Abstract
Dyskeratosis congenita (DC) is an inherited poikiloderma which in addition to the skin abnormalities is typically associated with nail dystrophy, leucoplakia, bone marrow failure, cancer predisposition and other features. Approximately 50% of DC patients remain genetically uncharacterized. All the DC genes identified to date are important in telomere maintenance. To determine the genetic basis of the remaining cases of DC, we undertook linkage analysis in 20 families and identified a common candidate gene region on chromosome 16 in a subset of these. This region included the C16orf57 gene recently identified to be mutated in poikiloderma with neutropenia (PN), an inherited poikiloderma displaying significant clinical overlap with DC. Analysis of the C16orf57 gene in our uncharacterized DC patients revealed homozygous mutations in 6 of 132 families. In addition, three of six families previously classified as Rothmund–Thomson syndrome (RTS—a poikiloderma that is sometimes confused with PN) were also found to have homozygous C16orf57 mutations. Given the role of the previous DC genes in telomere maintenance, telomere length was analysed in these patients and found to be comparable to age-matched controls. These findings suggest that mutations in C16orf57 unify a distinct set of families which clinically can be categorized as DC, PN or RTS. This study also highlights the multi-system nature (wider than just poikiloderma and neutropenia) of the clinical features of affected individuals (and therefore house-keeping function of C16orf57), a possible role for C16orf57 in apoptosis, as well as a distinct difference from previously characterized DC patients because telomere length was normal.
INTRODUCTION
The main problem facing any clinician is making an accurate diagnosis from the symptoms and signs presenting in a particular individual at a given time point. This is compounded when several diseases have many overlapping features. This is exemplified by the ‘inherited poikilodermas' dyskeratosis congenita (DC), poikiloderma with neutropenia (PN) and Rothmund–Thomson syndrome (RTS); the overlapping features, particularly the poikiloderma, can often make it difficult to accurately classify a patient into one of these diagnoses.
DC is often described as being a multi-system bone marrow failure syndrome characterized by abnormal skin pigmentation (poikiloderma), nail dystrophy and oral leucoplakia (1). It is genetically and clinically very heterogeneous with three modes of inheritance being described: X-linked recessive, autosomal dominant and autosomal recessive. The causative mutations have been identified in some, but not all families presenting with DC. Six genes have been implicated in causing DC, and all these are important in telomere maintenance either by telomere elongation (telomerase) or telomere protection (shelterin). Five genes (DKC1, TERC, TERT, NOP10 and NHP2) involved in the telomerase complex have been shown to be mutated in some patients with DC. Dyskerin (DKC1) is the main causative gene in the X-linked form of the disease (2). Autosomal dominant DC is caused by heterozygous mutations in the core components of telomerase, namely TERC (the RNA component) (3) and TERT (the enzymatic component) (4). Autosomal recessive DC is caused by biallelic mutations in NOP10, NHP2 (both components of the small nucleolar ribonucleoprotein particle) and TERT (5–7). Heterozygous mutations have been described in TINF2 (TIN2) of the shelterin complex (8). Although some of these TINF2 mutations are inherited in a dominant fashion, the majority are de novo (9). These six genes, however, do not account for all the patients with DC as ∼40–50% still remain uncharacterized at the genetic level.
RTS is another rare autosomal recessive poikiloderma. Generally, it presents in infancy with a characteristic facial rash (poikiloderma) and heterogeneous clinical features including short stature, juvenile cataracts, sparse hair, skeletal abnormalities and a predisposition to osteosarcoma. Abnormalities in gastrointestinal, respiratory and haematological systems have also been reported. Mutations in RECQL4 (ATP-dependent DNA helicase Q4) have been identified in ∼60% of patients with RTS (10,11). Two clinical subforms of RTS have been defined: RTS I is characterized by poikiloderma, ectodermal dysplasia and juvenile cataracts. RTS II is characterized by poikiloderma, congenital bone defects and an increased risk of osteosarcoma in childhood and skin cancer in later life. RTS II is caused by biallelic mutations in RECQL4, but the function of this gene in the disease still remains unclear. RTS I is transmitted in an autosomal recessive manner, but the genetic aetiology remains unknown (12).
PN, previously described as Navajo poikiloderma, is a third rare autosomal recessive disorder involving poikiloderma (13,14). In addition to the skin involvement, it is characterized by neutropenia, short stature, pachyonychia (nail dystrophy) and pulmonary disease. There is much debate in the literature as to the degree of separation between this and RTS which is the more common hereditary poikiloderma (15,16). Generally, the RECQL4 mutations that are common to RTS are absent in PN. Recently, however, the gene C16orf57 has been implicated to cause PN. C16orf57 is a gene that has no known function, but its encoded product has been suggested to be interconnected to RECQL4 via the SMAD4 protein (17).
The aim of this study was to identify further genes that are involved in the pathology of DC. In doing so, an unexpected link was identified between the three syndromes mentioned above; a subset of patients classified clinically as DC, PN and RTS are unified by C16orf57 mutations.
