Abstract
Upon entry of Francisella tularensis to macrophages, the Francisella-containing phagosome (FCP) is trafficked into an acidified late endosome-like phagosome with limited fusion to the lysosomes followed by rapid escape into the cytosol where the organism replicates. Although the Francisella Pathogenicity Island (FPI), which encodes a type VI-like secretion apparatus, is required for modulation of phagosome biogenesis and escape into the cytosol, the mechanisms involved are not known. To decipher the molecular bases of modulation of biogenesis of the FCP and bacterial escape into the macrophage cytosol, we have screened a comprehensive mutant library of F. tularensis subsp novicida for their defect in proliferation within human macrophages, followed by characterization of modulation of phagosome biogenesis and bacterial escape into the cytosol. Our data show that at least 202 genes are required for intracellular proliferation within macrophages. Among the 125 most defective mutants in intracellular proliferation, we show that the FCP of at least 91 mutants co-localize persistently with the late endosomal/lysosomal marker LAMP-1 and fail to escape into the cytosol, as determined by fluorescence-based phagosome integrity assays and transmission electron microscopy. At least 34 genes are required for proliferation within the cytosol but do not play a detectable role in modulation of phagosome biogenesis and bacterial escape into the cytosol. Our data indicate a tremendous adaptation and metabolic reprogramming by F. tularensis to adjust to the micro-environmental and nutritional cues within the FCP, and these adjustments play essential roles in modulation of phagosome biogenesis and escape into the cytosol of macrophages as well as proliferation in the cytosol. The plethora of the networks of genes that orchestrate F. tularensis-mediated modulation of phagosome biogenesis, phagosomal escape, and bacterial proliferation within the cytosol is novel, complex, and involves an unusually large portion of the genome of an intracellular pathogen.
Keywords: endosome, lysosome, phagosome, cytosol, intracellular, tularemia, Listeria
Introduction
Francisella tularensis is an intracellular bacterium that causes tularemia, a fatal zoonotic disease that infects small mammals and humans (Ellis et al., 2002; Pechous et al., 2009; Santic et al., 2010). There are four subspecies of F. tularensis, which are subsp tularensis, holarctica, mediasiatica and novicida (Keim et al., 2007; Nigrovic and Wingerter, 2008). Subspecies tularensis is most virulent, while subsp holarctica and mediasiatica cause a mild form of tularemia (Santic et al., 2006; Pechous et al., 2009). All subspecies share about 97% genome identities (Champion et al., 2009; Larsson et al., 2009). Recent studies have shown that the high virulence of subsp tularensis and holarctica may be due to loss of gene functions or an increase in the copy number of genes such as duplication of the Francisella Pathogenicity Island (FPI) (Champion et al., 2009; Larsson et al., 2009). Because of low infectivity, ease of dissemination, and high morbidity and mortality, F. tularensis is classified by the CDC as a category A select bioterrorism agent (Dennis et al., 2001).
Clinical manifestation of tularemia depends on the route of infection and it includes glandular, ulceroglandular, oculoglandular, oropharyngeal, pneumonic and typhoidal tularemia (Ellis et al., 2002). Tularemia often presents with nonspecific flu-like symptoms such as headache, fever, chills, nausea, diarrhea, and myalgia (Oyston et al., 2004; Nigrovic and Wingerter, 2008).
The ability of F. tularensis to cause disease is due to its capacity to replicate within cells (Oyston et al., 2004). Like other intracellular pathogens, F. tularensis must overcome the host innate immune response to successfully colonize the intracellular niche. The primary host defense is centered on the antimicrobial properties of the phagosome. Most successful intracellular pathogens either escape the phagosome or divert phagosome maturation to an idiosyncratic niche where they replicate. Bacteria that escape from the phagosome include Shigella flexneri and Listeria monocytogenes (Goebel and Kuhn, 2000; Ray et al., 2009). Escape of S. flexneri and L. monocytogenes from the acidified phagosome into the host cell cytosol is mediated by a pore-forming cytolysin and phospholipases (Ray et al., 2009). Similarly, the Francisella-containing phagosome (FCP) transiently acquires early and late endosomal markers as well as the vacuolar ATPase, which acidifies the phagosome followed by rapid escape of the bacteria into the cytosol within 30–60 min (Golovliov et al., 2003a; Clemens et al., 2004; Santic et al., 2005a; Santic et al., 2005b; Checroun et al., 2006; Santic et al., 2007; Bonquist et al., 2008; Santic et al., 2008; Qin et al., 2009). Mutants that are unable to escape into the cytosol do not replicate (Santic et al., 2005b; Bonquist et al., 2008; Qin et al., 2009) and are attenuated in animal models (Lauriano et al., 2004; Weiss et al., 2007; Mohapatra et al., 2008). Similar to trafficking within human macrophages, F. tularensis transiently occupies a late endosome-like phagosome in Drosophila melanogaster-derived S2 cells followed by rapid bacterial escape into the cytosol, where the bacteria proliferate robustly (Santic et al., 2009). This may suggest that common mechanisms are utilized by F. tularensis to modulate phagosome biogenesis, escape into the cytosol, and to proliferate within the cytosol of mammalian and arthropod-derived cells. F. tularensis subsp novicida is very similar to the virulent subspecies in modulation of phagosome biogenesis, phagosomal escape, proliferation within the cytosol of mammalian macrophages, and manifestation of disease in animal models (Santic et al., 2005a; Santic et al., 2005b; Santic et al., 2006; Santic et al., 2007; Santic et al., 2009; Pechous et al., 2009). These characteristics render F. tularensis subsp novicida a very useful model to dissect the molecular bases of intracellular proliferation of F. tularensis under BSL2 containment.
The Francisella Pathogenicity Island (FPI) encodes a type VI-like secretion apparatus, which is required for modulation of phagosome biogenesis and escape into the cytosol (Barker et al., 2009). The VgrG and IglI proteins are secreted into the host cell cytosol, and the translocation of IgII is FPI-dependent (Barker et al., 2009). The three FPI-encoded VgrG, IglI, and IglC proteins, the MglA global regulator, four acid phosphatases (AcpA, AcpB, AcpC and Hap) and a lipoprotein (FTT1103) have been shown to play important roles in phagosomal escape of F. tularensis into the cytosol of macrophages, but the molecular bases of phagosomal escape are not known (Barker et al., 2009; Golovliov et al., 2003b; Santic et al., 2005a; Santic et al., 2005b; Checroun et al., 2006; Santic et al., 2007; Bonquist et al., 2008; Santic et al., 2008; Qin et al., 2009). Interestingly, the two FPI genes iglC, iglD, and their regulator MglA are also required for intracellular proliferation within arthropod-derived cells (Santic et al., 2009). The phagosome containing the mglA and iglC mutants matures into a phagolysosome and bacteria fail to escape into the cytosol within macrophages and arthropod-derived cells (Santic et al., 2005b; Santic et al., 2009; Bonquist et al., 2008). In addition, genes involved in oxidative stress, protein turnover, capsule or lipopolysaccharide (LPS) biosynthesis, type IV pilin assembly, iron uptake, outer membrane channels, purine biosynthesis, and regulation through mglAB, sspA or pmrA are required for intracellular growth and virulence of F. tularensis (see (Pechous et al., 2009; Santic et al., 2010) for recent reviews).
Genome-wide screens using transposon-based mutagenesis have identified genes involved in various aspects of virulence or dissemination in animal models of tularemia (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009). An in vivo negative selection mutant screen has identified genes of F. tularensis subsp novicida required for virulence in the pulmonary routes of infection in the mice model (Kraemer et al., 2009). On the other hand, genes required for growth of Francisella in vivo by the subcutaneous route have been identified (Weiss et al., 2007). In vivo negative selection screen in mice using signature-tagged mutagenesis in the LVS strain has identified genes required for growth in the lung during respiratory tularemia (Su et al., 2007). Two other mutant screens have identified F. tularensis genes required for replication in macrophages and HepG2 cells, respectively, but the mutant libraries used in these screens are biased and do not cover the entire genome (Qin and Mann, 2006; Maier et al., 2007). Although modulation of phagosome biogenesis and escape into the cytosol are the two crucial steps in the intracellular infection and manifestation of disease, no studies have been reported to identify the genes repertoire involved in these crucial pathogenic processes. Therefore, the molecular bases of phagosome biogenesis and bacterial escape into the cytosol by F. tularensis remain unknown, and are the goals of this study.
To decipher the molecular bases of phagosome biogenesis and bacterial escape and proliferation within the cytosol, we utilized a comprehensive transposon insertion mutant library of F. tularensis subsp novicida (Gallagher et al., 2007). We identified 202 genes that contribute to intracellular growth in human macrophages. Among the mutants defective in replication in human macrophages, 137 of them are required for replication in D. melanogaster S2 cells (see accompanying manuscript). In contrast to the wild type strain that co-localize transiently with late endosomal/lysosomal markers prior to rapid escape into the cytosol, 91 of the mutants that are severely defective in intracellular growth in macrophages co-localize persistently with late endosome/lysosome markers and fail to escape into the cytosol. Another thirty four mutants severely defective in intra-macrophages growth but successfully escape into the cytosol fail to proliferate. Our findings are surprising, since phagosomal escape of other bacteria such as L. monocytogenes and S. flexineri is mediated by few loci (Ray et al., 2009), which indicates a novel molecular complexity governing phagosomal escape of F. tularensis.
Results
Replication of F. tularensis mutants in U937 macrophages
The ability of F. tularensis to cause tularemia is dependent on its proliferation within the macrophage cytosol after rapid phagosomal escape. To identify the bacterial genes involved in phagosomal escape, our experimental design was based on a two-step screen. First, we identified the mutants defective in intracellular proliferation; and second, these mutants were analyzed for phagosome biogenesis and phagosomal escape. Therefore, we performed a primary screen of a comprehensive library of 3,050 sequence-defined insertion mutants of F. tularensis subsp novicida corresponding to 1448 genes with a minimum of two mutant alleles for most genes (Gallagher et al., 2007) for their defect in intracellular proliferation in human-derived U937 macrophages. Infections were performed at MOI of 10 for 1 h followed by 1 h of gentamicin treatment. At 24 h post-infection, cells were lysed and serial dilutions were plated on agar plates for colony enumeration. To exclude the mutants with mild defective phenotype from our analyses, a mutant was considered defective in intracellular proliferation if it showed ≥102 fold reduction in intracellular growth compared to the wild type strain at 24 h post-infection.
Among the 3,050 mutants tested in the primary screen, we identified 425 mutant alleles with ≥102 fold reduction in the number of cfus recovered at 24h post-infection compared to the wild type strain. Since all the FPI genes have been reported to be involved in intracellular proliferation (Barker et al., 2009; Pechous et al., 2009; Santic et al., 2010), we focused our screen on the non-FPI genes. To confirm the phenotype of the primary screen, growth kinetics of the non-FPI mutants was re-examined twice. When the defect was re-examined, the OD of all the 425 mutants was determined after overnight culture in broth, and equivalent OD for all the mutant bacteria was used for infection to ensure equivalent input for all the 425 mutants. Our data confirmed that 271 mutant alleles corresponding to 202 genes showed a consistent ≥102 fold reduction in the number of cfus recovered at 24h post-infection compared to the wild type strain (Table 1). Remarkably, defect in at least 125 genes caused ≥103 fold reduction in the number of cfus recovered at 24h post-infection compared to the wild type strain, and these were selected for further analyses of phagosome biogenesis and bacterial escape into the cytosol (see below).
Table 1.
List of Growth-defective mutants of F. tularensis in U937 macrophages and S2 cells grouped according to function.
| List of growth defective mutants in both U937 and S2 Cells | |||||
|---|---|---|---|---|---|
| Strain Name | Locus Tag | Gene | Description | Log reduction in Growth relative to WT | |
| U937 | S2 | ||||
| Controls | |||||
| Wild type | 0 | 0 | |||
| Intracellular growth locus C | IglC | 5 | 5 | ||
| Proteins of unknown Function | |||||
| tnfn1_pw060323p08q148 | FTN_0027 | conserved protein of unknown function | 4 | 6* | |
| tnfn1_pw060510p03q161 | FTN_0027 | conserved protein of unknown function | 2 | 2* | |
| tnfn1_pw060323p03q103 | FTN_0041 | protein of unknown function | 5 | 2# | |
| tnfn1_pw060420p01q149 | FTN_0041 | protein of unknown function | 2 | 3# | |
| tnfn1_pw060420p04q143 | FTN_0149 | conserved protein of unknown function | 5 | 5 | |
| tnfn1_pw060323p02q193 | FTN_0275 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060419p03q124 | FTN_0275 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060510p02q121 | FTN_0275 | conserved protein of unknown function | 3 | 2# | |
| tnfn1_pw060420p04q134 | FTN_0297 | conserved protein of unknown function | 7 | 7 | |
| tnfn1_pw060328p05q119 | FTN_0444 | membrane protein of unknown function | 6 | 6# | |
| tnfn1_pw060420p03q175 | FTN_0444 | membrane protein of unknown function | 5 | 5# | |
| tnfn1_pw060323p07q141 | FTN_0788 | conserved protein of unknown function | 5 | 5 | |
| tnfn1_pw060420p04q176 | FTN_0855 | protein of unknown function | 5 | 2 | |
| tnfn1_pw060323p03q147 | FTN_0930 | protein of unknown function | 6 | 3# | |
| tnfn1_pw060323p05q150 | FTN_0930 | protein of unknown function | 6 | 3# | |
| tnfn1_pw060510p01q108 | FTN_0977 | conserved protein of unknown function | 7 | 7 | |
| tnfn1_pw060510p01q128 | FTN_1170 | conserved protein of unknown function | 2 | 3* | |
| tnfn1_pw060418p02q157 | FTN_1170 | conserved protein of unknown function | 2 | 4* | |
| tnfn1_pw060420p04q196 | FTN_1256 | membrane protein of unknown function | 4 | 5 | |
| tnfn1_pw060323p01q113 | FTN_1343 | conserved protein of unknown function | 4 | 4# | |
| tnfn1_pw060418p02q105 | FTN_1343 | conserved protein of unknown function | 4 | 4# | |
| tnfn1_pw060328p02q110 | FTN_1457 | protein of unknown function | 5 | 5# | |
| tnfn1_pw060420p02q183 | FTN_1457 | protein of unknown function | 6 | 6# | |
| tnfn1_pw060328p01q172 | FTN_1542 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060328p02q177 | FTN_1713 | protein of unknown function | 2 | 2 | |
| tnfn1_pw060328p06q155 | FTN_1764 | protein of unknown function | 6 | 7# | |
| Hypothetical Proteins | |||||
| tnfn1_pw060323p03q142 | FTN_0030 | hypothetical membrane protein | 4 | 3# | |
