Abstract
The complex interactions between proteins and small organic molecules (ligands) are intensively studied because they play key roles in biological processes and drug activities. Here, we present a novel approach to characterise and map the ligand-binding cavities of proteins without direct geometric comparison of structures, based on Principal Component Analysis of cavity properties (related mainly to size, polarity and charge). This approach can provide valuable information on the similarities, and dissimilarities, of binding cavities due to mutations, between-species differences and flexibility upon ligand-binding. The presented results show that information on ligand-binding cavity variations can complement information on protein similarity obtained from sequence comparisons. The predictive aspect of the method is exemplified by successful predictions of serine proteases that were not included in the model construction. The presented strategy to compare ligand-binding cavities of related and unrelated proteins has many potential applications within protein and medicinal chemistry, for example in the characterisation and mapping of “orphan structures”, selection of protein structures for docking studies in structure-based design and identification of proteins for selectivity screens in drug design programs.
Keywords: protein cavity comparison, physicochemical properties, alignment independent, SCREEN, principal component analysis, binding sites, medicinal chemistry, drug design, PCA clustering tree, bioinformatics
Introduction
Many biological processes, such as cell signalling and enzymatic reactions, are regulated by interactions between proteins and small organic molecules (ligands). Elucidating these interactions is of fundamental biological interest and essential for rational drug discovery. In most cases the binding of a ligand to a protein occurs on a solvent accessible surface formed as a cavity within the protein. Binding affinity depends on the physicochemical properties of both the protein (especially the cavity in which the interaction occurs) and the ligand. Here we present a novel approach for analyzing and mapping structural and physicochemical properties of such cavities without making direct geometric comparisons of the structures. The cavities are compared using a wider set of attributes (i.e. an extensive structural and physicochemical characterisation) than in existing work and the multivariate modelling enables for an interpretable overview of the property relatedness of cavities. The unique strategy of making all-against-all comparisons based on an exhaustive property characterisation without a geometric alignment give the presented work many potential applications. For instance in the characterisation and mapping of “orphan structures”, selection of protein structures for docking studies in structure-based design, investigation of similarity of binding cavities for multitarget-directed ligands, and identification of proteins for selectivity screens in drug design programs.
Two basic approaches have been developed to represent ligand-binding cavities for comparing diverse proteins, using either the positions of amino acids in the cavities, i.e. coordinate-based techniques,1–10 or defined surfaces to represent the cavities of interest, i.e. surface-based techniques.11–18 In both cases, the relevant cavities must be manually or automatically identified; in terms of included amino acids in the former, and the included surfaces in the latter. Comprehensive introductions of advances in pattern recognition and comparison of proteins have been presented by Kuhn et al.,9 Xie et al.,19 Chen et al.20 and Watson et al.21
In the coordinate-based approach, the positions of the amino acids are described using, for example, all atoms,1,2 only the side chains,3,4 pseudo-atoms,5,6 chemical groups,7 and/or pseudo-centers.8–10 Representations using side chains and pseudo-atoms have been utilized to identify common structural motifs, such as catalytic residues, in enzymes.3–5,10 The use of pseudo-centres to represent side chains is an extension of the pseudo-atom approach to include selected physicochemical properties of amino acids.8–10 In Cavbase, for instance, the physicochemical properties of the cavity-flanking residues are condensed into a restricted set of five generic pseudo-centres, corresponding to hydrogen-bond donor, acceptor, mixed donor/acceptor, hydrophobic aliphatic and aromatic.8 These rules have been modified to improve similarity searches for classifications9 and the recognition of functional sites.10 A comparable approach has been used to create similarity networks of binding sites22 and a method has recently been proposed for predicting protein-drug interactions based on the physicochemical attributes of all surface atoms in a cavity.1
In the surface-based approach, pre-defined surfaces of ligand-binding cavities are partitioned into smaller molecular surface patches, which are then further evaluated by comparison.11–18 A wide range of techniques for calculating surfaces has been described over the last few decades23–30 and the surface patch characterisations may contain both geometrical and physicochemical information about the proteins.11–13 Proteins have been mapped, and enzyme classes predicted by self-organizing neural networks, based on Connolly surface calculations and assignment of generalized atom types (i.e. aliphatic, hydrogen-bond donors/acceptors, aromatic-face/edge) to each point on a surface grid.11 In addition, a large-scale analysis has been presented recently of surface patches of different binding cavities using emergent self-organising maps, based on Cavbase pseudo-centres and wavelet-based shape descriptors.13 The objective was to describe functional properties of the cavities by identifying sets of similar substructures within them. In addition to the methods of representing ligand-binding cavities discussed above, probe spheres have been used to fill cavities and the shapes of ligand-binding cavities of different proteins have been described using spherical harmonic expansions.31 An alternative protein surface-based approach has been reported recently, in which surface representations are generated by the expansion of 3D functions into Zernike-Canterakis series with subsequent normalisation to obtain the rotational invariance.32,33 The shape, electrostatic potential and an approximate description of the hydrophobicity of protein surfaces obtained by this method have been used to compare the total surfaces of globins against a representative set of proteins. The same approach has also been used to compare the active sites of 19 TIM barrel enzymes.32 In addition, a program called SiteAlign has been used to compare proteins in the sc-PDB database by calculating fixed-length cavity “fingerprints” containing topological and chemical information (data on H-bond donors/acceptors, aromatic and aliphatic characters and charge) of the cavities; projecting these cavity descriptors to the centre of spheres; then aligning the spheres by exhaustive rotation/translation for comparison.18
Binding cavities can be compared with similarity searches using a query cavity,5,6,8,9,18,34 all-against-all comparisons1,2,4,22,31 and/or protein diversity mapping.11,13,22,31,32,35 In order to make such a comparison, the representations and characterisations (i.e. the geometric patterns and/or numerical vectors) of the cavities must be systematically applied to all proteins considered. The vast majority of these approaches first require a computationally expensive geometric alignment of the atoms, side chains, pseudo-atoms/centres or surface patches followed by the assignment of similarity scores. In general, the difficulty is to identify relevant substructures to overlap; which substructures in protein X are equivalent to which substructures in protein Y?36 Furthermore, selecting the appropriate geometric alignment method can be difficult and most, if not all, existing methods are heuristic.37 In similarity scoring, the relevance of the comparison of two cavities is highly dependent on the degree of spatial overlap of the aligned binding site representations, which is likely to be high for structurally similar cavities. However, in comparisons of dissimilar cavities the similarity scoring can be problematic since it is difficult to estimate the biological significance of a proposed geometric alignment.38 In the present work, we focus on cavity comparisons that are based on a structural and physicochemical characterisation of binding cavities, without making a geometric alignment. The approach of characterising and comparing ligand-binding cavities in a manner that is independent of the rotation of the proteins, but which is still based on 3D structures, facilitates the comparison of structurally diverse proteins.
This paper presents an approach to map, characterize and compare ligand-binding cavities of a diverse set of proteins while avoiding geometric comparisons. Descriptors were calculated by SCREEN (Surface Cavity REcognition and EvaluatioN)12 and the number of different structural and physicochemical properties (i.e. size, shape, polarity, charge, electrostatics, flexibility, secondary structure and hydrogen bonding capabilities) covered by the descriptors exceeds previously reported attempts to physicochemically describe cavities.1,8,9,11,13,18,32 The idea of describing cavities and analysing the cavity property relationships is comparable to the characterisation of small molecules, for example in investigations of molecular diversity39–42 and/or the creation of quantitative structure-activity relationship (QSAR) models.43,44 To explore the structural and physicochemical space of cavities, their key properties were extracted from the descriptor matrix using Principal Component Analysis (PCA). This method has been widely used to explore the properties of ligands,39,40 protein-ligand space (from a chemogenomics perspective),45 whole proteins46,47 and binding sites of closely related proteins,48 but to our knowledge this is the first time that PCA has been applied to explore and compare cavities of a diverse set of proteins, applying an all-against-all comparison and mapping the resulting principal properties of the cavities. Here, the structural and physicochemical similarities and dissimilarities between ligand-binding cavities are visualised in PCA clustering trees, then analyzed and considered in relation to their respective domain classifications in the structural classification of proteins (SCOP)49 database. Furthermore, the predictive aspect of the method is explored by predicting the similarity of ligand-binding cavities of a new set of proteins to the cavities included in the PCA model. Finally, information obtained in this exploration of the physicochemical and structural property space of cavities is related to information obtained in corresponding explorations of ligands, and suggestions for applications of the described method in protein structure-activity-function analysis, medicinal chemistry and structure-based design are given.
Method
Protein Preparation
The proteins used in this study were selected from the PDBbind database50,51 and are named by the Protein Data Bank (PDB) code as entered in the Research Collaborator for Structural Bioinformatics (RCSB) protein data bank. Proteins containing metals in the ligand-binding cavities were not included because SCREEN does not take cofactors and metals into account. Since SCREEN operates by mapping the solvent-accessible surface of proteins, some deep, enclosed cavities in the proteins were not recognized, and those proteins were not included. A filtration was applied to reduce the data set to a maximum of five randomly selected representatives of each SCOP classification protein domain (hereafter the word domain refer to the SCOP definition of a protein domain). Keeping all representatives for each domain from the original database would have led to an over-representation of some proteins, e.g. Human Immunodeficiency Virus (HIV)-1-Protease, in the PCA. However, retaining up to five versions of the same protein in the final selection was deemed to be potentially valuable to broaden the description of each protein and account for variations arising from differences in protein conformation in and around these cavities. The full, final set of proteins included in the study is presented in the Supporting Information.