RESULTS
Illumina™ 6k SNP chip analysis
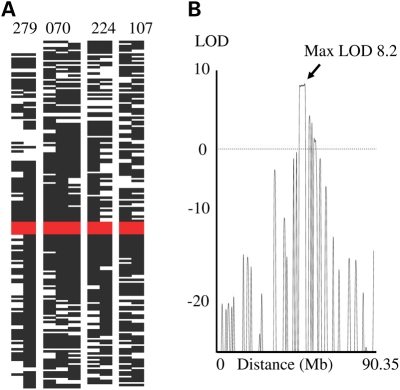
Previous analysis of 16 consanguineous families using microsatellite analysis showed that DC is a very heterogeneous disorder (5). In comparative terms, this method is crude in terms of the resolution obtained, so therefore we screened eight consanguineous multiplex families with DC using the Illumina 6k SNP chip as well as 12 sporadic individuals to identify possible smaller overlapping regions of homozygosity that could have been missed by the larger intervals of microsatellite mapping. Interestingly, one of the families (DCR070) though enrolled on the DCR in 1999 has subsequently been reported with a diagnosis of PN, Clericuzio type (18). All the autosomal SNP data were examined for homozygosity in the families using Genehunter. Although in each family several regions with significant LOD scores (1.5 or greater depending on family structure) were observed, there was only one overlapping region on chromosome 16 that was common to four families (DCR070, DCR107, DCR224 and DCR279, including a total of nine affected individuals). When the data from these four families were combined, a LOD score of 8.2 was achieved, which is highly significant. This interval was defined as being between SNPs rs1982395 and rs768462 (57668756–60069848bp on GRCh37; Fig. 1). On further scrutiny of the sporadic cases, it was noted that three individuals also had a significant block of homozygosity overlapping this interval (DCR106, DCR311 and DCR324). No other significant overlapping intervals were identified in the remaining families.
Figure 1.
Identification of a common region of homozygosity in four families with DC. (A) Homozygosity of all SNPs on chromosome 16. Black boxes indicate a homozygous call, white spaces heterozygous calls. The red bar indicates the overlapping region. (B) LOD score plot of families DCR279, DCR070, DCR224 and DCR107 showing the maximum LOD score of 8.2 on chromosome 16.
Mutation detection
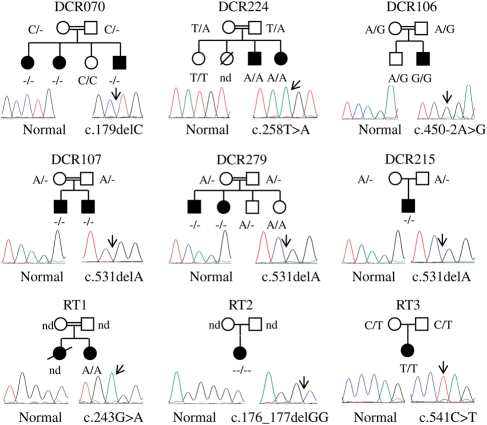
Using the UCSC genome bioinformatics site (http://genome.ucsc.edu), 22 genes were found to be in this region, none of which were obvious candidates. Out of these 22 genes, snoRNA46, snoRNA50, GINS3 and CSNK2A2 were sequenced, but no mutation was identified. A recent paper by Volpi et al. (17) described a homozygous splicing mutation in the orphan gene C16orf57, in a large kindred with PN. Owing to the clinical overlap between the published family and our DCR families, we sequenced this gene in the index case from DCR070, DCR107, DCR224 and DCR279 as well as the sporadic individuals from DCR106, DCR311 and DCR324 which also overlapped this interval. Homozygous mutations were identified in all patients except DCR311 and DCR324 (Table 1). Where a mutation was observed, the rest of the family was sequenced for that exon to confirm the presence of the mutation in the index case and to determine segregation (Fig. 2).
Table 1.
Mutations identified in C16orf57 in patients in the DCR and RTS patients
| Family | Exon | Nucleotide | Protein |
|---|---|---|---|
| 070 | 2 | c.179 del C | Pro60Leu fsX54a |
| 106 | 4 | c.450-2 A>G | Splice site |
| 107 | 5 | c.531 del A | His179Met fsX86 |
| 215 | 5 | c.531 del A | His179Met fsX86 |
| 224 | 2 | c.258 T>A | Tyr86X |
| 279 | 5 | c.531 del A | His179Met fsX86 |
| RTS 1 | 2 | c.243 G>A | Trp81Xb |
| RTS 2 | 2 | c.176,177 del GG | Gly59Ala fsX2 |
| RTS 3 | 5 | c.541 C>T | Gln181X |
Figure 2.
Pedigrees of families with mutations in C16orf57. Sequence panel on the left shows the normal trace while the right shows the observed mutation. The arrow highlights the point of mutation. c., coding nucleotide number; nd, sample unavailable for analysis; open shape, unaffected individual; filled shape, affected individual.