| tnfn1_pw060420p02q155 | FTN_0030 | hypothetical membrane protein | 3 | 3# | |
| tnfn1_pw060328p06q180 | FTN_0038 | hypothetical protein | 4 | 4# | |
| tnfn1_pw060419p02q127 | FTN_0038 | hypothetical protein | 2 | 2# | |
| tnfn1_pw060420p02q173 | FTN_0169 | conserved hypothetical membrane protein | 6 | 6* | |
| tnfn1_pw060510p01q193 | FTN_0169 | conserved hypothetical membrane protein | 5 | 5* | |
| tnfn1_pw060328p05q136 | FTN_0384 | conserved hypothetical protein | 4 | 7 | |
| tnfn1_pw060328p05q130 | FTN_0534 | conserved hypothetical membrane protein | 5 | 7 | |
| tnfn1_pw060418p01q143 | FTN_0556 | hypothetical protein | 7 | 7 | |
| tnfn1_pw060419p03q188 | FTN_0696 | hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060323p01q155 | FTN_0696 | hypothetical membrane protein | 5 | 3# | |
| tnfn1_pw060328p06q185 | FTN_0709 | hypothetical protein | 2 | 7 | |
| tnfn1_pw060323p07q129 | FTN_0759 | conserved hypothetical protein | 4 | 2 | |
| tnfn1_pw060419p02q102 | FTN_0792 | hypothetical protein | 5 | 6# | |
| tnfn1_pw060420p01q167 | FTN_0792 | hypothetical protein | 2 | 2# | |
| tnfn1_pw060323p02q140 | FTN_0895 | hypothetical protein | 2 | 2* | |
| tnfn1_pw060323p07q105 | FTN_0895 | hypothetical protein | 4 | 2* | |
| tnfn1_pw060328p08q188 | FTN_1098 | conserved hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060510p03q192 | FTN_1098 | conserved hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060510p04q192 | FTN_1098 | conserved hypothetical membrane protein | 7 | 6# | |
| tnfn1_pw060419p04q117 | FTN_1156 | hypothetical protein | 2 | 4 | |
| tnfn1_pw060328p02q129 | FTN_1612 | hypothetical protein | 2 | 2 | |
| Metabolic Proteins | |||||
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain | 5 | 7 |
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | riboflavin synthase beta-chain | 4 | 5 |
| tnfn1_pw060328p06q174 | FTN_0125 | ackA | propionate kinase 2/acetate kinase A | 4 | 4# |
| tnfn1_pw060418p03q133 | FTN_0199 | cyoE | heme O synthase | 2 | 4 |
| tnfn1_pw060323p04q102 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase | 1 | 1# |
| tnfn1_pw060418p03q177 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase | 3 | 4# |
| tnfn1_pw060418p01q187 | FTN_0319 | amino acid-polyamine-organocation family protein | 6 | 7 | |
| tnfn1_pw060323p06q113 | FTN_0420 | SAICAR synthetase/phosphoribosylamine-glycine ligase | 7 | 5 | |
| tnfn1_pw060323p05q182 | FTN_0504 | lysine decarboxylase | 4 | 4 | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 | 5 | 7 |
| tnfn1_pw060510p02q154 | FTN_0511 | shikimate 5-dehydrogenase | 2 | 2# | |
| tnfn1_pw060510p02q157 | FTN_0511 | shikimate 5-dehydrogenase | 6 | 6# | |
| tnfn1_pw060510p04q157 | FTN_0511 | shikimate 5-dehydrogenase | 3 | 2# | |
| tnfn1_pw060323p06q194 | FTN_0527 | thrC | threonine synthase | 7 | 7# |
| tnfn1_pw060510p01q172 | FTN_0527 | thrC | threonine synthase | 5 | 5# |
| tnfn1_pw060510p03q172 | FTN_0527 | thrC | threonine synthase | 2 | 2# |
| tnfn1_pw060323p03q127 | FTN_0567 | tRNA synthetase class II (D, K and N) | 5 | 2 | |
| tnfn1_pw060510p03q171 | FTN_0588 | asparaginase | 2 | 2 | |
| tnfn1_pw060419p03q116 | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit | 2 | 2 |
| tnfn1_pw060418p02q128 | FTN_0633 | katG | peroxidase/catalase | 7 | 7 |
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | quinolinate sythetase A | 3 | 2# |
| tnfn1_pw060419p04q164 | FTN_0692 | nadA | quinolinate sythetase A | 2 | 2# |
| tnfn1_pw060510p01q159 | FTN_0695 | add | deoxyadenosine deaminase/adenosine deaminase | 3 | 7 |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | biotin--acetyl-CoA-carboxylase ligase | 6 | 7 |
| tnfn1_pw060328p01q128 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) | 5 | 5 |
| tnfn1_pw060420p02q175 | FTN_0877 | cls | cardiolipin synthetase | 7 | 5 |
| tnfn1_pw060328p06q142 | FTN_0954 | histidine acid phosphatase | 4 | 4 | |
| tnfn1_pw060420p01q130 | FTN_0965 | metal-dependent exopeptidase | 3 | 3 | |
| tnfn1_pw060328p01q151 | FTN_0983 | bifunctional protein: glutaredoxin 3/ribonucleotide reductase beta subunit | 5 | 3# | |
| tnfn1_pw060328p06q189 | FTN_0995 | hslV | ATP-dependent protease HslVU, peptidase subunit | 2 | 2# |
| tnfn1_pw060420p04q195 | FTN_0995 | hslV | ATP-dependent protease HslVU, peptidase subunit | 2 | 2# |
| tnfn1_pw060510p02q187 | FTN_1018 | aldolase/adducin class II family protein | 3 | 3 | |
| tnfn1_pw060323p02q168 | FTN_1046 | wzb | low molecular weight (LMW) phosphotyrosine protein phosphatase | 2 | 2 |
| tnfn1_pw060328p06q184 | FTN_1061 | acid phosphatase, HAD superfamily protein | 2 | 2# | |
| tnfn1_pw060420p02q103 | FTN_1061 | acid phosphatase, HAD superfamily protein | 3 | 3# | |
| tnfn1_pw060510p04q113 | FTN_1121 | phrB | deoxyribodipyrimidine photolyase | 5 | 7 |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase | 6 | 6 |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase | 3 | 4# |
| tnfn1_pw060328p03q107 | FTN_1222 | kpsF | phosphosugar isomerase | 4 | 3 |
| tnfn1_pw060510p02q164 | FTN_1231 | gloA | lactoylglutathione lyase | 4 | 4* |
| tnfn1_pw060420p04q194 | FTN_1231 | gloA | lactoylglutathione lyase | 3 | 5* |
| tnfn1_pw060510p04q146 | FTN_1231 | gloA | lactoylglutathione lyase | 2 | 2* |
| tnfn1_pw060510p01q142 | FTN_1333 | tktA | transketolase I | 5 | 5 |
| tnfn1_pw060418p02q109 | FTN_1376 | disulfide bond formation protein, DsbB family | 4 | 4 | |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase | 4 | 7 |
| tnfn1_pw060419p01q104 | FTN_1523 | amino acid-polyamine-organocation family protein | 4 | 4# | |
| tnfn1_pw060328p02q165 | FTN_1523 | amino acid-polyamine-organocation family protein | 4 | 5# | |
| tnfn1_pw060419p02q191 | FTN_1523 | amino acid-polyamine-organocation family protein | 2 | 2# | |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase | 5 | 5# |
| tnfn1_pw060418p01q131 | FTN_1557 | oxidoreductase iron/ascorbate family protein | 7 | 7 | |
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | glycerol-3-phosphate dehydrogenase | 3 | 5 |
| tnfn1_pw060419p04q130 | FTN_1585 | glpK | glycerol kinase | 3 | 3 |
| tnfn1_pw060510p01q146 | FTN_1597 | prfC | peptide chain release factor 3 | 5 | 5 |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | cytochrome bd-II terminal oxidase subunit I | 5 | 7 |
| tnfn1_pw060328p02q105 | FTN_1620 | appB | cytochrome bd-II terminal oxidase subunit II | 6 | 3 |
| tnfn1_pw060418p04q111 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 3 | 3# | |
| tnfn1_pw060418p04q112 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 2 | 2# | |
| tnfn1_pw060420p04q169 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 4 | 4# | |
| tnfn1_pw060323p04q160 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C | 7 | 7# |
| tnfn1_pw060510p02q165 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C | 2 | 2# |
| Transporter Proteins | |||||
| tnfn1_pw060420p04q149 | FTN_0008 | 10 TMS drug/metabolite exporter protein | 4 | 4# | |
| tnfn1_pw060420p02q151 | FTN_0018 | sdaC | serine permease | 2 | 4 |
| tnfn1_pw060418p04q168 | FTN_0141 | ABC transporter, ATP-binding protein | 5 | 6# | |
| tnfn1_pw060418p03q147 | FTN_0299 | putP | proline:Na+ symporter | 2 | 2# |
| tnfn1_pw060510p02q139 | FTN_0299 | putP | proline:Na+ symporter | 2 | 2# |
| tnfn1_pw060323p03q141 | FTN_0619 | pseudogene: nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 3 | 3* | |
| tnfn1_pw060328p06q129 | FTN_0619 | pseudogene: nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | 5* | |
| tnfn1_pw060510p02q156 | FTN_0624 | serine permease | 2 | 2* | |
| tnfn1_pw060323p06q164 | FTN_0624 | serine permease | 2 | 2* | |
| tnfn1_pw060418p01q161 | FTN_0636 | glpT | glycerol-3-phosphate transporter | 7 | 7 |
| tnfn1_pw060419p04q142 | FTN_0687 | galP1 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 2 | 3* |
| tnfn1_pw060510p04q158 | FTN_0687 | galP1 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 2 | 2* |
| tnfn1_pw060328p06q132 | FTN_0728 | predicted Co/Zn/Cd cation transporter | 2 | 5 | |
| tnfn1_pw060418p03q103 | FTN_0739 | potG | ATP-binding cassette putrescine uptake system, ATP-binding protein | 2 | 2# |
| tnfn1_pw060328p08q153 | FTN_0739 | potG | ATP-binding cassette putrescine uptake system, ATP-binding protein | 2 | 5# |
| tnfn1_pw060510p04q103 | FTN_0799 | emrE | putative membrane transporter of cations and cationic drugs, multidrug resistance protein | 2 | 2 |
| tnfn1_pw060323p01q177 | FTN_0799 | emrE | putative membrane transporter of cations and cationic drugs, multidrug resistance protein | 4 | 3 |
| tnfn1_pw060328p04q109 | FTN_0885 | proton-dependent oligopeptide transporter (POT) family protein, di-or tripeptide:H+ symporter | 5 | 2 | |
| tnfn1_pw060328p04q167 | FTN_0997 | proton-dependent oligopeptide transporter (POT) family protein, di-or tripeptide:H+ symporter | 5 | 3 | |
| tnfn1_pw060323p05q110 | FTN_1215 | kpsC | capsule polysaccharide export protein KpsC | 2 | 5 |
| tnfn1_pw060323p07q172 | FTN_1344 | major facilitator superfamily (MFS) transport protein | 4 | 4* | |
| tnfn1_pw060420p04q148 | FTN_1344 | major facilitator superfamily (MFS) transport protein | 5 | 5* | |
| tnfn1_pw060323p01q175 | FTN_1441 | sugar porter (SP) family protein | 4 | 4# | |
| tnfn1_pw060420p02q182 | FTN_1441 | sugar porter (SP) family protein | 6 | 6# | |
| tnfn1_pw060419p02q126 | FTN_1581 | small conductance mechanosensitive ion channel (MscS) family protein | 3 | 3 | |
| tnfn1_pw060323p03q106 | FTN_1593 | oppA | ABC-type oligopeptide transport system, periplasmic component | 2 | 2* |
| tnfn1_pw060420p03q104 | FTN_1593 | oppA | ABC-type oligopeptide transport system, periplasmic component | 4 | 6* |
| tnfn1_pw060420p01q189 | FTN_1611 | major facilitator superfamily (MFS) transport protein | 7 | 5 | |
| tnfn1_pw060328p02q121 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2 | 1* |
| tnfn1_pw060420p02q159 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2 | 2* |
| tnfn1_pw060418p03q187 | FTN_1733 | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | 4 | |
| Transferases | |||||
| tnfn1_pw060323p02q177 | FTN_0019 | pyrB | aspartate carbamoyltransferase | 2 | 2# |
| tnfn1_pw060323p03q119 | FTN_0019 | pyrB | aspartate carbamoyltransferase | 2 | 2# |
| tnfn1_pw060510p01q103 | FTN_0063 | ilvE | branched-chain amino acid aminotransferase protein (class IV) | 3 | 5 |
| tnfn1_pw060323p03q121 | FTN_0343 | aminotransferase | 7 | 2 | |
| tnfn1_pw060328p03q179 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 4 | 4* | |
| tnfn1_pw060419p01q169 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 2 | 2* | |
| tnfn1_pw060323p06q168 | FTN_0545 | glycosyl transferase, group 2 | 4 | 4# | |
| tnfn1_pw060419p01q187 | FTN_0545 | glycosyl transferase, group 2 | 5 | 5# | |
| tnfn1_pw060328p01q142 | FTN_0928 | cysD | sulfate adenylyltransferase subunit 2 | 3 | 3# |
| tnfn1_pw060323p03q182 | FTN_1428 | wbtO | transferase | 3 | 2# |
| tnfn1_pw060510p01q119 | FTN_1428 | wbtO | transferase | 2 | 6# |
| DNA modifying | |||||
| tnfn1_pw060323p03q125 | FTN_0133 | ribonuclease II family protein | 2 | 2 | |
| tnfn1_pw060510p02q141 | FTN_0133 | ribonuclease II family protein | 5 | 5 | |
| tnfn1_pw060323p03q122 | FTN_0577 | mutL | DNA mismatch repair enzyme with ATPase activity | 7 | 6# |
| tnfn1_pw060510p01q148 | FTN_0577 | mutL | DNA mismatch repair enzyme with ATPase activity | 5 | 5# |
| tnfn1_pw060510p04q193 | FTN_0680 | uvrC | excinuclease ABC, subunit C | 6 | 3 |
| tnfn1_pw060328p04q156 | FTN_1027 | ruvC | holliday junction endodeoxyribonuclease | 3 | 4# |
| tnfn1_pw060510p01q132 | FTN_1027 | holliday junction endodeoxyribonuclease | 3 | 3# | |
| tnfn1_pw060510p01q114 | FTN_1073 | DNA/RNA endonuclease G | 5 | 6* | |
| tnfn1_pw060510p02q114 | FTN_1073 | DNA/RNA endonuclease G | 2 | 2* | |
| tnfn1_pw060510p01q153 | FTN_1154 | type I restriction-modification system, subunit S | 5 | 6 | |
| tnfn1_pw060323p03q167 | FTN_1197 | recR | RecFOR complex, RecR component | 2 | 4# |
| tnfn1_pw060510p02q106 | FTN_1197 | recR | RecFOR complex, RecR component | 3 | 3# |
| tnfn1_pw060328p06q158 | FTN_1293 | rnhB | ribonuclease HII | 2 | 5 |
| tnfn1_pw060323p07q175 | FTN_1487 | restriction endonuclease | 3 | 6 | |
| Cell Division | |||||
| tnfn1_pw060328p03q149 | FTN_0162 | ftsQ | cell division protein FtsQ | 2 | 2# |
| tnfn1_pw060328p01q167 | FTN_0330 | minD | septum formation inhibitor-activating ATPase | 2 | 2 |
| Type IV Pilin | |||||
| tnfn1_pw060323p03q109 | FTN_1137 | pilQ | Type IV pili secretin component | 2 | 2 |
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | Type IV pili secretin component | 4 | 4 |
| tnfn1_pw060323p06q157 | FTN_1139 | pilO | Type IV pili glycosylation protein | 2 | 2 |
| Others | |||||
| tnfn1_pw060323p06q138 | FTN_0107 | lepA | GTP-binding protein LepA | 2 | 4# |
| tnfn1_pw060418p02q123 | FTN_0107 | lepA | GTP-binding protein LepA | 2 | 4# |
| tnfn1_pw060420p04q150 | FTN_0155 | competence protein | 2 | 7* | |
| tnfn1_pw060510p04q189 | FTN_0155 | competence protein | 6 | 3* | |
| tnfn1_pw060418p04q181 | FTN_0338 | MutT/nudix family protein | 2 | 2 | |
| tnfn1_pw060328p06q137 | FTN_0465 | Sua5/YciO/YrdC family protein | 2 | 2# | |
| tnfn1_pw060323p03q111 | FTN_0465 | Sua5/YciO/YrdC family protein | 2 | 2# | |
| tnfn1_pw060323p06q115 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060420p03q193 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060510p01q120 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060328p06q167 | FTN_0985 | DJ-1/PfpI family protein | 6 | 6# | |
| tnfn1_pw060328p06q167 | FTN_0985 | DJ-1/PfpI family protein | 5 | 5# | |
| tnfn1_pw060420p04q127 | FTN_1031 | ftnA | ferric iron binding protein, ferritin-like | 2 | 6 |
| tnfn1_pw060419p02q137 | FTN_1034 | rnfB | iron-sulfur cluster-binding protein | 2 | 3 |
| tnfn1_pw060420p03q121 | FTN_1064 | PhoH family protein, putative ATPase | 2 | 4 | |
| tnfn1_pw060328p06q178 | FTN_1241 | DedA family protein | 4 | 5 | |
| tnfn1_pw060418p01q185 | FTN_1355 | regulatory factor, Bvg accessory factor family | 6 | 7 | |
| tnfn1_pw060328p03q154 | FTN_1453 | two-component regulator, sensor histidine kinase | 2 | 2 | |
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 2 | 2* |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 4 | 4* |
| Intergenic | |||||
| tnfn1_pw060323p03q164 | intergenic | 3 | 2 | ||
| tnfn1_pw060328p06q190 | intergenic | 3 | 3 | ||
| tnfn1_pw060419p03q131 | intergenic | 2 | 2 | ||
| tnfn1_pw060419p04q189 | intergenic | 5 | 3 | ||
| tnfn1_pw060323p08q139 | intergenic | 4 | 4 | ||
| List of growth defective mutants in only U937 Cells | |||||
| Proteins of unknown function | |||||
| tnfn1_pw060328p06q147 | FTN_0109 | protein of unknown function | 3# | ||
| tnfn1_pw060418p04q193 | FTN_0109 | protein of unknown function | 4# | ||
| tnfn1_pw060510p01q123 | FTN_0132 | protein of unknown function | 2 | ||
| tnfn1_pw060323p07q115 | FTN_0290 | protein of unknown function | 5 | ||
| tnfn1_pw060328p04q122 | FTN_0428 | protein of unknown function | 2* | ||
| tnfn1_pw060510p04q109 | FTN_0428 | protein of unknown function | 2* | ||
| tnfn1_pw060419p03q140 | FTN_0477 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060420p02q178 | FTN_0915 | conserved protein of unknown function | 7 | ||
| tnfn1_pw060419p04q188 | FTN_0925 | protein of unknown function | 4 | ||
| tnfn1_pw060420p02q181 | FTN_0933 | protein of unknown function | 7 | ||
| tnfn1_pw060419p04q118 | FTN_1172 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060420p01q127 | FTN_1175 | membrane protein of unknown function | 4 | ||
| tnfn1_pw060420p01q109 | FTN_1367 | protein of unknown function | 2 | ||
| tnfn1_pw060420p01q132 | FTN_1624 | conserved protein of unknown function | 4 | ||
| tnfn1_pw060420p02q184 | FTN_1696 | protein of unknown function | 7 | ||
