytic activity, and further, it can only be used for RNAs whose catalytic activity is relatively robust. Also, the method provides limited information on steps in folding and the structures of intermediates.
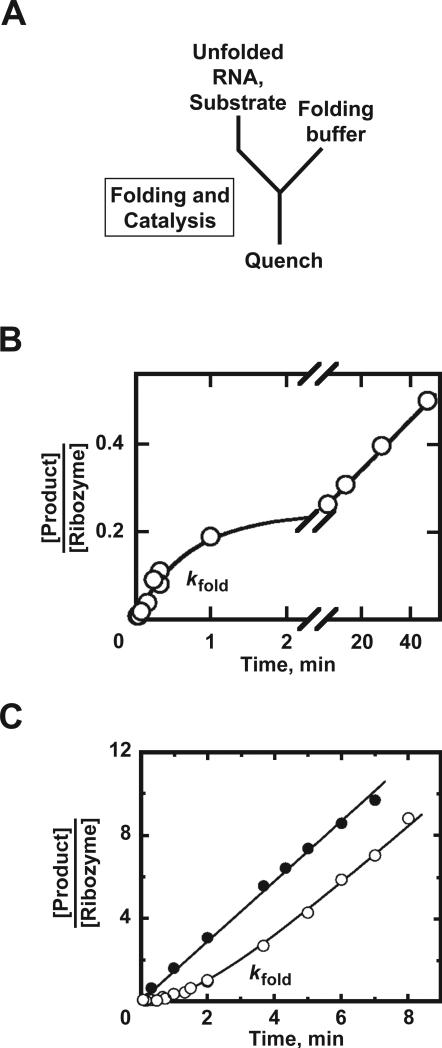
There are two general classes of catalytic activity assay, termed continuous and discontinuous assays (Fig. 1). In the continuous assay, folding and catalysis are monitored together, and the rate constant observed by monitoring the catalytic reaction reflects the folding process if folding is rate-limiting. In the discontinuous assay, folding is separated from the chemical reaction by performing the reaction in two stages, where the first stage allows folding but not catalysis, and the second stage allows catalysis but not further formation of native RNA.
Figure 1.
Schematics of continuous and discontinuous catalytic activity assays for folding. A, Continuous assay. The reaction is performed in a single stage, in which both folding and catalysis occur. Aliquots are quenched at various times t. B, Discontinuous assay. In the first stage, folding is allowed but the catalytic reaction is blocked. At various times t1, aliquots are transferred to stage 2. Here, the conditions allow robust catalysis while preventing further accumulation of native ribozyme, allowing determination of the fraction of native ribozyme at time t1.
In the sections below, we describe how to perform and interpret RNA folding experiments using each of these assays. We and others have used catalytic activity extensively to study ribozymes that cleave RNA, particularly group I introns (Zarrinkar and Williamson, 1994; Pan and Woodson, 1998; Russell and Herschlag, 1999; Russell and Herschlag, 2001; Russell et al., 2007). For simplicity we focus on these ribozymes in our descriptions, but the same methods are applicable to ribozymes that perform other types of reactions. Also for simplicity, we assume in the sections below that the reaction substrate is radiolabeled and is separated from the reaction product using polyacrylamide gel electrophoresis, but any method that separates reaction substrates and products quantitatively can be used. Our goal in the sections below is not to list detailed protocols, as the details will differ for different RNAs and different methods of labeling and detecting substrates, but rather to outline general strategies of experimental design and interpretation that allow catalytic activity to be an effective monitor of RNA folding.
2. PRELIMINARY MEASUREMENTS OF CATALYTIC REACTION
The only absolute requirement for using catalytic activity to monitor folding is that conditions exist under which the overall rate constant for the catalytic reaction, including all steps leading up to detectable product formation, exceeds the rate constant for folding. The degree to which catalysis must be faster than folding depends on a variety of factors, but as a general rule the difference must be at least 3 – 5-fold. As a first step, it is important to measure the catalytic reaction, independently from folding, and to survey conditions to maximize the fraction of active ribozyme and the catalytic rate constant. In practice, these measurements may be performed in parallel with trial folding measurements using the continuous assay described in section 3 below.
2.1. How to measure the catalytic reaction and establish pre-folding conditions
The basic experiment for monitoring the catalytic reaction is described in Fig. 2A. First, the RNA is pre-folded in the presence of Mg2+, typically at elevated temperature (10–50 mM Mg2+, 37–60 °C, 10–60 min) and in the absence of a substrate or cofactor. In practice, the pre-folding conditions will not yet be established for a new ribozyme, so the time and conditions of this incubation should be varied. Next, samples are adjusted to a constant set of conditions to measure the catalytic reaction rate, and the substrate is added. In an initial experiment, the substrate should be present in modest excess of the ribozyme (2–5-fold) so that the first round of the catalytic reaction can be observed and distinguished from subsequent rounds. Aliquots are quenched at various times by adding formamide (50–90% final concentration) and EDTA (≥2-fold in excess of Mg2+), and products are separated from substrate, typically by denaturing polyacrylamide gel electrophoresis (Zaug et al., 1988; Herschlag and Cech, 1990). The fraction of substrate converted to product is normalized by the substrate and the ribozyme concentrations (as determined by UV absorbance) and plotted against reaction time.
Figure 2.
Measurement of the catalytic reaction. A, Experimental scheme. RNA is pre-folded (typically 10–50 mM Mg2+, 37–60 °C, 10–60 min) and then mixed with substrate. B, Measurement of substrate cleavage by the Tetrahymena ribozyme (Russell and Herschlag, 1999). The reaction included 500 nM oligonucleotide substrate (with trace 5′-radiolabeled substrate) and 200 nM pre-folded ribozyme and was performed at 37 °C, pH 7.0, 10 mM Mg2+, with 1 mM guanosine. The cleavage reaction gave a burst of 0.8 product/ribozyme, indicating that most of the ribozyme was catalytically active. The rate constant was ≥8 min–1, faster than folding under these conditions. C, Simulated data showing biphasic kinetics from an internal equilibrium (KInt) between substrate cleavage and ligation. The approach to equilibrium occurs rapidly (lower dashed line at 0.6), and then dissociation of one of the products drives the reaction forward to a [product]/[ribozyme] value of 1 (upper dashed line, amplitude of 0.4 for the slow phase). From eq. 3 and 4, KInt = 1.5 (0.6/0.4) and the correction factor CF = 0.6. Panel B reprinted from Russell and Herschlag (1999) with permission from Elsevier.