Screening of the dyskeratosis congenita registry
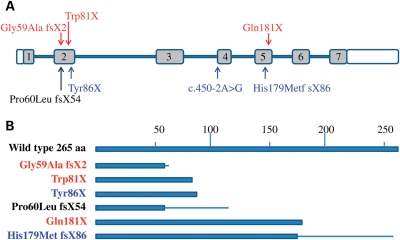
In light of the new C16orf57 mutations discovered in several of the families with DC, we then screened additional uncharacterized families entered into our dyskeratosis congenita registry (DCR) by heteroduplex analysis to determine whether any more individuals had mutations in this gene. The DCR is a large collection of over 350 families with DC from all over the world. The genetic basis of ∼40–50% still remains unknown, and it was these families that were targeted in the next screen. In addition, six individuals with a clinical diagnosis of RTS we had in our collection were also included in the screen, with two of the patients being normal for RECQL4 (RECQL4 mutation data were unavailable for the other patients). As mentioned earlier, RTS is very similar in presentation to PN and two additional mutations in C16orf57 were described by Volpi et al. in a family with atypical RTS (17,19). A total of 124 additional patients were analysed, and homozygous mutations were identified in 4 patients (1 with DC and 3 with RTS; Table 1 and Fig. 2). In total, seven homozygous missense, non-sense or frame-shift mutations were identified in nine families affecting 14 individuals (five of these are novel). One mutation was recurrent in three families, i.e. c.531delA, p.His179Met fsX86. Apart from one splice-site mutation, all involve the premature truncation or the generation of a partially scrambled protein (Fig. 3).
Figure 3.
Schematic structure of C16orf57. (A) Relative position of the mutations to the exons (grey boxes). (B) Relative protein lengths predicted to remain after mutation. Text colour relates to disease type: blue, DC; red, RTS; black, PN/DC: bar, correct amino acid sequence; line, mutant amino acid sequence before stop codon. The effect of the splice-site mutation is not shown because of the lack of appropriate material in which to determine the sequence of the aberrant transcript.
Clinical features
Table 2 lists all the various clinical features in the families with mutations in C16orf57. The most striking feature is the presentation of abnormal skin pigmentation (poikiloderma) at a very young age (all <6 years) and a low white blood cell count which is common to all. However, there is a large degree of overlap between all the patients. All the DCR patients fulfil the criteria for the diagnosis of DC as defined by Vulliamy et al. (20), namely the index case presents with the classical mucocutaneous triad of abnormal skin pigmentation, nail dystrophy and leucoplakia or has at least two of the mucocutaneous features combined with hypocellular bone marrow or two or more of the other somatic abnormalities known to occur in DC; even DCR070, which has been recently published as having PN, fulfils these criteria (18). The patients with RTS are slightly atypical in that they do have defects in the bone marrow compartment, but this is not a new observation (12,19). The overlap of features suggests that none of the disease classifications based on clinical criteria is completely satisfactory. The unifying feature of a mutation in C16orf57 now helps to define a tight subtype of patient profile that shares early-onset skin pigmentation abnormalities and haematological abnormalities as well as a range of other features.
Table 2.
Features of individuals with C16orf57 mutations
| Identification | 279 |
070 |
224 |
107 |
106 | 215 | RT 1 |
RT 2 | RT 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diagnosis (reference) | DC |
DC/PN (18,21) | DC |
DC |
DC | DC | RTS (28) | RTS | RTS | ||||||
| Mutation | H179M fsX86 |
P60L fsX55 |
Y86X |
H179M fsX86 |
c.450-2A>G | H179M fsX86 | ND | W81X | G59A fsX2 | Q181X | |||||
| Clinical feature | |||||||||||||||
| Abnormal skin (poikiloderma) | +(1y) | + (1y) | + | + | +(1y) | +(4y) | +(6y) | +(1y) | +(1y) | +(1y) | +(1y) | + | + | + (<2y) | + (6m) |
| Nail dystrophy | +(1y) | +(1y) | + | + | + | +(3y) | +(6y) | +(1y) | +(1y) | +(5y) | + | + | + | ||
| Leucoplakia | + | + | + | ||||||||||||
| Epiphora | + | + | + | + | |||||||||||
| Head abnormality | Tear duct | Tear duct | Tear duct | Abnormal ear, hearing loss | Abnormal ear | Hair loss | Sparse eyelashes | Nasal bridge | Prominent forehead | ||||||
| Dental problems | + | + | + | + | + | + | + | + | + | ||||||
| Skeletal | Osteoporosis | + | Osteoporosis | ||||||||||||
| Liver–spleen | Enlarged | Enlarged | |||||||||||||
| Phimosis | + | ||||||||||||||
| Pulmonary | + | + | + | + | + | + | + | ||||||||
| Cardiac | VSD | ||||||||||||||
| IUGR | + | ||||||||||||||
| Short stature | + | + | + | + | + | + | + | + | + | ||||||
| Low BW | + | + | + | + | |||||||||||
| Dev delay | + | + | + | + | + | ||||||||||
| Cancer | AML | AML | Skin | ||||||||||||
| Blood | |||||||||||||||
| Hb | N | Low | N | N | Low | Low | N | Low | N | Low | N | Low | N | ||
| WBC Neutrophils | Low | Low | Low | Low | Low | Low | Low | Low | Low | Low | Low | Died AML 1988 | Lowa | Lowa | Low |
| Platelets | Low | Low | Low | Low | Low | Variable | N | N | N | N | N | N | N | N | |
| Bone marrow | N | Hypo-cellular | Hypo-cellular | Hypo-cellular | MDS changes Myeloid maturation defect | MDS changes | MDS changes | MDS changes | MDS changes | Hypo-cellular | Hypo-cellular | ||||
| Immuno-globulins | ↑ | ↑ | ↑ | ||||||||||||
Mutations identified in nine families (14 affected individuals): five DC families (279, 224, 107, 106, 215), one DC/PN (070), three RTS families (RT1, RT2, RT3).