| Hypothetical Proteins | |||||
| tnfn1_pw060323p01q181 | FTN_0336 | hypothetical protein | 3 | ||
| tnfn1_pw060510p01q147 | FTN_0403 | hypothetical membrane protein | 4 | ||
| tnfn1_pw060323p01q163 | FTN_0727 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060418p03q110 | FTN_0847 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060510p02q108 | FTN_0847 | conserved hypothetical protein | 4# | ||
| tnfn1_pw060419p02q152 | FTN_0888 | hypothetical membrane protein | 2 | ||
| tnfn1_pw060418p01q191 | FTN_1349 | hypothetical protein | 4# | ||
| tnfn1_pw060328p06q182 | FTN_1395 | conserved hypothetical protein | 4 | ||
| tnfn1_pw060328p04q136 | FTN_1406 | conserved hypothetical membrane protein | 4 | ||
| tnfn1_pw060420p02q127 | FTN_1656 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060420p02q176 | FTN_1686 | hypothetical membrane protein | 5 | ||
| tnfn1_pw060418p03q159 | FTN_1736 | hypothetical protein | 2 | ||
| Metabolic Proteins | |||||
| tnfn1_pw060419p02q150 | FTN_0090 | acpA | acid phosphatase | 5 | |
| tnfn1_pw060419p03q169 | FTN_0218 | nfnB | dihydropteridine reductase | 2 | |
| tnfn1_pw060420p01q123 | FTN_0524 | asd | aspartate semialdehyde dehydrogenase | 5 | |
| tnfn1_pw060323p06q168 | FTN_0545 | glycosyl transferase, group 2 | 5# | ||
| tnfn1_pw060419p01q187 | FTN_0545 | glycosyl transferase, group 2 | 5# | ||
| tnfn1_pw060510p03q168 | FTN_0598 | tRNA-dihydrouridine synthase | 3 | ||
| tnfn1_pw060328p04q196 | FTN_0746 | alr | alanine racemase | 6# | |
| tnfn1_pw060420p04q108 | FTN_0822 | para-aminobenzoate synthase component I | 5 | ||
| tnfn1_pw060420p04q140 | FTN_0957 | short chain dehydrogenase | 4 | ||
| tnfn1_pw060420p02q174 | FTN_1233 | haloacid dehalogenase-like hydrolase | 6 | ||
| tnfn1_pw060420p04q116 | FTN_1421 | wbtH | glutamine amidotransferase/asparagine synthase | 3 | |
| tnfn1_pw060419p04q135 | FTN_1415 | thioredoxin | 6 | ||
| tnfn1_pw060510p04q185 | FTN_1701 | glutamate decarboxylase | 3 | ||
| tnfn1_pw060510p04q136 | FTN_1767 | rbsK | ribokinase, pfkB family | 3 | |
| tnfn1_pw060328p05q154 | FTN_1777 | trpG | anthranilate synthase component II | 2# | |
| Transporter Proteins | |||||
| tnfn1_pw060420p04q158 | FTN_0800 | ArsB arsenite/antimonite exporter | 2 | ||
| tnfn1_pw060510p01q152 | FTN_1711 | tyrP | tyrosine permease | 6 | |
| DNA Modification | |||||
| tnfn1_pw060419p04q116 | FTN_0287 | type I restriction-modification system, subunit R (restriction) | 2 | ||
| tnfn1_pw060420p03q134 | FTN_0710 | type I restriction-modification system, subunit R (restriction) | 4 | ||
| tnfn1_pw060510p04q179 | FTN_0838 | xthA | exodeoxyribonuclease III | 3 | |
| tnfn1_pw060419p04q152 | FTN_1017 | pseudogene: DNA-3-methyladenine glycosylase | 5 | ||
| tnfn1_pw060323p04q111 | FTN_1176 | uvrB | excinuclease ABC, subunit B | 2 | |
| Transferases | |||||
| tnfn1_pw060420p02q180 | FTN_0483 | bifunctional NMN adenylyltransferase/Nudix hydrolase | 7 | ||
| tnfn1_pw060510p01q158 | FTN_0988 | prmA | 50S ribosomal protein L11, methyltransferase | 7 | |
| tnfn1_pw060510p02q144 | FTN_1234 | queA | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | 6 | |
| Transcription/Translation | |||||
| tnfn1_pw060323p03q127 | FTN_0567 | tRNA synthetase class II (D, K and N) | 2 | ||
| tnfn1_pw060510p03q168 | FTN_0598 | tRNA-dihydrouridine synthase | 3 | ||
| tnfn1_pw060419p04q129 | FTN_1290 | mglA | macrophage growth locus, protein A | 3# | |
| Others | |||||
| tnfn1_pw060328p08q161 | - | isftu1 | isftu1 | 2 | |
| tnfn1_pw060510p04q176 | FTN_0182 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060323p08q110 | FTN_0286 | transposase | 3 | ||
| tnfn1_pw060420p01q168 | FTN_0646 | cscK | ROK family protein | 5 | |
| tnfn1_pw060328p04q123 | FTN_0672 | secA | preprotein translocase, subunit A (ATPase, RNA helicase) | 2 | |
| tnfn1_pw060328p04q112 | FTN_1002 | blaA | beta-lactamase class A | 2# | |
| tnfn1_pw060419p02q192 | FTN_1002 | blaA | beta-lactamase class A | 2# | |
| tnfn1_pw060420p02q177 | FTN_1145 | era | GTP-binding protein | 6 | |
| tnfn1_pw060418p03q107 | FTN_1217 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060328p06q171 | FTN_1263 | comL | competence lipoprotein ComL | 2# | |
| tnfn1_pw060420p02q179 | FTN_1263 | comL | competence lipoprotein ComL | 7# | |
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 2* | |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 4* | |
| Intergenic | |||||
| tnfn1_pw060328p03q108 | intergenic | 2 | |||
| tnfn1_pw060419p04q165 | intergenic | 5 | |||
| tnfn1_pw060510p01q102 | intergenic | 5 | |||
| tnfn1_pw060510p01q112 | intergenic | 4 | |||
| tnfn1_pw060510p01q135 | intergenic | 4 | |||
| List of growth defective mutants in only S2 cells according to their functions | |||||
| Proteins of unknown Function | |||||
| tnfn1_pw060419p01q176 | FTN_0043 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060418p01q155 | FTN_0044 | protein of unknown function | 3 | ||
| tnfn1_pw060418p02q158 | FTN_0050 | protein of unknown function | 4 | ||
| tnfn1_pw060328p08q104 | FTN_0051 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060420p01q142 | FTN_0052 | protein of unknown function | 2 | ||
| tnfn1_pw060419p04q191 | FTN_0077 | protein of unknown function | 3# | ||
| tnfn1_pw060323p06q122 | FTN_0077 | protein of unknown function | 2# | ||
| tnfn1_pw060510p04q143 | FTN_0099 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060418p04q193 | FTN_0109 | protein of unknown function | 4 | ||
| tnfn1_pw060418p04q117 | FTN_0207 | protein of unknown function containing a von Willebrand factor type A (vWA) domain | 2 | ||
| tnfn1_pw060328p04q119 | FTN_0325 | membrane protein of unknown function | 2 | ||
| tnfn1_pw060328p08q156 | FTN_0340 | protein of unknown function | 2 | ||
| tnfn1_pw060323p03q157 | FTN_0364 | conserved protein of unknown function | 2* | ||
| tnfn1_pw060418p04q136 | FTN_0364 | conserved protein of unknown function | 3* | ||
| tnfn1_pw060328p08q149 | FTN_0439 | protein of unknown function | 4# | ||
| tnfn1_pw060418p01q142 | FTN_0482 | protein of unknown function | 6 | ||
| tnfn1_pw060328p04q110 | FTN_0573 | protein of unknown function | 2 | ||
| tnfn1_pw060418p02q126 | FTN_0573 | protein of unknown function | 4 | ||
| tnfn1_pw060328p08q173 | FTN_0584 | araJ | conserved inner membrane protein of unknown function | 5 | |
| tnfn1_pw060510p04q147 | FTN_0599 | protein of unknown function | 2# | ||
| tnfn1_pw060328p06q173 | FTN_0599 | protein of unknown function | 2# | ||
| tnfn1_pw060510p01q183 | FTN_0782 | protein of unknown function | 5 | ||
| tnfn1_pw060323p03q129 | FTN_0786 | protein of unknown function | 7# | ||
| tnfn1_pw060323p05q127 | FTN_0791 | protein of unknown function | 3# | ||
| tnfn1_pw060419p03q107 | FTN_0791 | protein of unknown function | 2# | ||
| tnfn1_pw060418p01q141 | FTN_0817 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060328p05q126 | FTN_0828 | protein of unknown function | 5 | ||
| tnfn1_pw060510p04q111 | FTN_0861 | conserved protein of unknown function | 4 | ||
| tnfn1_pw060418p04q148 | FTN_0878 | protein of unknown function | 2 | ||
| tnfn1_pw060328p02q106 | FTN_0884 | drug/metabolite transporter superfamily protein | 2# | ||
| tnfn1_pw060328p03q163 | FTN_0884 | drug/metabolite transporter superfamily protein | 4# | ||
| tnfn1_pw060323p03q150 | FTN_0900 | protein of unknown function with predicted hydrolase and phosphorylase activity | 2# | ||
| tnfn1_pw060418p03q108 | FTN_0900 | protein of unknown function with predicted hydrolase and phosphorylase activity | 6# | ||
| tnfn1_pw060323p04q104 | FTN_0918 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060418p02q131 | FTN_0918 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060419p04q188 | FTN_0925 | protein of unknown function | 5 | ||
| tnfn1_pw060419p04q179 | FTN_1001 | protein of unknown function | 2# | ||
| tnfn1_pw060323p07q181 | FTN_1001 | protein of unknown function | 3# | ||
| tnfn1_pw060418p02q145 | FTN_1020 | conserved protein of unknown function | 5 | ||
| tnfn1_pw060419p01q172 | FTN_1044 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060420p01q111 | FTN_1053 | outer membrane protein of unknown function | 3 | ||
| tnfn1_pw060420p02q158 | FTN_1071 | protein of unknown function | 5 | ||
| tnfn1_pw060418p02q133 | FTN_1093 | protein of unknown function | 5 | ||
| tnfn1_pw060420p01q134 | FTN_1103 | protein of unknown function | 2# | ||
| tnfn1_pw060328p01q140 | FTN_1103 | protein of unknown function | 3# | ||
| tnfn1_pw060323p08q143 | FTN_1235 | protein of unknown function | 2 | ||
| tnfn1_pw060510p03q135 | FTN_1254 | protein of unknown function | 4 | ||
| tnfn1_pw060323p03q102 | FTN_1257 | membrane protein of unknown function | 3# | ||
| tnfn1_pw060419p03q150 | FTN_1257 | membrane protein of unknown function | 4# | ||
| tnfn1_pw060418p04q121 | FTN_1261 | protein of unknown function | 2 | ||
| tnfn1_pw060323p08q134 | FTN_1270 | conserved membrane protein of unknown function | 2 | ||
| tnfn1_pw060418p01q149 | FTN_1298 | GTPase of unknown function | 7 | ||
| tnfn1_pw060419p01q143 | FTN_1334 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060328p08q108 | FTN_1334 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060328p05q124 | FTN_1372 | protein of unknown function | 5 | ||
| tnfn1_pw060323p04q183 | FTN_1386 | protein of unknown function | 3 | ||
| tnfn1_pw060328p01q156 | FTN_1442 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060420p01q165 | FTN_1442 | conserved protein of unknown function | 4# | ||
| tnfn1_pw060418p02q186 | FTN_1448 | protein of unknown function | 3 | ||
| tnfn1_pw060328p02q116 | FTN_1449 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060419p03q173 | FTN_1449 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060323p07q176 | FTN_1534 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060328p02q177 | FTN_1713 | protein of unknown function | 3 | ||
| tnfn1_pw060328p05q185 | FTN_1734 | protein of unknown function | 5 | ||
| tnfn1_pw060328p08q107 | FTN_1774 | protein of unknown function | 3 | ||
| Hypothetical Protein | |||||
| tnfn1_pw060418p04q139 | FTN_0011 | hypothetical protein | 2# | ||
| tnfn1_pw060420p02q108 | FTN_0012 | hypothetical protein | 2 | ||
| tnfn1_pw060420p02q139 | FTN_0013 | hypothetical protein | 3 | ||
| tnfn1_pw060328p01q141 | FTN_0014 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060419p04q178 | FTN_0028 | conserved hypothetical membrane protein | 3# | ||
| tnfn1_pw060323p04q145 | FTN_0028 | conserved hypothetical membrane protein | 2# | ||
| tnfn1_pw060418p04q143 | FTN_0053 | hypothetical protein | 2 | ||
| tnfn1_pw060328p06q157 | FTN_0170 | conserved hypothetical membrane protein | 5 | ||
| tnfn1_pw060418p03q151 | FTN_0212 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060323p08q114 | FTN_0326 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060328p05q165 | FTN_0360 | hypothetical protein | 5# | ||
| tnfn1_pw060419p01q145 | FTN_0368 | hypothetical protein | 2 | ||
| tnfn1_pw060419p03q186 | FTN_0375 | hypothetical protein | 3 | ||
| tnfn1_pw060420p02q163 | FTN_0398 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060420p04q104 | FTN_0466 | conserved hypothetical protein | 4 | ||
| tnfn1_pw060328p08q148 | FTN_0548 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060418p04q176 | FTN_0548 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060328p06q164 | FTN_0630 | hypothetical protein | 5 | ||
| tnfn1_pw060328p05q141 | FTN_0701 | conserved hypothetical protein | 5 | ||
| tnfn1_pw060418p02q152 | FTN_0706 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060418p02q175 | FTN_0717 | conserved hypothetical membrane protein | 5 | ||
| tnfn1_pw060328p06q126 | FTN_0732 | hypothetical protein | 5 | ||
| tnfn1_pw060323p07q129 | FTN_0759 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060323p04q134 | FTN_0938 | hypothetical protein | 2# | ||
| tnfn1_pw060418p02q170 | FTN_0938 | hypothetical protein | 4# | ||
| tnfn1_pw060419p03q187 | FTN_1123 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060418p04q105 | FTN_1180 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060420p04q159 | FTN_1223 | conserved hypothetical membrane protein | 7 | ||
| tnfn1_pw060323p08q166 | FTN_1232 | conserved hypothetical membrane protein | 2 | ||
| tnfn1_pw060328p03q180 | FTN_1260 | hypothetical membrane protein | 2 | ||
| tnfn1_pw060510p01q184 | FTN_1299 | hypothetical protein | 5 | ||
| tnfn1_pw060419p04q127 | FTN_1342 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060328p05q157 | FTN_1379 | pseudogene: hypothetical membrane protein, fragment | 5 | ||
| tnfn1_pw060323p06q178 | FTN_1389 | conserved hypothetical membrane protein | 3# | ||
| tnfn1_pw060420p01q172 | FTN_1389 | conserved hypothetical membrane protein | 2# | ||
| tnfn1_pw060420p01q153 | FTN_1458 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060323p04q147 | FTN_1761 | pseudogene: hypothetical protein, fragment | 3 | ||
| tnfn1_pw060418p04q149 | FTN_1765 | conserved hypothetical protein | 2 | ||
| Metabolic | |||||
| tnfn1_pw060510p02q160 | FTN_0021 | carA | carbamoyl-phosphate synthase small chain | 2 | |
| tnfn1_pw060418p04q115 | FTN_0095 | nitroreductase | 7 | ||
| tnfn1_pw060420p02q191 | FTN_0113 | ribC | riboflavin synthase alpha chain | 6 | |
| tnfn1_pw060328p05q159 | FTN_0118 | serine peptidase, S49 family | 3# | ||
| tnfn1_pw060420p02q187 | FTN_0118 | serine peptidase, S49 family | 5# | ||
| tnfn1_pw060328p06q139 | FTN_0127 | gabD | succinate semialdehyde dehydrogenase (NAD(P)+ dependent) | 5 | |
| tnfn1_pw060510p01q130 | FTN_0154 | rimK | glutathione synthase/ribosomal protein S6 modification enzyme | 3 | |
| tnfn1_pw060328p01q150 | FTN_0168 | lysU | lysyl-tRNA synthetase | 2# | |
| tnfn1_pw060510p02q178 | FTN_0217 | L-lactate dehydrogenase | 2 | ||
| tnfn1_pw060323p07q113 | FTN_0362 | deoxyribodipyrimidine photolyase-related protein | 4 | ||
| tnfn1_pw060323p04q144 | FTN_0406 | sterol desaturase | 3# | ||
| tnfn1_pw060418p01q189 | FTN_0406 | sterol desaturase | 6# | ||
| tnfn1_pw060328p06q134 | FTN_0443 | maeA | NAD-dependent malic enzyme | 5# | |
| tnfn1_pw060328p06q125 | FTN_0496 | slt | soluble lytic murein transglycosylase | 3 | |
| tnfn1_pw060418p04q116 | FTN_0512 | glgX | pullulanase | 4 | |
| tnfn1_pw060510p03q154 | FTN_0516 | glgA | glycogen synthase | 7 | |
| tnfn1_pw060420p01q135 | FTN_0540 | pckA | phosphoenolpyruvate carboxykinase | 2 | |
| tnfn1_pw060419p04q153 | FTN_0597 | protein-disulfide isomerase | 2 | ||
| tnfn1_pw060510p02q110 | FTN_0603 | mutM | formamidopyrimidine-DNA glycosylase | 2 | |
| tnfn1_pw060328p02q139 | FTN_0621 | eno | enolase (2-phosphoglycerate dehydratase) | 2# | |
| tnfn1_pw060510p03q188 | FTN_0627 | chiA | chitinase, glycosyl hydrolase family 18 | 6 | |
| tnfn1_pw060418p01q120 | FTN_0651 | cdd | cytidine deaminase | 5# | |
| tnfn1_pw060419p01q168 | FTN_0651 | cdd | cytidine deaminase | 2# | |
| tnfn1_pw060328p04q151 | FTN_0661 | guaB | IMP dehydrogenase/GMP reductase | 6# | |
| tnfn1_pw060328p06q131 | FTN_0674 | glxK | glycerate kinase | 3 | |
| tnfn1_pw060420p01q148 | FTN_0694 | nadB | L-aspartate oxidase | 4 | |
| tnfn1_pw060323p06q103 | FTN_0711 | predicted metal-dependent hydrolase | 2 | ||
| tnfn1_pw060328p04q116 | FTN_0765 | choloylglycine hydrolase family protein | 2 | ||
| tnfn1_pw060510p03q119 | FTN_0806 | glycosyl hydrolase family 3 | 3 | ||
| tnfn1_pw060323p07q185 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase | 3# | |
| tnfn1_pw060419p02q138 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase | 3# | |
| tnfn1_pw060328p04q175 | FTN_0818 | lipase/esterase | 5 | ||
| tnfn1_pw060418p02q142 | FTN_0826 | aldo/keto reductase family protein | 3 | ||
| tnfn1_pw060328p08q145 | FTN_0907 | D-alanyl-D-alanine carboxypeptidase | 4# | ||
