Summary
Arthropod vectors are important vehicles for transmission of Francisella tularensis between mammals, but very little is known about the F. tularensis-arthropod vector interaction. Drosophila melanogaster has been recently developed as an arthropod vector model for F. tularensis. We have shown that intracellular trafficking of F. tularensis within human monocytes-derived macrophages and D. melanogaster-derived S2 cells is very similar. Within both evolutionarily distant host cells, the Francisella-containing phagosome matures to a late endosome-like phagosome with limited fusion to lysosomes followed by rapid bacterial escape into the cytosol where the bacterial proliferate. To decipher the molecular bases of intracellular proliferation of F. tularensis within arthropod-derived cells, we screened a comprehensive library of mutants of F. tularensis subsp novicida for their defect in intracellular proliferation within D. melanogaster-derived S2 cells. Our data show that 394 genes, representing 22% of the genome, are required for intracellular proliferation within D. melanogaster-derived S2 cells, including many of the Francisella Pathogenicity Island (FPI) genes that are also required for proliferation within mammalian macrophages. Functional gene classes that exhibit growth defect include metabolic (25%), FPI (2%), Type IV pili (1%), transport (16%) and DNA modification (5%). Among 168 most defective mutants in intracellular proliferation in S2 cells, 80 are defective in lethality and proliferation within adult D. melanogaster. The observation that only 135 of the 394 mutants that are defective in S2 cells are also defective in human macrophages indicates that F. tularensis utilize common as well as distinct mechanisms to proliferate within mammalian and arthropod cells. Our studies will facilitate deciphering the molecular aspects of F. tularensis-arthropod vector interaction and its patho-adaptation to infect mammals.
Keywords: S2 cells, intracellular, tularemia, Drosophila, macrophages
Introduction
Francisella tularensis is a Gram-negative intracellular bacterium that causes tularemia in small mammals and humans (Ellis et al., 2002; Hazlett and Cirillo, 2009; Santic et al., 2010). Because of low infectivity, ease of dissemination, and high morbidity and mortality, F. tularensis is classified by the CDC as a category A select bioterrorism agent (Dennis et al., 2001). The bacteria are maintained in nature primarily through infection of rodents and lagomorphs, and are transmitted to humans through insects bites, skin abrasions, inhalation, or ingestion (Ellis et al., 2002; Hazlett and Cirillo, 2009; Santic et al., 2010).
There are four subspecies of F. tularensis, which are subspecies tularensis, holarctica, mediasiatica and novicida (Keim et al., 2007; Nigrovic and Wingerter, 2008). All the four subspecies share about 97% genomic identities (Champion et al., 2009; Larsson et al., 2009). Human infections are mostly caused by subspecies tularensis and holarctica. F. tularensis subspecies tularensis is restricted to North America while the other species are distributed across the northern hemisphere (Keim et al., 2007; Nigrovic and Wingerter, 2008). Unlike the other subspecies, F. tularensis subsp. novicida is attenuated in humans, but it causes disease in animal models similar to the virulent subspecies (Santic et al., 2006; Santic et al., 2010). In addition, it replicates intracellularly within human and mouse macrophages, which is an important step in the disease process in mammals (Oyston et al., 2004).
Vector-borne transmission of tularemia to mammalian hosts has an important role in pathogenesis of the disease, and F. tularensis-arthropod vector interaction has likely played a major role in bacterial ecology and maintenance in the environment (Keim et al., 2007). Deer flies, horse flies, ticks and mosquitoes are common arthropod vectors of transmission between mammals (Keim et al., 2007). Although transmission via arthropod vectors may play an important role in the infectious life cycle of F. tularensis and subsequent pathogenesis to mammalian host, very little is known about the interaction of F. tularensis with the arthropod vector at the molecular, cellular, and organism level.
The well-studied and genetically tractable arthropod Drosophila melanogaster has been developed as an arthropod host model for the study of host-pathogen interactions at the organism level for fungi, gram positive and gram negative bacteria (Apidianakis and Rahme, 2009). Recent studies have shown that adult flies or Drosophila cell line could be used as a model system to study Francisella pathogenesis (Vonkavaara et al., 2008; Santic et al., 2009). D. melanogaster is an attractive model system for a number of reasons: 1) Studies of the signal transduction cascade underlying the innate immune system in D. melanogaster have revealed striking similarities to the mammalian innate immune response (Hoffmann et al., 1999; Anderson, 2000); 2) In insects and mammals, Toll family receptors (Hoffmann et al., 1999; Anderson, 2000) trigger host innate immune responses that are highly conserved; 3) Amenability for genetic manipulations; and most importantly 4) Arthropods are vectors for transmission of tularemia between mammals.
The genetic tractability of Drosophila has enabled unbiased approaches to the identification of host-encoded factors that impact the pathogen-host interface both at the cellular and molecular levels (D’Argenio et al., 2001; Dionne et al., 2003; Kurz et al., 2003; Needham et al., 2004; Kim et al., 2008). Although many studies using Drosophila S2 cells as a model system has focused on RNAi screening of host factors required for the pathogen-host interaction (Cherry, 2008), no comprehensive screen has been performed to identify genes of Francisella or any other intracellular pathogens required for proliferation in arthropod-derived cells.
The sualB cell line from Anopheles gambiae has also been used as a model to study intracellular replication of F. tularensis (Read et al., 2008). Interestingly, trafficking of F. tularensis subsp novicida in D. melanogaster-derived S2 cells is similar to mammalian macrophages (Santic et al., 2009). Within both host cells, F. tularensis transiently occupies a late endosome-like phagosome followed by rapid bacterial escape into the cytosol, where the bacteria proliferate robustly (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a; Santic et al., 2005b; Checroun et al., 2006; Santic et al., 2007; Bonquist et al., 2008; Chong et al., 2008; Santic et al., 2008; Qin et al., 2009; Wehrly et al., 2009). This may suggest that some common mechanisms are utilized by F. tularensis to modulate phagosome biogenesis, escape into the cytosol, and to proliferate within mammalian and arthropod-derived cells.
Intracellular trafficking and robust intracellular proliferation of F. tularensis subsp novicida within mammalian macrophages and S2 cells is very similar. In addition, the ability of F. tularensis subsp novicida to infect D. melanogaster and mice has made F. tularensis subsp novicida a very useful model to dissect molecular basis of the intracellular infection by F. tularensis under BSL2 containment (Santic et al., 2006; Santic et al., 2010). Several genes within the 30-Kb pathogenicity island have been shown to be required for intracellular replication of F. tularensis within macrophages (Baron and Nano, 1998; Santic et al., 2005b; Bonquist et al., 2008; Schmerk et al., 2009). Similar to macrophages, intracellular replication of F. tularensis in S2 and SualB cells has been shown to be dependent on MglA, MglB, IglA, IglC, IglD as well as PdpA and PdpB (Read et al., 2008; Vonkavaara et al., 2008; Santic et al., 2009). We have shown that replication of F. tularensis subsp novicida within human macrophages involve a large percentage of the bacterial genome (see accompanying manuscript).
Since the molecular bases of intracellular proliferation of F. tularensis in arthropod-derived cells are not known, we screened a comprehensive sequence-defined mutant library of F. tularensis subsp novicida containing 3,050 alleles corresponding to 1448 non-essential genes for mutants defective in intracellular proliferation (Gallagher et al., 2007). Our data show that 394 genes, representing 22% of the genome, are required for replication within D. melanogaster S2 cells. Among 168 most defective mutants in S2 cells, 80 are required for replication and lethality to D. melanogaster adult flies. Interestingly, 135 of the 394 mutants that are defective in S2 cells are also defective in macrophages (see accompanying manuscript). Therefore, F. tularensis might have acquired some of the mechanisms to proliferate within mammalian cells through patho-adaptation to the arthropod host. However, additional distinct molecular mechanisms are also required for proliferate within both evolutionarily distant hosts.
Results and Discussion
Replication of F. tularensis mutants in S2 Cells
Arthropod vectors are important vehicles for transmission of. F. tularensis between mammals, but knowledge on the interaction between the bacteria and the arthropod host is very limited. The well-studied and genetically tractable arthropod model D. melanogaster has been recently explored as an arthropod vector model for F. tularensis subsp holarctica-derived LVS strain and F. tularensis subsp novicida with similar findings for both subsp (Vonkavaara et al., 2008; Santic et al., 2009). There are approximately 1800 genes in the genome of F. tularensis subsp novicida, of which 312 genes are essential. To identify the repertoire of genes essential for intracellular replication of F. tularensis in S2 cells, we screened F. tularensis subsp novicida mutant library containing 3050 sequence-defined multiple-allele insertion mutants corresponding to the 1448 non-essential genes of the genome (Gallagher et al., 2007). The S2 cells were seeded at 1 × 106 cells ml−1 in 96 well plates. Infections were performed at MOI of 10 for 1 h followed by 1 h of gentamicin treatment to kill extracellular bacteria, which resulted in infection of ~25% of the cells with an average of 1 bacterium/cell. At 24 h post-infection, cells were lysed and serial dilutions were plated on agar plates for colony enumeration. The CFU for the wild type strain at 24h post-infection was ~ 1×108 with slight variations between multiple experiments.
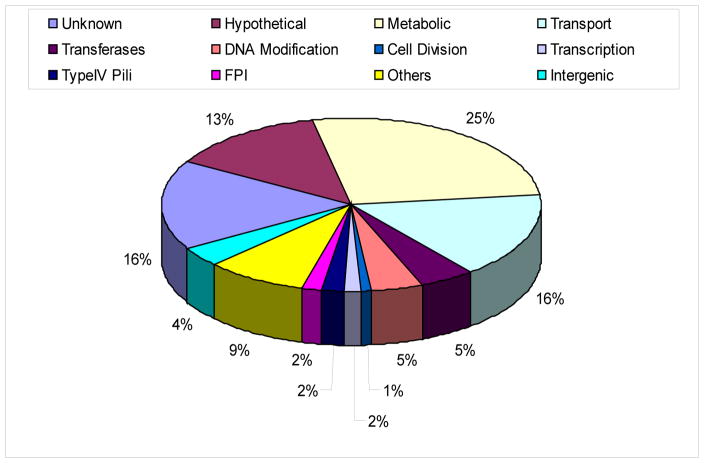
Our primary screen showed that 501 mutant alleles were defective in S2 cells, but the observed defect for some of the mutants may be to a defect in uptake, which was not accounted for in our large and comprehensive screen. To confirm findings of the primary screen, we re-screened the 501 mutants twice. For these infections, the OD of the bacteria for all the mutants was measured and adjusted to a similar OD for all infections, to ensure equivalent input. Our data confirmed that 476 alleles, corresponding to 394 genes, were consistently and reproducibly required for proliferation within S2 cells (Table 1 and 2). Remarkably, this represented about 22% of the genome. The defect in growth ranged from 10 fold reductions in growth for the less attenuated mutants to 107 fold reductions in growth for the severely attenuated mutants, compared to the wild type strain (Table 1). This indicates that while some of the mutants were completely attenuated for growth in S2 cells others exhibited only modest reduction in intracellular growth. About 38% of the mutants that were defective had insertions in either hypothetical proteins or proteins of unknown function (Fig. 1). Other functional gene classes that showed growth defect included metabolic (25%), FPI (2%), Type IV pili (1%), transport (16%), and DNA modification (5%) (Table 1 and Fig. 1). Clearly, there were a large number of genes that belong to proteins of unknown function or hypothetical proteins that are required for intracellular proliferation. It will be interesting to identify the functions of these proteins. This will facilitate deciphering the molecular mechanisms utilized by F. tularensis to proliferate in arthropod-derived cells. Many of the FPI genes were identified in our screen including iglC, iglD, pdpC and pdpD (Table 1 and 2).
Table 1.
List of growth-defective mutants of F. tularensis within S2 and U937 cells according to their functions.
| List of growth defective mutants in both U937 and S2 Cells | |||||
|---|---|---|---|---|---|
| Strain Name | Locus Tag | Gene | Description | Log reduction in Growth relative to WT | |
| Controls | |||||
| Wild type | 0 | 0 | |||
| Intracellular growth locus C | IglC | 5 | 5 | ||
| Proteins of unknown Function | |||||
| tnfn1_pw060323p08q148 | FTN_0027 | conserved protein of unknown function | 4 | 6* | |
| tnfn1_pw060510p03q161 | FTN_0027 | conserved protein of unknown function | 2 | 2* | |
| tnfn1_pw060323p03q103 | FTN_0041 | protein of unknown function | 5 | 2# | |
| tnfn1_pw060420p01q149 | FTN_0041 | protein of unknown function | 2 | 3# | |
| tnfn1_pw060420p04q143 | FTN_0149 | conserved protein of unknown function | 5 | 5 | |
| tnfn1_pw060323p02q193 | FTN_0275 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060419p03q124 | FTN_0275 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060510p02q121 | FTN_0275 | conserved protein of unknown function | 3 | 2# | |
| tnfn1_pw060420p04q134 | FTN_0297 | conserved protein of unknown function | 7 | 7 | |
| tnfn1_pw060328p05q119 | FTN_0444 | membrane protein of unknown function | 6 | 6# | |
| tnfn1_pw060420p03q175 | FTN_0444 | membrane protein of unknown function | 5 | 5# | |
| tnfn1_pw060323p07q141 | FTN_0788 | conserved protein of unknown function | 5 | 5 | |
| tnfn1_pw060420p04q176 | FTN_0855 | protein of unknown function | 5 | 2 | |
| tnfn1_pw060323p03q147 | FTN_0930 | protein of unknown function | 6 | 3# | |
| tnfn1_pw060323p05q150 | FTN_0930 | protein of unknown function | 6 | 3# | |
| tnfn1_pw060510p01q108 | FTN_0977 | conserved protein of unknown function | 7 | 7 | |
| tnfn1_pw060510p01q128 | FTN_1170 | conserved protein of unknown function | 3 | 3* | |
| tnfn1_pw060418p02q157 | FTN_1170 | conserved protein of unknown function | 5 | 6* | |
| tnfn1_pw060420p04q196 | FTN_1256 | membrane protein of unknown function | 4 | 5 | |
| tnfn1_pw060323p01q113 | FTN_1343 | conserved protein of unknown function | 4 | 4# | |
| tnfn1_pw060418p02q105 | FTN_1343 | conserved protein of unknown function | 4 | 4# | |
| tnfn1_pw060328p02q110 | FTN_1457 | protein of unknown function | 5 | 5# | |
| tnfn1_pw060420p02q183 | FTN_1457 | protein of unknown function | 6 | 6# | |
| tnfn1_pw060328p01q172 | FTN_1542 | conserved protein of unknown function | 2 | 2# | |
| tnfn1_pw060328p02q177 | FTN_1713 | protein of unknown function | 3 | 3 | |
| tnfn1_pw060328p06q155 | FTN_1764 | protein of unknown function | 6 | 7# | |
| Hypothetical Proteins | |||||
| tnfn1_pw060323p03q142 | FTN_0030 | hypothetical membrane protein | 4 | 3# | |
| tnfn1_pw060420p02q155 | FTN_0030 | hypothetical membrane protein | 3 | 3# | |
| tnfn1_pw060328p06q180 | FTN_0038 | hypothetical protein | 4 | 4# | |
| tnfn1_pw060419p02q127 | FTN_0038 | hypothetical protein | 2 | 2# | |
| tnfn1_pw060420p02q173 | FTN_0169 | conserved hypothetical membrane protein | 6 | 6* | |