AML, acute myeloid leucaemia; BM, bone marrow; ND, not determined; Hb, haemoglobin; MDS, myelodysplasia; VSD, ventricular septal defect; WBC, white blood cells; N, normal; plus, feature present; upward arrow, increased levels. Age in brackets is in months (m) or years (y).
aLow neutrophils and lymphocytes—in all other patients, the low WBC count was mainly due to the low neutrophil count. The pulmonary abnormalities included bronchiectasis (DCR279), chronic pulmonary infiltrates (DCR070), pulmonary cystic changes (DCR107), recurrent chest infections (DCR279, DCR070, RT1) and restrictive lung disease (DCR107). Imaging of the liver in DCR106 and DCR107 showed no evidence of cirrhosis. Bone marrows which showed hypocellularity were usually associated with some features of dysplasia. BM cytogenetic analyses were undertaken in some patients (DCR070, RT1 and RT2), and these showed no abnormal cytogenetic clones.
Telomeres are not short in patients with C16orf57 mutations
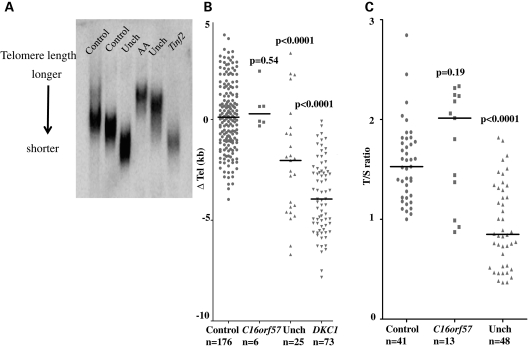
One of the main defining features of the genetically characterized DC is that most of the patients regardless of mutation tend to have short telomeres compared with age-matched controls. We therefore selected two methods to investigate this in our patient cohort. The first method used Southern blotting with a subtelomeric probe specific to chromosome 7. This method has been widely reported but does involve a large quantity of DNA being available. Recently, a new multiplexed PCR [monochrome multiplex quantitative PCR (MM-qPCR)]-based method has been described that compares the ratios of a single-copy gene to telomere length. The advantage of this method is that it needs much less DNA, so is more widely applicable to samples where DNA is in limited supply. From our patients with C16orf57 mutations, we only had enough DNA to perform Southern blot analysis on 6, whereas we were able to perform the MM-qPCR method on 13 patients. We also performed MM-qPCR analysis on 41 healthy controls to establish a normal range. For comparison, we analysed 48 of our uncharacterized DC patients by MM-qPCR, and included previously obtained age-adjusted (▵ Tel) lengths from Southern blots for healthy controls and patients with known DKC1 mutations. What is clear from both data sets is that telomere length is normal in all patients with C16orf57 mutations when compared with the healthy controls (Fig. 4), with no significant difference observed when either set is analysed using the Mann–Whitney U test. This group is also quite different from most other DC patients in terms of the telomere lengths whether the genetic basis is characterized or not. Although we do not know the function of C16orf57, it is clear that it is not involved in control of telomere length. It also strengthens the idea that C16orf57 mutations are responsible for a very specific subtype of disease whether it is classified as DC, RTS or PN.
Figure 4.
Telomere lengths are not short in patients with mutations in C16orf57 compared with healthy controls. (A) Sample of a Southern blot using the subtelomeric probe pTelBam8. This shows the variety of lengths obtained from different individuals both with and without disease. (B) Age-adjusted ▵ tel values for controls compared with patients with C16orf57 mutations, uncharacterized patients with DC and patients with known DKC1 mutations. (C) T/S ratio as a measure of telomere length comparing controls with patients with C16orf57 mutations and uncharacterized DC patients. AA, patient with aplastic anaemia; Unch, genetically uncharacterized DC patients; Tinf2, DC patient with TINF2 mutation; solid line, median length obtained for the sample group, P-values from the Mann–Whitney U test relative to controls.