A set of pre-incubation conditions should be selected that maximizes the fraction of active ribozyme, as determined from the amount of product in a burst or from the steady-state rate (see below). Once optimal pre-incubation conditions are established, this step is kept constant while the reaction conditions are surveyed (section 2.3).
2.2. Interpreting results from catalytic rate measurements
The method used to analyze the progress curve depends on the shape of the curve, which in turn depends on whether the reaction products are released faster or slower than they are produced. For a ribozyme that cleaves a substrate to generate two products, there are three regimes (Table 1; corresponding regimes for ribozymes that self-cleave or produce only one reaction product are also shown). If both products are released substantially faster than the substrate is cleaved (≥3–5-fold), a linear accumulation of product will be observed. Changes in the fraction of native ribozyme from different pre-folding conditions are reflected in corresponding changes in the reaction rate. If dissociation of one product is faster than cleavage and the other is slower, a rapid burst of product will be generated, which gives values both for the cleavage rate and for the amount of active ribozyme. If both products are released slower, a burst will be present, but the amplitude of the burst may correspond to less than the total amount of active ribozyme (see below).
Table 1.
Effects of product release rates on reaction and folding kinetics
| Relative product release rates† | Intramolecular cleavage | Intermolecular reaction 1 product | Intermolecular reaction 2 products (e.g. substrate cleavage) |
|---|---|---|---|
| Fast | Burst kinetics, amplitude corresponds to active ribozyme | Linear product accumulation, no burst. Rate-limiting folding detectable by lag kinetics | Linear product accumulation, no burst. Rate-limiting folding detectable by lag kinetics |
| One product fast, one product slow | N/A | N/A | Burst kinetics, amplitude corresponds to active ribozyme |
| Slow | Burst kinetics, amplitude may be reduced by reverse reaction | Burst kinetics, amplitude may be reduced by reverse reaction | Burst kinetics, amplitude may be reduced by reverse reaction |
Rate constants for product release are considered relative to those for earlier steps of the catalytic reaction for reactions with pre-folded ribozyme, and relative to the folding rate constant for experiments using the continuous activity assay (section 3).
If the progress curve shows a burst of product formation, the normalized data are fit by Eq. 1, in which A is the amplitude of the product burst, which reflects the amount of active ribozyme. Pre-folding conditions should be chosen to maximize this value. kobs is the rate constant for the catalytic reaction for steps leading up to product formation, s is the rate constant for the slower steady-state reaction, and t is the reaction time.
| (1) |
A cleavage reaction of the Tetrahymena ribozyme, for which one product is released rapidly and the other slowly (middle regime in Table 1, Herschlag and Cech, 1990), is shown in Fig. 2B. After folding (30 min, 50 °C, 10 mM Mg2+), the ribozyme rapidly performs a single round of oligonucleotide substrate cleavage at 37 °C and 10 mM Mg2+ (Herschlag and Cech, 1990; Zarrinkar and Williamson, 1994; Russell and Herschlag, 1999). The burst amplitude indicates that nearly all of the ribozyme is active. One of the products is then released slowly, as is often observed for RNA products that bind by base-pairing interactions, producing a slow steady-state phase.
If both products are released slowly, the burst amplitude may correspond to less than the total amount of active ribozyme because the cleavage products can re-ligate on the ribozyme (Scheme 1), reaching an internal equilibrium (Eq. 2) (Buzayan et al., 1986; Hegg and Fedor, 1995; Golden and Cech, 1996; Karbstein et al., 2002).
| (2) |
However, a small amplitude may indicate instead that the ribozyme is not fully folded to the native state or that some of the ribozyme is damaged and therefore inactive.
Scheme 1.

Although it is not trivial to distinguish between these possibilities, a strong indicator can come from the shape of the reaction progress curve. If the reaction is reversible and the products dissociate with distinct rate constants, the progress curve will give a double exponential within the first turnover, where the first phase, with amplitude A1, represents the approach to KInt and the second phase, with amplitude A2, represents dissociation of the first product (Fig. 2C). In contrast, the presence of inactive or misfolded ribozyme will lead to a single exponential followed by a linear phase. The amplitudes of the two exponential phases allow calculation of KInt (Eq. 3) and a correction factor, CF (Eq. 4). The measured burst amplitude is divided by CF to obtain the fraction of native ribozyme.
| (3) |
| (4) |
It should also be confirmed that one of the products is released with the rate constant of the slower phase by measuring its dissociation directly using pulse-chase procedures (Rose et al., 1974; Herschlag and Cech, 1990). To confirm the conclusion of an internal equilibrium, the reverse reaction should also be probed directly by monitoring reaction beginning from products (Hertel and Uhlenbeck, 1995; Karbstein et al., 2002).
2.3. Surveying conditions for use in folding experiments
After pre-incubation conditions are established, it is important to measure the catalytic rate under various conditions. Conditions that should be varied include monovalent and divalent cation concentrations, pH, and temperature. Substrate concentration should also be varied to establish saturation. The main goals are to identify conditions under which the catalytic rate exceeds the folding rate and to maximize this difference. Increases in pH and Mg2+ concentration increase the catalytic rates of many RNAs (Herschlag and Khosla, 1994; Hertel and Uhlenbeck, 1995; Narlikar and Herschlag, 1997), and a decrease in temperature may be advantageous because it may give a smaller decrease in the catalytic rate than the folding rate (Russell et al., 2007).