| tnfn1_pw060418p04q131 | FTN_0907 | D-alanyl-D-alanine carboxypeptidase | 4# | ||
| tnfn1_pw060418p04q167 | FTN_0935 | asnB | asparagine synthase | 2 | |
| tnfn1_pw060510p02q145 | FTN_0945 | rsuA | 16S rRNA pseudouridine synthase | 4 | |
| tnfn1_pw060328p08q120 | FTN_0987 | tRNA-dihydrouridine synthase | 3# | ||
| tnfn1_pw060323p08q141 | FTN_1015 | isochorismatase family protein | 3# | ||
| tnfn1_pw060420p01q129 | FTN_1015 | isochorismatase family protein | 2# | ||
| tnfn1_pw060323p05q141 | FTN_1033 | grxB | glutaredoxin 2 | 3# | |
| tnfn1_pw060420p01q193 | FTN_1033 | grxB | glutaredoxin 2 | 4# | |
| tnfn1_pw060418p01q153 | FTN_1055 | lon | DNA-binding, ATP-dependent protease La | 2 | |
| tnfn1_pw060328p06q184 | FTN_1061 | acid phosphatase, HAD superfamily protein | 3# | ||
| tnfn1_pw060420p02q103 | FTN_1061 | acid phosphatase, HAD superfamily protein | 7# | ||
| tnfn1_pw060510p04q113 | FTN_1121 | phrB | deoxyribodipyrimidine photolyase | 6 | |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase | 4 | |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase | 5# | |
| tnfn1_pw060328p08q131 | FTN_1174 | murI | glutamate racemase | 2# | |
| tnfn1_pw060419p03q164 | FTN_1186 | pepO | M13 family metallopeptidase | 7 | |
| tnfn1_pw060418p01q124 | FTN_1245 | iscS | cysteine desulfurase | 7# | |
| tnfn1_pw060323p04q139 | FTN_1264 | rluD | ribosomal large subunit pseudouridine synthase D | 2# | |
| tnfn1_pw060510p03q183 | FTN_1264 | rluD | ribosomal large subunit pseudouridine synthase D | 6# | |
| tnfn1_pw060328p06q166 | FTN_1273 | long chain fatty acid CoA ligase | 2 | ||
| tnfn1_pw060419p03q126 | FTN_1278 | nadE | NAD synthase | 5 | |
| tnfn1_pw060328p05q128 | FTN_1329 | fbaA | fructose bisphosphate aldolase Class II | 3 | |
| tnfn1_pw060323p06q195 | FTN_1390 | Zn-dependent hydrolase | 5 | ||
| tnfn1_pw060510p04q137 | FTN_1425 | wbtF | NAD dependent epimerase | 2 | |
| tnfn1_pw060419p03q166 | FTN_1431 | wbtA | dTDP-glucose 4,6-dehydratase | 2 | |
| tnfn1_pw060323p07q169 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | 4# | ||
| tnfn1_pw060418p02q122 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | 3# | ||
| tnfn1_pw060328p08q196 | FTN_1459 | short chain dehydrogenase | 5 | ||
| tnfn1_pw060328p06q128 | FTN_1530 | lysA | diaminopimelate decarboxylase | 6 | |
| tnfn1_pw060328p05q101 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) | 2# | |
| tnfn1_pw060419p04q163 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) | 6# | |
| tnfn1_pw060418p02q178 | FTN_1536 | amino acid-polyamine-organocation (APC) superfamily protein | 4 | ||
| tnfn1_pw060323p06q106 | FTN_1552 | acid phosphatase, PAP2 family | 5 | ||
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase | 2 | |
| tnfn1_pw060323p04q110 | FTN_1678 | nuoC | NADH dehydrogenase I, C subunit | 5# | |
| tnfn1_pw060328p05q160 | FTN_1729 | dapB | dihydrodipicolinate reductase | 4# | |
| tnfn1_pw060510p01q178 | FTN_1729 | dapB | dihydrodipicolinate reductase | 3# | |
| tnfn1_pw060328p04q104 | FTN_1730 | lysC | aspartate kinase III | 2 | |
| tnfn1_pw060328p03q174 | FTN_1768 | pepN | aminopeptidase N | 3 | |
| Transporter proteins | |||||
| tnfn1_pw060323p03q117 | FTN_0005 | corA | divalent inorganic cation transporter | 2# | |
| tnfn1_pw060420p01q131 | FTN_0005 | corA | divalent inorganic cation transporter | 3# | |
| tnfn1_pw060420p01q180 | FTN_0097 | hydroxy/aromatic amino acid permease (HAAAP) family protein | 4# | ||
| tnfn1_pw060419p03q162 | FTN_0115 | Na+/H+ antiporter | 4 | ||
| tnfn1_pw060323p08q162 | FTN_0151 | ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component | 2 | ||
| tnfn1_pw060419p01q165 | FTN_0183 | manganese/Zinc/Iron chelate uptake transporter family protein | 3# | ||
| tnfn1_pw060419p04q103 | FTN_0183 | manganese/Zinc/Iron chelate uptake transporter family protein | 2# | ||
| tnfn1_pw060510p02q174 | FTN_0184 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060323p03q161 | FTN_0276 | mviN | multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) transporter | 2* | |
| tnfn1_pw060510p02q151 | FTN_0276 | mviN | multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) transporter | 3* | |
| tnfn1_pw060323p08q118 | FTN_0345 | DNA uptake protein, SMF family | 2 | ||
| tnfn1_pw060419p03q195 | FTN_0363 | sodium bile acid symporter family protein | 4 | ||
| tnfn1_pw060420p03q115 | FTN_0566 | mechanosensitive ion channel protein | 3 | ||
| tnfn1_pw060328p08q167 | FTN_0579 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060419p04q167 | FTN_0620 | major facilitator superfamily (MFS) transport protein | 5 | ||
| tnfn1_pw060328p06q114 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 2# | ||
| tnfn1_pw060510p02q115 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 5# | ||
| tnfn1_pw060510p02q167 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 5# | ||
| tnfn1_pw060418p02q189 | FTN_0640 | dctA | C4-dicarboxylate transport protein | 3 | |
| tnfn1_pw060510p02q159 | FTN_0688 | galP2 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 3 | |
| tnfn1_pw060510p03q140 | FTN_0741 | proton-dependent oligopeptide transporter (POT) family protein, di-or tripeptide:H+ symporter | 5 | ||
| tnfn1_pw060328p05q107 | FTN_0767 | betT | betaine/carnitine/choline transporter (BCCT) family protein | 4 | |
| tnfn1_pw060420p03q116 | FTN_0824 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060510p04q173 | FTN_0872 | small conductance mechanosensitive ion channel (MscS) family protein | 5 | ||
| tnfn1_pw060328p06q175 | FTN_0875 | metabolite:H+ symporter (MHS) family | 2 | ||
| tnfn1_pw060328p02q106 | FTN_0884 | drug/metabolite transporter superfamily protein | 1# | ||
| tnfn1_pw060328p03q163 | FTN_0884 | drug/metabolite transporter superfamily protein | 4# | ||
| tnfn1_pw060328p01q188 | FTN_0910 | sugar:cation symporter family protein | 2# | ||
| tnfn1_pw060419p04q109 | FTN_0910 | sugar:cation symporter family protein | 2# | ||
| tnfn1_pw060419p01q175 | FTN_0984 | ABC transporter, ATP-binding protein | 2 | ||
| tnfn1_pw060419p01q170 | FTN_1006 | transporter-associated protein, HlyC/CorC family | 4 | ||
| tnfn1_pw060418p02q160 | FTN_1010 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060419p01q133 | FTN_1014 | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | ||
| tnfn1_pw060328p02q109 | FTN_1107 | metlQ | methionine uptake transporter (MUT) family protein, membrane and periplasmic protein | 2 | |
| tnfn1_pw060323p05q139 | FTN_1166 | metabolite:H+ symporter (MHS) family protein | 7 | ||
| tnfn1_pw060419p02q107 | FTN_1267 | ATP-binding Cassette (ABC) superfamily protein | 4 | ||
| tnfn1_pw060418p02q182 | FTN_1275 | drug:H+ antiporter-1 (DHA2) family protein | 5 | ||
| tnfn1_pw060420p04q186 | FTN_1404 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060510p02q118 | FTN_1409 | major facilitator superfamily (MFS) transport protein | 6 | ||
| tnfn1_pw060328p06q119 | FTN_1549 | drug:H+ antiporter-1 (DHA1) family protein | 3 | ||
| tnfn1_pw060419p02q126 | FTN_1581 | small conductance mechanosensitive ion channel (MscS) family protein | 2 | ||
| tnfn1_pw060418p01q150 | FTN_1586 | sugar transporter, MFS superfamily | 2 | ||
| tnfn1_pw060420p01q146 | FTN_1681 | fur | ferric uptake regulation protein | 2* | |
| tnfn1_pw060510p04q167 | FTN_1681 | fur | ferric uptake regulation protein | 2* | |
| tnfn1_pw060323p03q163 | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | 3* | ||
| tnfn1_pw060328p02q192 | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | 4* | ||
| tnfn1_pw060323p06q117 | FTN_1685 | drug:H+ antiporter-1 (DHA1) family protein | 3 | ||
| tnfn1_pw060418p02q140 | FTN_1685 | drug:H+ antiporter-1 (DHA1) family protein | 5 | ||
| tnfn1_pw060328p05q182 | FTN_1707 | nhaD | Na+:H+ antiporter | 5 | |
| tnfn1_pw060328p02q121 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2* | |
| tnfn1_pw060420p02q159 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2* | |
| tnfn1_pw060510p03q118 | FTN_1717 | kdpB | potassium-transporting ATPase B chain | 3 | |
| tnfn1_pw060420p01q113 | FTN_1752 | nhaA | Na+:H+ antiporter | 3 | |
| Transferase | |||||
| tnfn1_pw060510p04q127 | FTN_0071 | LPS fatty acid acyltransferase | 2# | ||
| tnfn1_pw060419p03q160 | FTN_0080 | SAM-dependent methyltransferase | 4 | ||
| tnfn1_pw060328p08q125 | FTN_0120 | rhodanese-related sulfurtransferase | 4 | ||
| tnfn1_pw060328p01q137 | FTN_0153 | RimI-like acetyltransferase | 3 | ||
| tnfn1_pw060418p01q110 | FTN_0200 | UDP-3-O-[3-fatty acid] glucosamine N-acyltransferase | 2# | ||
| tnfn1_pw060510p02q131 | FTN_0200 | UDP-3-O-[3-fatty acid] glucosamine N-acyltransferase | 2# | ||
| tnfn1_pw060420p02q146 | FTN_0300 | glycosyl transferase, group 2 | 5 | ||
| tnfn1_pw060328p03q179 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 4* | ||
| tnfn1_pw060419p01q169 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 2* | ||
| tnfn1_pw060420p01q152 | FTN_0453 | glycosyl transferase | 5 | ||
| tnfn1_pw060419p02q135 | FTN_0560 | ksgA | dimethyladenosine transferase | 3 | |
| tnfn1_pw060419p04q168 | FTN_1091 | aroA | 3-phosphoshikimate 1-carboxyvinyltransferase | 2 | |
| tnfn1_pw060418p03q185 | FTN_1400 | S-adenosylmethionine-dependent methyltransferase | 5 | ||
| tnfn1_pw060418p04q172 | FTN_1418 | manC | mannose-1-phosphate guanylyltransferase | 4 | |
| DNA Modification | |||||
| tnfn1_pw060510p04q169 | FTN_0122 | recA | recombinase A protein | 2 | |
| tnfn1_pw060328p06q179 | FTN_0492 | parC | DNA topoisomerase IV subunit A | 2# | |
| tnfn1_pw060510p04q168 | FTN_0666 | uvrA | excinuclease ABC, subunit A | 2 | |
| FTN_0704 | type I restriction-modification system, subunit M (methyltransferase) | 5 | |||
| tnfn1_pw060510p02q180 | FTN_0704 | type I restriction-modification system, subunit M (methyltransferase) | 2 | ||
| tnfn1_pw060510p03q158 | FTN_1294 | rRNA methylase, SpoU family | 2 | ||
| tnfn1_pw060510p02q176 | FTN_1413 | ATPase, AAA family, related to the helicase subunit of the Holliday junction resolvase | 2 | ||
| tnfn1_pw060328p08q179 | FTN_1491 | adenine specific DNA methylase | 2 | ||
| tnfn1_pw060328p06q176 | FTN_1544 | hemK | modification methylase, HemK family | 5 | |
| tnfn1_pw060328p05q164 | FTN_1594 | uvrD | DNA helicase II | 6 | |
| Cell Division | |||||
| tnfn1_pw060328p01q167 | FTN_0330 | minD | septum formation inhibitor-activating ATPase | 2 | |
| tnfn1_pw060323p08q146 | FTN_0331 | minC | septum formation inhibitor | 4# | |
| tnfn1_pw060420p02q170 | FTN_0331 | minC | septum formation inhibitor | 4# | |
| Transcription/Translation | |||||
| tnfn1_pw060328p06q196 | FTN_0552 | yhbY | RNA-binding protein | 5# | |
| tnfn1_pw060510p03q150 | FTN_0949 | rplI | 50S ribosomal protein L9 | 2 | |
| tnfn1_pw060328p06q170 | FTN_1099 | transcriptional regulator, LysR family | 7 | ||
| tnfn1_pw060419p03q165 | FTN_1300 | transcriptional regulator, LysR family | 2 | ||
| tnfn1_pw060328p02q148 | FTN_1393 | transcriptional regulator, ArsR family | 3# | ||
| tnfn1_pw060418p01q138 | FTN_1393 | transcriptional regulator, ArsR family | 2# | ||
| tnfn1_pw060419p02q151 | FTN_1628 | transcriptional regulator, LysR family | 2# | ||
| tnfn1_pw060510p03q194 | FTN_1628 | transcriptional regulator, LysR family | 2# | ||
| FPI | |||||
| tnfn1_pw060328p01q144 | FTN_1313 | hypothetical protein | 3 | ||
| tnfn1_pw060323p03q179 | FTN_1314 | conserved hypothetical protein | 1 | ||
| tnfn1_pw060328p06q163 | FTN_1315 | protein of unknown function | 5 | ||
| tnfn1_pw060328p06q115 | FTN_1322 | iglC | intracellular growth locus protein C | 5 | |
| tnfn1_pw060419p04q108 | FTN_1325 | pdpD | protein of unknown function | 2 | |
| Type IV Pili | |||||
| tnfn1_pw060418p04q123 | FTN_0070 | pilE | Type IV pili, pilus assembly protein | 3 | |
| tnfn1_pw060510p03q129 | FTN_0070 | pilE | Type IV pili, pilus assembly protein | 3 | |
| tnfn1_pw060323p06q179 | FTN_0305 | pilus assembly protein | 4 | ||
| tnfn1_pw060419p01q196 | FTN_0414 | Type IV pili, pilus assembly protein | 2 | ||
| tnfn1_pw060419p03q141 | FTN_0664 | fimT | Type IV pili, pilus assembly protein | 2 | |
| tnfn1_pw060328p05q146 | FTN_0946 | pilF | Type IV pili, pilus assembly protein | 5 | |
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | Type IV pili secretin component | 4 | |
| Others | |||||
| tnfn1_pw060328p08q161 | isftu1 | isftu1 | 2 | ||
| tnfn1_pw060323p03q115 | isftu3 | isftu3 | 1 | ||
| tnfn1_pw060328p04q157 | isftu2 | isftu2 | 1 | ||
| tnfn1_pw060510p02q150 | isftu6 | isftu6 | 2 | ||
| tnfn1_pw060328p01q179 | FTN_0010 | phage terminase, small subunit | 3 | ||
| tnfn1_pw060328p08q114 | FTN_0266 | htpG | chaperone Hsp90, heat shock protein HtpG | 2 | |
| tnfn1_pw060328p04q152 | FTN_0322 | VacJ like lipoprotein | 3# | ||
| tnfn1_pw060418p01q140 | FTN_0322 | VacJ like lipoprotein | 2# | ||
| tnfn1_pw060328p08q155 | FTN_0357 | pal | peptidoglycan-associated lipoprotein, OmpA family | 4* | |
| tnfn1_pw060419p01q158 | FTN_0357 | pal | peptidoglycan-associated lipoprotein, OmpA family | 2* | |
| tnfn1_pw060510p02q122 | FTN_0367 | phage integrase | 4 | ||
| tnfn1_pw060328p08q132 | FTN_0372 | regulatory protein, AlpA family | 4 | ||
| tnfn1_pw060323p07q171 | FTN_0585 | cutC | copper homeostasis protein CutC family protein | 2 | |
| tnfn1_pw060328p06q127 | FTN_0713 | ostA2 | organic solvent tolerance protein OstA | 5# | |
| tnfn1_pw060419p01q180 | FTN_0713 | ostA2 | organic solvent tolerance protein OstA | 4# | |
| tnfn1_pw060323p06q105 | FTN_0810 | ROK family protein | 4 | ||
| tnfn1_pw060419p01q139 | FTN_0836 | kinase-like protein | 2 | ||
| tnfn1_pw060418p04q134 | FTN_1051 | hfq | host factor I for bacteriophage Q beta replication | 2 | |
| tnfn1_pw060420p03q121 | FTN_1064 | PhoH family protein, putative ATPase | 4 | ||
| tnfn1_pw060328p05q177 | FTN_1192 | chitin-binding protein | 6 | ||
| tnfn1_pw060419p04q183 | FTN_1240 | BolA family protein | 4 | ||
| tnfn1_pw060418p02q190 | FTN_1242 | DedA family protein | 5 | ||
| tnfn1_pw060419p01q120 | FTN_1488 | prophage maintenance system killer protein (DOC) | 6 | ||
| tnfn1_pw060419p01q135 | FTN_1665 | magnesium chelatase | 2 | ||
| tnfn1_pw060419p04q180 | FTN_1682 | frgA | siderophore biosynthesis protein | 5 | |
| tnfn1_pw060510p04q122 | FTN_1698 | Dam-replacing family protein | 2 | ||
| Intergenic | |||||
| tnfn1_pw060418p01q125 | intergenic | 7 | |||
| tnfn1_pw060323p06q165 | intergenic | 5 | |||
| tnfn1_pw060323p08q117 | intergenic | 3 | |||
| tnfn1_pw060328p05q195 | intergenic | 6 | |||
| tnfn1_pw060419p01q148 | intergenic | 3 | |||
| tnfn1_pw060420p03q148 | intergenic | 3 | |||
| tnfn1_pw060420p01q164 | intergenic | 2 | |||
| tnfn1_pw060510p02q127 | intergenic | 2 | |||
| tnfn1_pw060328p08q109 | intergenic | 2 | |||
| tnfn1_pw060510p04q116 | intergenic | 2 | |||
Mutants for which all the mutant alleles showed similar growth defect
Mutants for which two out of three or three out of four alleles showed growth defect
There was a lack of consistent phenotype for the two mutant alleles for some of the mutants, which may be due to the site of the insertion that may generate a functional or partially functional protein in some of the mutants. The growth defect for most of the mutants identified was not due to a growth defect in vitro, since more than 98% of the mutants exhibited normal growth in vitro, compared to the wild type strain. It is likely that the defect for few of the mutants was due to a defect in attachment and/or entry into macrophages. It is likely that the reduction in intracellular growth for some of the mutants was due to or amplified by a polar effect of the transposon insertion on downstream genes. However, this would implicate the identified disrupted operon in intracellular proliferation. Similar findings were also observed in our screen for mutants defective in S2 cells (see accompanying manuscript).