C16orf57 knockdown studies
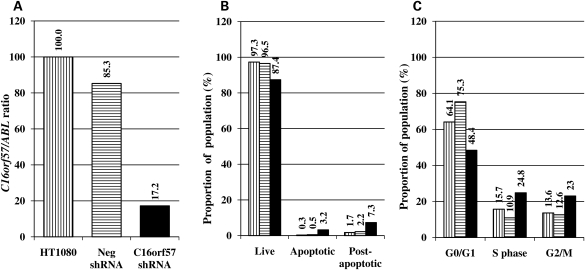
In light of the lack of functional information relating to the role of C16orf57 in the cell, we undertook a series of experiments to determine whether it had any role in cell biology either in terms of apoptosis or in the cell cycle. All experiments were performed on an unsorted population of HT1080 cells either 7 or 14 days after transduction with viral constructs encoding either a C16orf57 shRNA or a negative shRNA to observe the effects of knockdown of the C16orf57 transcript (Fig. 5A). Unmanipulated HT1080 cells were assayed at the same time. Apoptosis levels after knockdown with the C16orf57 shRNA showed a small increase in the proportion of cells that were either apoptotic or post-apoptotic relative to either the unmanipulated cells or the cells treated with the negative shRNA viral construct, suggesting the effect is specific to the action of reducing the C16orf57 levels in the cell (Fig. 5B). Repetition of these experiments show that this increase in the percentage of cells that are apoptotic following C16orf57 knockdown, although subtle, is statistically significant (Student's t-test P = 0.016, Supplementary Material, Table S3). Concomitant with this, the proportion of cells observed in the G2/M phase of the cell cycle was increased in the C16orf57 knockdown group (Fig. 5C) relative to the unmanipulated cells, but this failed to reach significance.
Figure 5.
Effects of knockdown of C16orf57 on apoptosis and cell cycle. (A) Levels of knockdown achieved in 7 days after treatment with lentiviral-shRNA. (B) Corresponding apoptosis levels in HT1080 cells. (C) Corresponding cell cycle analysis. Vertical stripes, unmanipulated cells; horizontal stripes, cells transduced with a negative shRNA lentiviral construct; solid black bars, cells transduced with a C16orf57 shRNA lentiviral construct. Numbers on graphs relate to the data point for each experiment.
DISCUSSION
The principal aim of this study was to determine the genetic basis of the uncharacterized cases of DC present on the DCR (London). Linkage analysis using the Illumina 6k SNP chip in 20 DC families (8 consanguineous multiplex families and 12 consanguineous families with sporadic cases) enabled a common candidate region of homozygosity to be established for a subgroup of these families on chromosome 16 between SNPs rs1982395 and rs768462 (Fig. 1). As well as providing strong evidence for a DC locus, this result also provided additional evidence for genetic heterogeneity as not all families shared this area of homozygosity.
A recent study by Volpi et al. identified a mutation in the orphan gene C16orf57 in a large family with PN. This gene was in our genetic interval of interest and the phenotype of PN overlaps significantly with DC. Sequence analysis of C16orf57 in our patients with overlapping genetic homozygosity revealed homozygous mutations in five of these families, with the mutations segregating in a recessive manner. Further screening of an additional 118 DC patients and 6 RTS patients identified an additional four homozygous mutations (one in DC and three in RTS). In total, seven mutations were identified, five of which were novel, including one recurrent mutation (c.531delA, p.His179MetfsX86). The other two mutations were reported during the preparation of this article: the first relates to the mutation in our family DCR070 which has been previously reported as having PN (18). This mutation published by Tanaka et al. (21) has been verified by this work as being c.179delC, p.Pro60Leu fsX54. The second mutation, c.243G>A, p.Trp81X, has recently been reported in a family with PN that is distinct from the family in which we observe this change (22).
The individuals with C16orf57 mutations have multi-system abnormalities, as summarized in Table 2. This includes the striking abnormal skin pigmentation/poikiloderma at a young age and a low white cell count (particularly the neutrophil count). In addition, affected individuals have a variable range of other features which are seen in classical cases of DC, PN and RTS. It is apparent from this table that a clinician could diagnose these patients as having any of these three syndromes particularly as the age of onset of some of these features is highly variable. Although C16orf57 mutations were first described in a family labelled as PN, it is clear from Table 2 that C16orf57 mutations produce a wider range of multi-system abnormalities and not just PN. The multi-system abnormalities are also more akin to DC and suggest that the C16orf57 gene is likely to have an important house-keeping role in the cell. This is also consistent with the ubiquitous expression profile of C16orf57 in different human tissues (http://www.genecards.org). Three out of the 14 of patients have developed a cancer at a young age (acute myeloid leucaemia in two, squamous cell carcinoma of the skin in one). Thus, like other cases of DC and RTS, patients with C16orf57 mutations have a predisposition to developing cancer.