For all reaction conditions, the ribozyme should be first pre-folded under the optimal set of conditions established above. This allows changes in observed reaction rate to be interpreted in terms of effects on the catalytic steps, not on the fraction of active ribozyme (assuming that the reaction conditions do not give loss of native ribozyme). In practice, reactions in which the ribozyme is not pre-folded are conveniently performed in parallel (section 3). This allows scanning of conditions to find those under which the catalytic rate exceeds the folding rate, allowing the continuous assay to be used. These conditions can also be used as the second stage of a discontinuous assay (section 4), allowing folding to be measured even under first-stage conditions that do not support robust activity, as well as allowing a broad set of applications (section 5).
3. FOLLOWING RNA FOLDING BY CONTINUOUS ACTIVITY ASSAY
This section describes a simple assay for measuring RNA folding in which the adoption of the native state is determined by onset of catalytic activity. RNA folding is initiated at the same time as the catalytic reaction and, provided that folding is the rate-limiting step, the observed rate constant for the catalytic activity reflects the rate constant for folding. The continuous assay is very simple, so it is a good first choice for ribozymes whose catalytic reactions are rapid. It is also useful for studying RNAs for which folding and cleavage steps cannot easily be separated, such as self-processing RNAs that do not require cofactors. The main limitations are that it cannot be used to follow folding under conditions where folding is faster than the catalytic reaction, and it cannot generally be used to track unfolding processes, with the fraction of native ribozyme decreasing over time.
3.1. How to perform experiments using the continuous assay
The overall technique for measuring the rate of RNA folding is very similar to that for measuring the rate of substrate cleavage. The only difference is that the substrate, in small excess relative to the ribozyme, is added as folding is initiated, typically by Mg2+ addition (Fig. 3A). As in section 2, aliquots are removed at various times and quenched.
Figure 3.
Monitoring RNA folding using the continuous assay. A, Experimental scheme. Folding is initiated in the presence of substrate, and folding and catalysis are quenched with formamide and EDTA. B, Example of burst kinetics for the wild-type Tetrahymena ribozyme (Russell and Herschlag, 1999). The observed rate constant reflects folding, and the slow, linear phase is limited by product release. The burst amplitude (0.27) is lower than in an equivalent reaction of pre-folded ribozyme (see Fig. 2B), indicating that most of the ribozyme did not reach the native state on this time scale. C, Lag kinetics for the EΔP5abc Tetrahymena variant ribozyme. The steady-state rate was the same as that for pre-folded ribozyme (see text). Panels B and C adapted from Russell and Herschlag (1999), with permission from Elsevier.
3.2. Interpreting results from the continuous assay
The results are interpreted in conjunction with catalytic rate measurements described in section 2. If the progress curves are the same with or without pre-folding of the ribozyme, folding is more rapid than cleavage and only a lower limit on the folding rate is obtained. On the other hand, if the reaction with co-initiation of folding and cleavage gives slower or less product formation, folding is limiting for at least a fraction of the population and further interpretation of the data is warranted.
There are two general types of behavior that may be observed. If the rate constant for release of at least one product is slower than folding and the catalytic reaction, product will be formed in an initial burst. In this case, the data are normalized for substrate and ribozyme concentration as in section 2 and fit by eq. 1. Here, kobs reflects the rate constant for folding (kfold), and the amplitude gives the fraction of ribozyme that folds to the native state in this transition. If the burst amplitude indicates less than stoichiometric formation of product, it suggests that some of the ribozyme does not reach the native state on this time scale (or that the reaction reaches an internal equiibrium before product dissociation, see Section 2.2). To rule out the possibility that the low value results from damaged ribozyme, the amplitude should be compared to that from a reaction with pre-folded ribozyme. Using this assay, it was shown that a small fraction of the Tetrahymena ribozyme reaches the native state with a rate constant of 1 min–1 (Fig. 3B), whereas the rest misfolds and remains non-native on the time scale of the experiment (Russell and Herschlag, 1999; Russell and Herschlag, 2001). In principle, the continuous assay can also reveal multiple folding pathways by the presence of additional exponential phases. However, in practice it can be difficult to these phases from continued cleavage by native ribozyme formed in the fastest phase of folding.
The second possible behavior is a steady-state accumulation of product without a burst, which may be preceded by a lag. A lag appears if the folding process is slower than the entire catalytic cycle, because the steady-state rate of product formation depends on the concentration of active ribozyme, and this concentration increases slowly as the native state accumulates in folding. An example of lag kinetics is shown in Fig. 3C (Russell and Herschlag, 1999). In this case, the data are fit by Eq. 5, in which s represents the rate constant for the steady-state phase and kobs is the rate constant for the lag, which is equal to the folding rate constant kfold. To maximize the signal from a lag, a large excess of substrate should be used relative to the ribozyme (≥10-fold) to increase the length and linearity of the steady-state phase.
| (5) |
To obtain a relative measure of the fraction of ribozyme present in the native state, the steady-state rate is compared to that for the pre-folded RNA. If the rate is lower without pre-folding, not all of the ribozyme has folded to the native state, whereas if they are the same, there is as much native ribozyme as in the pre-folded control. However, it is important that this comparison does not give an absolute measure of the fraction of native ribozyme, only a relative one. For example, a Tetrahymena ribozyme variant that lacks the P5abc peripheral element (EΔP5abc) gives the same steady-state rate with or without pre-folding (Fig. 3C, Russell and Herschlag, 1999). Nevertheless, this ribozyme equilibrates between the native and misfolded conformers, with the native state populated only by about 60% of the ribozyme even after pre-folding (Johnson et al., 2005). This incomplete occupancy was not detectable using the continuous assay.