All protein structures in the final selection were determined by X-ray crystallography with a resolution finer than 2.5 Å. Protein files were prepared by adding hydrogens, and selecting tautomers of Asn, Gln, His and orientations of the hydrogens of the functional groups SH and OH using Reduce software.52 Water, other inorganic molecules, cofactors and ligands were removed before calculations were made, and the atom names, arising from different atom identification conventions, were standardised for compatibility with the SCREEN application. The proteins were classified according to the SCOP classification system49 for comparison.
Protein Sets
The set of proteins described in the previous section was used as a training set to create the PCA of the cavity characterisation with subsequent all-against-all comparison and mapping. Two prediction sets of proteins, A and B (Table 1), were selected. Prediction set A consisted of one protein from each of twelve domains that had representatives in our subset of PDBbind. In addition, one protein with no domain representative included in the model was selected (A-1FDQ). Prediction set B included ten serine proteases belonging to two domains not present in the training set: five representatives of the Subtilisin domain and five from the Chymotrypsin(ogen) domain which were downloaded from the RCSB protein data bank.53
TABLE I.
Prediction Set Proteins
| Name# | Domain* | Family* |
|---|---|---|
| A-1AJP | Penicillin acylase | Penicillin acylase, catalytic domain |
| A-1AKW | Flavodoxin | Flavodoxin-related |
| A-1APW | Acid protease | Pepsin-like |
| A-1AX1 | Legume lectin | Legume lectins |
| A-1D1P | Tyrosine phosphatase | Low-molecular-weight phosphotyrosine protein |
| A-1ETT | Thrombin | Eukaryotic proteases |
| A-1FDQ | Brain fatty acid binding protein | Fatty acid binding protein-like |
| A-1GJ7 | Urokinase-type plasminogen activator (LMW U-PA) | Eukaryotic proteases |
| A-1GPN | Acetylcholinesterase | Acetylcholinesterase-like |
| A-1J16 | Trypsin(ogen) | Eukaryotic proteases |
| A-1NJD | Thymidylate synthase | Thymidylatesynthase/dCMP hydroxymethylase |
| A-1TCX | Simian immunodeficiency virus (SIV) protease | Retroviral protease (retropepsin) |
| A-2QWF | Influenza neuraminidase | Sialidases (neuraminidases) |
| B-1AB9 | (α, γ)-Chymotrypsin(ogen) | Eukaryotic proteases |
| B-1AFQ | (α, γ)-Chymotrypsin(ogen) | Eukaryotic proteases |
| B-1GG6 | (α, γ)-Chymotrypsin(ogen) | Eukaryotic proteases |
| B-1K2I | (α, γ)-Chymotrypsin(ogen) | Eukaryotic proteases |
| B-7GCH | (α, γ)-Chymotrypsin(ogen) | Eukaryotic proteases |
| B-1SBN | Subtilisin | Subtilases |
| B-1SBT | Subtilisin | Subtilases |
| B-1SUP | Subtilisin | Subtilases |
| B-1TM1 | Subtilisin | Subtilases |
| B-2SIC | Subtilisin | Subtilases |
prediction set A or B and PDB code.
As defined by the SCOP classification of proteins.
Property Calculations
The first two phases of SCREEN12 were modified for our purposes, i.e. detecting cavities and calculating their attributes, as follows. SCREEN discovers surface cavities by identifying "patches" of the solvent-accessible surface that are distant from the exterior of the investigated protein. Here, the solvent-accessible surface of each protein was computed using a 1.4 Å probe with a subroutine of GRASP2,54 which produced a series of triangles that closely approximate the analytical surface. An envelope surface, representing the exterior of the protein, was generated in a similar manner using a 5.0 Å probe. Analysing the position of each triangle, SCREEN identified distant surface triangles that were further than 2.0 Å from the envelope surface. Distant surface triangles that shared an edge were classified into patches that identified cavities on the protein surface.
In the second phase, SCREEN collected data for each individual patch according to 408 surface descriptors (see Nayal et al.12 for a complete list), which were separated into eight categorical and continuous groups (Table II). All descriptors were calculated by surveying atoms adjacent to each triangle in a patch and cataloguing the types of neighbouring amino acids, the types of nearby secondary structure, and the polarity (or non-polarity) of adjacent amino acids and nearby hydrogen bond donors and acceptors. The continuous groups of descriptors, including those related to properties such as electrostatic potential, shape, polarity (e.g. the atomic solvation parameter, ASP) and flexibility were each divided into 20 to 50 bins with narrow ranges, in which triangle points were assigned and counted. For instance, to map electrostatic potential (EP) distributions and surface curvature, the respective bins were incremented each time a triangle point within the encompassed range of EP and surface curvature was encountered. The cavity description vectors formed by the 408 attributes generated by SCREEN, including all categorical and continuous bins were scaled to zero mean (in terms of deviating from the mean) and unit variance (to account for the attributes’ different units) to yield a data matrix appropriate for PCA operations.55 PCA was then applied to the data to identify (and subsequently remove) noisy descriptors with low modelling influence, leaving only informative descriptors (see Supporting Information for a complete list). These steps are discussed in more detail in the Principal Component Analysis section.
TABLE II.
Examples of Descriptors Used in the Study, Divided into Eight Main Groups.
| Property | Descriptors | Name |
|---|---|---|
| Size | Number of atoms | Number_of_atoms |
| Area | Area | |
| Radius | Radius | |
| Diameter | Diameter | |
| Polarity | Proportion of non-polar atoms | NONPOL_atoms |
| Atomic solvation parameter | ASP | |
| Atom solvation energy | Atom_solvation_energy | |
| Transfer free energy | TFE | |
| Surface solvation energy | Surface_solvation energy | |
| Hydrogen bonds | Hydrogen bond density | HBond_Density |
| Proportions of atoms with or without hydrogen bond acceptor | Hbond_ACCEPT, Hbond_DONOR | |
| Amino acid content | Proportions of atoms that are parts of a specific amino acid | AA-ALA, AA-ARG |
| Shape | Curvature | Curvature |
| Curvedness | Curvedness | |
| Gaussian curvature | Gaussian | |
| Shape index | Shape_Index | |
| Depth | Depth | |
| Normalized cavity moment of inertia | MoiANorm | |
| Electrostatics | Charge | Charges |
| Electrostatic potential | EP | |
| Electrostatic field | Efield | |
| Flexibility | Side chain entropy distribution | Side_chain_entropy |
| Secondary structure | Proportions of atoms that are parts of α-helix-, β-sheet-, coil | HELIX, SHEET |
Calculation of Ligand Descriptors
Physicochemical 2D descriptors were calculated for the ligands of all proteins included in this study using MOE software.56 Ligand PDB codes and descriptors are available in the Supporting Information.
Principal Component Analysis
PCA55,57,58 was used to compress the systematic variation in the descriptor matrix X by extracting linear combinations of the descriptors so called principal components (PCs). The first PC was extracted by calculating an eigenvector oriented in the descriptor space that accounted for the largest variation in the data. All objects (modelled cavities) were projected onto this eigenvector, giving each object a score value (t), i.e. its eigenvector value, and a loading value (p) reflecting the contribution of specific descriptor(s) to the orientation of the new vector. Further PCs were iteratively calculated by placing a new eigenvector, orthogonal to those already plotted, oriented to account for as much as possible of the remaining variation. The decomposition of matrix X can be written as:
| (Eq. 1) |
where A is the number of extracted PCs and E is the residual matrix. Each PC was evaluated based on its eigenvalue, R2-, and Q2 values.55 The eigenvalues reflect the length of eigenvectors with an average eigenvalue equal to 1 in the case of a complete linear transformation (i.e. the number of extracted PCs equals the number of variables) of centred and unit variance scaled data (as in our case). An eigenvalue substantially larger than 1 indicates a clear structural variation in the data in the direction of that eigenvector, and hence a significant PC. For PCs with eigenvalues approaching 1, the differences in eigenvalues between two subsequent PCs were investigated in a SCREE plot where the eigenvalues were plotted against the PC number. PCs with nearly equal eigenvalues that were close to average indicated a random structure of the data (noise) and were hence not significant. The R2 value for each PC is a measure of how much of the total variation in the original data that is extracted for each linear combination. A complete linear transformation would give a total sum of the PCs’ R2 values of 1, which corresponds to 100 % explained variation. Cross-validation (Q2) was also applied to each PC according to the equation:
| (Eq. 2) |
where PRESS is the predicted residual sum of squares and SS the residual sum of squares of the previous PC. A PC was considered to be significant if its Q2 value exceeded 0.01. The cumulative Q2 value of the model was calculated according to:
| (Eq. 3) |
where is the product of the PRESS/SS values for each PC over the total number of A components. The cumulative Q2 value was used as in indicator of the robustness of the model and its ability of predicting objects not included in the model, and its value should be close to the model’s total R2 value. The results of the evaluation of the first 16 PCs are given in the Supporting Information.