As well as the neutropenia, approximately half of the patients also have abnormalities in red cell (haemoglobin) and/or platelet counts. In addition, several of the bone marrows from these patients suggested a global defect in haematopoiesis either in the form of myelodysplasia or as hypocellularity. Therefore, although there is a major impact on the myeloid lineage with the consequent neutropenia, there are features suggestive of a more global haematopoietic defect. In one RTS patient, with the appropriate sample being available (RT2), peripheral blood progenitor studies showed a reduction in both myeloid and erythroid progenitors (Supplementary Material, Table S2), again providing evidence for a more global haematopoietic defect. Further investigations are warranted to validate this observation as only the one patient sample was available.
It is notable that the mutations identified in these families all spare the N-terminus of the protein (Fig. 3B). The smallest mutant protein is predicted to have at least 61 of the N-terminal amino acids. This suggests that an important key function of the protein may be located within this part of the molecule. At present, the function of the gene remains to be elucidated. Our knockdown studies suggest a subtle but significant increase in the proportion of apoptotic cells after knockdown of C16orf57. These data indicate that C16orf57 might have some role in controlling apoptosis, although this effect may well be indirect, possibly via its proposed interaction with SMAD4 (17). Additional studies need to be performed to further characterize the function of C16orf57 particularly in primary cells and the effects of long-term knockdown. It will also be important to determine the precise effect each mutation has on the protein.
The previously characterized cases of DC (due to mutations in DKC1, TERC, TERT, NOP10, NHP2 and TINF2) are associated with short telomeres (as demonstrated in Fig. 4B, DKC1 lengths). Telomere length was measured in patients by either Southern blot or MM-qPCR. Telomere length measurements by any method are highly variable in terms of the range and a wide overlap can exist between different individuals, as shown in Fig. 4A. This example shows how lengths vary and how not all patients with DC have short telomeres. Telomere length was found to be comparable to age-matched controls by both methods and longer than groups of patients either uncharacterized or with DKC1 mutations. This observation suggests that patients presenting with features of DC due to C16orf57 mutations are distinct from those due to mutations in the previously characterized DC genes. This observation suggests that telomere length maintenance is unaffected in this group of patients. It does not however reject telomere dysfunction as the mechanism of disease. This idea has been suggested previously by Lamm et al. in their study of patients with Hoyeraal–Hreidarsson syndrome (23), in which they propose that telomere length-independent pathways can contribute to telomere dysfunction.
In summary, we report the identification of novel homozygous mutations in the C16orf57 gene in several families clinically classified as DC, PN and RTS. The multi-system abnormalities in the affected individuals include poikiloderma, haematological abnormalities, a range of other abnormalities and a predisposition to cancer. These observations suggest a house-keeping role for this gene, but the precise function at this stage remains elusive. C16orf57 mutations therefore unify a specific subset of patients with multi-system abnormalities observed in DC, PN and RTS but which are distinct from previously characterized DC patients as they have normal-length telomeres.
MATERIALS AND METHODS
Patient selection
All of the patients included in this study have been enrolled in the DCR initially at Hammersmith Hospital and relocated to Barts and The London in 2006. The clinical criteria used for entry into this registry have been described previously (20). In addition to patients with DC, we also included the six patients in our collection with RTS and any family members where appropriate. All samples were obtained with informed consent and with the approval of our local ethics committee.
SNP genotyping and linkage analysis
Genomic DNA samples, prepared from peripheral blood samples extracted using the Puregene DNA isolation kit (Gentra, Minneapolis, USA), from 8 consanguineous multiplex families and 12 consanguineous families with sporadic cases were analysed using the Illumina 6k SNP chip (service provided by The Genome Centre, Barts and The London School of Medicine). Of the 51 individuals analysed, 27 had a diagnosis of DC, 14 were unaffected parents and 10 were unaffected siblings. Multi-point LOD score analysis was performed on each of the families using Genehunter (24), assuming an autosomal recessive inheritance, 100% penetrance and a disease allele frequency of 0.005.
Mutation screening
Primers (Supplementary Material, Table S1) were designed to amplify the entire coding region (seven exons) of C16orf57 in the index cases of the families that had an overlapping LOD score on chromosome 16 (n = 4). These fragments were sequenced using Applied Biosystems Big dye V3.1 chemistry on a 3130XL DNA analyser (Applied Biosystems, Warrington, UK). An additional set of primers (available on request) were designed to screen 118 uncharacterized index case patient samples in the DCR by denaturing high-performance liquid chromatography (Transgenomic, Glasgow, UK) at a single temperature at which the fragment was at least 80% helical. Samples were pooled pairwise to identify any possible homozygous changes. Any pools which gave abnormal elution patterns were re-amplified separately and sequenced.
Telomere length measurements
Southern blotting
Genomic DNA was digested with BamH1 and analysed by Southern blot using a 0.75% agarose gel and the subtelomeric probe pTelBam8 (25,26). Telomere lengths were measured as the size of the fragment of peak signal intensity, which includes ∼8kb of subtelomeric DNA. Sizes were determined with reference to the same standards run on each gel using Image Quant software. Data from normal control samples, patients with C16orf57 mutations and age-adjusted telomere length (▵ Tel) values were obtained as described previously (3). Differences between the various groups were tested using the Mann–Whitney U test.