4. FOLLOWING RNA FOLDING BY DISCONTINOUS ACTIVITY ASSAY
The essence of the discontinuous assay is that the reaction is performed in two stages (see Fig. 1). In the first stage, folding is allowed to proceed but the catalytic reaction is blocked. In the second stage, the conditions are changed such that folding is arrested and the catalytic reaction is initiated, allowing determination of the fraction of native ribozyme that accumulated during the first stage. The discontinuous assay has several advantages over the continuous assay. By separating folding from the catalytic reaction, conditions can be chosen for stage 2 that maximize the catalytic rate, increasing the robustness of the signal. Indeed, using this assay it is even possible to measure the folding rate under conditions in stage one that do not give rapid catalysis, such as low Mg2+ concentration. Further, decreases in the fraction of native ribozyme can be determined, allowing unfolding processes to be followed.
There are also disadvantages and limitations to the discontinuous assay. It can only be used if conditions exist that arrest folding. Minimally, folding must become much slower than catalysis, and in practice it is valuable to establish conditions that arrest folding on the experimental time scale (minutes for experiments performed by hand). Another limitation is that the discontinuous assay requires more work than the continuous assay, because each folding time course consists of a series of reaction time courses. This disadvantage can be minimized by collecting only a small number of points during each cleavage reaction, which can be sufficient to measure reliably the fraction of native ribozyme if the reaction kinetics are well-understood.
4.1. How to design and perform experiments using the discontinuous assay
The key to using the discontinuous assay is to establish conditions for the two reaction stages (Fig. 4A). In the first stage, the catalytic reaction must be inhibited, most commonly by omitting the substrate or a necessary cofactor. In the second stage, folding must be arrested, or at least slowed to a lower rate than the catalytic reaction. This goal has been achieved by using high Mg2+ concentration (50–100 mM) and/or low temperature (Russell and Herschlag, 2001; Russell et al., 2006; Russell et al., 2007; Pan et al., 1999) (see section 2.3).
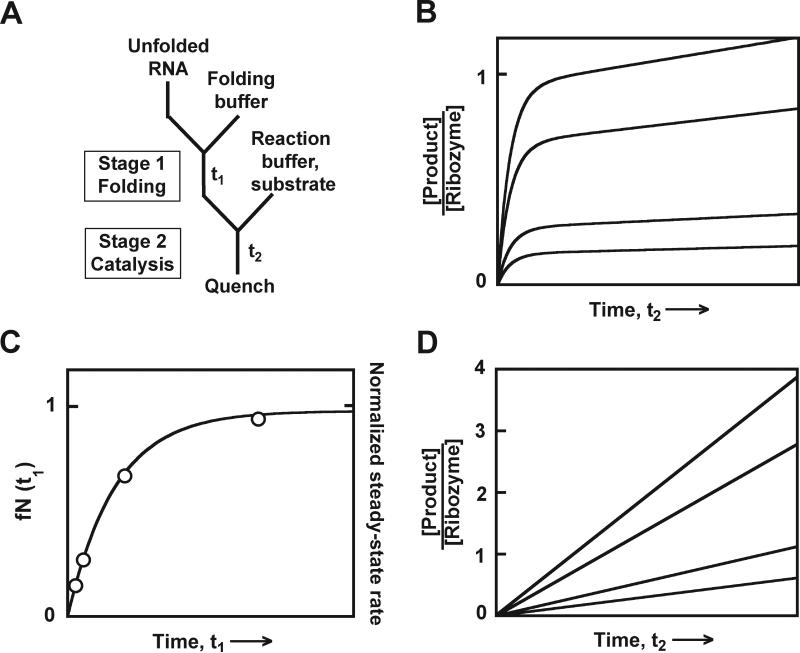
Figure 4.
Monitoring RNA folding by the discontinuous activity assay. A, Experimental design. Unfolded RNA is first mixed with Mg2+ to initiate folding in stage 1. After various times t1, aliquots are transferred to stage 2, blocking further native folding, and are mixed with substrate. Aliquots from stage 2 are quenched at various times t2 by formamide and EDTA. B, Simulated data for a ribozyme that gives burst kinetics. The four curves represent stage 2 reactions after different folding times t1. Each curve has the same rate constant, which reflects the catalytic reaction, followed by a steady-state increase that reflects subsequent turnovers. C, Simulated folding curve. Plotting the burst amplitudes from panel B against folding time t1 gives an exponential curve with rate constant kfold. D, Simulated product accumulation for four stage 2 reactions with a ribozyme that rapidly releases products. The slopes are also plotted in panel C and give the same folding curve as the burst amplitudes from panel B, except that here the amounts of native ribozyme are on a relative scale (alternate y-axis labels at right).
When conditions for the two stages are established, folding experiments can be performed. Folding is typically initiated in stage 1 by adding divalent ions (Mg2+). Aliquots are removed from the mixture at different times t1 and transferred to conditions for stage 2. This can be done in a single step or, if the folding reaction is sufficiently slow under stage 2 conditions, it can be performed in multiple steps. For example, the aliquot can first be transferred to low temperature and/or high Mg2+ conditions, and then substrate can be added. Because aliquots must be quenched at relatively defined times after substrate addition for a good determination of the burst amplitude or reaction rate, it is helpful to ‘unlink’ this reaction from the stage 1 reaction by adding substrate separately. Quenched aliquots are processed as above.
4.2. Interpreting results from the discontinuous assay
For each folding time t1, the fraction of cleaved substrate is normalized by the substrate and ribozyme concentrations and plotted against t2. Most commonly, the progress curve can be fit to a single exponential phase of product formation followed by a linear increase (Fig. 4B, eq. 1). Here the rate constant kobs reflects the catalytic reaction, as in section 2. The burst amplitude A reflects the fraction of native ribozyme at the time the aliquot was transferred from stage 1 to stage 2 (fN(t1)).
To obtain the folding rate, fN(t1) is plotted against folding time t1 (Fig. 4C). If a simple pathway is traversed, this curve will give a single exponential curve in which the rate constant reflects the observed folding process and the amplitude reflects the fraction of native ribozyme present at the endpoint. The amplitude value will be less than one if the ribozyme is not completely folded at equilibrium, if there are additional folding pathways that do not give native ribozyme on the time scale observed, or if some of the ribozyme is damaged. It may be necessary to include a constant term if additional folding pathways give rapid, unresolved phases of native ribozyme formation or to include additional exponential terms if there are multiple pathways that produce native ribozyme on the observed time scale (Russell et al., 2007).