| tnfn1_pw060510p01q193 | FTN_0169 | conserved hypothetical membrane protein | 5 | 5* | |
| tnfn1_pw060328p05q136 | FTN_0384 | conserved hypothetical protein | 4 | 7 | |
| tnfn1_pw060328p05q130 | FTN_0534 | conserved hypothetical membrane protein | 3 | 8 | |
| tnfn1_pw060418p01q143 | FTN_0556 | hypothetical protein | 7 | 7 | |
| tnfn1_pw060419p03q188 | FTN_0696 | hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060323p01q155 | FTN_0696 | hypothetical membrane protein | 5 | 3# | |
| tnfn1_pw060328p06q185 | FTN_0709 | hypothetical protein | 3 | 7 | |
| tnfn1_pw060323p07q129 | FTN_0759 | conserved hypothetical protein | 4 | 2 | |
| tnfn1_pw060419p02q102 | FTN_0792 | hypothetical protein | 5 | 6# | |
| tnfn1_pw060420p01q167 | FTN_0792 | hypothetical protein | 2 | 2# | |
| tnfn1_pw060323p02q140 | FTN_0895 | hypothetical protein | 2 | 2* | |
| tnfn1_pw060323p07q105 | FTN_0895 | hypothetical protein | 4 | 2* | |
| tnfn1_pw060328p08q188 | FTN_1098 | conserved hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060510p03q192 | FTN_1098 | conserved hypothetical membrane protein | 2 | 2# | |
| tnfn1_pw060510p04q192 | FTN_1098 | conserved hypothetical membrane protein | 7 | 6# | |
| tnfn1_pw060419p04q117 | FTN_1156 | hypothetical protein | 2 | 4 | |
| tnfn1_pw060328p02q129 | FTN_1612 | hypothetical protein | 2 | 2 | |
| Metabolic Proteins | |||||
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain | 5 | 7 |
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | riboflavin synthase beta-chain | 4 | 5 |
| tnfn1_pw060328p06q174 | FTN_0125 | ackA | propionate kinase 2/acetate kinase A | 4 | 4# |
| tnfn1_pw060418p03q133 | FTN_0199 | cyoE | heme O synthase | 2 | 4 |
| tnfn1_pw060323p04q102 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase | 1 | 1# |
| tnfn1_pw060418p03q177 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase | 3 | 4# |
| tnfn1_pw060418p01q187 | FTN_0319 | amino acid-polyamine-organocation family protein | 6 | 7 | |
| tnfn1_pw060323p06q113 | FTN_0420 | SAICAR synthetase/phosphoribosylamine-glycine ligase | 7 | 5 | |
| tnfn1_pw060323p05q182 | FTN_0504 | lysine decarboxylase | 4 | 4 | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 | 5 | 7 |
| tnfn1_pw060510p02q154 | FTN_0511 | shikimate 5-dehydrogenase | 2 | 2# | |
| tnfn1_pw060510p02q157 | FTN_0511 | shikimate 5-dehydrogenase | 6 | 6# | |
| tnfn1_pw060510p04q157 | FTN_0511 | shikimate 5-dehydrogenase | 3 | 2# | |
| tnfn1_pw060323p06q194 | FTN_0527 | thrC | threonine synthase | 7 | 7# |
| tnfn1_pw060510p01q172 | FTN_0527 | thrC | threonine synthase | 5 | 5# |
| tnfn1_pw060510p03q172 | FTN_0527 | thrC | threonine synthase | 2 | 2# |
| tnfn1_pw060323p03q127 | FTN_0567 | tRNA synthetase class II (D, K and N) | 6 | 2 | |
| tnfn1_pw060510p03q171 | FTN_0588 | asparaginase | 2 | 2 | |
| tnfn1_pw060419p03q116 | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit | 2 | 2 |
| tnfn1_pw060418p02q128 | FTN_0633 | katG | peroxidase/catalase | 7 | 7 |
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | quinolinate sythetase A | 3 | 2# |
| tnfn1_pw060419p04q164 | FTN_0692 | nadA | quinolinate sythetase A | 2 | 2# |
| tnfn1_pw060510p01q159 | FTN_0695 | add | deoxyadenosine deaminase/adenosine deaminase | 3 | 7 |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | biotin--acetyl-CoA-carboxylase ligase | 6 | 7 |
| tnfn1_pw060328p01q128 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) | 5 | 5 |
| tnfn1_pw060420p02q175 | FTN_0877 | cls | cardiolipin synthetase | 7 | 5 |
| tnfn1_pw060328p06q142 | FTN_0954 | histidine acid phosphatase | 4 | 4 | |
| tnfn1_pw060420p01q130 | FTN_0965 | metal-dependent exopeptidase | 3 | 3 | |
| tnfn1_pw060328p01q151 | FTN_0983 | bifunctional protein: glutaredoxin 3/ribonucleotide reductase beta subunit | 5 | 3# | |
| tnfn1_pw060328p06q189 | FTN_0995 | hslV | ATP-dependent protease HslVU, peptidase subunit | 2 | 2# |
| tnfn1_pw060420p04q195 | FTN_0995 | hslV | ATP-dependent protease HslVU, peptidase subunit | 4 | 4# |
| tnfn1_pw060510p02q187 | FTN_1018 | aldolase/adducin class II family protein | 3 | 3 | |
| tnfn1_pw060323p02q168 | FTN_1046 | wzb | low molecular weight (LMW) phosphotyrosine protein phosphatase | 3 | 2 |
| tnfn1_pw060328p06q184 | FTN_1061 | acid phosphatase, HAD superfamily protein | 2 | 2# | |
| tnfn1_pw060420p02q103 | FTN_1061 | acid phosphatase, HAD superfamily protein | 3 | 3# | |
| tnfn1_pw060510p04q113 | FTN_1121 | phrB | deoxyribodipyrimidine photolyase | 5 | 7 |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5- carboxylate dehydrogenase | 6 | 6 |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase | 3 | 4# |
| tnfn1_pw060328p03q107 | FTN_1222 | kpsF | phosphosugar isomerase | 4 | 3 |
| tnfn1_pw060510p02q164 | FTN_1231 | gloA | lactoylglutathione lyase | 4 | 4* |
| tnfn1_pw060420p04q194 | FTN_1231 | gloA | lactoylglutathione lyase | 3 | 5* |
| tnfn1_pw060510p04q146 | FTN_1231 | gloA | lactoylglutathione lyase | 2 | 2* |
| tnfn1_pw060510p01q142 | FTN_1333 | tktA | transketolase I | 5 | 5 |
| tnfn1_pw060418p02q109 | FTN_1376 | disulfide bond formation protein, DsbB family | 4 | 4 | |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase | 4 | 7 |
| tnfn1_pw060419p01q104 | FTN_1523 | amino acid-polyamine-organocation family protein | 4 | 4 | |
| tnfn1_pw060328p02q165 | FTN_1523 | amino acid-polyamine-organocation family protein | 4 | 5# | |
| tnfn1_pw060419p02q191 | FTN_1523 | amino acid-polyamine-organocation family protein | 2 | 2# | |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase | 5 | 5# |
| tnfn1_pw060418p01q131 | FTN_1557 | oxidoreductase iron/ascorbate family protein | 7 | 7 | |
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | glycerol-3-phosphate dehydrogenase | 3 | 5 |
| tnfn1_pw060419p04q130 | FTN_1585 | glpK | glycerol kinase | 3 | 3 |
| tnfn1_pw060510p01q146 | FTN_1597 | prfC | peptide chain release factor 3 | 5 | 5 |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | cytochrome bd-II terminal oxidase subunit I | 5 | 7 |
| tnfn1_pw060328p02q105 | FTN_1620 | appB | cytochrome bd-II terminal oxidase subunit II | 6 | 3 |
| tnfn1_pw060418p04q111 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 3 | 3# | |
| tnfn1_pw060418p04q112 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 2 | 2# | |
| tnfn1_pw060420p04q169 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 4 | 4# | |
| tnfn1_pw060323p04q160 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C | 7 | 7# |
| tnfn1_pw060510p02q165 | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C | 2 | 2# |
| Transporter Proteins | |||||
| tnfn1_pw060420p04q149 | FTN_0008 | 10 TMS drug/metabolite exporter protein | 4 | 4# | |
| tnfn1_pw060420p02q151 | FTN_0018 | sdaC | serine permease | 2 | 4 |
| tnfn1_pw060418p04q168 | FTN_0141 | ABC transporter, ATP-binding protein | 5 | 6# | |
| tnfn1_pw060418p03q147 | FTN_0299 | putP | proline:Na+ symporter | 2 | 2# |
| tnfn1_pw060510p02q139 | FTN_0299 | putP | proline:Na+ symporter | 2 | 2# |
| tnfn1_pw060323p03q141 | FTN_0619 | pseudogene: nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 3 | 3* | |
| tnfn1_pw060328p06q129 | FTN_0619 | pseudogene: nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | 5* | |
| tnfn1_pw060510p02q156 | FTN_0624 | serine permease | 2 | 2* | |
| tnfn1_pw060323p06q164 | FTN_0624 | serine permease | 2 | 2* | |
| tnfn1_pw060418p01q161 | FTN_0636 | glpT | glycerol-3-phosphate transporter | 7 | 7 |
| tnfn1_pw060419p04q142 | FTN_0687 | galP1 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 2 | 3* |
| tnfn1_pw060510p04q158 | FTN_0687 | galP1 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 2 | 2* |
| tnfn1_pw060328p06q132 | FTN_0728 | predicted Co/Zn/Cd cation transporter | 2 | 5 | |
| tnfn1_pw060418p03q103 | FTN_0739 | potG | ATP-binding cassette putrescine uptake system, ATP-binding protein | 2 | 2# |
| tnfn1_pw060328p08q153 | FTN_0739 | potG | ATP-binding cassette putrescine uptake system, ATP-binding protein | 2 | 5# |
| tnfn1_pw060510p04q103 | FTN_0799 | emrE | putative membrane transporter of cations and cationic drugs, multidrug resistance protein | 2 | 2 |
| tnfn1_pw060323p01q177 | FTN_0799 | emrE | putative membrane transporter of cations and cationic drugs, multidrug resistance protein | 4 | 3 |
| tnfn1_pw060328p04q109 | FTN_0885 | proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | 5 | 2 | |
| tnfn1_pw060328p04q167 | FTN_0997 | proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | 5 | 3 | |
| tnfn1_pw060323p05q110 | FTN_1215 | kpsC | capsule polysaccharide export protein KpsC | 2 | 5 |
| tnfn1_pw060323p07q172 | FTN_1344 | major facilitator superfamily (MFS) transport protein | 4 | 4* | |
| tnfn1_pw060420p04q148 | FTN_1344 | major facilitator superfamily (MFS) transport protein | 5 | 5* | |
| tnfn1_pw060323p01q175 | FTN_1441 | sugar porter (SP) family protein | 4 | 4# | |
| tnfn1_pw060420p02q182 | FTN_1441 | sugar porter (SP) family protein | 6 | 6# | |
| tnfn1_pw060419p02q126 | FTN_1581 | small conductance mechanosensitive ion channel (MscS) family protein | 3 | 3 | |
| tnfn1_pw060323p03q106 | FTN_1593 | oppA | ABC-type oligopeptide transport system, periplasmic component | 2 | 2* |
| tnfn1_pw060420p03q104 | FTN_1593 | oppA | ABC-type oligopeptide transport system, periplasmic component | 4 | 6* |
| tnfn1_pw060420p01q189 | FTN_1611 | major facilitator superfamily (MFS) transport protein | 7 | 5 | |
| tnfn1_pw060328p02q121 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2 | 1* |
| tnfn1_pw060420p02q159 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2 | 2* |
| tnfn1_pw060418p03q187 | FTN_1733 | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | 4 | |
| Transferases | |||||
| tnfn1_pw060323p02q177 | FTN_0019 | pyrB | aspartate carbamoyltransferase | 2 | 2# |
| tnfn1_pw060323p03q119 | FTN_0019 | pyrB | aspartate carbamoyltransferase | 2 | 2# |
| tnfn1_pw060510p01q103 | FTN_0063 | ilvE | branched-chain amino acid aminotransferase protein (class IV) | 3 | 5 |
| tnfn1_pw060323p03q121 | FTN_0343 | aminotransferase | 7 | 2 | |
| tnfn1_pw060328p03q179 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 4 | 4* | |
| tnfn1_pw060419p01q169 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 2 | 2* | |
| tnfn1_pw060323p06q168 | FTN_0545 | glycosyl transferase, group 2 | 4 | 4# | |
| tnfn1_pw060419p01q187 | FTN_0545 | glycosyl transferase, group 2 | 5 | 5# | |
| tnfn1_pw060328p01q142 | FTN_0928 | cysD | sulfate adenylyltransferase subunit 2 | 3 | 3# |
| tnfn1_pw060323p03q182 | FTN_1428 | wbtO | transferase | 3 | 2# |
| tnfn1_pw060510p01q119 | FTN_1428 | wbtO | transferase | 2 | 6# |
| DNA modifying | |||||
| tnfn1_pw060323p03q125 | FTN_0133 | ribonuclease II family protein | 2 | 2 | |
| tnfn1_pw060510p02q141 | FTN_0133 | ribonuclease II family protein | 5 | 5 | |
| tnfn1_pw060323p03q122 | FTN_0577 | mutL | DNA mismatch repair enzyme with ATPase activity | 7 | 6# |
| tnfn1_pw060510p01q148 | FTN_0577 | mutL | DNA mismatch repair enzyme with ATPase activity | 5 | 5# |
| tnfn1_pw060510p04q193 | FTN_0680 | uvrC | excinuclease ABC, subunit C | 6 | 3 |
| tnfn1_pw060328p04q156 | FTN_1027 | ruvC | holliday junction endodeoxyribonuclease | 3 | 4# |
| tnfn1_pw060510p01q132 | FTN_1027 | holliday junction endodeoxyribonuclease | 3 | 3# | |
| tnfn1_pw060510p01q114 | FTN_1073 | DNA/RNA endonuclease G | 5 | 6* | |
| tnfn1_pw060510p02q114 | FTN_1073 | DNA/RNA endonuclease G | 2 | 2* | |
| tnfn1_pw060510p01q153 | FTN_1154 | type I restriction-modification system, subunit S | 5 | 6 | |
| tnfn1_pw060323p03q167 | FTN_1197 | recR | RecFOR complex, RecR component | 2 | 4# |
| tnfn1_pw060510p02q106 | FTN_1197 | recR | RecFOR complex, RecR component | 3 | 3# |
| tnfn1_pw060328p06q158 | FTN_1293 | rnhB | ribonuclease HII | 2 | 5 |
| tnfn1_pw060323p07q175 | FTN_1487 | restriction endonuclease | 3 | 6 | |
| Cell Division | |||||
| tnfn1_pw060328p03q149 | FTN_0162 | ftsQ | cell division protein FtsQ | 2 | 2# |
| tnfn1_pw060328p01q167 | FTN_0330 | minD | septum formation inhibitor-activating ATPase | 2 | 2 |
| FPI Proteins | |||||
| Type IV Pilin | |||||
| tnfn1_pw060323p03q109 | FTN_1137 | pilQ | Type IV pili secretin component | 2 | 2 |
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | Type IV pili secretin component | 4 | 4 |
| tnfn1_pw060323p06q157 | FTN_1139 | pilO | Type IV pili glycosylation protein | 2 | 2 |
| Others | |||||
| tnfn1_pw060323p06q138 | FTN_0107 | lepA | GTP-binding protein LepA | 2 | 4# |
| tnfn1_pw060418p02q123 | FTN_0107 | lepA | GTP-binding protein LepA | 2 | 4# |
| tnfn1_pw060420p04q150 | FTN_0155 | competence protein | 2 | 7* | |
| tnfn1_pw060510p04q189 | FTN_0155 | competence protein | 6 | 3* | |
| tnfn1_pw060418p04q181 | FTN_0338 | MutT/nudix family protein | 2 | 2 | |
| tnfn1_pw060328p06q137 | FTN_0465 | Sua5/YciO/YrdC family protein | 2 | 2# | |
| tnfn1_pw060323p03q111 | FTN_0465 | Sua5/YciO/YrdC family protein | 2 | 2# | |
| tnfn1_pw060323p06q115 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060420p03q193 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060510p01q120 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3# |
| tnfn1_pw060328p06q167 | FTN_0985 | DJ-1/PfpI family protein | 6 | 6# | |
| tnfn1_pw060328p06q167 | FTN_0985 | DJ-1/PfpI family protein | 5 | 5# | |
| tnfn1_pw060420p04q127 | FTN_1031 | ftnA | ferric iron binding protein, ferritin-like | 2 | 6 |
| tnfn1_pw060419p02q137 | FTN_1034 | rnfB | iron-sulfur cluster-binding protein | 2 | 3 |
| tnfn1_pw060420p03q121 | FTN_1064 | PhoH family protein, putative ATPase | 2 | 4 | |
| tnfn1_pw060328p06q178 | FTN_1241 | DedA family protein | 4 | 5 | |
| tnfn1_pw060418p01q185 | FTN_1355 | regulatory factor, Bvg accessory factor family | 6 | 7 | |
| tnfn1_pw060328p03q154 | FTN_1453 | two-component regulator, sensor histidine kinase | 2 | 2 | |
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 2 | 2* |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 4 | 4* |
| Intergenic | |||||
| tnfn1_pw060323p03q164 | intergenic | 3 | 2 | ||
| tnfn1_pw060328p06q190 | intergenic | 3 | 3 | ||
| tnfn1_pw060419p03q131 | intergenic | 2 | 2 | ||
| tnfn1_pw060419p04q189 | intergenic | 5 | 3 | ||
| tnfn1_pw060323p08q139 | intergenic | 4 | 4 | ||
| List of growth defective mutants in only S2 cells according to their functions | |||||
| Proteins of unknown Function | |||||
| tnfn1_pw060419p01q176 | FTN_0043 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060418p01q155 | FTN_0044 | protein of unknown function | 3 | ||
| tnfn1_pw060418p02q158 | FTN_0050 | protein of unknown function | 4 | ||
| tnfn1_pw060328p08q104 | FTN_0051 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060420p01q142 | FTN_0052 | protein of unknown function | 2 | ||
| tnfn1_pw060419p04q191 | FTN_0077 | protein of unknown function | 3# | ||
| tnfn1_pw060323p06q122 | FTN_0077 | protein of unknown function | 2# | ||
| tnfn1_pw060510p04q143 | FTN_0099 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060418p04q193 | FTN_0109 | protein of unknown function | 4 | ||
| tnfn1_pw060418p04q117 | FTN_0207 | protein of unknown function containing a von Willebrand factor type A (vWA) domain | 2 | ||
| tnfn1_pw060328p04q119 | FTN_0325 | membrane protein of unknown function | 2 | ||
| tnfn1_pw060328p08q156 | FTN_0340 | protein of unknown function | 2 | ||
| tnfn1_pw060323p03q157 | FTN_0364 | conserved protein of unknown function | 2* | ||