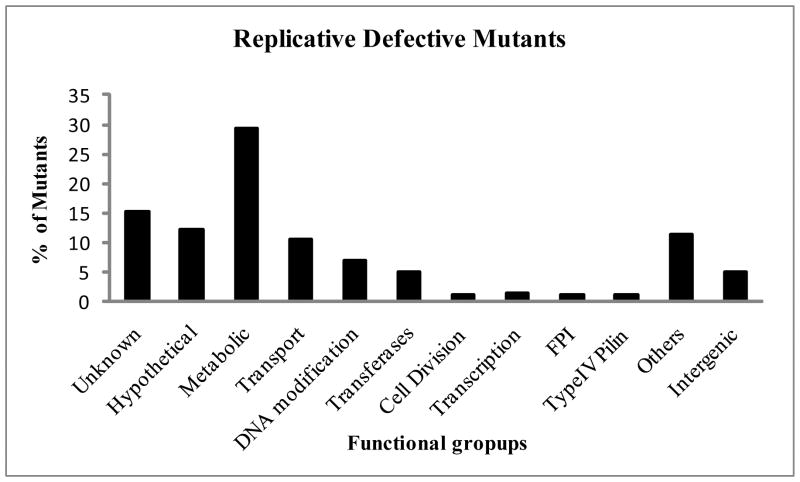
The 202 mutants defective in intracellular proliferation were not skewed to any particular protein functional group but rather distributed across several different functional groups (Fig. 1). The largest percentage of mutants comprising ~30% of all the mutants identified had mutations in metabolic genes (Fig. 1). Interestingly, about 10% of the mutants had insertions in genes required for the transport of nutrients (Fig. 1), which supports findings about the fastidious nature of F. tularensis. The metabolic genes were grouped according to their putative biochemical pathways. Our analysis shows that mutations in genes involved in carbohydrate, amino acid, and nucleotide metabolism are defective in intracellular replication (Table 2).
Fig. 1. Functional groups of mutants defective in intra-macrophage growth.
U937 macrophages were infected with each of the mutants of F. tularensis at MOI of 10 for 1 h followed by 1 h of gentamicin treatment. Growth of the mutants was compared to the wild type strain at 24 h post-infection, and the relative reduction in the number of cfu relative to the wild type strain was determined. After the primary screen, 425 mutants were tested twice in triplicate.
Table 2.
List of growth defective or dissemination defective mutants identified in previous screens and are defective in both U937 macrophages and S2 cells
| Strain Name | Locus Tag | Gene | Description |
|---|---|---|---|
| tnfn1_pw060323p03q172α | FTN_0008 | 10 TMS drug/metabolite exporter protein | |
| tnfn1_pw060420p02q151βγδ | FTN_0018 | sdaC | serine permease |
| tnfn1_pw060323p02q177βγδ | FTN_0019 | pyrB | aspartate carbamoyltransferase |
| tnfn1_pw060323p08q120βγδ | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060510p02q160βγδ | FTN_0021 | carA | carbamoyl-phosphate synthase small chain |
| tnfn1_pw060419p04q178α | FTN_0028 | conserved hypothetical membrane protein | |
| tnfn1_pw060418p04q123β | FTN_0070 | pilE | Type IV pili, pilus assembly protein |
| tnfn1_pw060420p01q180βγδπ | FTN_0097 | hydroxy/aromatic amino acid permease (HAAAP) family protein | |
| tnfn1_pw060419p01q106α | FTN_0111 | ribH | riboflavin synthase beta-chain |
| tnfn1_pw060510p04q169δ | FTN_0122 | recA | recombinase A protein |
| tnfn1_pw060510p01q123α | FTN_0132 | lpsA | protein of unknown function |
| tnfn1_pw060323p03q125α | FTN_0133 | ribonuclease II family protein | |
| tnfn1_pw060420p02q173α | FTN_0169 | conserved hypothetical membrane protein | |
| tnfn1_pw060418p03q133α | FTN_0199 | cyoE | heme O synthase |
| tnfn1_pw060323p04q102βγδ | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| tnfn1_pw060510p02q178α | FTN_0217 | L-lactate dehydrogenase | |
| tnfn1_pw060420p04q134α | FTN_0297 | conserved protein of unknown function | |
| tnfn1_pw060418p03q147α | FTN_0299 | putP | proline:Na+ symporter |
| tnfn1_pw060420p02q146βγδ | FTN_0300 | glycosyl transferase, group 2 | |
| tnfn1_pw060328p01q167βγδ | FTN_0330 | minD | septum formation inhibitor-activating ATPase |
| tnfn1_pw060323p08q146δ | FTN_0331 | minC | septum formation inhibitor |
| tnfn1_pw060328p08q156α | FTN_0340 | protein of unknown function | |
| tnfn1_pw060323p06q113βγδ | FTN_0420 | purCD | SAICAR synthetase/phosphoribosylamine-glycine ligase |
| tnfn1_pw060328p06q134βγδ | FTN_0443 | maeA | NAD-dependent malic enzyme |
| tnfn1_pw060328p05q119βγδπ | FTN_0444 | membrane protein of unknown function | |
| tnfn1_pw060323p05q182βγδ | FTN_0504 | cadC | lysine decarboxylase |
| tnfn1_pw060510p01q124βγδ | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 |
| tnfn1_pw060323p06q168βγδ | FTN_0545 | glycosyl transferase, group 2 | |
| tnfn1_pw060419p02q135β | FTN_0560 | ksgA | dimethyladenosine transferase |
| tnfn1_pw060419p03q116βγδ | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit |
| tnfn1_pw060510p04q147βγδ | FTN_0599 | protein of unknown function | |
| tnfn1_pw060323p06q164βγδ | FTN_0624 | serine permease | |
| tnfn1_pw060418p02q128δ | FTN_0633 | katG | peroxidase/catalase |
| tnfn1_pw060420p01q168α | FTN_0646 | cscK | ROK family protein |
| tnfn1_pw060419p01q168βγδ | FTN_0651 | cdd | cytidine deaminase |
| tnfn1_pw060510p04q168δ | FTN_0666 | uvrA | excinuclease ABC, subunit A |
| tnfn1_pw060328p04q123βγδ | FTN_0672 | secA | preprotein translocase, subunit A (ATPase, RNA helicase) |
| tnfn1_pw060420p03q134α | FTN_0710 | type I restriction-modification system, subunit R (restriction) | |
| tnfn1_pw060328p06q127α | FTN_0713 | ostA2 | organic solvent tolerance protein OstA |
| tnfn1_pw060328p06q132βγδ | FTN_0728 | predicted Co/Zn/Cd cation transporter | |
| tnfn1_pw060323p06q115α | FTN_0768 | tspO | tryptophan-rich sensory protein |
| tnfn1_pw060323p06q105α | FTN_0810 | ROK family protein | |
| tnfn1_pw060323p07q185βγδ | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase |
| tnfn1_pw060418p01q141β | FTN_0817 | conserved protein of unknown function | |
| tnfn1_pw060420p04q108βγδπ | FTN_0822 | para-aminobenzoate synthase component I | |
| tnfn1_pw060420p03q116α | FTN_0824 | major facilitator superfamily (MFS) transport protein | |
| tnfn1_pw060328p01q128α | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060420p04q176βγδ | FTN_0855 | protein of unknown function | |
| tnfn1_pw060420p02q175α | FTN_0877 | cls | cardiolipin synthetase |
| tnfn1_pw060323p04q104α | FTN_0918 | conserved protein of unknown function | |
| tnfn1_pw060419p04q188βγδ | FTN_0925 | protein of unknown function | |
| tnfn1_pw060420p02q181α | FTN_0933 | protein of unknown function | |
| tnfn1_pw060323p04q134α | FTN_0938 | hypothetical protein | |
| tnfn1_pw060419p01q170α | FTN_1006 | transporter-associated protein, HlyC/CorC family | |
| tnfn1_pw060323p08q141α | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060418p01q153α | FTN_1055 | lon | DNA-binding, ATP-dependent protease La |
| tnfn1_pw060510p01q114π | FTN_1073 | DNA/RNA endonuclease G | |
| tnfn1_pw060419p04q168βγδ | FTN_1091 | aroA | 3-phosphoshikimate 1-carboxyvinyltransferase |
| tnfn1_pw060328p08q188α | FTN_1098 | conserved hypothetical membrane protein | |
| tnfn1_pw060328p02q109α | FTN_1107 | metlQ | methionine uptake transporter (MUT) family protein, membrane and periplasmic protein |
| tnfn1_pw060328p02q175βγδ | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase |
| tnfn1_pw060418p03q107βγδ | FTN_1217 | ATP-binding cassette (ABC) superfamily protein | |
| tnfn1_pw060323p08q166α | FTN_1232 | conserved hypothetical membrane protein | |
| tnfn1_pw060328p06q178βγδ | FTN_1241 | DedA family protein | |
| tnfn1_pw060510p03q135β | FTN_1254 | protein of unknown function | |
| tnfn1_pw060420p04q196βγδπ | FTN_1256 | membrane protein of unknown function | |
| tnfn1_pw060323p03q102βγδ | FTN_1257 | membrane protein of unknown function | |
| tnfn1_pw060420p02q179βγδ | FTN_1263 | comL | competence lipoprotein ComL |
| tnfn1_pw060418p01q149βγδ | FTN_1298 | GTPase of unknown function | |
| tnfn1_pw060328p01q144βγδ | FTN_1313 | hypothetical protein | |
| tnfn1_pw060328p06q163β | FTN_1315 | protein of unknown function | |
| tnfn1_pw060510p01q110αβγδ | FTN_1321 | iglD | intracellular growth locus protein D |
| tnfn1_pw060328p06q115βγδ | FTN_1322 | iglC | intracellular growth locus protein C |
| tnfn1_pw060419p04q108βγδ | FTN_1325 | pdpD | protein of unknown function |
| tnfn1_pw060510p01q142βγδ | FTN_1333 | tktA | transketolase I |
| tnfn1_pw060418p01q191α | FTN_1349 | hypothetical protein | |
| tnfn1_pw060418p01q185α | FTN_1355 | regulatory factor, Bvg accessory factor family | |
| tnfn1_pw060418p02q109π | FTN_1376 | disulfide bond formation protein, DsbB family | |
| tnfn1_pw060418p03q185α | FTN_1400 | S-adenosylmethionine-dependent methyltransferase | |
| tnfn1_pw060419p04q135α | FTN_1415 | thioredoxin | |
| tnfn1_pw060420p04q116βγδ | FTN_1421 | wbtH | glutamine amidotransferase/asparagine synthase |
| tnfn1_pw060510p04q137βγδ | FTN_1425 | wbtF | NAD dependent epimerase |
| tnfn1_pw060419p03q166βγδπ | FTN_1431 | wbtA | dTDP-glucose 4,6-dehydratase |
| tnfn1_pw060418p02q122βγδ | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | |
| tnfn1_pw060328p08q196α | FTN_1459 | short chain dehydrogenase | |
| tnfn1_pw060323p06q110βγδ | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
| tnfn1_pw060328p06q128β | FTN_1530 | lysA | diaminopimelate decarboxylase |
| tnfn1_pw060323p07q176α | FTN_1534 | conserved protein of unknown function | |
| tnfn1_pw060418p02q178α | FTN_1536 | amino acid-polyamine-organocation (APC) superfamily protein | |
| tnfn1_pw060418p01q150γ | FTN_1586 | sugar transporter, MFS superfamily | |
| tnfn1_pw060510p01q146βγδ | FTN_1597 | prfC | peptide chain release factor 3 |
| tnfn1_pw060420p01q189α | FTN_1611 | major facilitator superfamily (MFS) transport protein | |
| tnfn1_pw060323p04q160βγδ | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C |
| tnfn1_pw060419p04q180δ | FTN_1682 | frgA | siderophore biosynthesis protein |
| tnfn1_pw060323p03q163βγδ | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | |
| tnfn1_pw060328p05q154βγδ | FTN_1777 | trpG | anthranilate synthase component II |
Approximately, 15% of the mutants had transposon insertion in genes encoding proteins of unknown function and 12% were hypothetical proteins (Fig. 1). Identifying the functions of these genes, which make up about 30% of all the genes identified, will shed more light on the molecular mechanism required for the intracellular infection of macrophages by F. tularensis.