If, under the conditions of the stage 2 catalytic reaction, the first turnover is not faster than subsequent turnovers, the product will not accumulate in a burst, but will instead accumulate linearly (Fig. 4D). This rate can be used to provide a relative measure of the fraction of native ribozyme, as it will be proportional to the fraction of active ribozyme. Therefore, the rate can be plotted against folding time t1 to monitor the progress of folding. However, in this case the fraction of native ribozyme cannot be determined absolutely, only relative to a pre-folded control. This situation is analogous to that of lag kinetics in the continuous assay (section 3.2).
For certain applications of the discontinuous assay, the substrate cleavage reaction has been performed under single turnover conditions, with ribozyme in excess of substrate (Pan et al., 1999; Russell and Herschlag, 2001; Russell et al., 2006; Russell et al., 2007; see Fig. 6 and Fig. 8). Here, trace substrate is added in stage 2, and either the observed rate constant (Pan et al., 1999) or the fraction of the substrate cleaved rapidly (Russell and Herschlag, 2001) is taken as a measure of the fraction of native ribozyme. This method has the advantage that the burst amplitude does not depend on substrate concentration, or on ribozyme concentration in the latter case, which can increase precision and reproducibility. However, the single turnover assay has more extensive and specific requirements for the relative rate constants of substrate binding, release, and catalysis, and it should only be used under well-defined circumstances.
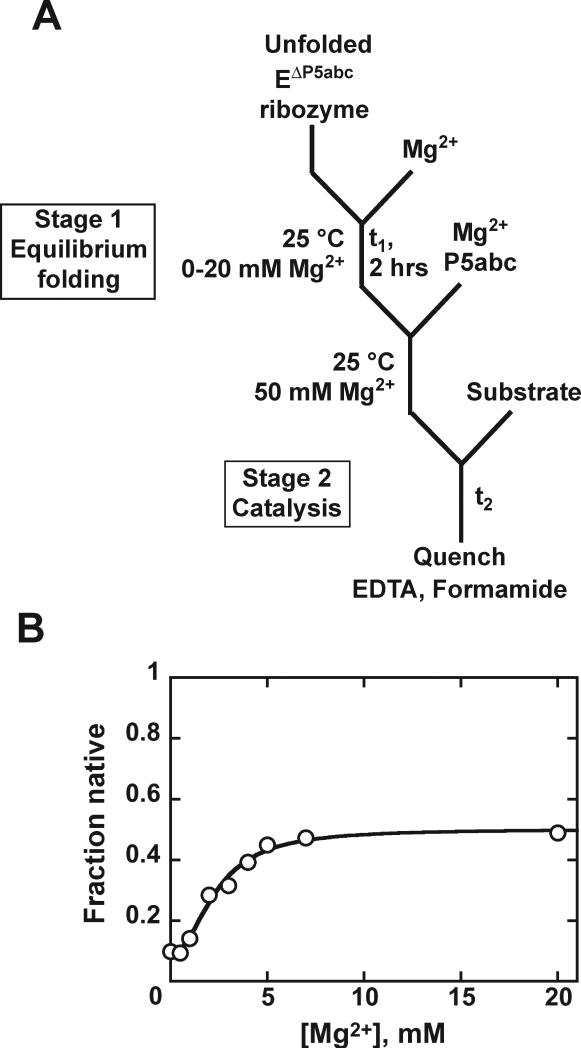
Figure 6.
Mg2+ concentration dependence of native ribozyme formation. A, Reaction scheme. The EΔP5abc Tetrahymena ribozyme was incubated in stage 1 with 0–20 mM Mg2+, allowing equilibration of the native RNA and folding intermediates. Additional Mg2+ (50 mM final) was then added, which induces folding predominantly to the long-lived misfolded form, and substrate was added to initiate the catalytic reaction in stage 2. B, Burst amplitude vs Mg2+ concentration in stage 1. The amplitude does not reach one because this RNA equilibrates between the native and misfolded conformations. Panel B adapted with permission from Russell et al. (2007), copyright 2007 American Chemical Society.
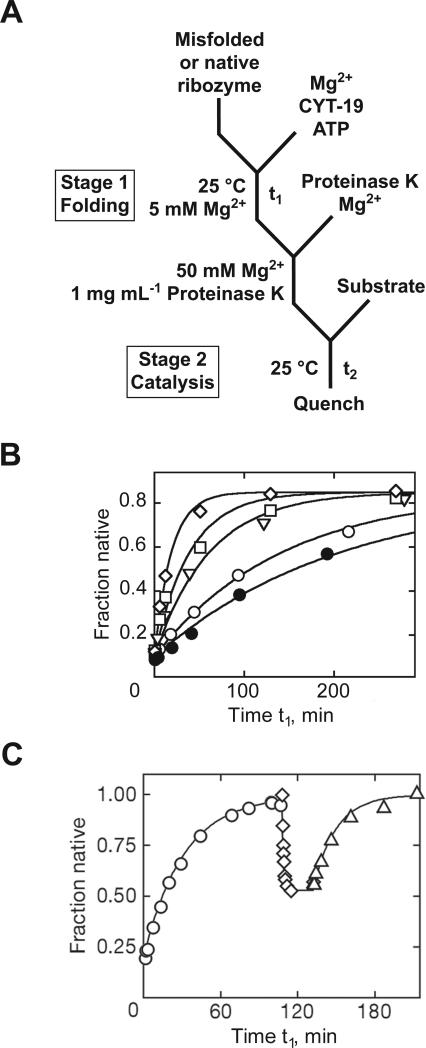
Figure 8.