| tnfn1_pw060418p04q136 | FTN_0364 | conserved protein of unknown function | 3* | ||
| tnfn1_pw060328p08q149 | FTN_0439 | protein of unknown function | 4# | ||
| tnfn1_pw060418p01q142 | FTN_0482 | protein of unknown function | 6 | ||
| tnfn1_pw060328p04q110 | FTN_0573 | protein of unknown function | 2 | ||
| tnfn1_pw060418p02q126 | FTN_0573 | protein of unknown function | 4 | ||
| tnfn1_pw060328p08q173 | FTN_0584 | araJ | conserved inner membrane protein of unknown function | 5 | |
| tnfn1_pw060510p04q147 | FTN_0599 | protein of unknown function | 2# | ||
| tnfn1_pw060328p06q173 | FTN_0599 | protein of unknown function | 2# | ||
| tnfn1_pw060510p01q183 | FTN_0782 | protein of unknown function | 5 | ||
| tnfn1_pw060323p03q129 | FTN_0786 | protein of unknown function | 7# | ||
| tnfn1_pw060323p05q127 | FTN_0791 | protein of unknown function | 3# | ||
| tnfn1_pw060419p03q107 | FTN_0791 | protein of unknown function | 2# | ||
| tnfn1_pw060418p01q141 | FTN_0817 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060328p05q126 | FTN_0828 | protein of unknown function | 5 | ||
| tnfn1_pw060510p04q111 | FTN_0861 | conserved protein of unknown function | 4 | ||
| tnfn1_pw060418p04q148 | FTN_0878 | protein of unknown function | 2 | ||
| tnfn1_pw060328p02q106 | FTN_0884 | drug/metabolite transporter superfamily protein | 2# | ||
| tnfn1_pw060328p03q163 | FTN_0884 | drug/metabolite transporter superfamily protein | 4# | ||
| tnfn1_pw060323p03q150 | FTN_0900 | protein of unknown function with predicted hydrolase and phosphorylase activity | 2# | ||
| tnfn1_pw060418p03q108 | FTN_0900 | protein of unknown function with predicted hydrolase and phosphorylase activity | 6# | ||
| tnfn1_pw060323p04q104 | FTN_0918 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060418p02q131 | FTN_0918 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060419p04q188 | FTN_0925 | protein of unknown function | 5 | ||
| tnfn1_pw060419p04q179 | FTN_1001 | protein of unknown function | 2# | ||
| tnfn1_pw060323p07q181 | FTN_1001 | protein of unknown function | 3# | ||
| tnfn1_pw060418p02q145 | FTN_1020 | conserved protein of unknown function | 5 | ||
| tnfn1_pw060419p01q172 | FTN_1044 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060420p01q111 | FTN_1053 | outer membrane protein of unknown function | 3 | ||
| tnfn1_pw060420p02q158 | FTN_1071 | protein of unknown function | 5 | ||
| tnfn1_pw060418p02q133 | FTN_1093 | protein of unknown function | 5 | ||
| tnfn1_pw060420p01q134 | FTN_1103 | protein of unknown function | 2# | ||
| tnfn1_pw060328p01q140 | FTN_1103 | protein of unknown function | 3# | ||
| tnfn1_pw060323p08q143 | FTN_1235 | protein of unknown function | 2 | ||
| tnfn1_pw060510p03q135 | FTN_1254 | protein of unknown function | 4 | ||
| tnfn1_pw060323p03q102 | FTN_1257 | membrane protein of unknown function | 3# | ||
| tnfn1_pw060419p03q150 | FTN_1257 | membrane protein of unknown function | 4# | ||
| tnfn1_pw060418p04q121 | FTN_1261 | protein of unknown function | 2 | ||
| tnfn1_pw060323p08q134 | FTN_1270 | conserved membrane protein of unknown function | 2 | ||
| tnfn1_pw060418p01q149 | FTN_1298 | GTPase of unknown function | 7 | ||
| tnfn1_pw060419p01q143 | FTN_1334 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060328p08q108 | FTN_1334 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060328p05q124 | FTN_1372 | protein of unknown function | 5 | ||
| tnfn1_pw060323p04q183 | FTN_1386 | protein of unknown function | 3 | ||
| tnfn1_pw060328p01q156 | FTN_1442 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060420p01q165 | FTN_1442 | conserved protein of unknown function | 4# | ||
| tnfn1_pw060418p02q186 | FTN_1448 | protein of unknown function | 3 | ||
| tnfn1_pw060328p02q116 | FTN_1449 | conserved protein of unknown function | 3# | ||
| tnfn1_pw060419p03q173 | FTN_1449 | conserved protein of unknown function | 2# | ||
| tnfn1_pw060323p07q176 | FTN_1534 | conserved protein of unknown function | 3 | ||
| tnfn1_pw060328p02q177 | FTN_1713 | protein of unknown function | 3 | ||
| tnfn1_pw060328p05q185 | FTN_1734 | protein of unknown function | 5 | ||
| tnfn1_pw060328p08q107 | FTN_1774 | protein of unknown function | 3 | ||
| Hypothetical Protein | |||||
| tnfn1_pw060418p04q139 | FTN_0011 | hypothetical protein | 2# | ||
| tnfn1_pw060420p02q108 | FTN_0012 | hypothetical protein | 2 | ||
| tnfn1_pw060420p02q139 | FTN_0013 | hypothetical protein | 3 | ||
| tnfn1_pw060328p01q141 | FTN_0014 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060419p04q178 | FTN_0028 | conserved hypothetical membrane protein | 3# | ||
| tnfn1_pw060323p04q145 | FTN_0028 | conserved hypothetical membrane protein | 2# | ||
| tnfn1_pw060418p04q143 | FTN_0053 | hypothetical protein | 2 | ||
| tnfn1_pw060328p06q157 | FTN_0170 | conserved hypothetical membrane protein | 5 | ||
| tnfn1_pw060418p03q151 | FTN_0212 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060323p08q114 | FTN_0326 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060328p05q165 | FTN_0360 | hypothetical protein | 5 | ||
| tnfn1_pw060419p01q145 | FTN_0368 | hypothetical protein | 2 | ||
| tnfn1_pw060419p03q186 | FTN_0375 | hypothetical protein | 3 | ||
| tnfn1_pw060420p02q163 | FTN_0398 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060420p04q104 | FTN_0466 | conserved hypothetical protein | 4 | ||
| tnfn1_pw060328p08q148 | FTN_0548 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060418p04q176 | FTN_0548 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060328p06q164 | FTN_0630 | hypothetical protein | 5 | ||
| tnfn1_pw060328p05q141 | FTN_0701 | conserved hypothetical protein | 5 | ||
| tnfn1_pw060418p02q152 | FTN_0706 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060418p02q175 | FTN_0717 | conserved hypothetical membrane protein | 5 | ||
| tnfn1_pw060328p06q126 | FTN_0732 | hypothetical protein | 5 | ||
| tnfn1_pw060323p07q129 | FTN_0759 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060323p04q134 | FTN_0938 | hypothetical protein | 2# | ||
| tnfn1_pw060418p02q170 | FTN_0938 | hypothetical protein | 4# | ||
| tnfn1_pw060419p03q187 | FTN_1123 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060418p04q105 | FTN_1180 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060420p04q159 | FTN_1223 | conserved hypothetical membrane protein | 7 | ||
| tnfn1_pw060323p08q166 | FTN_1232 | conserved hypothetical membrane protein | 2 | ||
| tnfn1_pw060328p03q180 | FTN_1260 | hypothetical membrane protein | 2 | ||
| tnfn1_pw060510p01q184 | FTN_1299 | hypothetical protein | 5 | ||
| tnfn1_pw060419p04q127 | FTN_1342 | conserved hypothetical protein | 3 | ||
| tnfn1_pw060328p05q157 | FTN_1379 | pseudogene: hypothetical membrane protein, fragment | 5 | ||
| tnfn1_pw060323p06q178 | FTN_1389 | conserved hypothetical membrane protein | 3# | ||
| tnfn1_pw060420p01q172 | FTN_1389 | conserved hypothetical membrane protein | 2# | ||
| tnfn1_pw060420p01q153 | FTN_1458 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060323p04q147 | FTN_1761 | pseudogene: hypothetical protein, fragment | 3 | ||
| tnfn1_pw060418p04q149 | FTN_1765 | conserved hypothetical protein | 2 | ||
| Metabolic | |||||
| tnfn1_pw060510p02q160 | FTN_0021 | carA | carbamoyl-phosphate synthase small chain | 2 | |
| tnfn1_pw060418p04q115 | FTN_0095 | nitroreductase | 7 | ||
| tnfn1_pw060420p02q191 | FTN_0113 | ribC | riboflavin synthase alpha chain | 6 | |
| tnfn1_pw060328p05q159 | FTN_0118 | serine peptidase, S49 family | 3# | ||
| tnfn1_pw060420p02q187 | FTN_0118 | serine peptidase, S49 family | 5# | ||
| tnfn1_pw060328p06q139 | FTN_0127 | gabD | succinate semialdehyde dehydrogenase (NAD(P)+ dependent) | 5 | |
| tnfn1_pw060510p01q130 | FTN_0154 | rimK | glutathione synthase/ribosomal protein S6 modification enzyme | 3 | |
| tnfn1_pw060328p01q150 | FTN_0168 | lysU | lysyl-tRNA synthetase | 2# | |
| tnfn1_pw060510p02q178 | FTN_0217 | L-lactate dehydrogenase | 2 | ||
| tnfn1_pw060323p07q113 | FTN_0362 | deoxyribodipyrimidine photolyase-related protein | 4 | ||
| tnfn1_pw060323p04q144 | FTN_0406 | sterol desaturase | 3# | ||
| tnfn1_pw060418p01q189 | FTN_0406 | sterol desaturase | 6# | ||
| tnfn1_pw060328p06q134 | FTN_0443 | maeA | NAD-dependent malic enzyme | 5# | |
| tnfn1_pw060328p06q125 | FTN_0496 | slt | soluble lytic murein transglycosylase | 3 | |
| tnfn1_pw060418p04q116 | FTN_0512 | glgX | pullulanase | 4 | |
| tnfn1_pw060510p03q154 | FTN_0516 | glgA | glycogen synthase | 7 | |
| tnfn1_pw060420p01q135 | FTN_0540 | pckA | phosphoenolpyruvate carboxykinase | 2 | |
| tnfn1_pw060419p04q153 | FTN_0597 | protein-disulfide isomerase | 2 | ||
| tnfn1_pw060510p02q110 | FTN_0603 | mutM | formamidopyrimidine-DNA glycosylase | 2 | |
| tnfn1_pw060328p02q139 | FTN_0621 | eno | enolase (2-phosphoglycerate dehydratase) | 2# | |
| tnfn1_pw060510p03q188 | FTN_0627 | chiA | chitinase, glycosyl hydrolase family 18 | 6 | |
| tnfn1_pw060418p01q120 | FTN_0651 | cdd | cytidine deaminase | 5# | |
| tnfn1_pw060419p01q168 | FTN_0651 | cdd | cytidine deaminase | 2# | |
| tnfn1_pw060328p04q151 | FTN_0661 | guaB | IMP dehydrogenase/GMP reductase | 6# | |
| tnfn1_pw060328p06q131 | FTN_0674 | glxK | glycerate kinase | 3 | |
| tnfn1_pw060420p01q148 | FTN_0694 | nadB | L-aspartate oxidase | 4 | |
| tnfn1_pw060323p06q103 | FTN_0711 | predicted metal-dependent hydrolase | 2 | ||
| tnfn1_pw060328p04q116 | FTN_0765 | choloylglycine hydrolase family protein | 2 | ||
| tnfn1_pw060510p03q119 | FTN_0806 | glycosyl hydrolase family 3 | 3 | ||
| tnfn1_pw060323p07q185 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase | 3# | |
| tnfn1_pw060419p02q138 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase | 3# | |
| tnfn1_pw060328p04q175 | FTN_0818 | lipase/esterase | 5 | ||
| tnfn1_pw060418p02q142 | FTN_0826 | aldo/keto reductase family protein | 3 | ||
| tnfn1_pw060328p08q145 | FTN_0907 | D-alanyl-D-alanine carboxypeptidase | 4# | ||
| tnfn1_pw060418p04q131 | FTN_0907 | D-alanyl-D-alanine carboxypeptidase | 4# | ||
| tnfn1_pw060418p04q167 | FTN_0935 | asnB | asparagine synthase | 2 | |
| tnfn1_pw060510p02q145 | FTN_0945 | rsuA | 16S rRNA pseudouridine synthase | 4 | |
| tnfn1_pw060328p08q120 | FTN_0987 | tRNA-dihydrouridine synthase | 3# | ||
| tnfn1_pw060323p08q141 | FTN_1015 | isochorismatase family protein | 3# | ||
| tnfn1_pw060420p01q129 | FTN_1015 | isochorismatase family protein | 2# | ||
| tnfn1_pw060323p05q141 | FTN_1033 | grxB | glutaredoxin 2 | 3# | |
| tnfn1_pw060420p01q193 | FTN_1033 | grxB | glutaredoxin 2 | 4# | |
| tnfn1_pw060418p01q153 | FTN_1055 | lon | DNA-binding, ATP-dependent protease La | 2 | |
| tnfn1_pw060328p06q184 | FTN_1061 | acid phosphatase, HAD superfamily protein | 3# | ||
| tnfn1_pw060420p02q103 | FTN_1061 | acid phosphatase, HAD superfamily protein | 7# | ||
| tnfn1_pw060510p04q113 | FTN_1121 | phrB | deoxyribodipyrimidine photolyase | 6 | |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5- carboxylate dehydrogenase | 4 | |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase | 5# | |
| tnfn1_pw060328p08q131 | FTN_1174 | murI | glutamate racemase | 2# | |
| tnfn1_pw060419p03q164 | FTN_1186 | pepO | M13 family metallopeptidase | 7 | |
| tnfn1_pw060418p01q124 | FTN_1245 | iscS | cysteine desulfurase | 7# | |
| tnfn1_pw060323p04q139 | FTN_1264 | rluD | ribosomal large subunit pseudouridine synthase D | 2# | |
| tnfn1_pw060510p03q183 | FTN_1264 | rluD | ribosomal large subunit pseudouridine synthase D | 6# | |
| tnfn1_pw060328p06q166 | FTN_1273 | long chain fatty acid CoA ligase | 2 | ||
| tnfn1_pw060419p03q126 | FTN_1278 | nadE | NAD synthase | 5 | |
| tnfn1_pw060328p05q128 | FTN_1329 | fbaA | fructose bisphosphate aldolase Class II | 3 | |
| tnfn1_pw060323p06q195 | FTN_1390 | Zn-dependent hydrolase | 5 | ||
| tnfn1_pw060510p04q137 | FTN_1425 | wbtF | NAD dependent epimerase | 2 | |
| tnfn1_pw060419p03q166 | FTN_1431 | wbtA | dTDP-glucose 4,6-dehydratase | 2 | |
| tnfn1_pw060323p07q169 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | 4# | ||
| tnfn1_pw060418p02q122 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | 3# | ||
| tnfn1_pw060328p08q196 | FTN_1459 | short chain dehydrogenase | 5 | ||
| tnfn1_pw060328p06q128 | FTN_1530 | lysA | diaminopimelate decarboxylase | 6 | |
| tnfn1_pw060328p05q101 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) | 2# | |
| tnfn1_pw060419p04q163 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) | 6# | |
| tnfn1_pw060418p02q178 | FTN_1536 | amino acid-polyamine-organocation (APC) superfamily protein | 4 | ||
| tnfn1_pw060323p06q106 | FTN_1552 | acid phosphatase, PAP2 family | 5 | ||
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase | 2 | |
| tnfn1_pw060323p04q110 | FTN_1678 | nuoC | NADH dehydrogenase I, C subunit | 5# | |
| tnfn1_pw060328p05q160 | FTN_1729 | dapB | dihydrodipicolinate reductase | 4# | |
| tnfn1_pw060510p01q178 | FTN_1729 | dapB | dihydrodipicolinate reductase | 3# | |
| tnfn1_pw060328p04q104 | FTN_1730 | lysC | aspartate kinase III | 2 | |
| tnfn1_pw060328p03q174 | FTN_1768 | pepN | aminopeptidase N | 3 | |
| Transporter proteins | |||||
| tnfn1_pw060323p03q117 | FTN_0005 | corA | divalent inorganic cation transporter | 2# | |
| tnfn1_pw060420p01q131 | FTN_0005 | corA | divalent inorganic cation transporter | 3# | |
| tnfn1_pw060420p01q180 | FTN_0097 | hydroxy/aromatic amino acid permease (HAAAP) family protein | 4# | ||
| tnfn1_pw060419p03q162 | FTN_0115 | Na+/H+ antiporter | 4 | ||
| tnfn1_pw060323p08q162 | FTN_0151 | ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component | 2 | ||
| tnfn1_pw060419p01q165 | FTN_0183 | manganese/Zinc/Iron chelate uptake transporter family protein | 3# | ||
| tnfn1_pw060419p04q103 | FTN_0183 | manganese/Zinc/Iron chelate uptake transporter family protein | 2# | ||
| tnfn1_pw060510p02q174 | FTN_0184 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060323p03q161 | FTN_0276 | mviN | multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) transporter | 2* | |
| tnfn1_pw060510p02q151 | FTN_0276 | mviN | multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) transporter | 3* | |
| tnfn1_pw060323p08q118 | FTN_0345 | DNA uptake protein, SMF family | 2 | ||
| tnfn1_pw060419p03q195 | FTN_0363 | sodium bile acid symporter family protein | 4 | ||
| tnfn1_pw060420p03q115 | FTN_0566 | mechanosensitive ion channel protein | 3 | ||
| tnfn1_pw060328p08q167 | FTN_0579 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060419p04q167 | FTN_0620 | major facilitator superfamily (MFS) transport protein | 5 | ||
| tnfn1_pw060328p06q114 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 2# | ||
| tnfn1_pw060510p02q115 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 5# | ||
| tnfn1_pw060510p02q167 | FTN_0631 | metabolite:H+ symporter (MHS) family protein | 5# | ||
| tnfn1_pw060418p02q189 | FTN_0640 | dctA | C4-dicarboxylate transport protein | 3 | |
| tnfn1_pw060510p02q159 | FTN_0688 | galP2 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 3 | |
| tnfn1_pw060510p03q140 | FTN_0741 | proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | 5 | ||
| tnfn1_pw060328p05q107 | FTN_0767 | betT | betaine/carnitine/choline transporter (BCCT) family protein | 4 | |
| tnfn1_pw060420p03q116 | FTN_0824 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060510p04q173 | FTN_0872 | small conductance mechanosensitive ion channel (MscS) family protein | 5 | ||