Monochrome multiplex quantitative PCR
Telomere lengths were measured using a multiplex real-time PCR method adapted from Cawthon (27). All samples were measured in triplicate. Briefly, each 15 µl of the reaction mixture contained 10 ng of genomic DNA and used the Roche SYBR Green Master I reaction mix (Roche Applied Science, Burgess Hill, UK). Telomere-specific primers were used at a concentration of 1 µm each and single-copy gene primers (specific to human beta-globin) at 200 nm each. Reactions were analysed on a Roche LightCycler 480 real-time thermal cycler (Roche Applied Science). A standard curve was established using a titration of a reference DNA sample and T/S ratios determined from the curve.
Interfering RNA study
C16orf57 mRNA transcript was knocked-down using a mission shRNA viral construct (TRCN0000137195; Sigma). HT1080 (human fibrosarcoma) cells were exposed to either the C16orf57 shRNA virus or a non-target shRNA virus for 3 days. Cells were subsequently cultured for 7 or 14 days before being assayed for levels of transcript reduction, apoptosis and cell cycle profiles relative to unmanipulated cells.
cDNA preparation
RNA was prepared from 0.5 × 106 cells from both types of viral manipulated cells and unmanipulated cells using the RNeasy mini kit (Qiagen). Following DNase1 (Invitrogen) treatment to remove any genomic DNA contamination, RNA was reverse transcribed into cDNA using M-MLV (Invitrogen) and random hexamer primers (Qiagen) in standard reactions according to manufacturer protocols.
Quantitative real-time PCR
RNA expression levels were determined by quantitative real-time PCR on an ABI prism 7500 real-time PCR system (Applied Biosystems). Twenty microlitre (final volume) reactions were performed using SYBR® Green PCR master mix (Applied Biosystems), 1 µl of cDNA and 25 nm forward and reverse primers. C16orf57 was measured using the Taqman gene expression assay Hs00984813_ml. The ABL gene expression was used as an endogenous control for normalization and expression calculated from standard curves as a C16orf57/ABL ratio.
Apoptosis
Apoptosis levels were determined using standard flow cytometry techniques. Briefly, this involved staining of the apoptotic membrane marker phosphatidylserine with a fluorescent annexin V–Alexafluor 647 conjugate and co-staining of nuclear DNA with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). These fluorophores were used to avoid interference from the GFP expressed by the viral shRNA constructs. After a 15 min incubation, cells were analysed using an LSRII four-laser flow cytometer (BD, Oxford, UK).
Cell cycle profile
G0/G1, S and G2M populations were determined using standard methods. A total of 0.2 × 106 cells were fixed overnight in 70% ethanol, washed twice in PBS and resuspended in 300 µl PBS with 10 µg/ml DAPI and 200 µg/ml RNase A. Cells were incubated at 37°C for 30 min before being analysed on the flow cytometer. Cells were pulse-width gated to exclude doublets.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by The Wellcome Trust (grant number 085937). Funding to pay the Open Access Charge was provided by a grant awarded to Queen Mary, University of London, by the Wellcome Trust.
ACKNOWLEDGEMENTS
We would like to thank all the clinicians and patients who have supported our research, particularly Professor Gluckman and Drs Dura, Guardiola, Jones, Kilic, Krahn and Sigaudy who provided the key patient samples for this study.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dokal I. Dyskeratosis congenita in all its forms. Br. J. Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. doi:10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 2.Heiss N.S., Knight S.W., Vulliamy T.J., Klauck S.M., Wiemann S., Mason P.J., Poustka A., Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. doi:10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 3.Vulliamy T., Marrone A., Goldman F., Dearlove A., Bessler M., Mason P.J., Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. doi:10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 4.Armanios M., Chen J.L., Chang Y.P., Brodsky R.A., Hawkins A., Griffin C.A., Eshleman J.R., Cohen A.R., Chakravarti A., Hamosh A., et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. doi:10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walne A.J., Vulliamy T., Marrone A., Beswick R., Kirwan M., Masunari Y., Al-Qurashi F.H., Aljurf M., Dokal I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007;16:1619–1629. doi: 10.1093/hmg/ddm111. doi:10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vulliamy T., Beswick R., Kirwan M., Marrone A., Digweed M., Walne A., Dokal I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. doi:10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marrone A., Walne A., Tamary H., Masunari Y., Kirwan M., Beswick R., Vulliamy T., Dokal I. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal–Hreidarsson syndrome. Blood. 