A descriptor was excluded from the model if its total R2 value, summarized over all included PCs, was less than 0.1. This procedure was repeated in an iterative manner to generate a new PCA model after every round of exclusions. Descriptors with negative Q2 values were also excluded. Each PC was interpreted by identifying the 20 descriptors with the highest and lowest loading values, i.e., the descriptors with greatest influence on determining the direction of that that eigenvector compared to the original descriptor space (40 in total for every PC). The dataset was also investigated for potential outliers by inspecting the cavity positions in the score plots and their distances to the model in X, DModX. No observation was found to be an extreme outlier, so no ligand-binding cavity was excluded for this reason. The DModX values of prediction sets A and B were carefully examined and all predicted observations were within the applicability domain of the model. Evince software was used for all of the PCA calculations.59
PCA Interpretation and PLS-DA
The relationships between the ligand-binding cavities, as described by the SCREEN descriptors, were explored by visual assessment of the PCA score plots (illustrating the groups and patterns amongst the cavities) and loading plots (indicating the descriptors of the cavities that contributed significantly to the observed patterns). More specifically, the similarity of the cavities was assessed by evaluating their proximity to one another along the PC axes, and the properties explaining their relationships were identified from the positions of the descriptors in the loading plots. To distinguish highly similar proteins (i.e. the Eukaryotic proteases), and to strengthen the PCA interpretations, a model was calculated using the supervised regression method, partial least squares-based projection to latent structure discriminant analysis (PLS-DA),58,60,61 using Evince.59 The procedure applied is briefly described in the Supporting Information.
PCA Clustering Tree
A matrix of Euclidean distances between all cavities in the PCA was calculated using the score values of significant PCs that were interpreted to described general trends. Pairs of proteins separated by small distances were also close in the multi-dimensional PCA space spanned by the PCs, and thus had similar properties. The cavities were clustered using the Euclidean distances and the Neighbour-Joining method of Saitou et al.62 implemented in PHYLIP63 software, to generate an un-rooted tree via randomised ordering of the cavities by successive clustering, displaying the similarities between the proteins without making assumptions of common origin. Branch lengths in the visualised tree were proportional to the distances between the cavities in the original multi-dimensional PCA score space. The tree was interpreted by identifying clusters of cavities and subsequent analysis of the PCA score and loading plots to identify properties shared by similar cavities (i.e. cavities in close proximity to each other in the tree). For clarity of presentation the tree presented here consists of a reduced set of proteins, selected by the inclusion of protein SCOP families consisting of three or more members, preserving the property diversity amongst the cavities. A complete list is provided in the Supporting Information. It should be noted that no cavities were deleted from the PCA.
Similarity Calculations
Similarities in the sequences of the examined proteins were calculated using the online resource ClustalW264 with all adjustable parameters set to default values, and results are presented as percent sequence identity. When several sequences were available for a protein, only those encoding parts of the ligand-binding cavities were considered. Ligand similarities, taking the volumes and the chemical group overlap of the two ligand molecules into account, were calculated using Comboscore in ROCS software.65 The score values ranged between 0 and 2, the latter indicating very high similarity.
Results and Discussion
The main principal properties for differentiating the 121 unique protein domains, representing 93 SCOP protein families were determined by PCA of 239 ligand-binding cavities, described by 264 structural and physicochemical properties. Generally, size and shape were the most important properties for differentiating the cavities, i.e. size and shape descriptors (Table II) had the strongest effects on PC1, followed by polarity (PC2), charges (PC3), depth/shape (PC4), electrostatic fields (PC5), and aromaticity/flexibility (PC6). These six PCs provided clear indications of the key physicochemical cavity properties of the set of proteins at a global level, and the most influential descriptors are considered more thoroughly in the following section. Note that 11 PCs were statistically significant and a SCREE-plot and statistical data are presented in the Supporting Information. PC7 to PC11 provided more detailed information about individual protein cavities and, although they were significant, these PCs were not included in this analysis of general trends. The PCA including 11 PCs described 54% of the original variation in the physicochemical descriptors (R2 = 0.54) and the cross-validation gave a cumulative Q2 value of 0.42. The statistical values for each of the six first PCs are given in Table III. The eigenvalues of the PCs were substantially higher than average, indicating that there was substantial systematic variation in the dataset and the relatively low R2 values indicated presence of data variation not contributing to the differentiation of protein cavities.
TABLE III.
Statistic Values for Each of the First Six PCs.
| PC* | R2 | Eigenvalue | Q2 |
|---|---|---|---|
| 1 | 0.134 | 32.1 | 0.121 |
| 2 | 0.101 | 24.2 | 0.104 |
| 3 | 0.067 | 16.0 | 0.072 |
| 4 | 0.046 | 11.0 | 0.051 |
| 5 | 0.040 | 9.6 | 0.043 |
| 6 | 0.031 | 7.3 | 0.033 |
Principal component.
The principal properties, derived through PCA, described the chemical space of the examined protein ligand-binding cavities, and enabled this space to be compared to the principal property space of ligands. Oprea et al.39 characterised the chemical space of drug-like ligands using ChemGPS to calculate descriptors and create a PCA model, which indicated that size and shape were the most significant properties (most strongly affecting PC1) followed by lipophilicity (PC2) and flexibility and polarity (PC3) of the ligands. This drug-like chemical space has recently been expanded to also include natural products (ChemGPS-NP).66 In ChemGPS-NP a different order of explained properties was revealed; lipophilicity was explained in PC3, aromaticity- and conjugation-related properties of the compounds in PCA2, and flexibility and rigidity properties were explained in PC4. Size, shape, polarity and flexibility were important properties for differentiating both the cavities and the ligands within their respective property spaces. Nevertheless, cavity size, shape and other characteristics can differ greatly from the corresponding ligand properties, in accordance with the proposal by Kahraman et al.31 that shape of cavities and the ligands they bind often do not correlate strongly. For most ligand/protein pairs, we observed little or no correlation between the main properties (size/shape and polarity) that differentiate the ligands and the cavities respectively. Correlation plots of score values between the first two PCs of ligands and cavities are presented in the Supporting Information. The lack of correlation is most likely due to the fact that cavities do not always embrace their ligands completely, and ligands often do not entirely fill their binding cavities. The fact that the property space of the binding cavities does not completely match the property space of the ligands has intriguing implications for ligand-based and structure-based design of novel ligands in drug discovery programs, since it indicates that ligands could be developed with very different binding modes from those of known ligands. One way to address this possibility would be to use fragment-based screening methods, either virtual (e.g., hot spot approaches) or experimental (e.g., nuclear magnetic resonance screening), to explore binding cavities and identify such potential ligands.67–69
Principal Property Identification
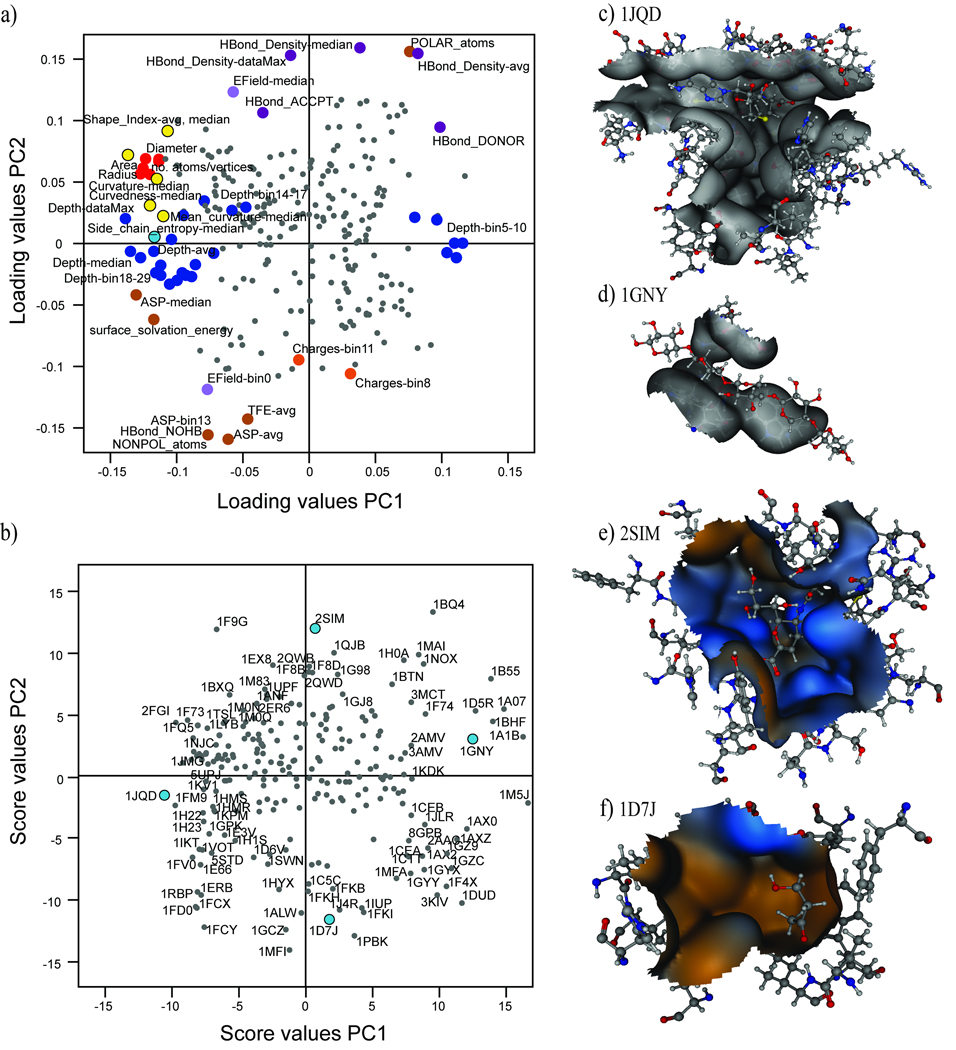
The PCA enabled an in-depth study of the properties possessed by the cavities by detailed study of the score-and loading plots. PC1 of the cavity PCA model was strongly associated with size and shape descriptors. It is evident in Figure 1 that PC1 was positively correlated to cavity depth, hydrophobicity, and flexibility (Figure 1a). PC2, containing the second most significant characteristics for discriminating between the cavities proteins, was dominated by properties such as polarity, electrostatic field (EField) and density of hydrogen-bond forming atoms (Figure 1a). Other descriptors exhibited less variation in the first two PCs.