| tnfn1_pw060328p06q175 | FTN_0875 | metabolite:H+ symporter (MHS) family | 2 | ||
| tnfn1_pw060328p02q106 | FTN_0884 | drug/metabolite transporter superfamily protein | 1# | ||
| tnfn1_pw060328p03q163 | FTN_0884 | drug/metabolite transporter superfamily protein | 4# | ||
| tnfn1_pw060328p01q188 | FTN_0910 | sugar:cation symporter family protein | 2# | ||
| tnfn1_pw060419p04q109 | FTN_0910 | sugar:cation symporter family protein | 2# | ||
| tnfn1_pw060419p01q175 | FTN_0984 | ABC transporter, ATP-binding protein | 2 | ||
| tnfn1_pw060419p01q170 | FTN_1006 | transporter-associated protein, HlyC/CorC family | 4 | ||
| tnfn1_pw060418p02q160 | FTN_1010 | major facilitator superfamily (MFS) transport protein | 2 | ||
| tnfn1_pw060419p01q133 | FTN_1014 | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | ||
| tnfn1_pw060328p02q109 | FTN_1107 | metlQ | methionine uptake transporter (MUT) family protein, membrane and periplasmic protein | 2 | |
| tnfn1_pw060323p05q139 | FTN_1166 | metabolite:H+ symporter (MHS) family protein | 7 | ||
| tnfn1_pw060419p02q107 | FTN_1267 | ATP-binding Cassette (ABC) superfamily protein | 4 | ||
| tnfn1_pw060418p02q182 | FTN_1275 | drug:H+ antiporter-1 (DHA2) family protein | 5 | ||
| tnfn1_pw060420p04q186 | FTN_1404 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060510p02q118 | FTN_1409 | major facilitator superfamily (MFS) transport protein | 6 | ||
| tnfn1_pw060328p06q119 | FTN_1549 | drug:H+ antiporter-1 (DHA1) family protein | 3 | ||
| tnfn1_pw060419p02q126 | FTN_1581 | small conductance mechanosensitive ion channel (MscS) family protein | 2 | ||
| tnfn1_pw060418p01q150 | FTN_1586 | sugar transporter, MFS superfamily | 2 | ||
| tnfn1_pw060420p01q146 | FTN_1681 | fur | ferric uptake regulation protein | 2* | |
| tnfn1_pw060510p04q167 | FTN_1681 | fur | ferric uptake regulation protein | 2* | |
| tnfn1_pw060323p03q163 | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | 3* | ||
| tnfn1_pw060328p02q192 | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | 4* | ||
| tnfn1_pw060323p06q117 | FTN_1685 | drug:H+ antiporter-1 (DHA1) family protein | 3 | ||
| tnfn1_pw060418p02q140 | FTN_1685 | drug:H+ antiporter-1 (DHA1) family protein | 5 | ||
| tnfn1_pw060328p05q182 | FTN_1707 | nhaD | Na+:H+ antiporter | 5 | |
| tnfn1_pw060328p02q121 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2* | |
| tnfn1_pw060420p02q159 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2* | |
| tnfn1_pw060510p03q118 | FTN_1717 | kdpB | potassium-transporting ATPase B chain | 3 | |
| tnfn1_pw060420p01q113 | FTN_1752 | nhaA | Na+:H+ antiporter | 3 | |
| Transferase | |||||
| tnfn1_pw060510p04q127 | FTN_0071 | LPS fatty acid acyltransferase | 2# | ||
| tnfn1_pw060419p03q160 | FTN_0080 | SAM-dependent methyltransferase | 4 | ||
| tnfn1_pw060328p08q125 | FTN_0120 | rhodanese-related sulfurtransferase | 4 | ||
| tnfn1_pw060328p01q137 | FTN_0153 | RimI-like acetyltransferase | 3 | ||
| tnfn1_pw060418p01q110 | FTN_0200 | UDP-3-O-[3-fatty acid] glucosamine N- acyltransferase | 2# | ||
| tnfn1_pw060510p02q131 | FTN_0200 | UDP-3-O-[3-fatty acid] glucosamine N- acyltransferase | 2# | ||
| tnfn1_pw060420p02q146 | FTN_0300 | glycosyl transferase, group 2 | 5 | ||
| tnfn1_pw060328p03q179 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 4* | ||
| tnfn1_pw060419p01q169 | FTN_0358 | tRNA-methylthiotransferase MiaB protein | 2* | ||
| tnfn1_pw060420p01q152 | FTN_0453 | glycosyl transferase | 5 | ||
| tnfn1_pw060419p02q135 | FTN_0560 | ksgA | dimethyladenosine transferase | 3 | |
| tnfn1_pw060419p04q168 | FTN_1091 | aroA | 3-phosphoshikimate 1-carboxyvinyltransferase | 2 | |
| tnfn1_pw060418p03q185 | FTN_1400 | S-adenosylmethionine-dependent methyltransferase | 5 | ||
| tnfn1_pw060418p04q172 | FTN_1418 | manC | mannose-1-phosphate guanylyltransferase | 4 | |
| DNA Modification | |||||
| tnfn1_pw060510p04q169 | FTN_0122 | recA | recombinase A protein | 2 | |
| tnfn1_pw060328p06q179 | FTN_0492 | parC | DNA topoisomerase IV subunit A | 2 | |
| tnfn1_pw060510p04q168 | FTN_0666 | uvrA | excinuclease ABC, subunit A | 2 | |
| tnfn1_pw060328p06q140 | FTN_0704 | type I restriction-modification system, subunit M (methyltransferase) | 5 | ||
| tnfn1_pw060510p02q180 | FTN_0704 | type I restriction-modification system, subunit M (methyltransferase) | 2 | ||
| tnfn1_pw060510p03q158 | FTN_1294 | rRNA methylase, SpoU family | 2 | ||
| tnfn1_pw060510p02q176 | FTN_1413 | ATPase, AAA family, related to the helicase subunit of the Holliday junction resolvase | 2 | ||
| tnfn1_pw060328p08q179 | FTN_1491 | adenine specific DNA methylase | 2 | ||
| tnfn1_pw060328p06q176 | FTN_1544 | hemK | modification methylase, HemK family | 5 | |
| tnfn1_pw060328p05q164 | FTN_1594 | uvrD | DNA helicase II | 6 | |
| Cell Division | |||||
| tnfn1_pw060328p01q167 | FTN_0330 | minD | septum formation inhibitor-activating ATPase | 2 | |
| tnfn1_pw060323p08q146 | FTN_0331 | minC | septum formation inhibitor | 4# | |
| tnfn1_pw060420p02q170 | FTN_0331 | minC | septum formation inhibitor | 4# | |
| Transcription/Translation | |||||
| tnfn1_pw060328p06q196 | FTN_0552 | yhbY | RNA-binding protein | 5# | |
| tnfn1_pw060510p03q150 | FTN_0949 | rplI | 50S ribosomal protein L9 | 2 | |
| tnfn1_pw060328p06q170 | FTN_1099 | transcriptional regulator, LysR family | 7 | ||
| tnfn1_pw060419p03q165 | FTN_1300 | transcriptional regulator, LysR family | 2 | ||
| tnfn1_pw060328p02q148 | FTN_1393 | transcriptional regulator, ArsR family | 3# | ||
| tnfn1_pw060418p01q138 | FTN_1393 | transcriptional regulator, ArsR family | 2# | ||
| tnfn1_pw060419p02q151 | FTN_1628 | transcriptional regulator, LysR family | 2# | ||
| tnfn1_pw060510p03q194 | FTN_1628 | transcriptional regulator, LysR family | 2# | ||
| FPI | |||||
| tnfn1_pw060328p01q144 | FTN_1313 | hypothetical protein | 3 | ||
| tnfn1_pw060323p03q179 | FTN_1314 | conserved hypothetical protein | 1 | ||
| tnfn1_pw060328p06q163 | FTN_1315 | protein of unknown function | 5 | ||
| tnfn1_pw060328p06q115 | FTN_1322 | iglC | intracellular growth locus protein C | 5 | |
| tnfn1_pw060419p04q108 | FTN_1325 | pdpD | protein of unknown function | 2 | |
| Type IV Pili | |||||
| tnfn1_pw060418p04q123 | FTN_0070 | pilE | Type IV pili, pilus assembly protein | 3 | |
| tnfn1_pw060510p03q129 | FTN_0070 | pilE | Type IV pili, pilus assembly protein | 3 | |
| tnfn1_pw060323p06q179 | FTN_0305 | pilus assembly protein | 4 | ||
| tnfn1_pw060419p01q196 | FTN_0414 | Type IV pili, pilus assembly protein | 2 | ||
| tnfn1_pw060419p03q141 | FTN_0664 | fimT | Type IV pili, pilus assembly protein | 2 | |
| tnfn1_pw060328p05q146 | FTN_0946 | pilF | Type IV pili, pilus assembly protein | 5 | |
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | Type IV pili secretin component | 4 | |
| Others | |||||
| tnfn1_pw060328p08q161 | isftu1 | isftu1 | 2 | ||
| tnfn1_pw060323p03q115 | isftu3 | isftu3 | 1 | ||
| tnfn1_pw060328p04q157 | isftu2 | isftu2 | 1 | ||
| tnfn1_pw060510p02q150 | isftu6 | isftu6 | 2 | ||
| tnfn1_pw060328p01q179 | FTN_0010 | phage terminase, small subunit | 3 | ||
| tnfn1_pw060328p08q114 | FTN_0266 | htpG | chaperone Hsp90, heat shock protein HtpG | 2 | |
| tnfn1_pw060328p04q152 | FTN_0322 | VacJ like lipoprotein | 3# | ||
| tnfn1_pw060418p01q140 | FTN_0322 | VacJ like lipoprotein | 2# | ||
| tnfn1_pw060328p08q155 | FTN_0357 | pal | peptidoglycan-associated lipoprotein, OmpA family | 4* | |
| tnfn1_pw060419p01q158 | FTN_0357 | pal | peptidoglycan-associated lipoprotein, OmpA family | 2* | |
| tnfn1_pw060510p02q122 | FTN_0367 | phage integrase | 4 | ||
| tnfn1_pw060328p08q132 | FTN_0372 | regulatory protein, AlpA family | 4 | ||
| tnfn1_pw060323p07q171 | FTN_0585 | cutC | copper homeostasis protein CutC family protein | 2 | |
| tnfn1_pw060328p06q127 | FTN_0713 | ostA2 | organic solvent tolerance protein OstA | 5# | |
| tnfn1_pw060419p01q180 | FTN_0713 | ostA2 | organic solvent tolerance protein OstA | 4# | |
| tnfn1_pw060323p06q105 | FTN_0810 | ROK family protein | 4 | ||
| tnfn1_pw060419p01q139 | FTN_0836 | kinase-like protein | 2 | ||
| tnfn1_pw060418p04q134 | FTN_1051 | hfq | host factor I for bacteriophage Q beta replication | 2 | |
| tnfn1_pw060420p03q121 | FTN_1064 | PhoH family protein, putative ATPase | 4 | ||
| tnfn1_pw060328p05q177 | FTN_1192 | chitin-binding protein | 6 | ||
| tnfn1_pw060419p04q183 | FTN_1240 | BolA family protein | 4 | ||
| tnfn1_pw060418p02q190 | FTN_1242 | DedA family protein | 5 | ||
| tnfn1_pw060419p01q120 | FTN_1488 | prophage maintenance system killer protein (DOC) | 6 | ||
| tnfn1_pw060419p01q135 | FTN_1665 | magnesium chelatase | 2 | ||
| tnfn1_pw060419p04q180 | FTN_1682 | frgA | siderophore biosynthesis protein | 5 | |
| tnfn1_pw060510p04q122 | FTN_1698 | Dam-replacing family protein | 2 | ||
| Intergenic | |||||
| tnfn1_pw060418p01q125 | intergenic | 7 | |||
| tnfn1_pw060323p06q165 | intergenic | 5 | |||
| tnfn1_pw060323p08q117 | intergenic | 3 | |||
| tnfn1_pw060328p05q195 | intergenic | 6 | |||
| tnfn1_pw060419p01q148 | intergenic | 3 | |||
| tnfn1_pw060420p03q148 | intergenic | 3 | |||
| tnfn1_pw060420p01q164 | intergenic | 2 | |||
| tnfn1_pw060510p02q127 | intergenic | 2 | |||
| tnfn1_pw060328p08q109 | intergenic | 2 | |||
| tnfn1_pw060510p04q116 | intergenic | 2 | |||
| List of growth defective mutants in only U937 Cells | |||||
| Proteins of unknown function | |||||
| tnfn1_pw060328p06q147 | FTN_0109 | protein of unknown function | 3# | ||
| tnfn1_pw060418p04q193 | FTN_0109 | protein of unknown function | 4# | ||
| tnfn1_pw060510p01q123 | FTN_0132 | protein of unknown function | 3 | ||
| tnfn1_pw060323p07q115 | FTN_0290 | protein of unknown function | 5 | ||
| tnfn1_pw060328p04q122 | FTN_0428 | protein of unknown function | 2* | ||
| tnfn1_pw060510p04q109 | FTN_0428 | protein of unknown function | 2* | ||
| tnfn1_pw060419p03q140 | FTN_0477 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060420p02q178 | FTN_0915 | conserved protein of unknown function | 7 | ||
| tnfn1_pw060419p04q188 | FTN_0925 | protein of unknown function | 4 | ||
| tnfn1_pw060420p02q181 | FTN_0933 | protein of unknown function | 7 | ||
| tnfn1_pw060419p04q118 | FTN_1172 | conserved protein of unknown function | 2 | ||
| tnfn1_pw060420p01q127 | FTN_1175 | membrane protein of unknown function | 4 | ||
| tnfn1_pw060420p01q109 | FTN_1367 | protein of unknown function | 2 | ||
| tnfn1_pw060420p01q132 | FTN_1624 | conserved protein of unknown function | 4 | ||
| tnfn1_pw060420p02q184 | FTN_1696 | protein of unknown function | 7 | ||
| Hypothetical Proteins | |||||
| tnfn1_pw060323p01q181 | FTN_0336 | hypothetical protein | 3 | ||
| tnfn1_pw060510p01q147 | FTN_0403 | hypothetical membrane protein | 4 | ||
| tnfn1_pw060323p01q163 | FTN_0727 | hypothetical membrane protein | 3 | ||
| tnfn1_pw060418p03q110 | FTN_0847 | conserved hypothetical protein | 2# | ||
| tnfn1_pw060510p02q108 | FTN_0847 | conserved hypothetical protein | 4# | ||
| tnfn1_pw060419p02q152 | FTN_0888 | hypothetical membrane protein | 2 | ||
| tnfn1_pw060418p01q191 | FTN_1349 | hypothetical protein | 4# | ||
| tnfn1_pw060328p06q182 | FTN_1395 | conserved hypothetical protein | 4 | ||
| tnfn1_pw060328p04q136 | FTN_1406 | conserved hypothetical membrane protein | 4 | ||
| tnfn1_pw060420p02q127 | FTN_1656 | conserved hypothetical protein | 2 | ||
| tnfn1_pw060420p02q176 | FTN_1686 | hypothetical membrane protein | 5 | ||
| tnfn1_pw060418p03q159 | FTN_1736 | hypothetical protein | 2 | ||
| Metabolic Proteins | |||||
| tnfn1_pw060419p02q150 | FTN_0090 | acpA | acid phosphatase | 5 | |
| tnfn1_pw060419p03q169 | FTN_0218 | nfnB | dihydropteridine reductase | 2 | |
| tnfn1_pw060420p01q123 | FTN_0524 | asd | aspartate semialdehyde dehydrogenase | 5 | |
| tnfn1_pw060323p06q168 | FTN_0545 | glycosyl transferase, group 2 | 5# | ||
| tnfn1_pw060419p01q187 | FTN_0545 | glycosyl transferase, group 2 | 5# | ||
| tnfn1_pw060510p03q168 | FTN_0598 | tRNA-dihydrouridine synthase | 3 | ||
| tnfn1_pw060328p04q196 | FTN_0746 | alr | alanine racemase | 6# | |
| tnfn1_pw060420p04q108 | FTN_0822 | para-aminobenzoate synthase component I | 5 | ||
| tnfn1_pw060420p04q140 | FTN_0957 | short chain dehydrogenase | 4 | ||
| tnfn1_pw060420p02q174 | FTN_1233 | haloacid dehalogenase-like hydrolase | 6 | ||
| tnfn1_pw060420p04q116 | FTN_1421 | wbtH | glutamine amidotransferase/asparagine synthase | 3 | |
| tnfn1_pw060419p04q135 | FTN_1415 | thioredoxin | 6 | ||
| tnfn1_pw060510p04q185 | FTN_1701 | glutamate decarboxylase | 3 | ||
| tnfn1_pw060510p04q136 | FTN_1767 | rbsK | ribokinase, pfkB family | 3 | |
| tnfn1_pw060328p05q154 | FTN_1777 | trpG | anthranilate synthase component II | 2# | |
| Transporter Proteins | |||||
| tnfn1_pw060420p04q158 | FTN_0800 | ArsB arsenite/antimonite exporter | 2 | ||
| tnfn1_pw060510p01q152 | FTN_1711 | tyrP | tyrosine permease | 6 | |
| DNA Modification | |||||
| tnfn1_pw060419p04q116 | FTN_0287 | type I restriction-modification system, subunit R (restriction) | 2 | ||
| tnfn1_pw060420p03q134 | FTN_0710 | type I restriction-modification system, subunit R (restriction) | 4 | ||
| tnfn1_pw060510p04q179 | FTN_0838 | xthA | exodeoxyribonuclease III | 3 | |
| tnfn1_pw060419p04q152 | FTN_1017 | pseudogene: DNA-3-methyladenine glycosylase | 5 | ||
| tnfn1_pw060323p04q111 | FTN_1176 | uvrB | excinuclease ABC, subunit B | 2 | |
| Transferases | |||||
| tnfn1_pw060420p02q180 | FTN_0483 | bifunctional NMN adenylyltransferase/Nudix hydrolase | 7 | ||
| tnfn1_pw060510p01q158 | FTN_0988 | prmA | 50S ribosomal protein L11, methyltransferase | 7 | |
| tnfn1_pw060510p02q144 | FTN_1234 | queA | S-adenosylmethionine:tRNA ribosyltransferase- isomerase | 6 | |
| Transcription/Translation | |||||
| tnfn1_pw060323p03q127 | FTN_0567 | tRNA synthetase class II (D, K and N) | 2 | ||
| tnfn1_pw060510p03q168 | FTN_0598 | tRNA-dihydrouridine synthase | 3 | ||
| tnfn1_pw060419p04q129 | FTN_1290 | mglA | macrophage growth locus, protein A | 3# | |
| Others | |||||
| tnfn1_pw060328p08q161 | - | isftu1 | isftu1 | 2 | |
| tnfn1_pw060510p04q176 | FTN_0182 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060323p08q110 | FTN_0286 | transposase | 3 | ||
| tnfn1_pw060420p01q168 | FTN_0646 | cscK | ROK family protein | 5 | |
| tnfn1_pw060328p04q123 | FTN_0672 | secA | preprotein translocase, subunit A (ATPase, RNA helicase) | 2 | |
| tnfn1_pw060328p04q112 | FTN_1002 | blaA | beta-lactamase class A | 2# | |
| tnfn1_pw060419p02q192 | FTN_1002 | blaA | beta-lactamase class A | 2# | |
| tnfn1_pw060420p02q177 | FTN_1145 | era | GTP-binding protein | 6 | |
| tnfn1_pw060418p03q107 | FTN_1217 | ATP-binding cassette (ABC) superfamily protein | 2 | ||
| tnfn1_pw060328p06q171 | FTN_1263 | comL | competence lipoprotein ComL | 2# | |
| tnfn1_pw060420p02q179 | FTN_1263 | comL | competence lipoprotein ComL | 7# | |
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 2* | |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 4* | |
| Intergenic | |||||
| tnfn1_pw060328p03q108 | intergenic | 2 | |||
| tnfn1_pw060419p04q165 | intergenic | 5 | |||
| tnfn1_pw060510p01q102 | intergenic | 5 | |||
| tnfn1_pw060510p01q112 | intergenic | 4 | |||
| tnfn1_pw060510p01q135 | intergenic | 4 | |||
- Mutants for which all the alleles showed growth defect
- Mutants for which two out of three or three out of four alleles showed growth defect
Table 2.