2007;110:4198–4205. doi: 10.1182/blood-2006-12-062851. doi:10.1182/blood-2006-12-062851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage S.A., Giri N., Baerlocher G.M., Orr N., Lansdorp P.M., Alter B.P. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. doi:10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walne A.J., Vulliamy T., Beswick R., Kirwan M., Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. doi:10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. doi:10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 11.Wang L.L., Gannavarapu A., Kozinetz C.A., Levy M.L., Lewis R.A., Chintagumpala M.M., Ruiz-Maldanado R., Contreras-Ruiz J., Cunniff C., Erickson R.P., et al. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund–Thomson syndrome. J. Natl Cancer Inst. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. doi:10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 12.Larizza L., Roversi G., Volpi L. Rothmund–Thomson syndrome. Orphanet. J. Rare Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. doi:10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clericuzio C.L., Hoyme H.E., Aase J.M. Immune deficient poikiloderma: a new genodermatosis. Am. J. Hum. Genet. 1991;49(Suppl. 1):131. [Google Scholar]
- 14.Erickson R.P. Southwestern Athabaskan (Navajo and Apache) genetic diseases. Genet. Med. 1999;1:151–157. doi: 10.1097/00125817-199905000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Wang L.L., Gannavarapu A., Clericuzio C.L., Erickson R.P., Irvine A.D., Plon S.E. Absence of RECQL4 mutations in poikiloderma with neutropenia in Navajo and non-Navajo patients. Am. J. Med. Genet. 2003;118A:299–301. doi: 10.1002/ajmg.a.10057. doi:10.1002/ajmg.a.10057. [DOI] [PubMed] [Google Scholar]
- 16.Van Hove J.L., Jaeken J., Proesmans M., Boeck K.D., Minner K., Matthijs G., Verbeken E., Demunter A., Boogaerts M. Clericuzio type poikiloderma with neutropenia is distinct from Rothmund–Thomson syndrome. Am. J. Med. Genet. A. 2005;132A:152–158. doi: 10.1002/ajmg.a.30430. doi:10.1002/ajmg.a.30430. [DOI] [PubMed] [Google Scholar]
- 17.Volpi L., Roversi G., Colombo E.A., Leijsten N., Concolino D., Calabria A., Mencarelli M.A., Fimiani M., Macciardi F., Pfundt R., et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. doi:10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mostefai R., Morice-Picard F., Boralevi F., Sautarel M., Lacombe D., Stasia M.J., McGrath J., Taieb A. Poikiloderma with neutropenia, Clericuzio type, in a family from Morocco. Am. J. Med. Genet. A. 2008;146A:2762–2769. doi: 10.1002/ajmg.a.32524. doi:10.1002/ajmg.a.32524. [DOI] [PubMed] [Google Scholar]
- 19.Pianigiani E., De Aloe G., Andreassi A., Rubegni P., Fimiani M. Rothmund–Thomson syndrome (Thomson-type) and myelodysplasia. Pediatr. Dermatol. 2001;18:422–425. doi: 10.1046/j.1525-1470.2001.01971.x. doi:10.1046/j.1525-1470.2001.01971.x. [DOI] [PubMed] [Google Scholar]
- 20.Vulliamy T.J., Marrone A., Knight S.W., Walne A., Mason P.J., Dokal I. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. doi:10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka A., Morice-Picard F., Lacombe D., Nagy N., Hide M., Taieb A., McGrath J. Identification of a homozygous deletion mutation in C16orf57 in a family with Clericuzio-type poikiloderma with neutropenia. Am. J. Med. Genet. A. 2010;152A:1347–1348. doi: 10.1002/ajmg.a.33455. [DOI] [PubMed] [Google Scholar]
- 22.Arnold A.W., Itin P.H., Pigors M., Kohlhase J., Bruckner-Tuderman L., Has C. Poikiloderma with neutropenia: a novel C16orf57 mutation and clinical diagnostic criteria. Br. J. Dermatol. 2010 doi: 10.1111/j.1365-2133.2010.09929.x. in press. DOI:10.111/j1365-2133.2010.09929.x. [DOI] [PubMed] [Google Scholar]
- 23.Lamm N., Ordan E., Shponkin R., Richler C., Aker M., Tzfati Y. Diminished telomeric 3′ overhangs are associated with telomere dysfunction in Hoyeraal–Hreidarsson syndrome. PLoS ONE. 2009;4:e5666. doi: 10.1371/journal.pone.0005666. doi:10.1371/journal.pone.0005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruglyak L., Daly M.J., Reeve-Daly M.P., Lander E.S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 25.Brown W.R., MacKinnon P.J., Villasante A., Spurr N., Buckle V.J., Dobson M.J. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. doi:10.1016/0092-8674(90)90293-N. [DOI] [PubMed] [Google Scholar]
- 26.Notaro R., Cimmino A., Tabarini D., Rotoli B., Luzzatto L. In vivo telomere dynamics of human hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:13782–13785. doi: 10.1073/pnas.94.25.13782. doi:10.1073/pnas.94.25.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawthon R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. doi:10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porter W.M., Hardman C.M., Abdalla S.H., Powles A.V. Haematological disease in siblings with Rothmund–Thomson syndrome. Clin. Exp. Dermatol. 1999;24:452–454. doi: 10.1046/j.1365-2230.1999.00530.x. doi:10.1046/j.1365-2230.1999.00530.x. [DOI] [PubMed] [Google Scholar]