Chaperone-mediated RNA folding monitored by a discontinuous activity assay. A, Reaction scheme. In stage 1, CYT-19 protein was added with ATP to populations of native or misfolded Tetrahymena ribozyme. At various times, further transitions between the native and misfolded conformations were blocked by Proteinase K and 50 mM Mg2+, and the fraction of native ribozyme was determined by adding substrate for 1 min (stage 2). B, Progress curves without CYT-19 (○), with 500 nM CYT-19 without ATP (•), or with ATP and 100 nM (▽), 200 nM (□), or 500 nM (◇) CYT-19. C, Unfolding of native ribozyme by CYT-19. Using a tertiary contact mutant, the experiment monitored initial folding to the native state (○), unfolding of native ribozyme by CYT-19 (◇) and subsequent re-folding after Proteinase K digestion of CYT-19 in stage 1 (△). Y-axis values are larger in panel C because data were normalized for 10-20% damaged ribozyme. Panel B adapted with permission from Tijerina et al. (2006), copyright 2006 National Academy of Sciences, U.S.A. Panel C reprinted from Bhaskaran and Russell (2007) with permission from Macmillan Publishers Ltd: Nature, copyright 2007.
5. OTHER APPLICATIONS OF CATALYTIC ACTIVITY AS A PROBE OF FOLDING
The preceding sections describe how to use catalytic activity to follow folding of structured RNAs. The same basic approach has been used in creative ways to gain additional information about RNA folding processes. As described below, the continuous assay has been applied to follow assembly of RNA-protein complexes. The discontinuous assay has been used to measure the equilibrium of native RNA formation and to follow folding co-transcriptionally or in the presence of chaperones.
5.1. Assembly of RNA-protein complexes
In addition to monitoring intramolecular folding events, the continuous activity assay has been used to follow assembly of a catalytic RNA with protein (Webb et al., 2001) or an activating RNA (Russell and Herschlag, 1999). The most general requirement here is that the catalytic reaction must be faster than the assembly process, which can be modulated by changing the concentrations of the interacting molecules.
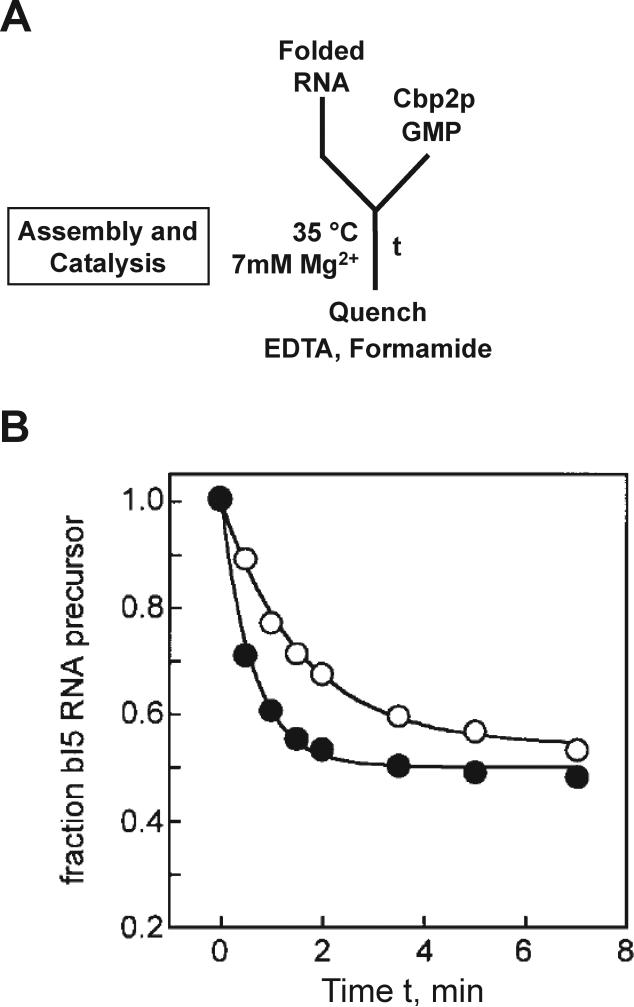
Weeks and colleagues used this method to measure association and assembly of a group I intron RNA with its cofactor protein Cbp2p (Webb et al., 2001). They first pre-incubated the intron to allow formation of a near-native collapsed state and then added Cbp2p to allow assembly and subsequent splicing (Fig. 5A). The observed rate constant was compared to that from a reaction in which Cbp2p and the intron were pre-incubated to allow complex formation, and then GMP was added to initiated splicing (analogous to pre-folded reactions of section 2). The observed rate constant was smaller for the reaction initiated with Cbp2p, indicating that complex assembly was rate-limiting for splicing under the experimental conditions (Fig. 5B).
Figure 5.
Continuous activity assay to follow RNA-protein complex assembly. A, Experimental design. Radiolabeled precursor of the bI5 group I intron was mixed with protein Cbp2p and cofactor GMP to initiate the folding and cleavage assay. B, This continuous assay gave a rate constant of 0.5 min–1 (○). A control reaction in which the RNA was pre-incubated with Cbp2p to allow complex formation and then mixed with GMP to initiate cleavage gave a larger rate constant, 1.6 min–1 (•). Panel B reprinted from Webb et al. (2001) with permission from Elsevier.
5.2. Native folding at equilibrium
The discontinuous assay has been used to measure the Mg2+-dependent equilibrium for native RNA folding of the Tetrahymena EΔP5abc ribozyme (Russell et al., 2007). In stage 1 the ribozyme was incubated at various Mg2+ concentrations to allow equilibration between intermediates and the native state, and then the fraction of native ribozyme was determined in stage 2 by adding 50 mM Mg2+ and P5abc RNA and then substrate. These additions allowed catalytic activity, while trapping most of the ribozyme that was non-native in stage 1 as a long-lived misfolded intermediate (Fig. 6A). A small fraction of the ribozyme did fold to the native state during stage 2 prior to substrate addition (10%, Fig. 6B), as expected from earlier results (Russell and Herschlag, 1999). Because this fraction was small and expected to be constant, it could be subtracted to calculate the fraction of native ribozyme in the stage 1 incubation (not shown).