List of growth defective or dissemination defective mutants in S2 and U937 cells identified in previous screens
| Strain Name | Locus Tag | Gene | Description |
|---|---|---|---|
| tnfn1_pw060323p03q172α | FTN_0008 | 10 TMS drug/metabolite exporter protein | |
| tnfn1_pw060420p02q151βγδ | FTN_0018 | sdaC | serine permease |
| tnfn1_pw060323p02q177 βγδ | FTN_0019 | pyrB | aspartate carbamoyltransferase |
| tnfn1_pw060323p08q120 βγδ | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060510p02q160 βγδ | FTN_0021 | carA | carbamoyl-phosphate synthase small chain |
| tnfn1_pw060419p04q178 α | FTN_0028 | conserved hypothetical membrane protein | |
| tnfn1_pw060418p04q123β | FTN_0070 | pilE | Type IV pili, pilus assembly protein |
| tnfn1_pw060420p01q180 βγδπ | FTN_0097 | hydroxy/aromatic amino acid permease (HAAAP) family protein | |
| tnfn1_pw060419p01q106 α | FTN_0111 | ribH | riboflavin synthase beta-chain |
| tnfn1_pw060510p04q169δ | FTN_0122 | recA | recombinase A protein |
| tnfn1_pw060510p01q123 α | FTN_0132 | lpsA | protein of unknown function |
| tnfn1_pw060323p03q125 α | FTN_0133 | ribonuclease II family protein | |
| tnfn1_pw060420p02q173 α | FTN_0169 | conserved hypothetical membrane protein | |
| tnfn1_pw060418p03q133 α | FTN_0199 | cyoE | heme O synthase |
| tnfn1_pw060323p04q102 βγδ | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| tnfn1_pw060510p02q178 α | FTN_0217 | L-lactate dehydrogenase | |
| tnfn1_pw060420p04q134 α | FTN_0297 | conserved protein of unknown function | |
| tnfn1_pw060418p03q147 α | FTN_0299 | putP | proline:Na+ symporter |
| tnfn1_pw060420p02q146 βγδ | FTN_0300 | glycosyl transferase, group 2 | |
| tnfn1_pw060328p01q167 βγδ | FTN_0330 | minD | septum formation inhibitor-activating ATPase |
| tnfn1_pw060323p08q146δ | FTN_0331 | minC | septum formation inhibitor |
| tnfn1_pw060328p08q156 α | FTN_0340 | protein of unknown function | |
| tnfn1_pw060323p06q113 βγδ | FTN_0420 | purCD | SAICAR synthetase/phosphoribosylamine- glycine ligase |
| tnfn1_pw060328p06q134 βγδ | FTN_0443 | maeA | NAD-dependent malic enzyme |
| tnfn1_pw060328p05q119 βγδπ | FTN_0444 | membrane protein of unknown function | |
| tnfn1_pw060323p05q182 βγδ | FTN_0504 | cadC | lysine decarboxylase |
| tnfn1_pw060510p01q124 βγδ | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 |
| tnfn1_pw060323p06q168 βγδ | FTN_0545 | glycosyl transferase, group 2 | |
| tnfn1_pw060419p02q135β | FTN_0560 | ksgA | dimethyladenosine transferase |
| tnfn1_pw060419p03q116 βγδ | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit |
| tnfn1_pw060510p04q147 βγδ | FTN_0599 | protein of unknown function | |
| tnfn1_pw060323p06q164 βγδ | FTN_0624 | serine permease | |
| tnfn1_pw060418p02q128δ | FTN_0633 | katG | peroxidase/catalase |
| tnfn1_pw060420p01q168 α | FTN_0646 | cscK | ROK family protein |
| tnfn1_pw060419p01q168 βγδ | FTN_0651 | cdd | cytidine deaminase |
| tnfn1_pw060510p04q168δ | FTN_0666 | uvrA | excinuclease ABC, subunit A |
| tnfn1_pw060328p04q123 βγδ | FTN_0672 | secA | preprotein translocase, subunit A (ATPase, RNA helicase) |
| tnfn1_pw060420p03q134 α | FTN_0710 | type I restriction-modification system, subunit R (restriction) | |
| tnfn1_pw060328p06q127 α | FTN_0713 | ostA2 | organic solvent tolerance protein OstA |
| tnfn1_pw060328p06q132 βγδ | FTN_0728 | predicted Co/Zn/Cd cation transporter | |
| tnfn1_pw060323p06q115 α | FTN_0768 | tspO | tryptophan-rich sensory protein |
| tnfn1_pw060323p06q105 α | FTN_0810 | ROK family protein | |
| tnfn1_pw060323p07q185 βγδ | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase |
| tnfn1_pw060418p01q141β | FTN_0817 | conserved protein of unknown function | |
| tnfn1_pw060420p04q108 βγδπ | FTN_0822 | para-aminobenzoate synthase component I | |
| tnfn1_pw060420p03q116 α | FTN_0824 | major facilitator superfamily (MFS) transport protein | |
| tnfn1_pw060328p01q128 α | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060420p04q176 βγδ | FTN_0855 | protein of unknown function | |
| tnfn1_pw060420p02q175 α | FTN_0877 | cls | cardiolipin synthetase |
| tnfn1_pw060323p04q104 α | FTN_0918 | conserved protein of unknown function | |
| tnfn1_pw060419p04q188 βγδ | FTN_0925 | protein of unknown function | |
| tnfn1_pw060420p02q181 α | FTN_0933 | protein of unknown function | |
| tnfn1_pw060323p04q134 α | FTN_0938 | hypothetical protein | |
| tnfn1_pw060419p01q170 α | FTN_1006 | transporter-associated protein, HlyC/CorC family | |
| tnfn1_pw060323p08q141 α | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060418p01q153 α | FTN_1055 | lon | DNA-binding, ATP-dependent protease La |
| tnfn1_pw060510p01q114π | FTN_1073 | DNA/RNA endonuclease G | |
| tnfn1_pw060419p04q168 βγδ | FTN_1091 | aroA | 3-phosphoshikimate 1- carboxyvinyltransferase |
| tnfn1_pw060328p08q188 α | FTN_1098 | conserved hypothetical membrane protein | |
| tnfn1_pw060328p02q109 α | FTN_1107 | metlQ | methionine uptake transporter (MUT) family protein, membrane and periplasmic protein |
| tnfn1_pw060328p02q175 βγδ | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase |
| tnfn1_pw060418p03q107 βγδ | FTN_1217 | ATP-binding cassette (ABC) superfamily protein | |
| tnfn1_pw060323p08q166 α | FTN_1232 | conserved hypothetical membrane protein | |
| tnfn1_pw060328p06q178 βγδ | FTN_1241 | DedA family protein | |
| tnfn1_pw060510p03q135 β | FTN_1254 | protein of unknown function | |
| tnfn1_pw060420p04q196 βγδπ | FTN_1256 | membrane protein of unknown function | |
| tnfn1_pw060323p03q102 βγδ | FTN_1257 | membrane protein of unknown function | |
| tnfn1_pw060420p02q179 βγδ | FTN_1263 | comL | competence lipoprotein ComL |
| tnfn1_pw060418p01q149 βγδ | FTN_1298 | GTPase of unknown function | |
| tnfn1_pw060328p01q144 βγδ | FTN_1313 | hypothetical protein | |
| tnfn1_pw060328p06q163β | FTN_1315 | protein of unknown function | |
| tnfn1_pw060510p01q110 αβγδ | FTN_1321 | iglD | intracellular growth locus protein D |
| tnfn1_pw060328p06q115 βγδ | FTN_1322 | iglC | intracellular growth locus protein C |
| tnfn1_pw060419p04q108 βγδ | FTN_1325 | pdpD | protein of unknown function |
| tnfn1_pw060510p01q142 βγδ | FTN_1333 | tktA | transketolase I |
| tnfn1_pw060418p01q191 α | FTN_1349 | hypothetical protein | |
| tnfn1_pw060418p01q185 α | FTN_1355 | regulatory factor, Bvg accessory factor family | |
| tnfn1_pw060418p02q109π | FTN_1376 | disulfide bond formation protein, DsbB family | |
| tnfn1_pw060418p03q185 α | FTN_1400 | S-adenosylmethionine-dependent methyltransferase | |
| tnfn1_pw060419p04q135 α | FTN_1415 | thioredoxin | |
| tnfn1_pw060420p04q116 βγδ | FTN_1421 | wbtH | glutamine amidotransferase/asparagine synthase |
| tnfn1_pw060510p04q137 βγδ | FTN_1425 | wbtF | NAD dependent epimerase |
| tnfn1_pw060419p03q166 βγδπ | FTN_1431 | wbtA | dTDP-glucose 4,6-dehydratase |
| tnfn1_pw060418p02q122 βγδ | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | |
| tnfn1_pw060328p08q196 α | FTN_1459 | short chain dehydrogenase | |
| tnfn1_pw060323p06q110 βγδ | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
| tnfn1_pw060328p06q128β | FTN_1530 | lysA | diaminopimelate decarboxylase |
| tnfn1_pw060323p07q176 α | FTN_1534 | conserved protein of unknown function | |
| tnfn1_pw060418p02q178 α | FTN_1536 | amino acid-polyamine-organocation (APC) superfamily protein | |
| tnfn1_pw060418p01q150γ | FTN_1586 | sugar transporter, MFS superfamily | |
| tnfn1_pw060510p01q146 βγδ | FTN_1597 | prfC | peptide chain release factor 3 |
| tnfn1_pw060420p01q189 α | FTN_1611 | major facilitator superfamily (MFS) transport protein | |
| tnfn1_pw060323p04q160 βγδ | FTN_1655 | rluC | ribosomal large subunit pseudouridine synthase C |
| tnfn1_pw060419p04q180δ | FTN_1682 | frgA | siderophore biosynthesis protein |
| tnfn1_pw060323p03q163 βγδ | FTN_1683 | drug:H+ antiporter-1 (DHA1) family protein | |
| tnfn1_pw060328p05q154 βγδ | FTN_1777 | trpG | anthranilate synthase component II |
Fig 1. Functional groups of mutants defective in intracellular growth.
The S2 cells were infected with mutants of F. tularensis subsp novicida at MOI of 10 for 1 h followed by 1 h of gentamicin treatment. Growth of the mutants was compared to the wild type strain at 24 h post-infection and the relative reduction in growth relative to the wild type strain was determined. Mutants were considered defective if they exhibited ≥10 fold reduction in growth. The growth defective mutants were grouped according to the function of the genes.
The metabolic genes were grouped according to the putative biochemical pathway. Our analysis showed that a large number of the metabolic genes that were required for replication in S2 cells were involved in both amino acid and carbohydrate metabolism or involved in the synthesis of co-enzymes or cofactors required for carbohydrate and amino acid metabolic pathways (Table 3). At least 11 genes required for nucleotide metabolism were required for intracellular proliferation (Table 3). Therefore, inability of F. tularensis to efficiently metabolize amino acids, carbohydrates, and nucleotides affected its ability to replicate within S2 cells. This indicates that the intracellular environment does not have sufficient nutrients required for intracellular bacterial proliferation and the bacteria require de novo synthesis of metabolic intermediates to support intracellular proliferation. Remarkably, the distribution of the metabolic genes across the various functional groups is similar for S2 cells and human macrophages (see accompanying manuscript), indicating similar metabolic requirements for F. tularensis to proliferate within evolutionarily distant host cells.
Table 3.