Figure 1.
Example cavities with extreme score values in PC1 and PC2 verify the analysis of dominating structural and physicochemical properties in the data set. The ligand-binding cavity of the Histamine methyltransferase 1JQD (Figure 1b and 1c) is an example of a large, deep cavity, while the cavity of the Xylan-binding module 1GNY is an example of a small and shallow cavity (Figure 1b and 1d). The first PC also demonstrated that, amongst the studied proteins, small ligand-binding cavities are quite polar, as indicated by the high values for polarity descriptors such as polar atoms and hydrogen bond donors. Small cavities were often shallow, indicating that small cavities were often not deeper than 5 Å, and were thus situated close to the surface of the protein.
The cavity of the Salmonella sialidase 2SIM (Figure 1b and 1e) is an example of a polar cavity that contains many polar atoms and charged amino acids, while the FK-506 binding protein 1D7J (Figure 1b and 1f) is representative of proteins that contain very hydrophobic ligand-binding cavities. The most important descriptors for hydrophobicity can be seen in the loading plot of PC2 (Figure 1a), and include non-polar atoms, non-hydrogen bonding atoms, high values of the average ASP and a high frequency in ASP-bin13 (i.e. energy of hydration of carbon atoms). The value of the average transfer free energy parameter (TFE) was high, and the EField-bin0 descriptor was highly populated, indicating that electric fields were very weak or non-existent in the cavities.
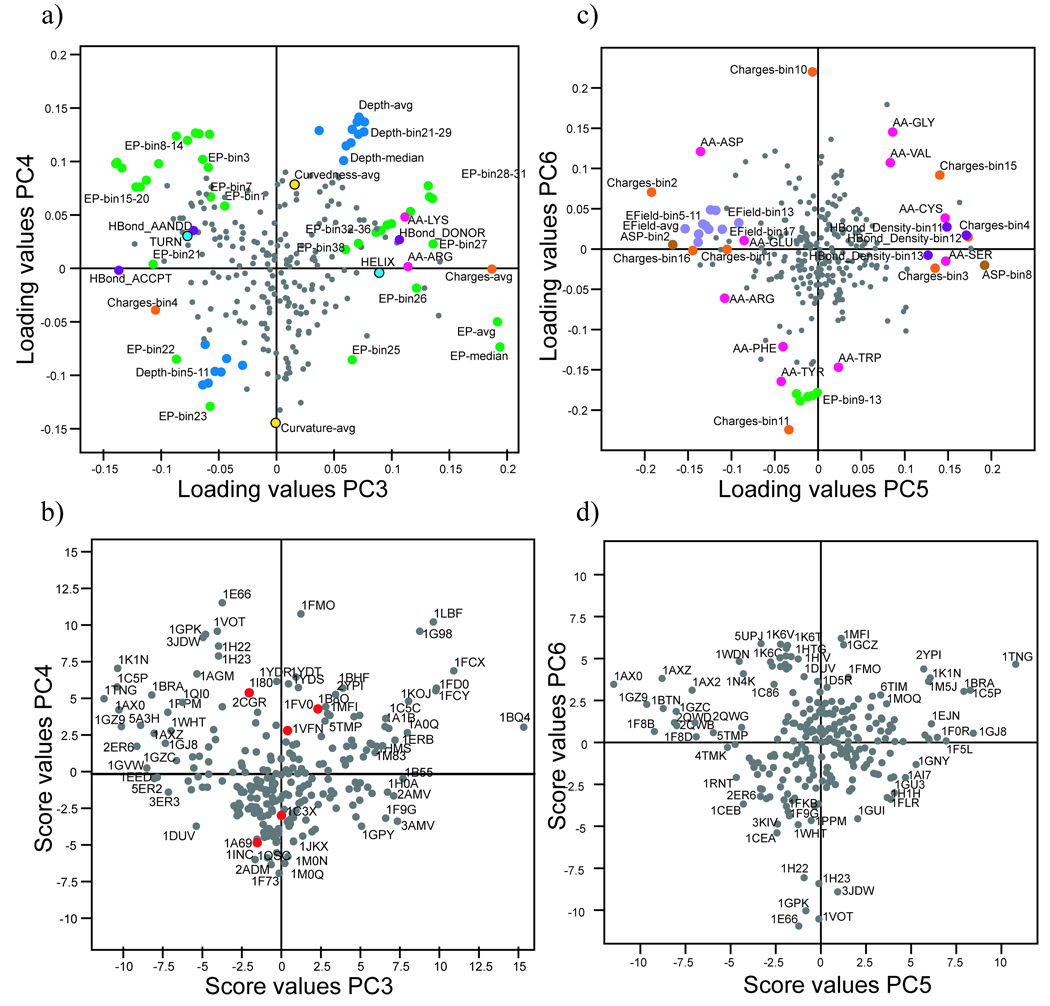
In PC3, the charge, EP and hydrogen-bond donor/acceptor capability of the cavities were the most significant properties. Cavities that were previously identified as polar in PC2 were separated by PC3 according to their charge. Descriptors such as Charges-avg, EP-avg and the presence of the amino acids Arg and Lys were highly influential and the presence of hydrogen bond acceptors was significant (Figure 2a). Cavities such as the Phosphoglycerate mutase 1BQ4, which contained a high percentage of amino acids with hydrogen-bond donor capability (e.g. positively charged amino acids such as Lys and Arg), had high score values (Figure 2b). Attributes associated with amino acids with low EPs, and thus negative charges (Charges-bin4) and hydrogen bond acceptors, had low loading values (Figure 2a). Ligand-binding cavities with a high percentage of amino acids with negatively charged, electron-donating atoms were abundant in this region of PC3, as exemplified by the Endothiapepsin 2ER6 (Figure 2b).
Figure 2.
The fourth most significant property for differentiating proteins based on cavity characteristics was the shape of the cavities (Figure 2a and b), described by depth, curvedness and curvature. The descriptors which exerted the greatest influence on PC4 were Depth-avg, Depth-median and bins that represented high depth values. In addition, the shape descriptor curvedness was correlated with depth, confirming the expectation that deep pockets are generally far from flat (i.e. they have a high value of curvedness-avg).
In the fifth PC, proteins were mainly separated by differences in the EField of their cavities (Figure 2c and d). Descriptors of negative and positive charge, EField and ASP were the most significant for this PC. The presence of several specific amino acids was also influential; charged amino acids (Arg, Asp and Glu) had low loading values, while Cys and Ser had high loading values. Hence, PC5 reflected variations in proportions of polar amino acids and their solvation (ASP-bin2 and bin8), charge and hydrogen-bond forming capability.
The presence of uncharged aromatic or aliphatic amino acids was the primary characteristic separating proteins in the sixth PC (Figure 2c and 2d). The descriptors Charge-bin10 and bin11 both described proportions of uncharged atoms and indicated the presence of aromatic amino acids (Tyr, Trp and Phe). Consequently, proteins with high proportions of aromatic amino acids and few charged amino acids were separated from proteins with high proportions of small, aliphatic, uncharged amino acids in their ligand-binding cavities (Figure 2d).
Mapping of ligand-binding cavities
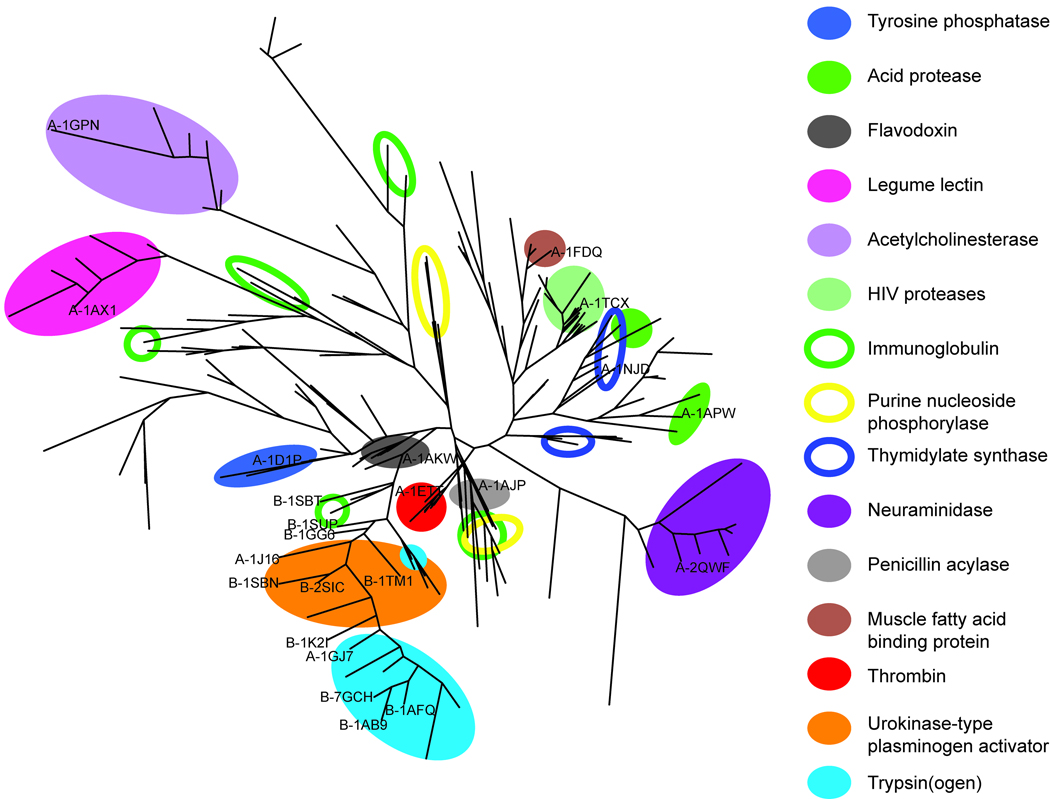
In order to map and compare the 239 protein cavities in the data set, a PCA clustering tree was created, based on their first six principal properties (Figure 3). The same tree, but with all PDB codes visible, is presented in the Supporting Information. The tree in Figure 3 illustrates the differences and similarities of protein cavities, both within and between domains (15 examples of SCOP protein domains are highlighted in Figure 3) based on their structural and physicochemical properties. These analyses of the underlying structural and physicochemical properties responsible for the functional variations in protein cavities clearly provide information that is complementary to the information yielded by structural classification systems such as SCOP49 or CATH.70 Furthermore, such mapping could be valuable in molecular modelling efforts, such as docking or virtual screens, where the choice of protein crystal structure can have a profound impact on the results.71
Figure 3.