Approximately, 25% of the mutants that we identified in our screen have been identified in other screens for various aspects of virulence of F. tularensis (Table 3). This indicates the power of our comprehensive screen that was aimed at identification of genetic loci required for modulation of phagosome biogenesis and escape into the cytosol of human macrophages, and both of these processes are essential for subsequent proliferation within the cytosol. Interestingly, 83% of the identified genes in our screen are conserved in the virulent F. tularensis subsp tularensis. This indicates that most of the genes that are necessary for intracellular replication in human macrophages are common to the highly virulent subsp of F. tularensis. This indicate that adaptation to the intracellular life in mammalian cells occurred before the subspecies diverged (Champion et al., 2009; Larsson et al., 2009).
Table 3.
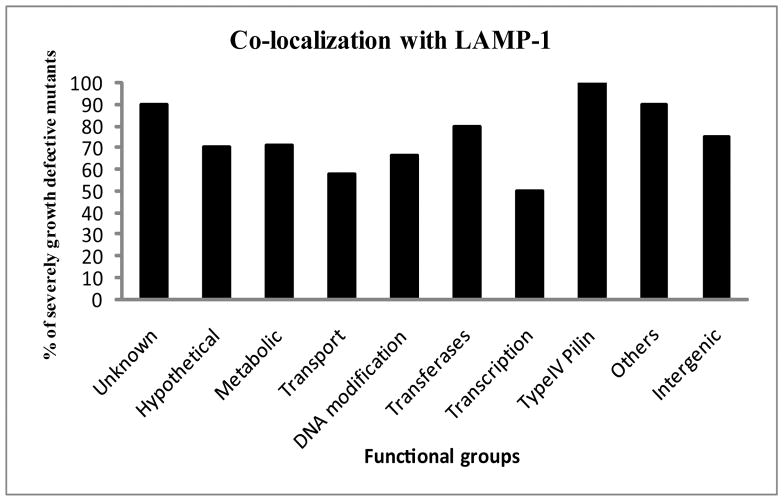
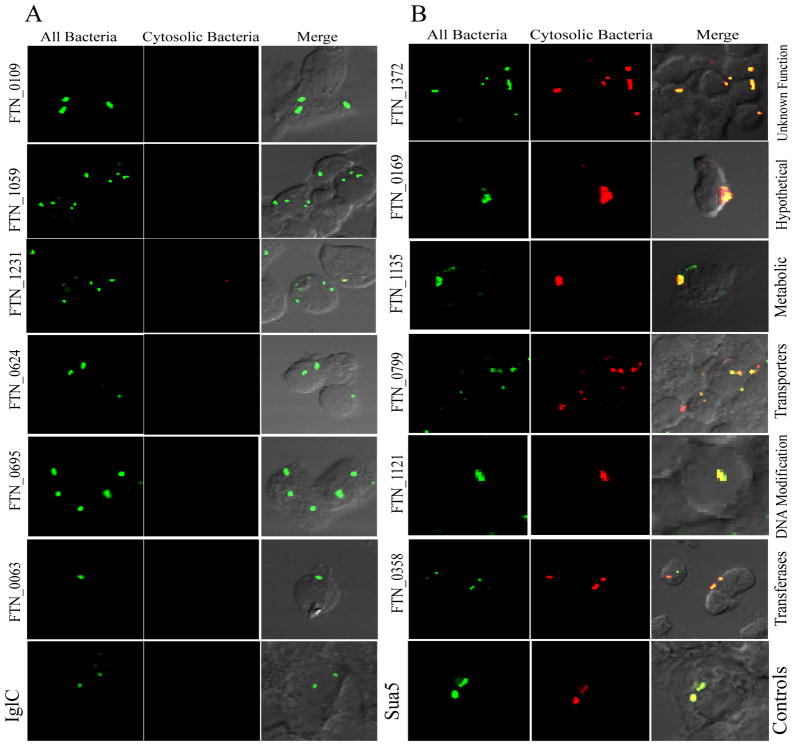
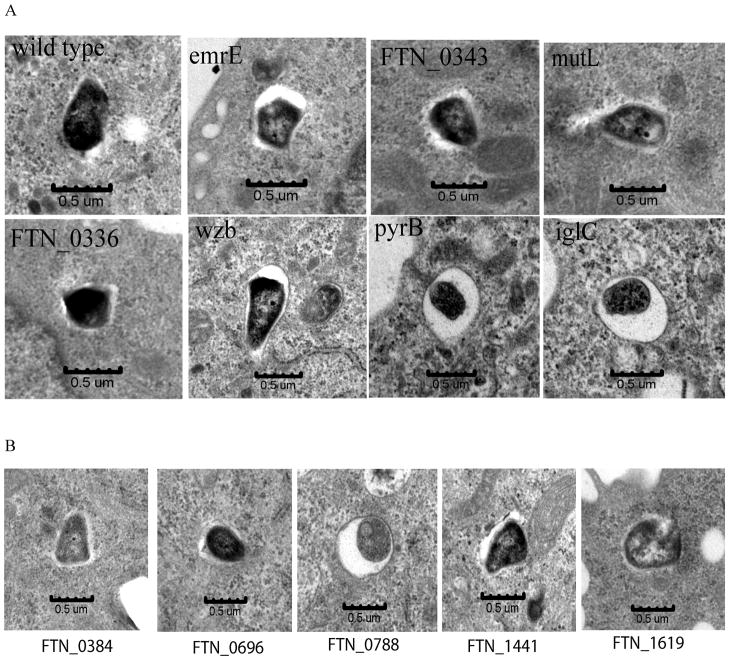
List of 6 growth-defective mutants of F. tularensis and the co-localization of the FCP with LAMP-1 in hMDMs
| Strain Name | Locus Tag | Gene | %Co-localization with Lamp1 | Log reduction in Growth relative to WT |
|---|---|---|---|---|
| Control | ||||
| Sua5/YciO/YrdC family protein | 17 | 0 | ||
| Intracellular growth locus C | IglC | 88 | 6 | |
| Selected Mutants | ||||
| tnfn1_pw060328p05q136 | FTN_0384 | 20 | 2 | |
| tnfn1_pw060323p07q141 | FTN_0788 | 75 | 4 | |
| tnfn1_pw060323p01q177 | FTN_0799 | emrE | 26 | 3 |
| tnfn1_pw060510p01q158 | FTN_0988 | prmA | 84 | 4 |
| tnfn1_pw060420p02q179 | FTN_1263 | comL | 78 | 5 |
| tnfn1_pw060323p01q113 | FTN_1343 | 72 | 2 | |
To confirm that the defect in replication was not limited to the U937 macrophage cell line, six mutants were selected randomly and tested in human monocytes-derived macrophages (hMDMs). Our data showed that all six mutants exhibited similar defective phenotypes in both U937 macrophages and hMDMs (Table 3). This indicates that the growth defect observed for the mutants is not restricted to the U937 human macrophage cell line but is also exhibited in primary human-derived macrophages.
Modulation of phagosome biogenesis by the mutants defective in intracellular replication
Since modulation of phagosome biogenesis and bacterial escape into the cytosol is essential for intra-macrophage proliferation, we decided to focus our additional studies on the most defective mutants in intra-macrophage proliferation to analyze the biogenesis of the FCP and bacterial escape into the cytosol. We determined whether the defect in intra-macrophage growth of the mutants with ≥103 fold reduction in intracellular growth was due to a defect in modulation in phagosome biogenesis. Since the FCP transiently co-localizes with the LAMP-1 late endosomal/lysosomal marker for ~ 30 min after which this co-localization is rapidly lost by 1–4h, we determined whether the FCP of the 125 most defective mutants co-localized transiently or persistently with LAMP-1 at 6h post-infection. We selected this time point because most FCPs harboring the wild type strain do not co-localize with LAMP-1 at this time points. This would exclude mutants that are mildly defective in modulation of phagosome biogenesis and would allow us to focus on the most defective mutants. We noticed that growth of the kanr mutant bacteria in presence of the kanamycin antibiotic before infection caused a significant increase in co-localization with LAMP-1 compared to growth in the absence of the antibiotic. Since the wild type strain is sensitive to kanamycin, we used the kanr Sua5 mutant as our negative control, since this mutant had no detectable defect in intracellular proliferation, co-localization with late endosomal/lysosomal markers, or phagosomal escape. As our positive control, we used the iglC mutant. To exclude the mildly defective mutants, a mutant was considered persistently co-localized with LAMP-1 if it exhibited more than 50% co-localization with LAMP-1 at 6h post infection, which is significantly different from the positive and negative controls (Student t-test, p<0.001). In contrast to the wild type strain that co-localized transiently with LAMP-1, 91 of the 125 defective mutants co-localized persistently with LAMP-1, similar to the iglC mutant positive control (Table 5). Most of these 91 mutants exhibited 65–75% co-localization with LAMP-1 (Fig. 2 and 3), while the negative control showed only 24% co-localization (Fig. 3 and Table 5) (Santic et al., 2005a; Santic et al., 2005b; Santic et al., 2007; Santic et al., 2008). The mutants that showed aberrant trafficking were distributed across several functional categories (Fig. 3 and Table 5).
Table 5.
Vacuolar vs. cytosolic localization of the mutants within U937 cells
| Strain Name | Locus Tag | Gene | %Co-localization with Lamp1 | Localization |
|---|---|---|---|---|
| Control | ||||
| Sua5/YciO/YrdC family protein | 24 | Cytoplasmic | ||
| Intracellular growth locus C | IglC | 81 | Phagosomal | |
| Proteins of unknown function | ||||
| tnfn1_pw060323p08q148 | FTN_0027 | 76 | Phagosomal | |
| tnfn1_pw060323p03q103 | FTN_0041 | 26 | Cytoplasmic | |
| tnfn1_pw060418p04q193 | FTN_0109 | 70 | Phagosomal | |
| tnfn1_pw060420p04q143 | FTN_0149 | 67 | Phagosomal | |
| tnfn1_pw060420p04q134 | FTN_0297 | 64 | Phagosomal | |
| tnfn1_pw060328p05q119 | FTN_0444 | 81 | Phagosomal | |
| tnfn1_pw060323p07q141 | FTN_0788 | 68 | Phagosomal* | |
| tnfn1_pw060420p04q176 | FTN_0855 | 89 | Phagosomal | |
| tnfn1_pw060420p02q178 | FTN_0915 | 70 | Phagosomal | |
| tnfn1_pw060419p04q188 | FTN_0925 | 72 | Phagosomal | |
| tnfn1_pw060323p03q147 | FTN_0930 | 55 | Phagosomal | |
| tnfn1_pw060420p02q181 | FTN_0933 | 66 | Phagosomal | |
| tnfn1_pw060510p01q108 | FTN_0977 | 69 | Phagosomal | |
| tnfn1_pw060420p01q127 | FTN_1175 | 80 | Phagosomal | |
| tnfn1_pw060420p04q196 | FTN_1256 | 83 | Phagosomal | |
| tnfn1_pw060323p01q113 | FTN_1343 | 60 | Phagosomal | |
| tnfn1_pw060328p02q110 | FTN_1457 | 32 | Cytoplasmic | |
| tnfn1_pw060420p01q132 | FTN_1624 | 70 | Phagosomal | |
| tnfn1_pw060420p02q184 | FTN_1696 | 45 | Cytoplasmic | |
| tnfn1_pw060328p06q155 | FTN_1764 | 76 | Phagosomal | |
| Hypothetical Proteins | ||||
| tnfn1_pw060323p03q142 | FTN_0030 | 63 | Phagosomal | |
| tnfn1_pw060328p06q180 | FTN_0038 | 79 | Phagosomal | |
| tnfn1_pw060420p02q173 | FTN_0169 | 32 | Cytoplasmic | |
| tnfn1_pw060323p01q181 | FTN_0336 | 40 | Cytoplasmic | |
| tnfn1_pw060328p05q136 | FTN_0384 | 28 | Cytoplasmic* | |
| tnfn1_pw060510p01q147 | FTN_0403 | 77 | Phagosomal | |
| tnfn1_pw060419p03q188 | FTN_0696 | 45 | Cytoplasmic* | |
| tnfn1_pw060323p01q163 | FTN_0727 | 68 | Phagosomal | |
| tnfn1_pw060419p02q102 | FTN_0792 | 80 | Phagosomal | |
| tnfn1_pw060510p02q108 | FTN_0847 | 64 | Phagosomal | |
| tnfn1_pw060323p07q105 | FTN_0895 | 30 | Cytoplasmic | |
| tnfn1_pw060510p03q192 | FTN_1098 | 75 | Phagosomal | |
| tnfn1_pw060418p01q191 | FTN_1349 | 56 | Phagosomal | |
| tnfn1_pw060328p06q182 | FTN_1395 | 80 | Phagosomal | |
| tnfn1_pw060328p04q136 | FTN_1406 | 84 | Phagosomal | |
| tnfn1_pw060328p02q129 | FTN_1612 | 69 | Phagosomal | |
| tnfn1_pw060420p02q176 | FTN_1686 | 89 | Phagosomal | |
| Metabolic Proteins | ||||
| tnfn1_pw060419p02q150 | FTN_0090 | 52 | Phagosomal | |
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | 45 | Cytoplasmic |
| tnfn1_pw060328p06q174 | FTN_0125 | ackA | 80 | Phagosomal |
| tnfn1_pw060323p06q113 | FTN_0420 | 37 | Cytoplasmic | |
| tnfn1_pw060323p05q182 | FTN_0504 | 67 | Phagosomal | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | 28 | Cytoplasmic |
| tnfn1_pw060510p02q157 | FTN_0511 | 72 | Phagosomal | |
| tnfn1_pw060420p01q123 | FTN_0524 | asd | 64 | Phagosomal |
| tnfn1_pw060323p06q194 | FTN_0527 | thrC | 58 | Phagosomal |
| tnfn1_pw060510p03q168 | FTN_0598 | 42 | Cytoplasmic | |
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | 68 | Phagosomal |
| tnfn1_pw060328p04q196 | FTN_0746 | alr | 72 | Phagosomal |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | 78 | Phagosomal |
| tnfn1_pw060420p04q108 | FTN_0822 | 76 | Phagosomal | |
| tnfn1_pw060420p03q153 | FTN_0840 | mdaB | 81 | Phagosomal |
| tnfn1_pw060420p02q175 | FTN_0877 | cls | 70 | Phagosomal |
| tnfn1_pw060328p06q142 | FTN_0954 | 85 | Phagosomal | |
| tnfn1_pw060420p04q140 | FTN_0957 | 24 | Cytoplasmic | |
| tnfn1_pw060420p01q130 | FTN_0965 | 83 | Phagosomal | |
| tnfn1_pw060328p01q151 | FTN_0983 | 66 | Phagosomal | |
| tnfn1_pw060328p06q184 | FTN_1061 | 70 | Phagosomal | |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 18 | Cytoplasmic |
| tnfn1_pw060328p03q107 | FTN_1222 | kpsF | 68 | Phagosomal |
| tnfn1_pw060420p04q194 | FTN_1231 | gloA | 87 | Phagosomal |
| tnfn1_pw060420p02q174 | FTN_1233 | 34 | Cytoplasmic | |
| tnfn1_pw060510p01q142 | FTN_1333 | tktA | 85 | Phagosomal |
| tnfn1_pw060418p02q109 | FTN_1376 | 78 | Phagosomal | |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | 87 | Phagosomal |
| tnfn1_pw060328p02q165 | FTN_1523 | 46 | Cytoplasmic | |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | 90 | Phagosomal |
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | 23 | Cytoplasmic |
| tnfn1_pw060510p01q146 | FTN_1597 | prfC | 80 | Phagosomal |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | 42 | Cytoplasmic* |
| tnfn1_pw060420p04q169 | FTN_1621 | 70 | Phagosomal | |
| tnfn1_pw060323p04q160 | FTN_1655 | rluC | 72 | Phagosomal |
| Transporter Proteins | ||||
| tnfn1_pw060420p04q149 | FTN_0008 | 35 | Cytoplasmic | |
| tnfn1_pw060418p04q168 | FTN_0141 | 25 | Cytoplasmic | |
| tnfn1_pw060323p03q141 | FTN_0619 | 26 | Cytoplasmic | |
| tnfn1_pw060323p06q164 | FTN_0624 | 83 | Phagosomal | |
| tnfn1_pw060328p06q132 | FTN_0728 | 65 | Phagosomal | |
| tnfn1_pw060323p01q177 | FTN_0799 | emrE | 34 | Cytoplasmic |
| tnfn1_pw060328p04q109 | FTN_0885 | 76 | Cytoplasmic | |
| tnfn1_pw060328p04q167 | FTN_0997 | 80 | Phagosomal | |
| tnfn1_pw060323p07q172 | FTN_1344 | 70 | Phagosomal | |
| tnfn1_pw060420p02q182 | FTN_1441 | 24 | Cytoplasmic* | |
| tnfn1_pw060420p01q189 | FTN_1611 | 87 | Phagosomal | |
| tnfn1_pw060510p01q152 | FTN_1711 | tyrP | 72 | Phagosomal |
| DNA Modification | ||||
| tnfn1_pw060510p02q141 | FTN_0133 | 81 | Phagosomal | |
| tnfn1_pw060323p03q122 | FTN_0577 | mutL | 31 | Cytoplasmic |
| tnfn1_pw060510p04q193 | FTN_0680 | uvrC | 60 | Phagosomal |
| tnfn1_pw060420p03q134 | FTN_0710 | 75 | Phagosomal | |
| tnfn1_pw060419p04q152 | FTN_1017 | 23 | Cytoplasmic | |
| tnfn1_pw060328p04q156 | FTN_1027 | ruvC | 60 | Phagosomal |
| tnfn1_pw060510p01q114 | FTN_1073 | 86 | Phagosomal | |
| tnfn1_pw060510p01q153 | FTN_1154 | 69 | Phagosomal | |
| tnfn1_pw060323p03q167 | FTN_1197 | recR | 27 | Cytoplasmic |
| Transferases | ||||
| tnfn1_pw060323p03q119 | FTN_0019 | pyrB | 63 | Phagosomal |
| tnfn1_pw060510p01q103 | FTN_0063 | ilvE | 68 | Phagosomal |
| tnfn1_pw060323p03q121 | FTN_0343 | 26 | Cytoplasmic | |
| tnfn1_pw060328p03q179 | FTN_0358 | 28 | Cytoplasmic | |
| tnfn1_pw060420p02q180 | FTN_0483 | 77 | Phagosomal | |
| tnfn1_pw060323p06q168 | FTN_0545 | 68 | Phagosomal | |
| tnfn1_pw060510p01q158 | FTN_0988 | prmA | 78 | Phagosomal |
| tnfn1_pw060510p02q144 | FTN_1234 | queA | 67 | Phagosomal |
| tnfn1_pw060418p04q172 | FTN_1418 | manC | 68 | Phagosomal |
| tnfn1_pw060510p01q119 | FTN_1428 | wbtO | 82 | Phagosomal |
| Transcription/Translation | ||||
| tnfn1_pw060510p03q168 | FTN_0598 | 42 | Cytoplasmic | |
| tnfn1_pw060419p04q129 | FTN_1290 | mglA | 71 | Phagosomal |
| Type IV Pilin | ||||
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | 66 | Phagosomal |
| tnfn1_pw060323p06q157 | FTN_1139 | pilO | 67 | Phagosomal |
| Others | ||||
| tnfn1_pw060323p08q110 | FTN_0286 | 73 | Phagosomal | |
| tnfn1_pw060420p01q168 | FTN_0646 | cscK | 77 | Phagosomal |
| tnfn1_pw060323p06q115 | FTN_0768 | tspO | 74 | Phagosomal |
| tnfn1_pw060328p06q167 | FTN_0985 | 37 | Cytoplasmic | |
| tnfn1_pw060419p02q137 | FTN_1034 | rnfB | 78 | Phagosomal |
| tnfn1_pw060420p02q177 | FTN_1145 | era | 84 | Phagosomal |
| tnfn1_pw060328p06q178 | FTN_1241 | 76 | Phagosomal | |
| tnfn1_pw060420p02q179 | FTN_1263 | comL | 79 | Phagosomal |
| tnfn1_pw060328p03q154 | FTN_1453 | 73 | Phagosomal | |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | 81 | Phagosomal |
| Intergenic | ||||
| tnfn1_pw060323p03q164 | intergenic | 28 | Cytoplasmic | |
| tnfn1_pw060323p08q139 | intergenic | 80 | Phagosomal | |
| tnfn1_pw060328p06q190 | intergenic | 35 | Cytoplasmic | |
| tnfn1_pw060419p04q165 | intergenic | 65 | Phagosomal | |
| tnfn1_pw060419p04q189 | intergenic | 89 | Phagosomal | |
| tnfn1_pw060510p01q102 | intergenic | 90 | Phagosomal | |
| tnfn1_pw060510p01q112 | intergenic | 78 | Phagosomal | |
| tnfn1_pw060510p01q135 | intergenic | 87 | Phagosomal | |
Mutants with discrepancy between LAMP-1 co-localization and Phagosomal integrity assay.