Metabolic genes required for intracellular proliferation of F. tularensis within S2 cells grouped according to metabolic pathways
| Amino acid metabolism | |||
|---|---|---|---|
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060510p02q160 | FTN_0021 | carA | carbamoyl-phosphate synthase small chain |
| tnfn1_pw060328p05q159 | FTN_0118 | serine peptidase, S49 family | |
| tnfn1_pw060420p02q187 | FTN_0118 | serine peptidase, S49 family | |
| tnfn1_pw060328p06q174 | FTN_0125 | ackA | propionate kinase 2/acetate kinase A |
| tnfn1_pw060323p05q182 | FTN_0504 | lysine decarboxylase | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 |
| tnfn1_pw060510p02q154 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060510p02q157 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060510p04q157 | FTN_0511 | shikimate 5-dehydrogenase | |
| tnfn1_pw060323p06q194 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060510p01q172 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060510p03q172 | FTN_0527 | thrC | threonine synthase |
| tnfn1_pw060420p01q135 | FTN_0540 | pckA | phosphoenolpyruvate carboxykinase |
| tnfn1_pw060510p03q171 | FTN_0588 | asparaginase | |
| tnfn1_pw060418p04q167 | FTN_0935 | asnB | asparagine synthase |
| tnfn1_pw060328p06q142 | FTN_0954 | histidine acid phosphatase | |
| tnfn1_pw060328p02q175 | FTN_1131 | putA | bifunctional proline dehydrogenase, pyrroline-5- carboxylate dehydrogenase |
| tnfn1_pw060328p08q131 | FTN_1174 | murI | glutamate racemase |
| tnfn1_pw060328p06q128 | FTN_1530 | lysA | diaminopimelate decarboxylase |
| tnfn1_pw060328p05q101 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) |
| tnfn1_pw060419p04q163 | FTN_1532 | gdhA | glutamate dehydrogenase (NADP+) |
| tnfn1_pw060328p05q160 | FTN_1729 | dapB | dihydrodipicolinate reductase |
| tnfn1_pw060510p01q178 | FTN_1729 | dapB | dihydrodipicolinate reductase |
| tnfn1_pw060328p04q104 | FTN_1730 | lysC | aspartate kinase III |
| tnfn1_pw060328p04q116 | FTN_9765 | choloylglycine hydrolase family protein | |
| Carbohydrate metabolism | |||
| tnfn1_pw060328p06q139 | FTN_0127 | gabD | succinate semialdehyde dehydrogenase (NAD(P)+ dependent) |
| tnfn1_pw060510p02q178 | FTN_0217 | L-lactate dehydrogenase | |
| tnfn1_pw060328p06q134 | FTN_0443 | maeA | NAD-dependent malic enzyme |
| tnfn1_pw060418p04q116 | FTN_0512 | glgX | pullulanase |
| tnfn1_pw060510p03q154 | FTN_0516 | glgA | glycogen synthase |
| tnfn1_pw060419p03q116 | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit |
| tnfn1_pw060328p02q139 | FTN_0621 | eno | enolase (2-phosphoglycerate dehydratase) |
| tnfn1_pw060510p03q188 | FTN_0627 | chiA | chitinase, glycosyl hydrolase family 18 |
| tnfn1_pw060328p06q131 | FTN_0674 | glxK | glycerate kinase |
| tnfn1_pw060510p03q119 | FTN_0806 | glycosyl hydrolase family 3 | |
| tnfn1_pw060510p02q187 | FTN_1018 | aldolase/adducin class II family protein | |
| tnfn1_pw060328p03q107 | FTN_1222 | kpsF | phosphosugar isomerase |
| tnfn1_pw060328p05q128 | FTN_1329 | fbaA | fructose bisphosphate aldolase Class II |
| tnfn1_pw060510p04q137 | FTN_1425 | wbtF | NAD dependent epimerase |
| tnfn1_pw060419p03q166 | FTN_1431 | wbtA | dTDP-glucose 4,6-dehydratase |
| tnfn1_pw060323p07q169 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | |
| tnfn1_pw060418p02q122 | FTN_1438 | bifunctional protein: 3-hydroxacyl-CoA dehydrogenase/acyl-CoA-binding protein | |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase |
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | glycerol-3-phosphate dehydrogenase |
| tnfn1_pw060419p04q130 | FTN_1585 | glpK | glycerol kinase |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | cytochrome bd-II terminal oxidase subunit I |
| tnfn1_pw060328p02q105 | FTN_1620 | appB | cytochrome bd-II terminal oxidase subunit II |
| tnfn1_pw060510p04q136 | FTN_1767 | rbsK | ribokinase, pfkB family |
| Nucleotide metabolism | |||
| tnfn1_pw060323p08q120 | FTN_0020 | carB | carbamoyl-phosphate synthase large chain |
| tnfn1_pw060510p02q160 | FTN_0021 | carA | carbamoyl-phosphate synthase small chain |
| tnfn1_pw060418p03q133 | FTN_0199 | cyoE | heme O synthase |
| tnfn1_pw060323p06q113 | FTN_0420 | SAICAR synthetase/phosphoribosylamine-glycine ligase | |
| tnfn1_pw060419p01q168 | FTN_0651 | cdd | cytidine deaminase |
| tnfn1_pw060328p04q151 | FTN_0661 | guaB | IMP dehydrogenase/GMP reductase |
| tnfn1_pw060510p01q159 | FTN_0695 | add | deoxyadenosine deaminase/adenosine deaminase |
| tnfn1_pw060328p01q151 | FTN_0983 | bifunctional protein: glutaredoxin 3/ribonucleotide reductase beta subunit | |
| tnfn1_pw060419p04q135 | FTN_1415 | thioredoxin | |
| tnfn1_pw060419p04q181 | FTN_1415 | thioredoxin | |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase |
| Reductive Metabolism | |||
| tnfn1_pw060418p04q115 | FTN_0095 | nitroreductase | |
| tnfn1_pw060510p01q130 | FTN_0154 | rimK | glutathione synthase/ribosomal protein S6 modification enzyme |
| tnfn1_pw060419p04q153 | FTN_0597 | protein-disulfide isomerase | |
| tnfn1_pw060418p02q128 | FTN_0633 | katG | peroxidase/catalase |
| tnfn1_pw060418p02q142 | FTN_0826 | aldo/keto reductase family protein | |
| tnfn1_pw060328p01q128 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060420p03q153 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) |
| tnfn1_pw060323p05q141 | FTN_1033 | grxB | glutaredoxin 2 |
| tnfn1_pw060420p01q193 | FTN_1033 | grxB | glutaredoxin 2 |
| tnfn1_pw060420p04q194 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060510p02q164 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060510p04q146 | FTN_1231 | gloA | lactoylglutathione lyase |
| tnfn1_pw060328p08q196 | FTN_1459 | short chain dehydrogenase | |
| tnfn1_pw060418p01q131 | FTN_1557 | oxidoreductase iron/ascorbate family protein | |
| tnfn1_pw060418p04q112 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | |
| tnfn1_pw060420p04q169 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | |
| cholesterol Metabolism | |||
| tnfn1_pw060323p04q144 | FTN_0406 | sterol desaturase | |
| tnfn1_pw060418p01q189 | FTN_0406 | sterol desaturase | |
| Lipid Metabolism | |||
| tnfn1_pw060328p04q175 | FTN_0818 | lipase/esterase | |
| tnfn1_pw060420p02q175 | FTN_0877 | cls | cardiolipin synthetase |
| tnfn1_pw060328p06q166 | FTN_1273 | long chain fatty acid CoA ligase | |
| conenzyme synthesis | |||
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | quinolinate sythetase A |
| tnfn1_pw060419p04q164 | FTN_0692 | nadA | quinolinate sythetase A |
| tnfn1_pw060420p01q148 | FTN_0694 | nadB | L-aspartate oxidase |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | biotin--acetyl-CoA-carboxylase ligase |
| tnfn1_pw060323p07q185 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase |
| tnfn1_pw060419p02q138 | FTN_0814 | bioF | 8-amino-7-oxononanoate synthase |
| tnfn1_pw060418p01q124 | FTN_1245 | iscS | cysteine desulfurase |
| tnfn1_pw060419p03q126 | FTN_1278 | nadE | NAD synthase |
| tnfn1_pw060323p04q110 | FTN_1678 | nuoC | NADH dehydrogenase I, C subunit |
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | riboflavin synthase beta-chain |
| tnfn1_pw060420p02q191 | FTN_0113 | ribC | riboflavin synthase alpha chain |
| Peptidoglycan biosynthesis | |||
| tnfn1_pw060328p06q125 | FTN_0496 | slt | soluble lytic murein transglycosylase |
| tnfn1_pw060323p04q102 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| tnfn1_pw060418p03q177 | FTN_0211 | pcp | pyrrolidone carboxylylate peptidase |
| Aromatic compound biosynthesis | |||
| tnfn1_pw060323p08q141 | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060420p01q129 | FTN_1015 | isochorismatase family protein | |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase |
| ppGpp biosynthesis | |||
| tnfn1_pw060323p06q110 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase |
For some mutants defective in intracellular proliferation, the two or more mutant alleles resulted in different defective phenotype (Table 1). The lack of consistent phenotype for the two mutant alleles for some of the mutants may be due to the site of the insertion, which may generate a functional or partially functional protein. The intracellular growth defect for most of the mutant alleles that exhibited growth defect was not due to a general defect in growth, since more than 98% of the mutants exhibited normal growth in vitro, compared to the wild type strain. The in vitro growth was analyzed by measuring the OD of all the 501 mutants identified in the primary screen and the wild type strain after overnight growth in broth. It is possible the defect in some of the mutants may be due to a defect in attachment and/or invasion. It is likely that the reduction in intracellular growth for some of the mutants was due to a polar effect of the transposon insertion on downstream genes. However, the possible polar effect would implicate the identified disrupted operon in intracellular proliferation.
Interestingly, 75 of the 394 mutants that showed defect in intracellular proliferation within S2 cells, have been shown in previous screens to exhibit growth defect for various aspects of virulence of F. tularensis in mammals (Table 2) (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009). Importantly, ~90% of F. tularensis subsp novicida genes that affect intracellular proliferation in S2 cells are conserved in the virulent F. tularensis subsp tularensis, which validates the use of subsp novicida as a model system.
Comparison of growth-defective mutants in S2 cells to the phenotype in human macrophages
A concurrent study from our lab has identified mutants that exhibit growth defect in human macrophages (see accompanying manuscript). Infections of U937 human macrophages were performed similar to S2 cells. To assess whether similar molecular mechanisms are utilized by F. tularensis to infect arthropod-derived cells and human macrophages, we compared the mutants identified to be defective in S2 cells to the mutants identified to be defective in human macrophages. Among the 394 mutants defective in replication in D. melanogaster S2 cells 135 of them were also required for replication in human macrophages (Fig. 2). This large number (259) of loci indicates that some common mechanisms are utilized by F. tularensis to proliferate within both insect-derived cells and human macrophages.
Fig 2. Distribution of functional categories of mutants defective for growth in S2 cells compared to mutants defective in both human U937 macrophages and S2 cells.
Growth of the mutants was compared to the wild type strain and the relative reduction in cfu relative to the wild type strain at 24 h post-infection was determined. The mutants were divided into two groups depending on whether they showed growth defect in S2 cells only or both S2 cells and U937 macrophages. The defective mutants in each group were categorized according to the function of the genes.
However, there is a significant difference in the molecular mechanisms required for replication in the two evolutionarily distinct host cells, since 259 of the genes identified to be required for intracellular replication are specific to S2 cells. Similarly, among 202 mutants defective in replication in human macrophages more than 143 were specific to U937 cells, indicating common as well as distinct mechanisms required for the infection of the two evolutionarily distant host cells. However, we can’t exclude the possibility that some of the differences in the phenotypes of the mutants between the two host cells may be due to the fact that simply different cell lines were used and the differences might not be host-specific for some of the mutants. We also can’t exclude the possibility that some of the differences of the phenotypes of the mutants between U937 and S2 cells may be due to the fact that the infection of both host cells was done at different temperatures where the S2 cells must be incubated below 30°C. The U937 cells are PMA-differentiated human macrophage-like cells, while S2 are embryonic macrophage-like cells of an insect. The distribution of the mutants across the different functional categories in the S2 cells was similar to mutants that were defective in both S2 cells and human macrophages and mutants defective in S2 cells only (Fig. 2). Among 70% of the mutants that were attenuated in both S2 cells and human macrophages, the level of attenuation of growth was similar in both S2 cells and human macrophages. However, the other 30% of the mutants showed up to 103 fold difference in the levels of attenuation between S2 cells and human macrophages (Table 1).
Previous studies have shown that in both arthropod cells and human macrophages, F. tularensis transiently occupies a late endosome-like phagosome followed by rapid bacterial escape into the cytosol, where the bacteria proliferate robustly (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005a; Santic et al., 2005b; Checroun et al., 2006; Santic et al., 2007; Bonquist et al., 2008; Chong et al., 2008; Santic et al., 2008; Qin et al., 2009; Wehrly et al., 2009). In our concurrent study, we showed that of the 135 genes required for intracellular proliferation in both macrophages and S2 cells, 59 loci were required for modulation of phagosome biogenesis and bacterial escape into the cytosol of macrophages (see accompanying manuscript). Since we have shown that modulation of phagosome biogenesis and escape into the cytosol by F. tularensis is similar in both arthropod cells and macrophages (Vonkavaara et al., 2008; Santic et al., 2009), it is most likely that these 59 loci are also required for modulation of phagosome biogenesis and phagosomal escape in S2 cells, similar to their role in macrophages. This indicates that conserved as well as distinct molecular mechanisms are utilized by F. tularensis to escape from the phagosome of evolutionarily distant host cells. It is likely that interaction of F. tularensis with arthropods has played a role in its ecology and patho-adaptation to infect mammals. This may not be surprising, since there are numerous evolutionary conserved pathways between the two species, such as innate immune responses and intracellular trafficking events.
Lethality of F. tularensis mutants to D. melanogaster adult flies
Previous in vivo studies on replication of F. tularensis in adult D. melanogaster has shown that some mutants, which are attenuated in human cells or in other mammalian models of infection, are also attenuated in growth and are less lethal to D. melanogaster adult flies (Vonkavaara et al., 2008; Santic et al., 2009). Thus, we determined whether mutants that were attenuated in S2 cells were also attenuated in virulence in adult flies. A total of 168 most defective mutants in intracellular proliferation in S2 cells were selected to be tested in the adult flies for lethality first, then for proliferation (Table 4). To obtain the survival ratio of infected flies, the number of flies that survived the infection at day 15 was divided by the number of flies immediately after infection. We calculated from 13 independent experiments that the average percent survival of wild type strain-infected flies by day 15 was ~22% whereas the percentage of survival of flies infected with the iglC mutant was 85.5% (p < 0.001, unpaired student t-test). We considered a mutant to be attenuated if flies infected with that mutant showed ≥50% survival rate at 15 days post-infection, which is statistically different from the wild type strain-infected flies for the same period (p< 0.0012, unpaired student t-test). A 50–74% survival rate for infected flies was considered moderate attenuation for the mutant and 75–100% survival rate for the flies represented highly attenuated mutant strain. From the 168 mutants tested, a total of 80 mutants were attenuated in lethality to D. melanogaster (Table 4). Twenty two of 80 mutant strains were highly attenuated for growth in D. melanogaster and 58 mutant strains exhibited moderate attenuation (Table 4 and Fig 3). We classified the mutants that showed attenuation in D. melanogaster to groups of functional categories. Higher percentages (60%) of mutants from the “Others” and “unknown function” functional categories were found to be attenuated in lethality to adult D. melanogaster (Table 4), which indicates that further studies of these novel genes might reveal unique mechanism that F. tularensis utilizes for interaction with the arthropod host. For the rest of the functional categories, the percentage of mutants attenuated in lethality to D. melanogaster was 35–45% (Table 4). Among mutants defective in intracellular growth in S2 cells that were tested, 21 of them were also found in other screens to be attenuated in vivo or in-vitro growth of mammalian models of tularemia (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009). We observed that 10 of these loci were also associated with less lethality to D. melanogaster in our screen. Mutants that had differential effects in D. melanogaster and the mammalian host may perhaps highlight the differences of these hosts during F. tularensis infection and warrant further studies.
Table 4.
List of F. tularensis mutants defective in growth and lethality to D. melanogaster grouped according to function.
| Strain Name | Locus Tag | Gene | Description | Log Reduction in Growth Relative to WT | Percent survival | Mut/WT cfu ratio lls | |