Three types of domain distributions were observed in the tree, which we defined as tight, loosely coherent and segregated. To illustrate these patterns, and other features of the data, the distributions of representatives of five of the 15 domains highlighted in Figure 3 (the HIV proteases, Acetylcholinesterases (AChE), Immunoglobulins (Ig), Purine nucleoside phosphorylases (PNP) and Thymidylate synthases (TS)) are briefly discussed below.
The HIV proteases had a tight domain distribution, indicating that their respective ligand-binding cavities were very similar and their crystal structures do not differ significantly in our data set. There are known, significant differences between the ligated and native forms of these proteins,72 especially in their flap-regions. However, since all HIV proteases included in this study were ligated, no differences were identified here in the flap-region.
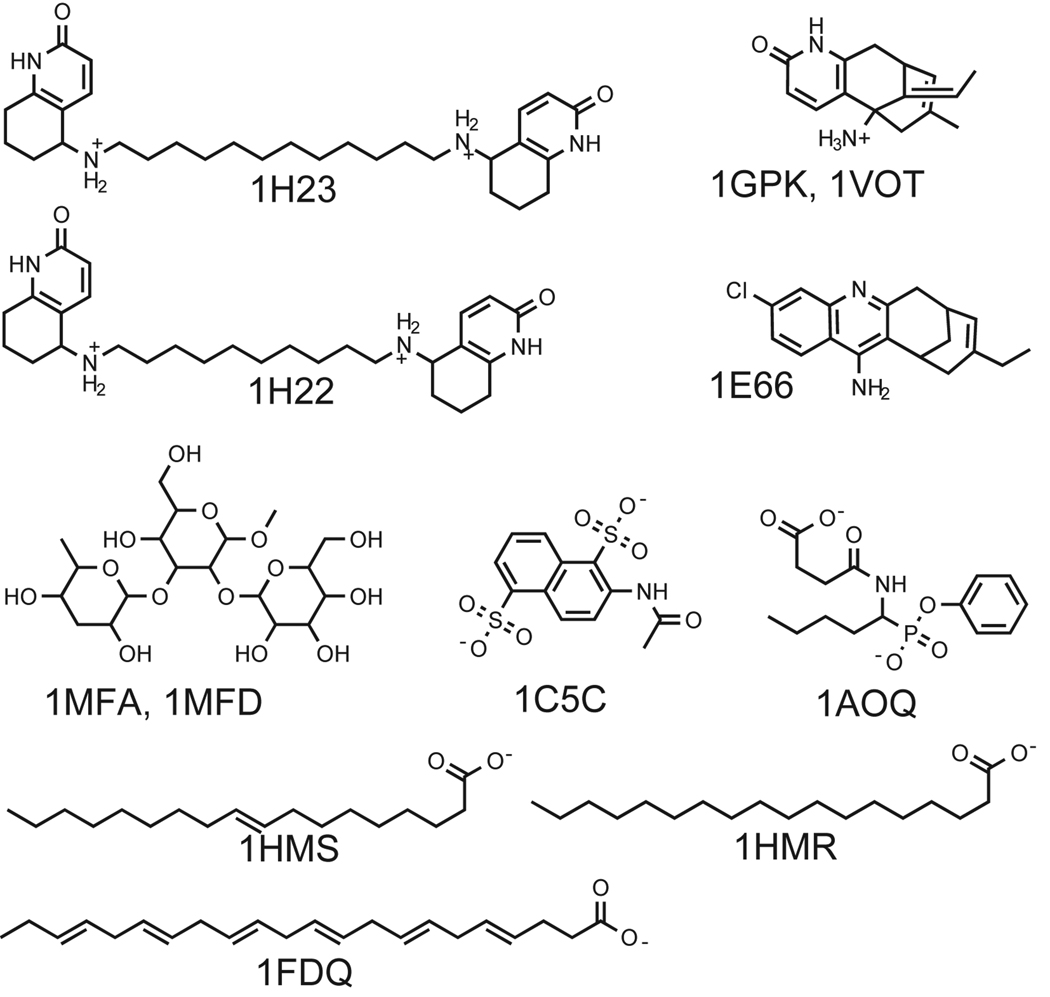
The AChEs were loosely coherent (Figure 3) and differed more widely within their domain than the HIV proteases, notably in shape (PC4) and aromaticity (PC6). All the AChE cavities included both the peripheral anionic site and the catalytic site (where most differences were found). Accordingly, three groups within the AChE domain were identified, with corresponding groups amongst their respective ligands; as shown in Figure 4, 1GPK and 1VOT bind the same ligand, while 1H22 and 1H23 have very similar ligands and 1E66 has a third, different type of ligand. Some of the hydrophobic residues in the active site of AChE are known to very flexible. Phe330 is one of those residues described by Xu et al.73 and a shift in the position of this side chain, exposing an adjacent small pocket, was the cause for the property-differences in the cavities of 1H22/1H23 compared to 1E66.
Figure 4.
The Ig domain was found to be segregated, appearing in five widely separated parts of the tree (Figure 3), indicating that there are large differences between their ligand-binding cavities, in accordance with expectations since the Ig antigen binding-cavity is known to be extremely variable in 3D structure.74 The examined Igs differed in most of the calculated properties (e.g. size, polarity and charge distribution). However, the two Igs that bind the same ligand (1MFA and 1MFD; Figure 4) also appeared close to each other in the tree, and thus have similar ligand-binding cavities. Interestingly, 1C5C and 1A0Q also appeared close together, although their respective ligands (Figure 4) have only a modest similarity (Comboscore, 0.9). Their ligands both have two acidic functional groups, i.e. phosphonic, carboxylic or sulfonic acids, which induce the rotation of positively charged Arg side chains into the cavity, thus causing these two cavities to be more similar. The Ig and AChE findings illustrate how our method, not dependant on relevant geometric alignments, enables the study of the property differences arising due to conformational differences between the cavities induced by ligands.
The tree in Figure 3 also shows the segregation of the PNP domain which is divided into two groups, differing (according to the PCA score and loading plots) in depth and shape; the significant properties for PC4 (Figure 2b). PNP proteins 1A69 and 1C3X have cavities that are shallower and flatter (with low average depth and high average curvature) than those of 1B8O, 1I80 and 1VFN. Further visual inspection of the data for PNP ligand-binding cavities (Figure 5a and 5b) indicates that there are large conformational differences within the PNP domain. The ligand-binding cavities of 1B8O, 1I80 and 1VFN are deeper and more enclosed than those of 1A69 and 1C3X, mainly because of conformational differences in three loop regions which restrict them. However, the protein sequences of these chains show different similarity patterns (Table IV). The most similar are 1B8O and 1VFN (99%), which also have physicochemically similar cavities. Interestingly, 1A69 and 1C3X have similar cavities, but very low sequence identity, demonstrating that cavities can be similar even if their overall protein sequences differ substantially.
Figure 5.
TABLE IV.
Sequence Identity Between Protein Chains of PNP.
| 1C3X* | 1A69# | 1VFN | 1B8O# | 1I80 | |
|---|---|---|---|---|---|
| 1C3X* | 100 | ||||
| 1A69# | 5 | 100 | |||
| 1VFN | 33 | 8 | 100 | ||
| 1B8O# | 33 | 8 | 99 | 100 | |
| 1I80 | 57 | 13 | 35 | 35 | 100 |
Chain A.
Chain B.
TS is another protein domain that was segregated, being split into two groups (Figure 3), differing (according to the PCA score and loading plots) in both size (PC1) and charge (PC3). A score plot of PC1 versus PC3 is presented in the Supporting Information. The TS proteins 1JMG, 1NJC and 1TSL are larger and shallower than 1F4F and 1F4G (Figures 5c and 5d). These groups of proteins originate from two different bacteria, 1F4F and 1F4G are from Escherichia coli,75 while 1JMG, 1NJC and 1TSL are from Lactobacillus casei,76–78 and have sequence differences that influence their cavity properties. The cavities of 1TSL and 1NJC that were identified as very similar bind their respective ligands at different binding sites within the cavity. A comparison of these cavities based solely on residues in the immediate vicinity of the ligands would fail to recognise their similarity because the two ligands have different surroundings. In contrast, the method used in the present study, which considers the entire cavity in which the ligands bind, recognises the similarity of such structures.
In addition to gene origin, ligand induced flexibility, and sequence differences, other factors such as protein crystallisation conditions and the resolution of the crystals could also influence the intra-domain separation of proteins, especially when the cavity is part of a flexible and/or poorly characterised region of the protein. An analysis of the cavity similarities as the one presented here is thus an important step in the choice of target for different kinds of molecular modelling strategies. In this study an automatic cavity detection method was employed, without including ligands to define the pockets. Depending on purpose of using the method, this approach may or may not be optimal. If it is not, the cavity surface of interest (e.g. hot spots) could be identified for example, by using ligands and could subsequently be used to calculate descriptors and finally to create PCA clustering trees.