5.3. Co-transcriptional folding
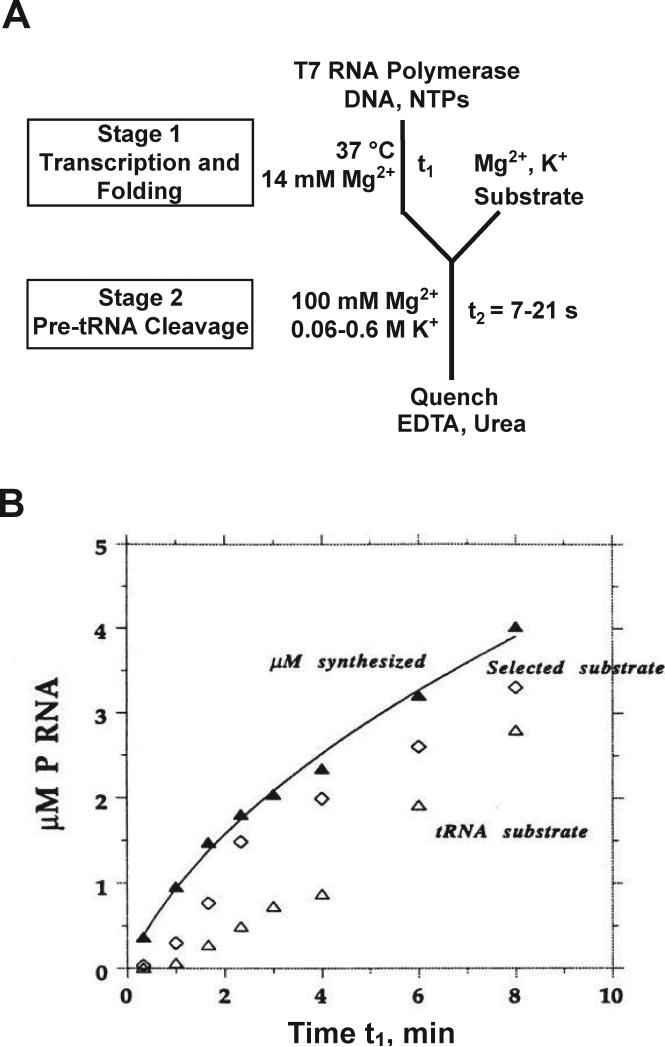
A discontinuous activity assay has been used by Pan, Sosnick and colleagues to follow folding of the RNase P RNA during in vitro transcription (Pan et al., 1999; Wong et al., 2005; Wong et al., 2007). In this work, stage 1 included a DNA template, T7 RNA polymerase, and NTPs to allow transcription. At various times, aliquots were transferred to a stage 2 reaction that included a tRNA precursor substrate and high ionic strength conditions (100 mM MgCl2 and 0.06 – 0.6 M KCl), permitting cleavage of the substrate but blocking transcription initiation (Fig. 7). The activity assay was used to monitor active RNase P RNA formed over time in the stage 1 reaction, and the amount of RNA synthesized was determined from incorporation of radiolabeled nucleotide. The lag between the accumulation of large RNA and the appearance of cleavage activity provided evidence for slow co-transcriptional folding of the RNase P RNA, and further work showed that pausing of RNA polymerase during transcription accelerated folding.
Figure 7.
Co-transcriptional folding of RNase P RNA monitored by a discontinuous activity assay. A, Reaction scheme. In stage 1, T7 RNA polymerase was incubated with template DNA and NTPs (including [α-32P]CTP) at 37 °C and pH 8.1 to initiate transcription and concurrent folding. At various times t1, transcription was arrested by transferring aliquots to high ionic strength conditions (stage 2), and the catalytic reaction was initiated by adding substrate, either a pre-tRNA or an oligonucleotide substrate obtained by in vitro selection that is cleaved efficiently by the catalytic domain of RNase P RNA (Pan and Jakacka, 1996). Stage 2 reactions were quenched after 7-21 s, allowing determination of the relative amount of native RNase P RNA. B, The amount of RNA synthesized (▲) was measured by incorporation of labeled CTP, while native folding was monitored by cleavage of the tRNA precursor substrate (△) or the selected substrate for folding of the catalytic domain (◇). Panel B reprinted with permission from Pan et al. (1999), copyright 1999 National Academy of Sciences, U.S.A.
The use of RNA catalysis to follow co-transcriptional folding was taken a step further by Fedor and colleagues, who studied the hairpin self-cleaving RNA in vivo using a continuous assay (Mahen et al., 2005). Starting from a steady state of precursors and RNA cleavage products, transcription was shut off using a glucose-sensitive promoter, and the rate of the subsequent loss of precursor RNA was used to determine the efficiency of folding to the native state vs alternative structures.
5.4. RNA chaperone-mediated folding and unfolding
Certain DEAD-box ‘RNA helicase’ proteins possess RNA chaperone activity and function during folding of several group I introns (Mohr et al., 2002; Huang et al., 2005). Our group has applied the discontinuous assay to study re-folding of the Tetrahymena ribozyme in the presence of the CYT-19 DEAD-box protein (Tijerina et al., 2006; Bhaskaran and Russell, 2007). The ribozyme was first misfolded by incubation with Mg2+ (Fig. 8A). Then, CYT-19 and ATP were added in stage 1. In stage 2, Proteinase K was added to inactivate CYT-19, and the Mg2+ concentration was increased to 50 mM to block further re-folding while allowing robust catalytic activity. By this assay, CYT-19 was shown to accelerate re-folding in a concentration-dependent manner (Fig. 8B).