|---|---|---|---|---|---|---|---|
| S2 | U937 | ||||||
| Controls | |||||||
| Wild type strain | WT | ||||||
| Intracellular growth locus | IglC | ||||||
| Cell Division | |||||||
| tnfn1_pw060328p01q167 | FTN_0330 | minD | septum formation inhibitor-activating ATPase | 2 | 2 | 13% | 24.6E-03 |
| DNA Modification | |||||||
| tnfn1_pw060510p01q114 | FTN_1073 | DNA/RNA endonuclease G | 6 | 5 | 90% | 1.2E-02 | |
| tnfn1_pw060328p04q156 | FTN_1027 | ruvC | holliday junction endodeoxyribonuclease | 4 | 3 | 60% | 4.7E-01 |
| tnfn1_pw060328p08q179 | FTN_1491 | adenine specific DNA methylase | 2 | 50% | 6.5E-01 | ||
| tnfn1_pw060323p03q167 | FTN_1197 | recR | RecFOR complex, RecR component | 4 | 2 | 44% | 6.7E-04 |
| tnfn1_pw060328p06q158 | FTN_1293 | rnhB | ribonuclease HII | 5 | 3 | 40% | 1.4E+00 |
| tnfn1_pw060328p05q121 | FTN_1487 | restriction endonuclease | 1 | 1 | 40% | 9.0E+00 | |
| tnfn1_pw060510p04q193 | FTN_0680 | uvrC | excinuclease ABC, subunit C | 3 | 6 | 20% | 1.0E-01 |
| tnfn1_pw060510p01q153 | FTN_1154 | type I restriction-modification system, subunit S | 6 | 5 | 20% | 3.2E-01 | |
| tnfn1_pw060323p03q122 | FTN_0577 | mutL | DNA mismatch repair enzyme with ATPase activity | 5 | 5 | 0% | 2.5E-02 |
| FPI | |||||||
| tnfn1_pw060418p04q106 | FTN_1319 | pdpC | hypothetical protein | 5 | 3 | 90% | 1.2E-04 |
| tnfn1_pw060510p01q110 | FTN_1321 | iglD | intracellular growth locus protein D | 6 | 4 | 60% | 2.1E-01 |
| tnfn1_pw060328p06q115 | FTN_1322 | iglC | intracellular growth locus protein C | 5 | 33% | ||
| Hypothetical Protein | |||||||
| tnfn1_pw060420p04q159 | FTN_1223 | conserved hypothetical membrane protein | 7 | 90% | 3.1E-02 | ||
| tnfn1_pw060419p03q188 | FTN_0696 | hypothetical membrane protein | 2 | 2 | 78% | 2.1E-02 | |
| tnfn1_pw060323p07q129 | FTN_0759 | conserved hypothetical protein | 2 | 4 | 71% | 2.1E+00 | |
| tnfn1_pw060328p06q180 | FTN_0038 | hypothetical protein | 4 | 4 | 70% | 3.5E-04 | |
| tnfn1_pw060323p03q142 | FTN_0030 | hypothetical membrane protein | 3 | 4 | 63% | 2.2E-03 | |
| tnfn1_pw060328p05q136 | FTN_0384 | conserved hypothetical protein | 7 | 4 | 62% | 2.3E-02 | |
| tnfn1_pw060328p06q126 | FTN_0732 | hypothetical protein | 5 | 60% | 1.6E+01 | ||
| tnfn1_pw060328p02q129 | FTN_1612 | hypothetical protein | 2 | 2 | 50% | 2.3E-02 | |
| tnfn1_pw060328p04q136 | FTN_1406 | conserved hypothetical membrane protein | 3 | 5 | 50% | 9.0E-02 | |
| tnfn1_pw060328p08q101 | FTN_0054 | hypothetical protein | 1 | 1 | 40% | 3.1E-07 | |
| tnfn1_pw060418p01q191 | FTN_1349 | hypothetical protein | 6 | 6 | 40% | 2.0E-03 | |
| tnfn1_pw060323p07q105 | FTN_0895 | hypothetical protein | 2 | 4 | 38% | 4.4E-02 | |
| tnfn1_pw060420p02q139 | FTN_0013 | hypothetical protein | 3 | 33% | 2.3E+02 | ||
| tnfn1_pw060418p02q175 | FTN_0717 | conserved hypothetical membrane protein | 5 | 30% | 2.3E-02 | ||
| tnfn1_pw060420p02q173 | FTN_0169 | conserved hypothetical membrane protein | 6 | 6 | 30% | 1.9E+03 | |
| tnfn1_pw060323p06q178 | FTN_1389 | conserved hypothetical membrane protein | 3 | 25% | 9.6E-05 | ||
| tnfn1_pw060419p04q117 | FTN_1156 | hypothetical protein | 4 | 4 | 25% | 3.0E+01 | |
| tnfn1_pw060419p02q102 | FTN_0792 | hypothetical protein | 6 | 5 | 22% | 2.3E+01 | |
| tnfn1_pw060419p03q187 | FTN_1123 | conserved hypothetical protein | 3 | 20% | 1.0E-01 | ||
| tnfn1_pw060420p04q104 | FTN_0466 | conserved hypothetical protein | 4 | 20% | 1.3E+00 | ||
| tnfn1_pw060328p06q185 | FTN_0709 | hypothetical protein | 7 | 3 | 20% | 1.3E+00 | |
| tnfn1_pw060510p04q192 | FTN_1098 | conserved hypothetical membrane protein | 6 | 7 | 11% | 2.7E-02 | |
| tnfn1_pw060510p01q184 | FTN_1299 | hypothetical protein | 5 | 10% | 1.3E-01 | ||
| tnfn1_pw060418p03q151 | FTN_0212 | hypothetical membrane protein | 3 | 10% | 3.3E+00 | ||
| Intergenic | |||||||
| tnfn1_pw060328p06q190 | intergenic | 3 | 3 | 89% | 9.9E-03 | ||
| tnfn1_pw060420p03q148 | intergenic | 5 | 80% | 5.0E-02 | |||
| tnfn1_pw060323p08q139 | intergenic | 4 | 4 | 44% | 1.6E-02 | ||
| tnfn1_pw060419p03q131 | intergenic | 2 | 2 | 40% | 5.5E-01 | ||
| tnfn1_pw060419p04q189 | intergenic | 3 | 5 | 22% | 9.8E-02 | ||
| Metabolic | |||||||
| tnfn1_pw060420p04q105 | FTN_1584 | glpD | glycerol-3-phosphate dehydrogenase | 5 | 3 | 100% | 5.6E-04 |
| tnfn1_pw060323p05q182 | FTN_0504 | lysine decarboxylase | 4 | 4 | 100% | 6.7E-04 | |
| tnfn1_pw060510p03q154 | FTN_0516 | glgA | glycogen synthase | 4 | 90% | 3.6E-04 | |
| tnfn1_pw060328p06q142 | FTN_0954 | histidine acid phosphatase | 4 | 4 | 90% | 9.8E-03 | |
| tnfn1_pw060328p06q130 | FTN_0692 | nadA | quinolinate sythetase A | 2 | 3 | 90% | 2.3E-01 |
| tnfn1_pw060328p02q174 | FTN_1135 | aroB | 3-dehydroquinate synthetase | 4 | 3 | 80% | 3.5E-04 |
| tnfn1_pw060328p06q150 | FTN_1494 | aceE | pyruvate dehydrogenase complex, E1 component, pyruvate dehydrogenase | 1 | 4 | 70% | 4.7E-01 |
| tnfn1_pw060418p02q109 | FTN_1376 | disulfide bond formation protein, DsbB family | 4 | 4 | 70% | 6.1E+02 | |
| tnfn1_pw060420p04q169 | FTN_1621 | predicted NAD/FAD-dependent oxidoreductase | 4 | 4 | 67% | 4.2E-01 | |
| tnfn1_pw060510p01q124 | FTN_0507 | gcvP1 | glycine cleavage system P protein, subunit 1 | 7 | 5 | 67% | 4.0E+00 |
| tnfn1_pw060323p06q106 | FTN_1552 | acid phosphatase, PAP2 family | 4 | 60% | 2.8E-04 | ||
| tnfn1_pw060419p01q106 | FTN_0111 | ribH | riboflavin synthase beta-chain | 5 | 4 | 60% | 5.6E-03 |
| tnfn1_pw060328p08q131 | FTN_1174 | murI | glutamate racemase | 5 | 60% | 2.3E-01 | |
| tnfn1_pw060419p03q116 | FTN_0593 | sucD | succinyl-CoA synthetase, alpha subunit | 2 | 2 | 60% | 2.6E+01 |
| tnfn1_pw060323p04q110 | FTN_1678 | nuoC | NADH dehydrogenase I, C subunit | 2 | 55% | 7.9E-04 | |
| tnfn1_pw060323p03q127 | FTN_0567 | tRNA synthetase class II (D, K and N) | 2 | 6 | 55% | 5.6E-03 | |
| tnfn1_pw060510p03q171 | FTN_0588 | asparaginase | 2 | 1 | 50% | 3.0E-02 | |
| tnfn1_pw060510p03q188 | FTN_0627 | chiA | chitinase, glycosyl hydrolase family 18 | 2 | 50% | 1.4E+00 | |
| tnfn1_pw060510p01q142 | FTN_1333 | tktA | transketolase I | 5 | 5 | 45% | 1.8E-06 |
| tnfn1_pw060510p02q178 | FTN_0217 | L-lactate dehydrogenase | 2 | 44% | 4.1E+01 | ||
| tnfn1_pw060323p06q195 | FTN_1390 | Zn-dependent hydrolase | 3 | 40% | 1.2E-01 | ||
| tnfn1_pw060419p01q168 | FTN_0651 | cdd | cytidine deaminase | 6 | 38% | 3.5E+00 | |
| tnfn1_pw060510p02q187 | FTN_1018 | aldolase/adducin class II family protein | 3 | 3 | 33% | 3.0E-03 | |
| tnfn1_pw060328p02q105 | FTN_1620 | appB | cytochrome bd-II terminal oxidase subunit II | 3 | 6 | 33% | 4.4E-02 |
| tnfn1_pw060323p06q103 | FTN_0711 | predicted metal-dependent hydrolase | 4 | 30% | 2.4E-04 | ||
| tnfn1_pw060328p08q145 | FTN_0907 | D-alanyl-D-alanine carboxypeptidase | 3 | 30% | 1.7E-01 | ||
| tnfn1_pw060419p04q130 | FTN_1585 | glpK | glycerol kinase | 3 | 3 | 29% | 4.4E-01 |
| tnfn1_pw060328p01q151 | FTN_0983 | bifunctional protein: glutaredoxin 3/ribonucleotide reductase beta subunit | 3 | 5 | 25% | 1.9E-03 | |
| tnfn1_pw060419p02q112 | FTN_1619 | appC | cytochrome bd-II terminal oxidase subunit I | 7 | 5 | 22% | 4.9E-02 |
| tnfn1_pw060323p07q167 | FTN_1518 | relA | GDP pyrophosphokinase/GTP pyrophosphokinase | 4 | 4 | 22% | 3.6E-01 |
| tnfn1_pw060420p02q175 | FTN_0877 | cls | cardiolipin synthetase | 5 | 7 | 20% | 6.8E-02 |
| tnfn1_pw060328p03q174 | FTN_1768 | pepN | aminopeptidase N | 2 | 20% | 8.0E+00 | |
| tnfn1_pw060418p02q122 | FTN_1438 | bifunctional protein: 3-hydroxacyl- CoA dehydrogenase/acyl-CoA- binding protein | 4 | 20% | 8.8E+01 | ||
| tnfn1_pw060323p06q113 | FTN_0420 | SAICAR synthetase/phosphoribosylamine- glycine ligase | 5 | 7 | 13% | 2.0E-01 | |
| tnfn1_pw060510p01q146 | FTN_1597 | prfC | peptide chain release factor 3 | 5 | 5 | 13% | 6.8E+02 |
| tnfn1_pw060510p01q118 | FTN_1553 | nudH | dGTP pyrophosphohydrolase | 5 | 5 | 11% | 1.3E-01 |
| tnfn1_pw060328p06q156 | FTN_0811 | birA | biotin--acetyl-CoA-carboxylase ligase | 7 | 6 | 11% | 4.5E-02 |
| tnfn1_pw060328p08q120 | FTN_0987 | tRNA-dihydrouridine synthase | 4 | 11% | 2.7E+00 | ||
| tnfn1_pw060420p03q153 | FTN_0840 | mdaB | NADPH-quinone reductase (modulator of drug activity B) | 5 | 5 | 10% | 2.9E-01 |
| tnfn1_pw060420p01q130 | FTN_0965 | metal-dependent exopeptidase | 3 | 3 | 9% | 1.2E+00 | |
| tnfn1_pw060419p03q164 | FTN_1186 | pepO | M13 family metallopeptidase | 2 | 0% | 4.0E-01 | |
| Others | |||||||
| tnfn1_pw060418p02q123 | FTN_0107 | lepA | GTP-binding protein LepA | 4 | 4 | 90% | 9.4E-02 |
| tnfn1_pw060323p06q105 | FTN_0810 | ROK family protein | 4 | 70% | 2.4E-04 | ||
| tnfn1_pw060328p08q114 | FTN_0266 | htpG | chaperone Hsp90, heat shock protein HtpG | 2 | 67% | 5.8E-01 | |
| tnfn1_pw060419p01q120 | FTN_1488 | prophage maintenance system killer protein (DOC) | 6 | 60% | 7.2E-01 | ||
| tnfn1_pw060328p06q178 | FTN_1241 | DedA family protein | 5 | 4 | 55% | 2.8E-04 | |
| tnfn1_pw060418p04q181 | FTN_0338 | MutT/nudix family protein | 2 | 1 | 50% | 1.3E-01 | |
| tnfn1_pw060420p03q193 | FTN_0768 | tspO | tryptophan-rich sensory protein | 3 | 3 | 50% | 1.4E-01 |
| tnfn1_pw060419p02q137 | FTN_1034 | rnfB | iron-sulfur cluster-binding protein | 3 | 2 | 50% | 1.2E+00 |
| tnfn1_pw060323p03q111 | FTN_0465 | Sua5/YciO/YrdC family protein | 2 | 2 | 33% | 2.8E+00 | |
| tnfn1_pw060328p06q167 | FTN_0985 | DJ-1/PfpI family protein | 6 | 2 | 20% | 5.2E-01 | |
| tnfn1_pw060420p03q121 | FTN_1064 | PhoH family protein, putative ATPase | 4 | 1 | 11% | 1.2E-01 | |
| tnfn1_pw060328p03q154 | FTN_1453 | two-component regulator, sensor histidine kinase | 2 | 2 | 11% | 1.0E-02 | |
| tnfn1_pw060420p04q127 | FTN_1031 | ftnA | ferric iron binding protein, ferritin- like | 6 | 2 | 0% | 3.0E+00 |
| Proteins of unknown function | |||||||
| tnfn1_pw060420p01q142 | FTN_0052 | protein of unknown function | 2 | 100% | 1.6E-04 | ||
| tnfn1_pw060420p04q176 | FTN_0855 | protein of unknown function | 2 | 5 | 100% | 6.9E-01 | |
| tnfn1_pw060323p06q122 | FTN_0077 | protein of unknown function | 2 | 91% | 3.2E-05 | ||
| tnfn1_pw060418p02q186 | FTN_1448 | protein of unknown function | 6 | 6 | 90% | 8.8E-01 | |
| tnfn1_pw060418p03q108 | FTN_0900 | protein of unknown function with predicted hydrolase and phosphorylase activity | 6 | 78% | 1.3E-03 | ||
| tnfn1_pw060418p04q193 | FTN_0109 | protein of unknown function | 4 | 70% | 2.9E-01 | ||
| tnfn1_pw060328p06q173 | FTN_0599 | protein of unknown function | 2 | 67% | 3.3E-03 | ||
| tnfn1_pw060328p08q104 | FTN_0051 | conserved protein of unknown function | 3 | 67% | 2.4E-02 | ||
| tnfn1_pw060328p08q149 | FTN_0439 | protein of unknown function | 4 | 60% | 1.2E-03 | ||
| tnfn1_pw060328p01q172 | FTN_1542 | conserved protein of unknown function | 2 | 2 | 60% | 5.5E-03 | |
| tnfn1_pw060323p05q127 | FTN_0791 | protein of unknown function | 1 | 60% | 8.0E-03 | ||
| tnfn1_pw060418p04q121 | FTN_1261 | protein of unknown function | 2 | 60% | 7.7E-01 | ||
| tnfn1_pw060418p02q157 | FTN_1170 | conserved protein of unknown function | 6 | 5 | 60% | 2.6E+01 | |
| tnfn1_pw060418p04q117 | FTN_0207 | protein of unknown function containing a von Willebrand factor type A (vWA) domain | 2 | 56% | 5.0E+00 | ||
| tnfn1_pw060510p04q111 | FTN_0861 | conserved protein of unknown function | 4 | 55% | 4.2E+02 | ||
| tnfn1_pw060323p04q183 | FTN_1386 | protein of unknown function | 3 | 50% | 1.2E-03 | ||
| tnfn1_pw060418p01q142 | FTN_0482 | protein of unknown function | 6 | 50% | 4.8E-03 | ||
| tnfn1_pw060328p05q124 | FTN_1372 | protein of unknown function | 5 | 50% | 1.5E+00 | ||
| tnfn1_pw060323p07q176 | FTN_1534 | conserved protein of unknown function | 3 | 50% | 2.7E+00 | ||
| tnfn1_pw060418p02q105 | FTN_1343 | conserved protein of unknown function | 4 | 4 | 50% | 6.9E+00 | |
| tnfn1_pw060323p03q157 | FTN_0364 | conserved protein of unknown function | 2 | 45% | 6.2E-01 | ||
| tnfn1_pw060328p06q155 | FTN_1764 | protein of unknown function | 7 | 6 | 40% | 3.5E-06 | |
| tnfn1_pw060323p08q148 | FTN_0027 | conserved protein of unknown function | 6 | 4 | 40% | 2.7E-02 | |
| tnfn1_pw060418p02q145 | FTN_1020 | conserved protein of unknown function | 5 | 40% | 6.0E+00 | ||
| tnfn1_pw060420p02q158 | FTN_1071 | protein of unknown function | 5 | 33% | 2.6E+02 | ||
| tnfn1_pw060328p08q107 | FTN_1774 | protein of unknown function | 3 | 33% | 3.6E-01 | ||
| tnfn1_pw060419p04q110 | FTN_0048 | conserved protein of unknown function | 2 | 2 | 29% | 4.2E+01 | |
| tnfn1_pw060419p04q188 | FTN_0925 | protein of unknown function | 5 | 22% | 5.4E+00 | ||
| tnfn1_pw060323p07q141 | FTN_0788 | conserved protein of unknown function | 5 | 5 | 22% | 5.4E+00 | |
| tnfn1_pw060328p02q110 | FTN_1457 | protein of unknown function | 5 | 5 | 20% | 1.5E+00 | |
| tnfn1_pw060420p04q143 | FTN_0149 | conserved protein of unknown function | 5 | 5 | 20% | 3.2E+00 | |
| tnfn1_pw060418p01q155 | FTN_0044 | protein of unknown function | 3 | 10% | 3.5E+01 | ||
| tnfn1_pw060328p02q177 | FTN_1713 | protein of unknown function | 3 | 3 | 0% | 7.0E-04 | |
| Transcription/Translation | |||||||
| tnfn1_pw060510p03q150 | FTN_0949 | rplI | 50S ribosomal protein L9 | 2 | 75% | 2.8E-05 | |
| tnfn1_pw060328p02q148 | FTN_1393 | transcriptional regulator, ArsR family | 3 | 30% | 2.3E-01 | ||
| Transferase | |||||||
| tnfn1_pw060323p06q168 | FTN_0545 | glycosyl transferase, group 2 | 4 | 4 | 87% | 2.7E-03 | |
| tnfn1_pw060328p01q142 | FTN_0928 | cysD | sulfate adenylyltransferase subunit 2 | 3 | 3 | 57% | 9.6E-02 |
| tnfn1_pw060510p01q119 | FTN_1428 | wbtO | transferase | 6 | 2 | 50% | 4.1E-01 |
| tnfn1_pw060420p02q146 | FTN_0300 | glycosyl transferase, group 2 | 5 | 30% | 6.8E-02 | ||
| tnfn1_pw060323p03q121 | FTN_0343 | aminotransferase | 2 | 7 | 20% | 1.9E+01 | |
| tnfn1_pw060418p01q110 | FTN_0200 | UDP-3-O-[3-fatty acid] glucosamine N-acyltransferase | 2 | 2 | 20% | 2.4E+02 | |
| tnfn1_pw060510p01q103 | FTN_0063 | ilvE | branched-chain amino acid aminotransferase protein (class IV) | 5 | 3 | 13% | 3.1E-01 |
| tnfn1_pw060418p04q172 | FTN_1418 | manC | mannose-1-phosphate guanylyltransferase | 4 | 10% | 8.3E-01 | |
| Transporter proteins | |||||||
| tnfn1_pw060420p01q180 | FTN_0097 | hydroxy/aromatic amino acid permease (HAAAP) family protein | 4 | 4 | 89% | 3.0E-05 | |
| tnfn1_pw060420p01q189 | FTN_1611 | - | major facilitator superfamily (MFS) transport protein | 5 | 7 | 89% | 9.2E-02 |
| tnfn1_pw060420p03q104 | FTN_1593 | oppA | ABC-type oligopeptide transport system, periplasmic component | 6 | 4 | 70% | 8.4E-04 |
| tnfn1_pw060323p06q117 | FTN_1685 | drug:H+ antiporter-1 (DHA1) family protein | 4 | 70% | 9.5E-02 | ||
| tnfn1_pw060418p04q168 | FTN_0141 | ABC transporter, ATP-binding protein | 6 | 5 | 70% | 1.1E-01 | |
| tnfn1_pw060328p06q119 | FTN_1549 | drug:H+ antiporter-1 (DHA1) family protein | 6 | 62% | 1.3E-01 | ||
| tnfn1_pw060510p03q140 | FTN_0741 | proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | 3 | 60% | 3.7E-02 | ||
| tnfn1_pw060419p01q175 | FTN_0984 | ABC transporter, ATP-binding protein | 2 | 56% | 5.0E-01 | ||
| tnfn1_pw060323p05q110 | FTN_1215 | kpsC | capsule polysaccharide export protein KpsC | 5 | 2 | 55% | 8.7E-07 |
| tnfn1_pw060323p01q175 | FTN_1441 | sugar porter (SP) family protein | 4 | 4 | 50% | 9.5E-05 | |
| tnfn1_pw060328p04q167 | FTN_0997 | proton-dependent oligopeptide transporter (POT) family protein, di- or tripeptide:H+ symporter | 3 | 5 | 50% | 2.4E-03 | |
| tnfn1_pw060419p01q170 | FTN_1006 | transporter-associated protein, HlyC/CorC family | 2 | 50% | 6.7E-01 | ||
| tnfn1_pw060328p06q132 | FTN_0728 | predicted Co/Zn/Cd cation transporter | 5 | 2 | 50% | 2.1E+00 | |
| tnfn1_pw060328p08q167 | FTN_0579 | major facilitator superfamily (MFS) transport protein | 3 | 44% | 1.6E+00 | ||
| tnfn1_pw060419p03q162 | FTN_0115 | Na+/H+ antiporter | 4 | 4 | 40% | 4.4E-02 | |
| tnfn1_pw060419p02q126 | FTN_1581 | small conductance mechanosensitive ion channel (MscS) family protein | 3 | 3 | 40% | 1.6E-01 | |
| tnfn1_pw060418p03q187 | FTN_1733 | - | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 4 | 2 | 40% | 3.4E+00 |
| tnfn1_pw060418p02q182 | FTN_1275 | drug:H+ antiporter-1 (DHA2) family protein | 4 | 40% | 4.4E+00 | ||
| tnfn1_pw060510p02q159 | FTN_0688 | galP2 | galactose-proton symporter, major facilitator superfamily (MFS) transport protein | 3 | 40% | 2.2E+04 | |
| tnfn1_pw060323p07q172 | FTN_1344 | major facilitator superfamily (MFS) transport protein | 4 | 4 | 37% | 4.3E-05 | |
| tnfn1_pw060420p02q159 | FTN_1716 | kdpC | potassium-transporting ATPase C chain | 2 | 2 | 33% | 2.0E+03 |
| tnfn1_pw060419p01q133 | FTN_1014 | nicotinamide ribonucleoside (NR) uptake permease (PnuC) family protein | 2 | 30% | 3.8E+00 | ||
| tnfn1_pw060510p02q118 | FTN_1409 | major facilitator superfamily (MFS) transport protein | 2 | 20% | 4.8E-01 | ||
| tnfn1_pw060323p05q139 | FTN_1166 | metabolite:H+ symporter (MHS) family protein | 1 | 20% | 1.4E+00 | ||
| tnfn1_pw060323p03q117 | FTN_0005 | corA | divalent inorganic cation transporter | 3 | 12% | 6.9E+03 | |
| tnfn1_pw060328p03q163 | FTN_0884 | drug/metabolite transporter superfamily protein | 4 | 4 | 11% | 1.6E-03 | |
| tnfn1_pw060510p04q167 | FTN_1681 | fur | ferric uptake regulation protein | 2 | 10% | 3.5E-01 | |
| tnfn1_pw060420p02q151 | FTN_0018 | sdaC | serine permease | 4 | 1 | 10% | 9.9E+02 |
| Type IV pili | |||||||
| tnfn1_pw060418p02q167 | FTN_1137 | pilQ | Type IV pili secretin component | 4 | 60% | 1.5E+03 | |
Fig 3. Lethality to D. melanogaster by F. tularensis subsp novicida mutants.