Fig. 2. Co-localization of the FCP of selected mutants with LAMP-1 in U937 macrophages.
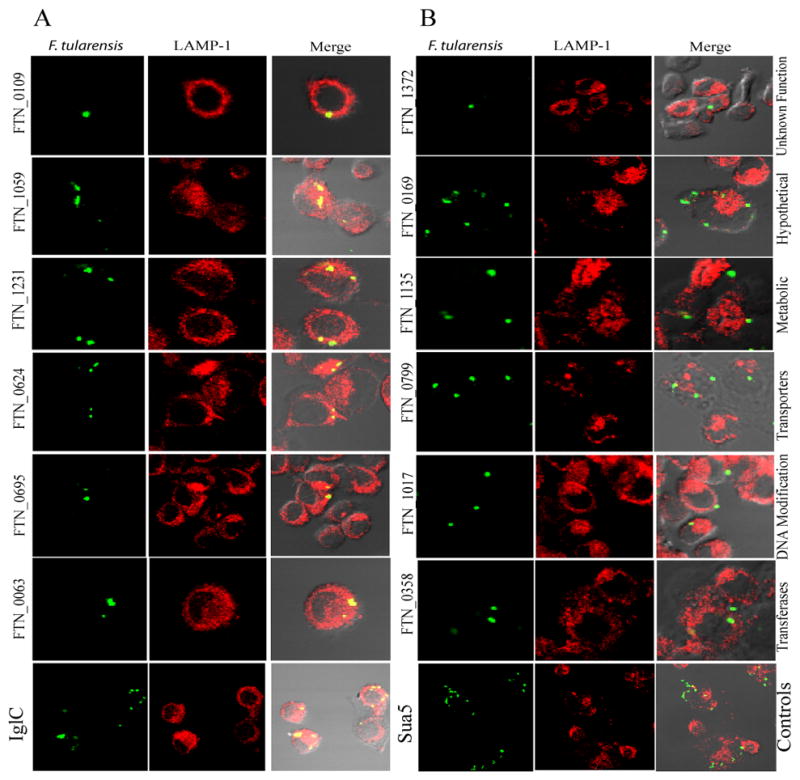
The cells were infected with each of the 125 mutants defective in intra-macrophage proliferation to determine co-localization of the phagosome with LAMP-1 at 6h post-infection. Bacteria were labeled with goat polyclonal antibody (green) and LAMP-1 was labeled with mouse monoclonal antibody (red). Representative confocal images of phagosomes harboring representative mutants from each functional group showing co-localization with Lamp-1 similar to the IglC mutant (A) or no co-localization, similar to the Sua5 WT-like phenotype (B). Data analyses were based on 100 infected cells analyzed from two different coverslips and the data were reproducible in two independent experiments.
Fig. 3. Functional categories of mutants that co-localize with LAMP-1.
U937 macrophages were infected with each of the 125 mutants severely defective in intra-macrophage proliferation to determine co-localization of the phagosome with LAMP-1 at 6h post-infection. Mutants were grouped according to the function of the mutated genes. Percentages of mutants in each functional group that co-localized persistently with LAMP-1 are shown.
To confirm that our observations of alterations in trafficking was not limited to the U937 macrophage cell line, the six mutants that were tested for intracellular proliferation in human monocytes-derived macrophages (hMDMs) were also examined for co-localization of the FCP with LAMP-1. Our data showed that the FCP of all the six mutants co-localized with LAMP-1 at a similar level in both U937 macrophages and hMDMs (Table 3). This indicates that the observed phenotypes for the mutants is not restricted to the U937 human macrophage cell line but is also exhibited in primary human-derived macrophages. These results indicate that most of the mutants defective in intracellular replication exhibit aberrant trafficking within human macrophages. Our findings indicate the complexity of the regulatory mechanisms that control modulation of biogenesis of the FCP.
Escape of the defective mutants into the host cell cytosol
To decipher the molecular bases of phagosomal escape, we examined the 125 mutants with ≥103 fold reduction in CFU at 24h post-infection in human-derived macrophages to examine their ability to escape into the macrophage cytosol. We utilized the fluorescence-based phagosome integrity assay to differentially label bacteria that are cytosolic/or within a compromised phagosome and those enclosed within an intact phagosome. This is achieved by loading the host cell cytosol with anti-F. tularensis antibody after preferential permeabilization of the plasma membrane, as previously described (Checroun et al., 2006; Santic et al., 2008). Among the 125 tested mutants, 31 bound the polyclonal anti-F. tularensis antibody, indicating their escape into the cytosol or localization in a compromised phagosome, similar to the positive control (Table 5 and Fig. 4). The other 94 mutants were not labeled with the anti-F. tularensis antibody loaded into the macrophage cytosol, indicating they were localized within intact phagosomes that were not permeable to the polyclonal antibody (Table 5). However, it is possible that the epitope recognized by the antibody might be altered, masked, or absent in few of the mutants that successfully escaped into the cytosol and were accessible to the antibody (see below). Results of the phagosomal escape assays were further confirmed by TEM for 6 randomly selected mutants, 4 of which escaped similar to the wild type strain while 2 (pyrB and wzb) were defective in phagosomal escape, similar to the iglC mutant (Fig 5). It is most likely that not all the genes are directly involved in phagosomal escape but are required for general bacterial fitness and adaptation to the acidified micro-environment within the FCP. Our data indicate that tremendous metabolic reprogramming is exhibited by F. tularensis during its short residence within the FCP. Overall, the results indicate a remarkable molecular complexity governs phagosomal escape of F. tularensis.
Fig. 4. Representative mutants analyzed for their escape into the cytosol of U937 macrophages.
U937 macrophages were infected with each of the 125 mutants of F. tularensis defective in intra-macrophage proliferation. Cytosolic bacteria were labeled with goat polyclonal antibody (red) loaded into the macrophage cytosol followed by permeabilization of all cellular membranes and labeling of all intracellular bacteria using mouse monoclonal antibody (green). Representative confocal images of F. tularensis mutant defective in phagosomal escape similar to the iglC mutant (A) and mutants that exhibit wild type-like Sua5 phenotype (B). Data analyses were based on 100 infected cells analyzed from two different coverslips and the data were reproducible in two independent experiments.
Fig. 5. Ultra-structural characterization of phagosomal escape.
U937 macrophages were infected with F. tularensis for 1 h followed by 1 h of gentamicin treatment. After a total of 6 h, the infected cells were analyzed by TEM to determine whether bacteria were within intact or disrupted phagosomes. A) Six mutants were selected randomly to determine their phagosomal escape; B) Five mutants with discrepancy in the results of the fluorescence-based phagosome integrity assays and LAMP-1 co-localization were examined by TEM for their phagosomal escape.
Surprisingly, there was discrepancy in the result of the LAMP-1 co-localization and the phagosome integrity assay for 5 mutants. One of the mutants (FTN_0788) that co-localized persistently with LAMP-1 had disrupted phagosome, as determined by fluorescence analyses. In contrast, the FCPs of the other 4 mutants (FTN_0384, FTN_0696, FTN_1619, and FTN_FTN_1441) were intact but were LAMP-1 negative (Table 5). Therefore, TEM was performed on the 5 mutants to decipher the reason for this discrepancy at the ultra-structural level. Our results showed that the four mutants that did not co-localize with LAMP-1 escaped into the cytosol, when examined by TEM (Fig. 5B). The other mutant that co-localized with LAMP-1 was found within intact phagosome. This indicated that our findings by TEM were more consistent with our finding related to LAMP-1 co-localization for these 5 mutants. Thus, at least 91 loci are required for modulation of phagosome biogenesis and escape of F. tularensis into the cytosol and at least 34 loci are required for proliferation within the cytosol but play no detectable role in phagosome biogenesis.
Comparison of the phenotype of the intra-macrophage growth-defective mutants to the phenotype within arthropod-derived cells
A concurrent study from our lab identified mutants that exhibited growth defect in D. melanogaster-derived S2 cells (see accompanying manuscript). To assess whether similar molecular mechanisms are utilized by F. tularensis to infect arthropod-derived cells and human macrophages, we compared the mutants identified to be defective in human macrophages to the mutants identified to be defective in D. melanogaster S2 cells. Among the 202 mutants defective in replication in human macrophages 135 of them were also required for replication in D. melanogaster S2 cells (see accompanying manuscript). This large number of loci indicates that common loci are utilized by F. tularensis to proliferate within both human macrophages and insect-derived cells. However, there are distinct molecular mechanisms required for replication in the two evolutionarily-distant host cells as more than 30% of the genes required for intracellular replication are specific to human macrophages.
Discussion
Most intracellular pathogens either escape the phagosome or divert phagosome maturation to an idiosyncratic niche where they replicate. Like other cytosolic bacteria, F. tularensis escapes from the phagosome into the cytosol where it replicates (Golovliov et al., 2003b; Santic et al., 2005a; Santic et al., 2005b; Checroun et al., 2006; Santic et al., 2007; Bonquist et al., 2008; Santic et al., 2008; Qin et al., 2009). Previous studies have shown that phagosomal escape is indispensable for the pathogenesis of tularemia (Santic et al., 2005b; Bonquist et al., 2008; Qin et al., 2009). Various mutagenesis screens have been performed to identify genes that are essential for the replication of F. tularensis in mouse models, in macrophages and hepatic cells (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009), but none identified mutants that fail to modulate phagosome biogenesis or fail to escape into the cytosol, which are the two major steps in the ability of F. tularensis to proliferate intracellularly and cause disease. Previous studies have shown that the three FPI gene products IglC, VgrG, and IglI, the MglA regulator, four acid phosphatases, and a lipoprotein are required for escape of F. tularensis subsp novicida into the host cell cytosol but the mechanism is not known (Barker et al., 2009; Santic et al., 2005b; Bonquist et al., 2008; Mohapatra et al., 2008; Qin et al., 2009). The FPI encodes a type VI-like secretion apparatus, and at least VgrG and IglI are secreted into the host cell cytosol, and the translocation of IglI is FPI-dependent (Barker et al., 2009). Since the FPI-encoded type VI-like secretion apparatus is essential for phagosome biogenesis and bacterial escape into the cytosol, it is most likely that the bacterial effectors directly involved in modulation of phagosome biogenesis and lysis of the phagosomal membranes are translocated by the type VI-like secretion apparatus encoded by the FPI.
Since mutants defective in phagosomal escape do not replicate in macrophages, our primary screen focused on identification of mutants that do not replicate in human macrophages followed by studies on phagosome biogenesis and a fluorescence-based phagosome integrity assay to identify the mutants that fail to escape into the cytosol. Transposon insertion in 202 genes result in ≥100 fold reduction in the number of cfus at 24h post-infection, compared to the wild type strain. Our data do not exclude mutants defective in attachment/entry into macrophages as well as polar effects of the insertion on downstream genes. Remarkably, the genes required for intracellular replication are involved in various physiological functions particularly in metabolic activities and nutrient transport. Interestingly, a large number of the identified genes encode proteins of unknown function or hypothetical proteins. Analysis of the function of these genes would shed light on the mechanisms of intracellular proliferation of F. tularensis. Importantly, only 25% of the genes we identified have been identified in other screens to be required for various aspects of virulence of F. tularensis (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009). This indicates the comprehensiveness and the power of our screen that is aimed at identification of bacterial loci required for phagosome biogenesis and escape into the cytosol, which are the two major steps essential for subsequent proliferation and manifestation of disease.
The FCP co-localizes transiently with late endosomal/lysosomal markers before bacterial escape into the cytosol (Pechous et al., 2009; Santic et al., 2010). Although the FCP harboring the mglA, iglC and FTT1103 mutants co-localize persistently with LAMPs and mature into a phagolysosome (Santic et al., 2005b; Bonquist et al., 2008; Qin et al., 2009), it is not known whether these mutants are defective in evasion of lysosomal fusion or escape into the cytosol. Our results show that the FCP of 91/125 mutants severely defective in intra-macrophage growth co-localize persistently with the late endosomal marker LAMP-1, and these mutants fail to escape into the cytosol of human macrophages. However, it is unlikely that all these genes are directly involved in bacterial escape into the cytosol but are required for general bacterial fitness and adaptation to the acidic micro-environment within the FCP, which is essential for modulation of phagosome biogenesis and bacterial escape into the cytosol (Santic et al., 2005b; Bonquist et al., 2008; Qin et al., 2009). Indeed, depending on the availability of pyrimidine nucleotide, auxotrophic mutants (carA, carB and pyrB) of F. tularensis subsp holarctica-derived LVS strain can either escape from the phagosome and grow in the cytosol or remain trapped in the phagosome (Schulert et al., 2009). Similar to LVS, it is most likely that similar mechanisms of nutritional and micro-environmental stresses within the FCP affect phagosomal escape of F. tularensis subsp novicida. The large numbers of various functional groups of mutants defective in escape into the cytosol suggest that the phagosome is not a hospitable environment for the growth of F. tularensis and that tremendous adaptation to the phagosomal micro-environment along with extensive metabolic reprogramming is required for successful phagosomal escape.
Interestingly, 4 of the mutants that have been shown to be defective in phagosomal escape, based on the fluorescence-based phagosome integrity assays, do not co-localize persistently with LAMP-1. In addition, one of the mutants that escape into the cytosol co-localize with LAMP-1. However, ultra-structural studies have clearly shown that this mutant is localized to an intact FCP while the other 4 mutants that are LMAP-1 negative are cytosolic, which is consistent for all the 5 mutants for correlation of LAMP-1 co-localization with the FCP. It is likely that in the phagosome integrity assay, failure of the antibody to bind cytosolic bacteria is due to alteration, masking, or absence of the epitope recognized by the antibody.