Prediction of Ligand-binding Cavity Similarities
A PCA model based on properties of protein cavities can be utilized to predict the similarity of cavities of “unknown” domains to those included in the model. Such predictions may be useful, for instance, in the characterisation and mapping of “orphan structures”. Mapping of the two prediction sets A and B using the PCA model based on the 239 protein cavities demonstrated that the model provides good predictability within the spanned property space. Prediction set A contained representatives of a wide variety of proteins and the result of these predictions is visualized in Figure 3. All of the proteins in set A (Table I) that had domain representatives in the PCA model were correctly predicted to be similar to their respective domains in the clustering tree (Figure 3). The protein in the prediction set that had no domain representative in the model was a brain fatty acid binding protein (A-1FDQ), and had predicted cavity similarity to 1HMR and 1HMS, both of which are muscle fatty acid binding proteins (ligands are presented in Figure 4). 1FDQ shares 66% sequence identity with 1HMR/1HMS, and these three proteins have the same fold according to SCOP and CATH. However, there was a small separation between 1FDQ and 1HMR/1HMS in PC5/PC6, indicating differences in the proportions of polar and/or aliphatic amino acids in their cavities. Accordingly, a closer inspection the cavities confirmed that there were differences in amino acids at four positions in their ligand-binding cavities, for instance, 1FDQ has a VAL and a GLY instead of THR and ALA at corresponding positions in 1HMR.
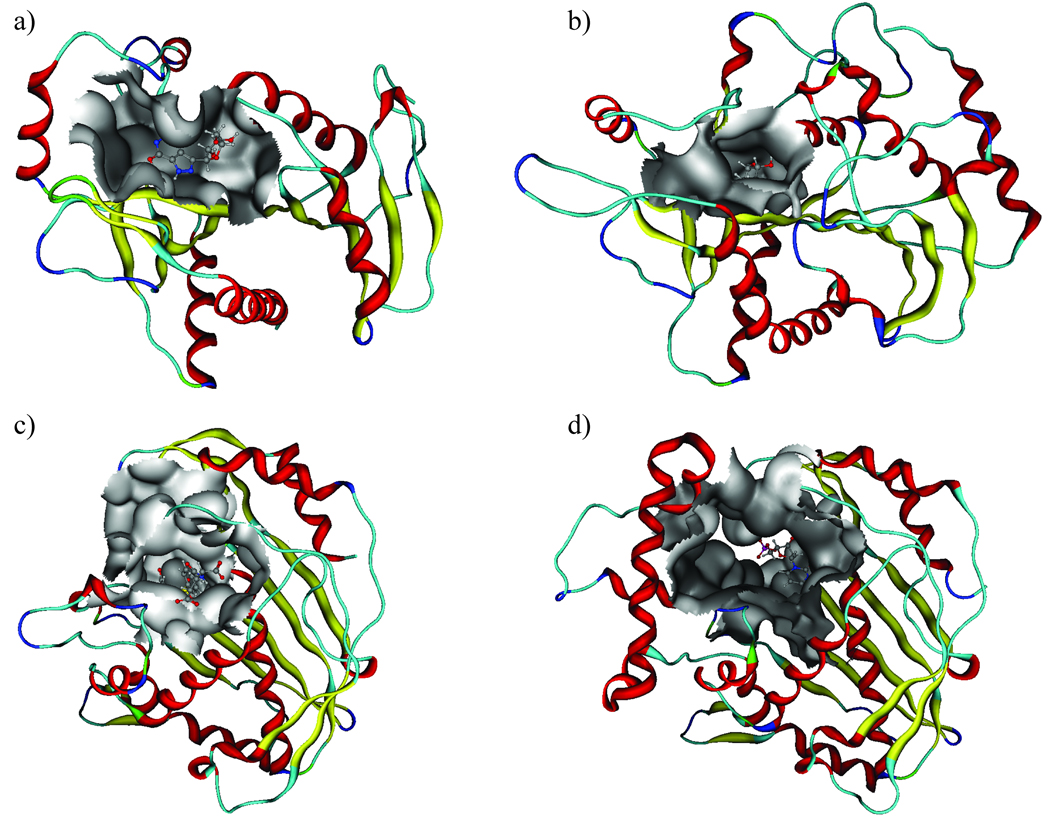
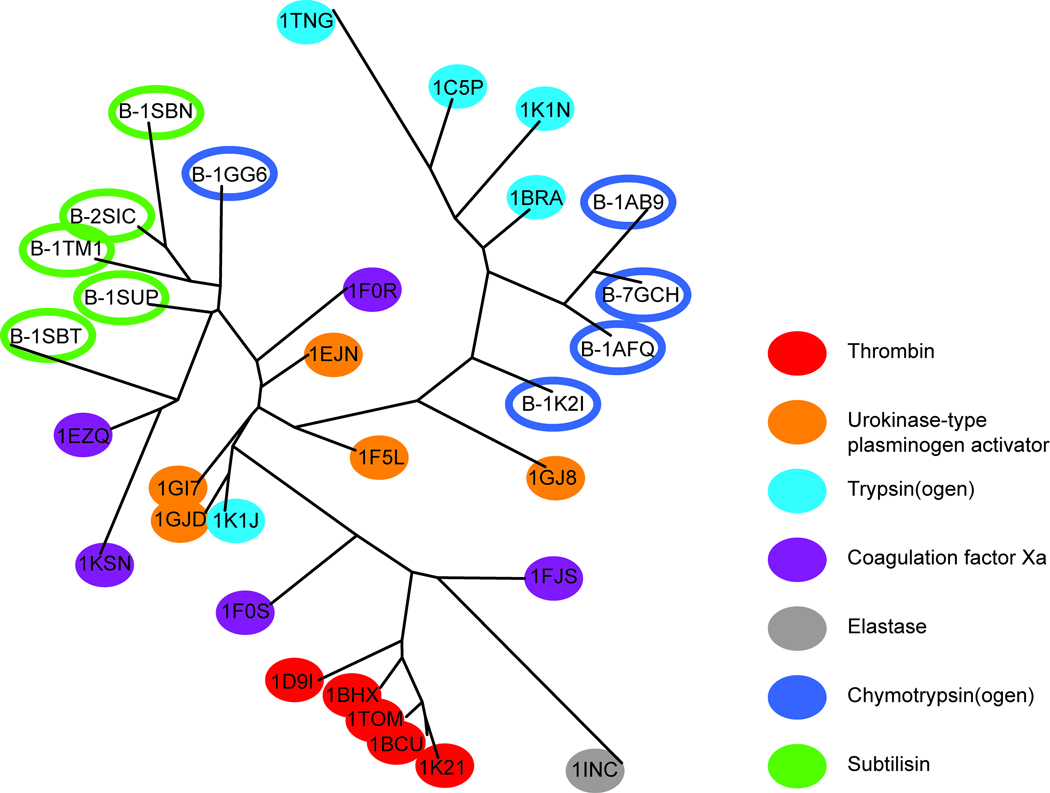
The prediction set B (Table I) included the serine proteases Chymotrypsin(ogen) and Subtilisin. Several other serine proteases, belonging to the Eukaryotic proteases, were included in the training set of proteins, but these particular domains were not represented. Chymotrypsin(ogen) has the same fold as the serine proteases included in the model, while Subtilisin has a completely different fold (but, nevertheless, a similar function). The proteins included in prediction set B were correctly predicted as being similar to the Eukaryotic proteases included in the training set, and appeared on the branch containing the Trypsin(ogen) and Urokinase-type plasminogen activators (UPA) domains (Figure 3).
To further examine the ability of the PCA model to predict and detect similarities and dissimilarities of the ligand-binding cavities, a more detailed tree of the Eukaryotic protease family and prediction set B was created (still based on the PCA for all proteins in the training set), as presented in Figure 6. An interpretation of prediction set B in Figure 6 revealed that ligand-binding cavities of Chymotrypsin(ogen) were predicted to be most similar to those of Trypsin(ogen) and UPA, but less similar to Thrombin and Elastase. The cavity in Subtilisin was predicted to be similar to cavities of the UPA and some Xa Coagulation factors, but less similar to Thrombin and Elastase.
Figure 6.
Furthermore, we concluded from Figure 6 that the training set proteins were roughly grouped into their respective domains. The ligand-binding cavities of Thrombin and Trypsin(ogen) were clearly separated in the tree. A ligand-induced single residue conformational change in the cavity of Trypsin(ogen) 1K1J caused this cavity to become more similar to the UPA cavities, especially in terms of size and shape. Cavities such as those of the Coagulation factor Xa and UPAs with more similar properties could not be easily distinguished in this summarized view of all PCs (i.e., plot of the positions of cavities in chemical space based on all of the considered structural and physicochemical properties). However, a closer inspection of the underlying PCs revealed more detailed information about these domains. For instance, the main differences between Trypsin(ogen) and Thrombin lie in PC1 and PC4, indicating differences in size and depth of these ligand-binding cavities. Furthermore, Coagulation factor Xa could be separated from UPA, in PC4, PC5 and PC6, corresponding to depth/shape, Ser/Cys amino acid content and aromaticity, respectively (see the score plot presented in the Supporting Information). A supervised regression (PLS-DA) model based on the original structural and physicochemical properties of the cavities, and classification of the cavities belonging to Trypsin(ogen) and Thrombin, and Coagulation factor Xa and UPA, as response factors, supported these findings, confirming that the descriptors contained physicochemical information that can be used to distinguish all domains in the Eukaryotic protease family (see Supporting Information for details). The predictive ability of the model to calculate the similarity between “new” cavities of proteins and those in the model has many potential applications, e.g. the identification of proteins with similar binding cavities for inclusion in selectivity screens to avoid undesirable off-target interactions in drug discovery programs. Analogously to the procedure outlined above, the PCA trees examined in such cases could be used to find nearest neighbours to a target of interest, and these proteins could then be further analysed. In addition, targets found to be similar could be candidates for a multitarget-directed ligand, i.e. a low-affinity ligand designed to interact with several targets in attempts to inhibit (or enhance) several associated biological processes.