D. melanogaster was infected and survival curves of representative F. tularensis subsp novicida mutants are shown. Wild type F. tularensis subsp novicida U112 and its isogenic iglC mutant are positive and negative controls, respectively. Data are represented as a daily mean % survival of 20 flies divided into two groups of 10. Error bar are standard deviation (SD) of the two groups.
Proliferation of F. tularensis mutants within D. melanogaster adult flies
We examined proliferation of the attenuated mutants within adult flies to determine if the defect in S2 cells correlated with reduced proliferation within D. melanogaster. To examine bacterial growth within the adult fly, 15 flies were infected with each mutant strain. A total of five flies were used for determination of average CFU at day 0 post infection and 10 flies used for the day 3 post-infection. The average CFU on day 15 was divided by the average cfu on day 0 to obtain the relative bacterial growth rate of each mutant in the fly. Furthermore, to normalize bacterial growth rate of all mutants in order to compare them to each other, the results are expressed as the ratio of mutant bacterial load to wild type (control) bacterial load. The averages cfu within the same test group were calculated using a trimmean, which excluded outliers (~20%) from each end of a data set. The ratio of the wild type strain of 1 was considered a reference. A mutant was considered to be attenuated if it showed 10 or more fold reduction in growth compared to the wild type bacteria. The average cfu of inoculums recovered from flies immediately after pricking considered day 0, was ~30 cfu per fly. By day 3 post infection, the wild type bacterial growth increased by ~108 cfu, whereas the iglC mutant (control) increased by ~103 cfu. Some mutants such as FTN_0054 (hypothetical), FTN_0097 (transporter), and FTN_0516 (metabolic) were severely attenuated in proliferation within D. melanogaster (Table 4). Other mutants such as FTN_0984 (transport) exhibited only a slight reduction in growth whereas some mutants such as FTN_0688 (transport) showed higher levels of growth compared to the wild type strain (Table 4). Thirteen out of 24 mutants from the “hypothetical proteins” functional category were found to be attenuated in replication within D. melanogaster adults. Mutants in the small category “intergenic” are interesting, 4 out 5 of these mutants, showed less proliferation within D. melanogaster. For the remaining categories, a fair percentage (~40%) of mutants tested showed defect in growth within D. melanogaster (Table 4).
The difference observed between intracellular growth in S2 cells and proliferation of F. tularensis within the fly might indicate that other organs, tissues or cells other than the macrophage-like cells might have a crucial role in replication of F. tularensis in arthropods. Importantly, previous findings have shown that, albeit hemocytes that are macrophage-like cells being important for infection, F. tularensis also multiply within extracellular spaces in the head, legs and wings of D. melanogaster (Vonkavaara et al., 2008). Other pathogens such as Listeria monocytogenes also spread through D. melanogaster tissues such as the fat body and epithelium (Mansfield et al., 2003). Mutants with the lowest bacterial load (more than 103 fold less than the wild type strain) were not necessarily the least lethal mutants to D. melanogaster, as only 13/40 of these caused ≤25% lethality to infected flies (Table 4). This finding indicates that the bacterial factors defective in these mutants have additional roles that modulate virulence within D. melanogaster. Among mutants defective in intracellular growth in S2 cells, 21 of them were involved in other aspects of virulence or dissemination in animal models of tularemia (Table 2) (Qin and Mann, 2006; Maier et al., 2007; Su et al., 2007; Weiss et al., 2007; Kraemer et al., 2009), 7 of these mutated genes were also associated with less lethality to D. melanogaster in our screen. Mutants that have differential effects in D. melanogaster and the mammalian host may highlight host-specific differences during infection by F. tularensis of evolutionarily distant hosts
When we classified our data according to lethality effects on D. melanogaster, we uncovered a correlation between the number of bacteria in the fly and the survival of the fly (Table 4). Mutants that exhibited low CFU resulted in higher survival rate for the infected flies. In addition, D. melanogaster infected with mutants that proliferated to high rates were less likely to survive (Table 4). Conversely, the least lethal a mutant was to D. melanogaster, the more likely it was to be attenuated in intracellular proliferation (Table 4). Approximately 64% of the mutants that caused up to 25% lethality in D. melanogaster exhibited at least 103 fold less CFU than wild type strain. Overall, 86% of these mutants had at least 10 fold less bacterial load than the wild type strain.
Combined, these observations indicate that decreased bacterial load offers survival advantage in most cases. Thus, screening F. tularensis strains for lethality to D. melanogaster might be a reasonable approach to identify important bacterial factors involved in arthropod-Francisella interaction. Although maintenance of F. tularensis in mammals is thought to be associated mainly with cottontail rabbits (Farlow et al., 2005; Keim et al., 2007; Goethert et al., 2009), recent cases of tularemia are mainly associated with arthropod vectors (2002; Goethert et al., 2009). In addition, many different kinds of hematophagous arthropods are vectors for transmission of tularemia, including tabanid flies and mosquitoes (Goethert et al., 2009). The mode of perpetuation seems to involve both horizontal (feeding of an arthropod on an infected mammal) and vertical (inheritance of infection by arthropod progeny) transmission (Hopla, 1974).
The association between F. tularensis and arthropod has most likely resulted in patho-adaptation that has enabled the bacteria to survive within the natural environment, and has likely equipped F. tularensis with molecular mechanisms for survival in mammalian cells. This is supported by our observations that a large number of the F. tularensis genes that are required for intracellular proliferation within human macrophages are also required for proliferation within S2 cells, for lethality to and proliferation within adult flies. Importantly, we have recently shown that modulation of phagosome biogenesis and phagosomal escape of F. tularensis into the cytosol are similar in both arthropod cells and macrophages (Santic et al., 2009). Since 59 bacterial loci are required for phagosomal escape in macrophages (see accompanying manuscript), it is most likely that these loci are also required for phagosomal escape in S2 cells. This indicates conserved as well as distinct molecular mechanisms utilized by F. tularensis to escape from the phagosome of evolutionarily distant host cells. Taken together, it is likely that interaction of F. tularensis with arthropods has played a factor in its ecology and patho-adaptation to infect mammals, since there are numerous evolutionary conserved pathways between the two species, such as innate immune responses and intracellular trafficking events. This is the first comprehensive molecular identification of the genetic loci of an intracellular pathogen required for intracellular proliferation within arthropod-derived cells and adult arthropod model system. Our studies will facilitate deciphering the molecular aspects of F. tularensis-arthropod vector interaction and its role in bacterial ecology and patho-adaptation to infect mammals.
Experimental Procedure
Bacterial strains, U937 macrophages, S2 cells and Media
F. tularensis subsp. novicida strain U112 and its isogenic mutants mglA and iglC have been described previously (Lauriano et al., 2004). The construction of the F. tularensis subsp. novicida mutants library which was obtained from Biodefence and Emerging Infections Resource Repository (http://www.beiresources.org) has been described previously (Gallagher et al., 2007). F. tularensis subsp. novicida strain U112 and its isogenic iglC mutant harboring the plasmid pKK214, which encodes gfp, were used in fly experiments as positive and negative controls, respectively, and have been described elsewhere (Abd et al., 2003; Lauriano et al., 2004; Santic et al., 2005b; Santic et al., 2007). All F. tularensis subsp. novicida strains were grown on tryptic soy agar (TSA) plates or in tryptic soy broth (TSB) supplemented with 0.1% cysteine and 10 mg/ml of kanamycin for 2 days or overnight respectively. Drosophila S2 cells has been described previously (Santic et al., 2009). For recovering bacteria from flies, tryptic soy agar (TSA) plates supplemented with 0.1 % cysteine, 10 mg ml−1 tetracycline or 10 mg ml−1 kanamycin were used as described previously (Santic et al., 2009).
Intracellular survival assays in S2 cells
Intracellular growth kinetics of F. tularensis in S2 cells was performed as previously described (Santic et al., 2009) with minor modifications. Briefly, S2 cells were seeded at 1 × 106 cells ml−1 in 96 well plates pre-coated with 0.5 mg ml−1 concavalin A (ConA) and incubated at 28°C with SDM containing 10% FBS (SDM-10) overnight. Cells were washed 3 X with SDM-10 and infected in duplicates with all the alleles of each mutant as well as wild type F. tularensis subsp. novicida and its isogenic iglC mutant at a MOI of 10. MOI was determined for each of the mutant strains. Infected cells were incubated for 1h at 28°C followed by treatment with 50μg ml−1 gentamicin for 1 h to kill extracellular bacteria. Infected cells were subsequently washed three times and incubated with fresh SDM-10 for 22 h for a total of 24 h of infection. The culture supernatant was removed and the cells were lysed by the addition of 200μl of sterile water for 10 minutes. The culture supernatant and lysate were combined and the number of bacterial CFUs in each well was determined by plating serial dilutions on TSA agar plates supplemented with 0.1 % cysteine, 10 μg ml−1 tetracycline or 10 μg ml−1 kanamycin for colony enumeration.
F. tularensis infection of D. melanogaster adult flies
Infections of D. melanogaster adult fly were performed as previously described (Santic et al., 2009) with minor changes. 7–14 days old flies were used in all our experiments. For D. melanogaster infections, adult female flies were anesthetized with CO2, pricked in the dorsal thorax with a 30-gauge needle dipped into 1ml of bacteria culture diluted to 2×106 ml−1 in TSB-C broth, then transferred to their designated food vial. F. tularensis subsp novicida and its isogenic mutant iglC were used as positive and negative control, respectively. To screen for lethality effects of each mutant of interest upon infection of the adult flies, 20 flies were infected and housed in two different vials and their survival observed over a period of 15 days. The percent survival of flies was calculated as a ratio of flies surviving by day 15 to flies that were alive by day 1 post inoculation (day 15/day 1). To screen for bacterial growth, we performed viable cell count using 5 infected flies at day 0 and 10 infected flies at day 3 post-infection as previously described (Santic et al., 2009). Bacterial growth rate was calculated by dividing day 3 CFUs by day 0 CFUs. Groups of mutants were tested in independent experiments and wild type F tularensis subsp novicida was used as a control in all experiments. Thus to normalize bacterial growth rate of all mutants in order to compare them to each other, the results are expressed as the ratio of mutant bacterial load to wild type (control) bacterial load. The averages for bacterial cfu within the same test group were averaged using a trimmean, which excluded outliers (~20%) from each end of a data set.
Statistical Analyses
To analyze for statistically significant differences between two sets of data, student’s t-test was used and the p value was obtained. GraphPad Prism 5 was used for statistical analysis.
Acknowledgments
YAK is supported by Public Health Service Awards R01AI43965 and R01AI069321 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund.
References
- Tularemia--United States, 1990–2000. MMWR Morb Mortal Wkly Rep. 2002;51:181–184. [PubMed] [Google Scholar]
- Abd H, Johansson T, Golovliov I, Sandstrom G, Forsman M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl Environ Microbiol. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KV. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- Apidianakis Y, Rahme LG. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat Protoc. 2009;4:1285–1294. doi: 10.1038/nprot.2009.124. [DOI] [PubMed] [Google Scholar]
- Baron GS, Nano FE. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol Microbiol. 1998;29:247–259. doi: 10.1046/j.1365-2958.1998.00926.x. [DOI] [PubMed] [Google Scholar]
- Bonquist L, Lindgren H, Golovliov I, Guina T, Sjostedt A. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun. 2008;76:3502–3510. doi: 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MD, Zeng Q, Nix EB, Nano FE, Keim P, Kodira CD, et al. Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 2009;5:e1000459. doi: 10.1371/journal.ppat.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S. Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr Opin Microbiol. 2008;11:262–270. doi: 10.1016/j.mib.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, Celli J. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio DA, Gallagher LA, Berg CA, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2001;183:1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect Immun. 2003;71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow J, Wagner DM, Dukerich M, Stanley M, Chu M, Kubota K, et al. Francisella tularensis in the United States. Emerg Infect Dis. 2005;11:1835–1841. doi: 10.3201/eid1112.050728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, Manoil C. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethert HK, Saviet B, Telford SR., 3rd Metapopulation structure for perpetuation of Francisella tularensis tularensis. BMC Microbiol. 2009;9:147. doi: 10.1186/1471-2180-9-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjostedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Cirillo KA. Environmental adaptation of Francisella tularensis. Microbes Infect. 2009 doi: 10.1016/j.micinf.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- Hopla CE. The ecology of tularemia. Adv Vet Sci Comp Med. 1974;18:25–53. [PubMed] [Google Scholar]
- Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- Kim SH, Park SY, Heo YJ, Cho YH. Drosophila melanogaster-based screening for multihost virulence factors of Pseudomonas aeruginosa PA14 and identification of a virulence-attenuating factor, HudA. Infect Immun. 2008;76:4152–4162. doi: 10.1128/IAI.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer PS, Mitchell A, Pelletier MR, Gallagher LA, Wasnick M, Rohmer L, et al. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun. 2009;77:232–244. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz CL, Chauvet S, Andres E, Aurouze M, Vallet I, Michel GP, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22:1451–1460. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, et al. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:e1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, Klose KE. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier TM, Casey MS, Becker RH, Dorsey CW, Glass EM, Maltsev N, et al. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect Immun. 2007;75:5376–5389. doi: 10.1128/IAI.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- Needham AJ, Kibart M, Crossley H, Ingham PW, Foster SJ. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology. 2004;150:2347–2355. doi: 10.1099/mic.0.27116-0. [DOI] [PubMed] [Google Scholar]
- Nigrovic LE, Wingerter SL. Tularemia. Infect Dis Clin North Am. 2008;22:489–504. ix. doi: 10.1016/j.idc.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Oyston PC, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A, Scott DW, Thompson JA, Mann BJ. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77:152–161. doi: 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read A, Vogl SJ, Hueffer K, Gallagher LA, Happ GM. Francisella genes required for replication in mosquito cells. J Med Entomol. 2008;45:1108–1116. doi: 10.1603/0022-2585(2008)45[1108:fgrfri]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Abu Kwaik Y. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell Microbiol. 2005a;7:957–967. doi: 10.1111/j.1462-5822.2005.00529.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Al-Khodor S, Abu Kwaik Y. Cell biology and molecular ecology of Francisella tularensis. Cell Microbiol. 2010;12:129–139. doi: 10.1111/j.1462-5822.2009.01400.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Abu Kwaik Y. Francisella tularensis travels a novel, twisted road within macrophages. Trends Microbiol. 2006;14:37–44. doi: 10.1016/j.tim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Klose KE, Jones S, Kwaik YA. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell Microbiol. 2005b;7:969–979. doi: 10.1111/j.1462-5822.2005.00526.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Akimana C, Asare R, Kouokam JC, Atay S, Kwaik YA. Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ Microbiol. 2009;11:1473–1481. doi: 10.1111/j.1462-2920.2009.01875.x. [DOI] [PubMed] [Google Scholar]
- Santic M, Molmeret M, Barker JR, Klose KE, Dekanic A, Doric M, Abu Kwaik Y. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol. 2007;9:2391–2403. doi: 10.1111/j.1462-5822.2007.00968.x. [DOI] [PubMed] [Google Scholar]
- Schmerk CL, Duplantis BN, Howard PL, Nano FE. A Francisella novicida pdpA mutant exhibits limited intracellular replication and remains associated with the lysosomal marker LAMP-1. Microbiology. 2009;155:1498–1504. doi: 10.1099/mic.0.025445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonkavaara M, Telepnev MV, Ryden P, Sjostedt A, Stoven S. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis. Cell Microbiol. 2008;10:1327–1338. doi: 10.1111/j.1462-5822.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- Wehrly TD, Chong A, Virtaneva K, Sturdevant DE, Child R, Edwards JA, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;11:1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, Monack DM. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]