Many other bacteria, including L. monocytogenes, S. flexneri, B. pseudomallei and Rickettsia spp. escape into the host cell cytosol where they replicate (Ray et al., 2009). The mechanisms of phagosomal escape are best studied in L. monocytogenes which requires the activity of listeriolysin as well as two other phospholipases to escape into the cytosol (Ray et al., 2009). Similarly, F. tularensis resides in phagosomes that transiently acquire endosomal markers and become acidified within 15–30 min prior to rapid escape into the cytosol by 30–60 min (Pechous et al., 2009; Santic et al., 2010). The MglA regulator, four acid phosphatases and a lipoprotein are required for escape of F. tularensis subsp novicida in addition to the 3 FPI-encoded proteins IglC, VgrG, and IglI, but the mechanism is not known (Barker et al., 2009; Pechous et al., 2009; Santic et al., 2010). However, none of these proteins possess any properties of a cytoylsin. Although F. tularensis subsp novicida exhibits hemolytic activity (Lai et al., 2003), no hemolysin homologues have been identified in any of the F. tularensis subspecies whose genomes have been sequenced (Larsson et al., 2005; Petrosino et al., 2006; Beckstrom-Sternberg et al., 2007; Chaudhuri et al., 2007; Rohmer et al., 2007). Importantly, 3 out of the 4 acid phosphatase identified previously (Mohapatra et al., 2008) have been found to be required for phagosomal escape in the present study. The diversity of genes that affect trafficking and phagosomal escape shows that F. tularensis is unlike other cytosolic pathogens that utilize cytolysins, pore-forming toxins, or hydrolytic enzymes to escape into the host cell cytosol. It rather requires a complex array of network of genes involved in various physiological functions to orchestrate its fitness to the acidic micro-environment in response to the environmental and nutritional cues within the FCP to enable efficient phagosomal escape. Importantly the type VI-like secretion apparatus encoded by the FPI seems to be essential for translocation of some secreted proteins (Barker et al., 2009) and is required for phagosomal escape (Barker et al., 2009; Pechous et al., 2009; Santic et al., 2010), suggesting that the effector involved in escape is likely to be translocated through this system.
Importantly, 34 of the 125 mutants with severe defect in intracellular replication escape into the cytosol. The growth defect of some of these mutants in the cytosol may be due to increased sensitivity to host antimicrobial cytosolic factors, inability to acquire nutrients, failure to modulated host cytosolic processes, or failure to adapt to the cytosolic micro-environment.
Interestingly, among 135 common F. tularensis loci identified to be required for intracellular proliferation within both human macrophages and D. melanogaster-derived S2 cells, 59 are required for phagosomal escape in human-derived macrophages (see accompanying manuscript). These data suggest that some common molecular mechanisms are utilized by F. tularensis to escape from the phagosome in mammalian and arthropod-derived cells. This may not be surprising, considering our recent findings that phagosome biogenesis and bacterial escape into the cytosol are very similar in both evolutionarily-distant host cells (Santic et al., 2009). However, our data clearly show that distinct mechanisms are also employed by F. tularensis to escape into the cytosol of the two evolutionarily distant host cells.
Our results indicate that F. tularensis requires a plethora of networks of genes to orchestrate its fitness during transient residence within the acidified FCP for efficient phagosomal escape and subsequent replication within the host cell cytosol, and that at least 34 loci are indispensable for proliferation within the cytosol. These networks of genes are potential targets for therapy and vaccination against tularemia, since phagosomal escape is the major step in the ability of this pathogen to proliferate intracellularly and cause disease.
Experimental procedures
Bacterial strains, tissue culture and Media
F. tularensis subsp. novicida strain U112 and its isogenic mutants mglA and iglC have been described previously (Lauriano et al., 2004). The construction of the F. tularensis subsp. novicida mutants library which was obtained from Biodefence and Emerging Infections Resource Repository (http://www.beiresources.org) has been described previously (Gallagher et al., 2007). All F. tularensis subsp novicida strains were grown on tryptic soy agar (TSA) plates for 2 days or in tryptic soy broth (TSB) supplemented with 0.1% cysteine and 10 mg/ml of kanamycin overnight. U937 macrophages were maintained at 37°C and 5% CO2 in RPMI-1640 tissue culture medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco BRL). The U937 macrophages were differentiated for 48 h using 50μg/ml of phorbol 12-myristate 13-acetate (PMA). The hMDMs were obtained and maintained as we described previously
Intracellular growth in U937 Macrophages
Infection of U937 macrophages with F. tularensis subsp. novicida was performed as described previously (Santic et al., 2005a). Briefly, U937 cells were seeded in 96-well plate at a concentration of 1×106 cells/ml of RPMI and differentiated for 48 h with PMA. Differentiated cells were infected with F. tularensis subsp. novicida and its isogenic mutants at MOI of 10 (1×107 bacteria/ml of RPMI) for 1 h followed by 1 h of gentamicin (50μgml−1) treatment. MOI was determined for each infecting strain by measuring absorbance of bacterial suspension at 550 nm. Infections were done in duplicate for each strain. To synchronize the infection, infected cells were centrifuged at 150 x g for 5 min before incubation at 37°C in 5% CO2. Cells were then incubated with fresh RPMI for 22 hrs. The supernatant was removed and infected cells were lysed with 200μl of sterile water. The supernatant was recombined with the lysate and serial dilutions were plated on agar plates for colony enumeration. The experiment was done twice to confirm the results.
Co-localization of F. tularensis with LAMP-1 in U937 Cells
Infections were performed as described above in 24-well plate with glass coverslips. At 6 h post-infection, cells were fixed with 3.7% formaldehyde for 30 min. After washing 3 times with 1X PBS, cells were permeabilized with 0.1% triton X 100 for 15 min on ice followed by 3 times wash with 1X PBS. Cells were subsequently treated with goat polyclonal anti F. tularensis subsp. novicida antibody (Genscript) at 1:4000 dilution and mouse monoclonal anti LAMP-1 antibody (Hybridoma library, University of Iowa) at 1:500 dilution. After 1 hr incubation the cells were washed 3 times and treated with Alexa fluor 488-conjugated anti goat antibody and Alexa fluor 555-conjugated anti mouse antibody (Molecular probes) at 1:4000 dilution. Co-localization of bacteria with LAMP-1 was analyzed with FV1000 Olympus confocal microscope as described previously (Santic et al., 2005a; Santic et al., 2005b; Santic et al., 2008). At least 100 infected cells from more than 10 different fields were analyzed. Experiments were done in duplicate for each strain.
Phagosome integrity assays
We utilized fluorescence-based phagosome integrity assay to determine phagosomal escape into the macrophage cytosol, as described previously (Checroun et al., 2006; Santic et al., 2008). Infections were performed as described for the LAMP-1 experiment above. At 6 h post-infection, infected macrophages washed quickly with 1X PBS and permeabilized with 80μg/ml digitonin in 1X PBS containing. 1:1000 dilution of goat polyclonal anti F. tularensis subsp novicida antibody (Genscript) for 10 min. Subsequently, 1:1000 dilution of goat polyclonal anti F. tularensis subsp. novicida antibody was loaded into the macrophage cytosol for another 40 min., as we described previously (Santic et al., 2005a; Santic et al., 2005b; Santic et al., 2008). The infected cells were was 3 times with 1X PBS and fixed for 30 min with 3.7% formaldehyde. The cells were washed 3 times with 1X PBS and permeabilized with 0.1% triton X 100. After washing 3 times with 1X PBS, the cells were treated with mouse monoclonal anti F. tularensis subsp. novicida antibody (a gift from John Gunn, Ohio State University) at 1:200 dilution followed by 3 times washing with 1X PBX and treatment with Alexa fluor 555-conjugated anti goat antibody and Alexa fluor 488-conjugated anti mouse antibody at 1:4000 dilution. Localization of bacteria was analyzed with FV1000 Olympus confocal microscope at our imaging suite. At least 100 infected cells from more than 10 different fields were analyzed.
Transmission Electron Microscopy
The use of Transmission Electron Microscope (TEM) to analyze escape of F. tularensis subsp. novicida into the cytosol of U937 macrophages has been described previously (Santic et al., 2005a). Briefly, monolayers of U937 macrophages were infected with F. tularensis subsp. novicida in 12-well plates at an MOI of 10 for 1 h followed by gentamicin treatment. At 6 h, post-infection, the infected U937 macrophages were processed and sections were stained with Uranyl Acetate and lead citrate and examined with a Hitachi H-7000/STEM electron microscope at 80 kV as described previously (Santic et al., 2005a).
Table 4.
Classification of metabolic genes according to metabolic pathways
| Amino acid metabolism | |||
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060328p06q174 | FTN_0125 | ackA | propionate kinase 2/acetate kinase A |
| tnfn1_pw060323p05q182 | FTN_0504 | lysine decarboxylase | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 |
| tnfn1_pw060510p02q154 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060510p02q157 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060510p04q157 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060510p04q157 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060323p06q194 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060510p01q172 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060510p03q172 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060510p03q171 | FTN_0588 | asparaginase | |
| tnfn1_pw060328p04q196 | FTN_0746 | alr | alanine racemase |
| tnfn1_pw060328p06q142 | FTN_0954 | histidine acid phosphatase | |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase |
| tnfn1_pw060328p04q116 | FTN_9765 | choloylglycine hydrolase family protein | |
| tnfn1_pw060510p04q185 | FTN_1701 | glutamate decarboxylase | |
| Carbohydrate metabolism | |||
| tnfn1_pw060420p01q123 | FTN_0524 | asd | aspartate semialdehyde dehydrogenase |
| tnfn1_pw060419p03q116 | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit |
| tnfn1_pw060328p02q139 | FTN_0621 | eno | enolase (2-phosphoglycerate dehydratase) |
| tnfn1_pw060510p02q187 | FTN_1018 | aldolase/adducin class II family protein | |
| tnfn1_pw060328p03q107 | FTN_1222 | kpsF | phosphosugar isomerase |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase |
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | glycerol-3-phosphate dehydrogenase |
| tnfn1_pw060419p04q130 | FTN_1585 | glpK | glycerol kinase |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | cytochrome bd-II terminal oxidase subunit I |
| tnfn1_pw060328p02q105 | FTN_1620 | appB | cytochrome bd-II terminal oxidase subunit II |
| tnfn1_pw060510p04q136 | FTN_1767 | rbsK | ribokinase, pfkB family |
| Nucleotide metabolism | |||
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060510p02q160 | FTN_0021 | carA | carbamoyl-phosphate synthase small chain |
| tnfn1_pw060418p03q133 | FTN_0199 | cyoE | heme O synthase |
| tnfn1_pw060323p06q113 | FTN_0420 | SAICAR synthetase/phosphoribosylamine-glycine ligase | |
| tnfn1_pw060510p01q159 | FTN_0695 | add | deoxyadenosine deaminase/adenosine deaminase |
| tnfn1_pw060328p01q151 | FTN_0983 | bifunctional protein: glutaredoxin 3/ribonucleotide reductase beta subunit | |
| tnfn1_pw060419p04q135 | FTN_1415 | thioredoxin | |
| tnfn1_pw060419p04q181 | FTN_1415 | thioredoxin | |
| tnfn1_pw060420p04q116 | FTN_1421 | wbtH | glutamine amidotransferase/asparagine synthase |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase |
| tnfn1_pw060323p04q160 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C |
| tnfn1_pw060510p02q165 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C |
| Reductive Metabolism | |||
| tnfn1_pw060419p03q169 | FTN_0218 | nfnB | dihydropteridine reductase |
| tnfn1_pw060418p02q128 | FTN_0633 | katG | peroxidase/catalase |
| tnfn1_pw060328p01q128 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060420p03q153 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060420p04q194 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060510p02q164 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060510p04q146 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060418p01q131 | FTN_1557 | oxidoreductase iron/ascorbate family protein | |
| tnfn1_pw060418p04q111 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | |
| tnfn1_pw060418p04q112 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | |
| tnfn1_pw060420p04q169 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | |
| Lipid Metabolism | |||
| tnfn1_pw060420p02q175 | FTN_0877 | cls | cardiolipin synthetase |
| tnfn1_pw060420p04q140 | FTN_0957 | short chain dehydrogenase | |
| conenzyme synthesis | |||
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | quinolinate sythetase A |
| tnfn1_pw060419p04q164 | FTN_0692 | nadA | quinolinate sythetase A |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | biotin--acetyl-CoA-carboxylase ligase |
| tnfn1_pw060418p01q124 | FTN_1245 | iscS | cysteine desulfurase |
| tnfn1_pw060419p03q126 | FTN_1278 | nadE | NAD synthase |
| tnfn1_pw060323p04q110 | FTN_1678 | nuoC | NADH dehydrogenase I, C subunit |
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | riboflavin synthase beta-chain |
| tnfn1_pw060420p02q191 | FTN_0113 | ribC | riboflavin synthase alpha chain |
| Peptidoglycan biosynthesis | |||
| tnfn1_pw060323p04q102 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| tnfn1_pw060418p03q177 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| Aromatic compound biosynthesis | |||
| tnfn1_pw060420p04q108 | FTN_0822 | para-aminobenzoate synthase component I | |
| tnfn1_pw060323p08q141 | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060420p01q129 | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase |
| ppGpp biosynthesis | |||
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
Acknowledgments
YAK is supported by Public Health Service Awards R01AI43965 and R01AI069321 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund.
References
- Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, Liu J, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstrom-Sternberg SM, Auerbach RK, Godbole S, Pearson JV, Beckstrom-Sternberg JS, Deng Z, et al. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS ONE. 2007;2:e947. doi: 10.1371/journal.pone.0000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun. 2008;76:3502–3510. doi: 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, et al. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Ren CP, Desmond L, Vincent GA, Silman NJ, Brehm JK, et al. Genome sequencing shows that European isolates of Francisella tularensis subspecies tularensis are almost identical to US laboratory strain Schu S4. PLoS ONE. 2007;2:e352. doi: 10.1371/journal.pone.0000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W, Kuhn M. Bacterial replication in the host cell cytosol. Curr Opin Microbiol. 2000;3:49–53. doi: 10.1016/s1369-5274(99)00050-8. [DOI] [PubMed] [Google Scholar]
- Golovliov I, Sjostedt A, Mokrievich A, Pavlov V. A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett. 2003a;222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003b;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, et al. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun. 2009;77:232–244. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai XH, Wang SY, Edebro H, Sjostedt A. Francisella strains express hemolysins of distinct characteristics. FEMS Microbiol Lett. 2003;224:91–95. doi: 10.1016/S0378-1097(03)00431-2. [DOI] [PubMed] [Google Scholar]
- Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, et al. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Oyston PC, Chain P, Chu MC, Duffield M, Fuxelius HH, et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, et al. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect Immun. 2007;75:5376–5389. doi: 10.1128/IAI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra NP, Soni S, Reilly TJ, Liu J, Klose KE, Gunn JS. Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect Immun. 2008;76:3690–3699. doi: 10.1128/IAI.00262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Santic M, Asare R, Carabeo RA, Abu Kwaik Y. Rapid escape of the dot/icm mutants of Legionella pneumophila into the cytosol of mammalian and protozoan cells. Infect Immun. 2007;75:3290–3304. doi: 10.1128/IAI.00292-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigrovic LE, Wingerter SL. Tularemia. Infect Dis Clin North Am. 2008;22:489–504. ix. doi: 10.1016/j.idc.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- Pechous RD, McCarthy TR, Zahrt TC. Working toward the future: insights into Francisella tularensis pathogenesis and vaccine development. Microbiol Mol Biol Rev. 2009;73:684–711. doi: 10.1128/MMBR.00028-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino JF, Xiang Q, Karpathy SE, Jiang H, Yerrapragada S, Liu Y, et al. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J Bacteriol. 2006;188:6977–6985. doi: 10.1128/JB.00506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Thompson JA, Mann BJ. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77:152–161. doi: 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Marteyn B, Sansonetti PJ, Tang CM. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol. 2009;7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, Radey M, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Abu Kwaik Y. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol. 2005a;7:957–967. doi: 10.1111/j.1462-5822.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Al-Khodor S, Abu Kwaik Y. Cell biology and molecular ecology of Francisella tularensis. Cell Microbiol. 2010;12:129–139. doi: 10.1111/j.1462-5822.2009.01400.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Abu Kwaik Y. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006;14:37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005b;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Akimana C, Asare R, Kouokam JC, Atay S, Kwaik YA. Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ Microbiol. 2009;11:1473–1481. doi: 10.1111/j.1462-2920.2009.01875.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Barker JR, Klose KE, Dekanic A, Doric M, Abu Kwaik Y. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol. 2007;9:2391–2403. doi: 10.1111/j.1462-5822.2007.00968.x. [DOI] [PubMed] [Google Scholar]
- Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, Jones BD, Allen LA. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect Immun. 2009;77:1324–1336. doi: 10.1128/IAI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]