Conclusion
The analysis of similarities between ligand-binding cavities in a large collection of protein crystal structures presented here yielded a global perspective of the property space spanned by the included cavities. The approach of using numerical vectors of equal length for all proteins describing the physicochemical properties of the cavities, provides the means to compare cavities of proteins with little or no sequence identity, avoiding uncertainties in geometrical alignment. The cavities were thoroughly described by calculating a multitude of physicochemical properties using SCREEN software, in the absence of ligands. Hence, the entire cavities were studied rather than merely the surroundings of the ligands, enabling similar pockets to be identified, including those in which ligands bind in different areas.
In order to elucidate the relationships between the proteins based on physicochemical features of their ligand-binding cavities, PCA was employed to reduce the dimensionality and visualise general trends in the data. Analysis of the PCA score and loading plots indicated that the most important general features for differentiating the protein ligand-binding cavities were, in order of significance: size/shape, polarity, charge distribution, depth/shape, electrostatic field and aromaticity. The PCA analysis showed that many of these properties are correlated, and provided details about the nature of the correlations. In addition, comparison of the cavity property space with that of drug-like ligands indicated that size, shape and polarity are the most important properties for differentiating objects within both of these spaces, but the two spaces are not directly correlated.
Using the score vectors from the PCA a clustering tree was created to map and compare the investigated proteins based on their ligand-binding cavity properties. Coherent protein domains were identified and segregated domains were furthered analysed. Analysis of the score and loading plots provided valuable information of structural and physicochemical properties responsible for the distribution of the cavities. Hence, the presented method not only provided a good overview of potential targets, but also information on the variability of binding cavities due to mutations, between-species differences and flexibility upon ligand-binding. Knowledge of the variations in cavities is of high potential value when choosing targets for molecular modelling studies and for designing ligands based on 3D structures of a target. Furthermore, the distribution of the domains in the chemical space explored demonstrated that ligand-binding cavities of proteins can be very similar even when their sequences differ greatly, as in the cases of PNP proteins 1C3X and 1A69, or Subtilisin compared to other serine proteases. Finally, the principal properties of ligand-binding cavities of proteins that were not included in the model were found to be predicted accurately, and their similarity to cavities within the global property space could be established. The predictive capability of the method offers the possibility to predict the similarity of cavities from orphan protein structures cavities to proteins with known function. Mapping of proteins according to the structural and physicochemical properties of their ligand-binding cavities using PCA can provide complementary information to similarity data obtained using, for example, SCOP, CATH or ClustalW.
Supplementary Material
Acknowledgements
This work was funded by grants from the Swedish Research Council and financial support from AstraZeneca R&D Mölndal, Sweden. We are grateful to Gianlucas Rossi for preparing the protein files for the SCREEN calculations.
Footnotes
Supporting Information Available: Table of proteins used in the study, list of descriptors included in the final PCA model, table of proteins used in the PCA clustering tree, information of the PCA of ligand features, correlation plots between ligand properties and ligand-binding cavity properties, tables of ligand descriptors and ligand PCA score values, statistical values and a SCREE-plot for the total ligand-binding cavity PCA, score plot including Eukaryotic proteases and prediction set B, score plot highlighting the TS proteins, PCA clustering tree containing all PDB code and information regarding the PLS-DA. This material is available free of charge via the Internet at http://www3.interscience.wiley.com/journal/36176/home.
References
- 1.Minai R, Matsuo Y, Onuki H, Hirota H. Method for comparing the structures of protein ligand-binding sites and application for predicting protein-drug interactions. Proteins: Struct, Funct, Bioinf. 2008;72(1):367–381. doi: 10.1002/prot.21933. [DOI] [PubMed] [Google Scholar]
- 2.Gold ND, Jackson RM. Fold independent structural comparisons of protein-ligand binding sites for exploring functional relationships. J Mol Biol. 2006;355(5):1112–1124. doi: 10.1016/j.jmb.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 3.Wallace AC, Borkakoti N, Thornton JM. TESS: A geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 1997;6(11):2308–2323. doi: 10.1002/pro.5560061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamelryck T. Efficient identification of side-chain patterns using a multidimensional index tree. Proteins: Struct, Funct, Bioinf. 2003;51(1):96–108. doi: 10.1002/prot.10338. [DOI] [PubMed] [Google Scholar]
- 5.Artymiuk PJ, Poirrette AR, Grindley HM, Rice DW, Willett P. A graph-theoretic approach to the identification of 3-dimentional patterns of amino-acid side-chains in protein structures. J Mol Biol. 1994;243(2):327–344. doi: 10.1006/jmbi.1994.1657. [DOI] [PubMed] [Google Scholar]
- 6.Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285(4):1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 7.Jambon M, Imberty A, Deleage G, Geourjon C. A new bioinformatic approach to detect common 3D sites in protein structures. Proteins: Struct, Funct, Bioinf. 2003;52(2):137–145. doi: 10.1002/prot.10339. [DOI] [PubMed] [Google Scholar]
- 8.Schmitt S, Kuhn D, Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J Mol Biol. 2002;323(2):387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn D, Weskamp N, Schmitt S, Hullermeier E, Klebe G. From the similarity analysis of protein cavities to the functional classification of protein families using Cavbase. J Mol Biol. 2006;359(4):1023–1044. doi: 10.1016/j.jmb.2006.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shulman-Peleg A, Nussinov R, Wolfson HJ. Recognition of functional sites in protein structures. J Mol Biol. 2004;339(3):607–633. doi: 10.1016/j.jmb.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahl M, Taroni C, Schneider G. Mapping of protein surface cavities and prediction of enzyme class by a self-organizing neural network. Protein Eng, Des Sel. 2000;13(2):83–88. doi: 10.1093/protein/13.2.83. [DOI] [PubMed] [Google Scholar]
- 12.Nayal M, Honig B. On the nature of cavities on protein surfaces: Application to the identification of drug-binding sites. Proteins: Struct, Funct, Bioinf. 2006;63(4):892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- 13.Kupas K, Ultsch A, Klebe G. Large scale analysis of protein-binding cavities using self-organizing maps and wavelet-based surface patches to describe functional properties, selectivity discrimination, and putative cross-reactivity. Proteins: Struct, Funct, Bioinf. 2008;71(3):1288–1306. doi: 10.1002/prot.21823. [DOI] [PubMed] [Google Scholar]
- 14.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7(9):1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31(13):3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binkowski TA, Freeman P, Liang J. pvSOAR: Detecting similar surface patterns of pocket and void surfaces of amino acid residues on proteins. Nucleic Acids Res. 2004;32:W555–W558. doi: 10.1093/nar/gkh390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008;8:23. doi: 10.1186/1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schalon C, Surgand JS, Kellenberger E, Rognan D. A simple and fuzzy method to align and compare druggable ligand-binding sites. Proteins: Struct, Funct, Bioinf. 2008;71(4):1755–1778. doi: 10.1002/prot.21858. [DOI] [PubMed] [Google Scholar]
- 19.Xie L, Bourne PE. A robust and efficient algorithm for the shape description of protein structures and its application in predicting ligand binding sites. BMC Bioinf. 2007;8:S9. doi: 10.1186/1471-2105-8-S4-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen BY, Fofanov VY, Bryant DH, Dodson BD, Kristensen DM, Lisewski AM, Kimmel M, Lichtarge O, Kavraki LE. The MASH pipeline for protein function prediction and an algorithm for the geometric refinement of 3D motifs. J Comput Biol. 2007;14(6):791–816. doi: 10.1089/cmb.2007.R017. [DOI] [PubMed] [Google Scholar]
- 21.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15(3):275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhang ZD, Grigorov MG. Similarity networks of protein binding sites. Proteins: Struct, Funct, Bioinf. 2006;62(2):470–478. doi: 10.1002/prot.20752. [DOI] [PubMed] [Google Scholar]
- 23.Richards FM. Areas, volumes, packing, and protein-structure. Ann Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 24.Edelsbrunner H, Mucke EP. 3-Dimensional alpha-shapes. ACM TransactGraph. 1994;13(1):43–72. [Google Scholar]