The discontinuous assay was also used to determine whether CYT-19 mediates unfolding of the native ribozyme (Bhaskaran and Russell, 2007). The motivation was to test whether CYT-19 acts as a general chaperone without recognizing structural features of misfolded RNA. Here, the ribozyme was first pre-folded to the native state, and the stage 1 reaction was designed to destabilize the native structure, either with low Mg2+ concentration or with destabilizing mutations in the RNA. In stage 2, Proteinase K and 50 mM Mg2+ were added and the assay was performed exactly as described above.
Using this assay, cycles of folding and unfolding of the ribozyme were observed (Fig. 8C). Initial folding gave native ribozyme formation, and then after addition of CYT-19, the cleavage burst amplitude decreased, indicating loss of the native ribozyme. After CYT-19 proteolysis, the ribozyme returned to the native state. Note that it would not be possible to use the continuous assay to monitor the unfolding transition because the substrate would already be cleaved prior to CYT-19 addition. However, it is straightforward with the discontinuous assay, requiring only that unfolding be faster than folding in stage 1, so that net unfolding occurs, and that the stage 2 conditions permit catalytic activity and block re-folding of RNA that unfolded in stage 1.
ACKNOWLEDGMENTS
We thank members of the Russell lab for comments on the manuscript. Research in the Russell lab is funded by grants from the NIH (GM 070456), the Welch Foundation (F-1563), and the Advanced Research Program (003658-0242-2007).
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KM, Hampel KJ. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry. 2009;48:5669–5678. doi: 10.1021/bi900183r. [DOI] [PubMed] [Google Scholar]
- Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. Ribozymes, the first 20 years. Biochem. Soc. Trans. 2002;30:1162–1166. doi: 10.1042/bst0301162. [DOI] [PubMed] [Google Scholar]
- Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li N, Ellington AD. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007;4:633–655. doi: 10.1002/cbdv.200790055. [DOI] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell. Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr. Opin. Struct. Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Emerick VL, Pan J, Woodson SA. Analysis of rate-determining conformational changes during self-splicing of the Tetrahymena intron. Biochemistry. 1996;35:13469–13477. doi: 10.1021/bi960865i. [DOI] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR. RNase P: interface of the RNA and protein worlds. Trends Biochem. Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu. Rev. Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- Golden BL, Cech TR. Conformational switches involved in orchestrating the successive steps of group I RNA splicing. Biochemistry. 1996;35:3754–3763. doi: 10.1021/bi952599z. [DOI] [PubMed] [Google Scholar]
- Hegg LA, Fedor MJ. Kinetics and thermodynamics of intermolecular catalysis by hairpin ribozymes. Biochemistry. 1995;34:15813–15828. doi: 10.1021/bi00048a027. [DOI] [PubMed] [Google Scholar]
- Held WA, Nomura M. Rate determining step in the reconstitution of Escherichia coli 30S ribosomal subunits. Biochemistry. 1973;12:3273–3281. doi: 10.1021/bi00741a020. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Khosla M. Comparison of pH dependencies of the Tetrahymena ribozyme reactions with RNA 2'-substituted and phosphorothioate substrates reveals a rate-limiting conformational step. Biochemistry. 1994;33:5291–5297. doi: 10.1021/bi00183a036. [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Uhlenbeck OC. The internal equilibrium of the hammerhead ribozyme reaction. Biochemistry. 1995;34:1744–1749. doi: 10.1021/bi00005a031. [DOI] [PubMed] [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. U.S.A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Carroll KS, Herschlag D. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry. 2002;41:11171–11183. doi: 10.1021/bi0202631. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T, Quaas R, Hahn U, Schmid FX. Folding of ribonuclease T1. 1. Existence of multiple unfolded states created by proline isomerization. Biochemistry. 1990;29:3053–3061. doi: 10.1021/bi00464a023. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T. Protein folding kinetics. Methods Mol. Biol. 1995;40:313–341. doi: 10.1385/0-89603-301-5:313. [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Szostak JW. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature. 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Moore PB, Steitz TA, Nissen P, Hansen J, Ban N. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Herschlag D. Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 1997;66:19–59. doi: 10.1146/annurev.biochem.66.1.19. [DOI] [PubMed] [Google Scholar]
- Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- Pan T, Jakacka M. Multiple substrate binding sites in the ribozyme from Bacillus subtilis RNase P. EMBO J. 1996;15:2249–2255. [PMC free article] [PubMed] [Google Scholar]
- Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose IA, O'Connell EL, Litwin S. Determination of the rate of hexokinase-glucose dissociation by the isotope-trapping methods. J. Biol. Chem. 1974;249:5163–5168. [PubMed] [Google Scholar]
- Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J. Mol. Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: Commitment to form the native conformation is late in the folding pathway. J. Mol. Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- Russell R, Das R, Suh H, Travers K, Laederach A, Engelhardt M, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J. Mol. Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. Time-resolved synchrotron X-ray “footprinting”, a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J. Mol. Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- Su LJ, Brenowitz M, Pyle AM. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol. 2003;334:639–652. doi: 10.1016/j.jmb.2003.09.071. [DOI] [PubMed] [Google Scholar]
- Teixeira A, Tahiri-Alaoui A, West S, Thomas B, Ramadass A, et al. Autocatalytic RNA cleavage in the human beta-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. Non-specific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J. Mol. Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Webb AE, Rose MA, Westhof E, Weeks KM. Protein-dependent transition states for ribonucleoprotein assembly. J. Mol. Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]
- Wong T, Sosnick TR, Pan T. Mechanistic insights on the folding of a large ribozyme during transcription. Biochemistry. 2005;44:7535–7542. doi: 10.1021/bi047560l. [DOI] [PubMed] [Google Scholar]
- Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA, Cech TR. Alternative secondary structures in the 5' exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- Xiao M, Li T, Yuan X, Shang Y, Wang F, Chen S, Zhang Y. A peripheral element assembles the compact core structure essential for group I intron self-splicing. Nucleic Acids Res. 2005;33:4602–4611. doi: 10.1093/nar/gki770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- Zarrinkar PP, Williamson JR. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat. Struct. Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]