- 25.Laskowski RA. Surfnet - A program for visualizing molecular-surfaces, cavities and intermolecular interactions. J Mol Graph. 1995;13(5):323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 26.Kinoshita K, Nakamura H. Identification of protein biochemical functions by similarity search using the molecular surface database eF-site. Protein Sci. 2003;12(8):1589–1595. doi: 10.1110/ps.0368703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connolly ML. Solvent-accessible surfaces of proteins and nucleic-acids. Science. 1983;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 28.Poirrette AR, Artymiuk PJ, Rice DW, Willett P. Comparison of protein surfaces using a genetic algorithm. J Comput-Aided Mol Des. 1997;11(6):557–569. doi: 10.1023/a:1007966011516. [DOI] [PubMed] [Google Scholar]
- 29.Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem Cent J. 2007;1:7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholls A, Sharp KA, Honig B. Protein folding and association - insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct, Funct, Bioinf. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.Kahraman A, Morris RJ, Laskowski RA, Thornton JM. Shape variation in protein binding pockets and their ligands. J Mol Biol. 2007;368(1):283–301. doi: 10.1016/j.jmb.2007.01.086. [DOI] [PubMed] [Google Scholar]
- 32.Sael L, La D, Li B, Rustamov R, Kihara D. Rapid comparison of properties on protein surface. Proteins: Struct, Funct, Bioinf. 2008;73(1):1–10. doi: 10.1002/prot.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins: Struct, Funct, Bioinf. 2008;72(4):1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
- 34.Chen BY, Bryant DH, Fofanov VY, Kristensen DM, Cruess AE, Kimmel M, Lichtarge O, Kayraki LE. Cavity scaling: Automated refinement of cavity aware motifs in protein function prediction. J Bioinf Comput Biol. 2007;5(2a):353–382. doi: 10.1142/s021972000700276x. [DOI] [PubMed] [Google Scholar]
- 35.Cui JA, Han LY, Lin HH, Tang ZQ, Ji ZL, Cao ZW, Li YX, Chen YZ. Advances in exploration of machine learning methods for predicting functional class and interaction profiles of proteins and peptides irrespective of sequence homology. Curr Bioinf. 2007;2(2):95–112. [Google Scholar]
- 36.Holm L, Sander C. Mapping the protein universe. Science. 1996;273(5275):595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 37.Eidhammer I, Jonassen I, Taylor WR. Structure comparison and structure patterns. J Comput Biol. 2000;7(5):685–716. doi: 10.1089/106652701446152. [DOI] [PubMed] [Google Scholar]
- 38.Kellenberger E, Schalon C, Rognan D. How to measure the similarity between protein ligand-binding sites? Curr Comput-Aided Drug Des. 2008;4(3):209–220. [Google Scholar]
- 39.Oprea TI, Gottfries J. ChemGPS: A chemical space navigation tool. Rat Appr Drug Des. 2001:437–446. [Google Scholar]
- 40.Larsson J, Gottfries J, Muresan S, Bohlin L, Backlund A. ChemGPS-NP - tuned for navigation in biologically relevant chemical space. Planta Medica. 2006;72(11):1020–1021. doi: 10.1021/np070002y. [DOI] [PubMed] [Google Scholar]
- 41.Maldonado AG, Doucet JP, Petitjean M, Fan BT. Molecular similarity and diversity in chemoinformatics: From theory to applications. Mol Diversity. 2006;10(1):39–79. doi: 10.1007/s11030-006-8697-1. [DOI] [PubMed] [Google Scholar]
- 42.Gorse AD. Diversity in medicinal chemistry space. Curr Top Med Chem. 2006;6(1):3–18. doi: 10.2174/156802606775193310. [DOI] [PubMed] [Google Scholar]
- 43.Wold S, Dunn WJ. Multivariate quantitative structure activity relationships (QSAR) - conditions for their applicability. J Chem Inf Comput Sci. 1983;23(1):6–13. [Google Scholar]
- 44.Mager DE. Quantitative structure-pharmacokinetic/pharmacodynamic relationships. Adv Drug Delivery Rev. 2006;58(12–13):1326–1356. doi: 10.1016/j.addr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Strombergsson H, Kleywegt GJ. A chemogenomics view on protein-ligand spaces. BMC Bioinf. 2009;10 doi: 10.1186/1471-2105-10-S6-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gunnarsson I, Andersson P, Wikberg J, Lundstedt T. Multivariate analysis of G protein-coupled receptors. J Chemom. 2003;17(1):82–92. [Google Scholar]
- 47.Lindstrom A, Pettersson F, Linusson A. Quantitative protein descriptors for secondary structure characterization and protein classification. Chemom Intell Lab Syst. 2009;95(1):74–85. [Google Scholar]
- 48.Naumann T, Matter H. Structural classification of protein kinases using 3D molecular interaction field analysis of their ligand binding sites: Target family landscapes. J Med Chem. 2002;45(12):2366–2378. doi: 10.1021/jm011002c. [DOI] [PubMed] [Google Scholar]
- 49.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP - a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Fang X, Lu Y, Wang S. The PDBbind Database: Collection of Binding Affinities for Protein-Ligand Complexes With Known Three-Dimensional Structures. J Med Chem. 2004;47(12):2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 51.Wang R, Fang X, Lu Y, Yang C-Y, Wang S. The PDBbind database: Methodologies and Updates. J Med Chem. 2005;48(12):4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 52.Reduce. 3.03. Durham NC: The Richardson Laboratory, Duke University; [Google Scholar]
- 53.RCSB Protein data Bank. [Accessed 2009-08-18]; http://www.rcsb.org.
- 54.Petrey D, Xiang ZX, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh IYY, Alexov E, Honig B. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins: Struct, Funct, Bioinf. 2003;53(6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 55.Jackson JE. A user's guide to principal components. New York: John Wiley & sons, Inc.; 1991. [Google Scholar]
- 56.MOE (Molecular Operating Environment) Version 2008.10. Suite 910 – 1010 Sherbrooke St. W, Montreal, Quebec. Canada H3A 2R7: Chemical Computing Group Inc.; [Google Scholar]
- 57.Wold S, Esbensen K, Geladi P. Principal Component Analysis. Chemom Intell Lab Syst. 1987;2(1–3):37–52. [Google Scholar]
- 58.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikström C, Wold S. Multi- and megavariate data analysis - basic principles and applications, part 1. Umeå: Umetrics AB. 2006 [Google Scholar]
- 59.Evince. 2.2.2. Umbio AB. Box 7980, 90719 Umeå, Sweden: [Google Scholar]
- 60.Stahle L, Wold S. Partial least squares analysis with cross-validation for the two-class problem: a monte carlo study. J Chemom. 1987;1:185–196. [Google Scholar]
- 61.Wold S, Sjöström M, Eriksson L. PLS-Regression: a Basic Tool of Chemometrics. Chemom Intel Lab Sys. 2001;58(2):109–130. [Google Scholar]
- 62.Saitou N, Nei M. The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 63.Felsenstein J. PHYLIP (Phylogeny Interference Package). version 3.6. 2004 http://evolution.genetics.washington.edu/phylip.html. [Google Scholar]
- 64.Lloyd A. ClustalW2. 1997. [Accessed 2008-08-06];1997 http://www.ebi.ac.uk/Tools/clustalw2/index.html.
- 65.ROCS. 2.2. Openeye scientific software Inc. 3600 Cerrillos Road, Suite 1107, Santa Fe, NM 87507: [Google Scholar]
- 66.Larsson J, Gottfries J, Muresan S, Backlund A. ChemGPS-NP: Tuned for navigation in biologically relevant chemical space. J Nat Prod. 2007;70(5):789–794. doi: 10.1021/np070002y. [DOI] [PubMed] [Google Scholar]
- 67.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51(13):3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 68.Law R, Barker O, Barker JJ, Hesterkamp T, Godemann R, Andersen O, Fryatt T, Courtney S, Hallett D, Whittaker M. The multiple roles of computational chemistry in fragment-based drug design. J Comput Aided Mol Des. 2009;23(8):459–473. doi: 10.1007/s10822-009-9284-1. [DOI] [PubMed] [Google Scholar]
- 69.Fontaine F, Cross S, Plasencia G, Pastor M, Zamora I. SHOP: a method for structure-based fragment and scaffold hopping. ChemMedChem. 2009;4(3):427–439. doi: 10.1002/cmdc.200800355. [DOI] [PubMed] [Google Scholar]
- 70.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH - a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 71.Sheridan RP, McGaughey GB, Cornell WD. Multiple protein structures and multiple ligands: effects on the apparent goodness of virtual screening results. J Comput Aided Mol Des. 2008;22(3–4):257–265. doi: 10.1007/s10822-008-9168-9. [DOI] [PubMed] [Google Scholar]
- 72.Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SBH, Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3-a resolution. Science. 1989;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 73.Xu YC, Colletier JP, Weik M, Jiang HL, Moult J, Silman I, Sussman JL. Flexibility of aromatic residues in the active-site gorge of acetylcholinesterase: X-ray versus molecular dynamics. Biophys J. 2008;95(5):2500–2511. doi: 10.1529/biophysj.108.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bork P, Holm L, Sander C. The immunoglobulin fold - structural classification, sequence patterns and common core. J Mol Biol. 1994;242(4):309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 75.Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, Wells JA. Site-directed ligand discovery. Proc Natl Acad Sci U S A. 2000;97(17):9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finer-Moore JS, Liu L, Birdsall DL, Brem R, Apfeld J, Santi DV, Stroud RM. Contributions of orientation and hydrogen bonding to catalysis in Asn229 mutants of thymidylate synthase. J Mol Biol. 1998;276(1):113–129. doi: 10.1006/jmbi.1997.1495. [DOI] [PubMed] [Google Scholar]
- 77.Finer-Moore JS, Liu L, Schafmeister CE, Birdsall DL, Mau T, Santi DV, Stroud RM. Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: Aspargine-229 in thymidylate synthase. Biochemistry. 1996;35(16):5125–5136. doi: 10.1021/bi952751x. [DOI] [PubMed] [Google Scholar]
- 78.Stout TJ, Tondi D, Rinaldi M, Barlocco D, Pecorari P, Santi DV, Kuntz ID, Stroud RM, Shoichet BK, Costi MP. Structure-based design of inhibitors specific for bacterial thymidylate synthase. Biochemistry. 1999;38(5):1607–1617. doi: 10.1021/bi9815896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.