Abstract
PTPN2 (protein tyrosine phosphatase non-receptor type 2, also known as TC-PTP) is a cytosolic tyrosine phosphatase that functions as a negative regulator of a variety of tyrosine kinases and other signaling proteins.1–3 In agreement with its role in the regulation of the immune system, PTPN2 was identified as a susceptibility locus for autoimmune diseases.4,5 In this work, we describe the identification of focal deletions of PTPN2 in human T-cell acute lymphoblastic leukemia (T-ALL). Deletion of PTPN2 was specifically found in T-ALLs with aberrant expression of the TLX1 transcription factor oncogene,6 including four cases also expressing the NUP214-ABL1 tyrosine kinase.7 Knockdown of PTPN2 expression increased the proliferation and cytokine sensitivity of T-ALL cells. In addition, PTPN2 was identified as a negative regulator of NUP214-ABL1 kinase activity. Our study provides genetic and functional evidence for a tumor suppressor role of PTPN2, and suggests that expression levels of PTPN2 may modulate response to treatment.
Keywords: tumor suppressor gene, cancer, phosphorylation
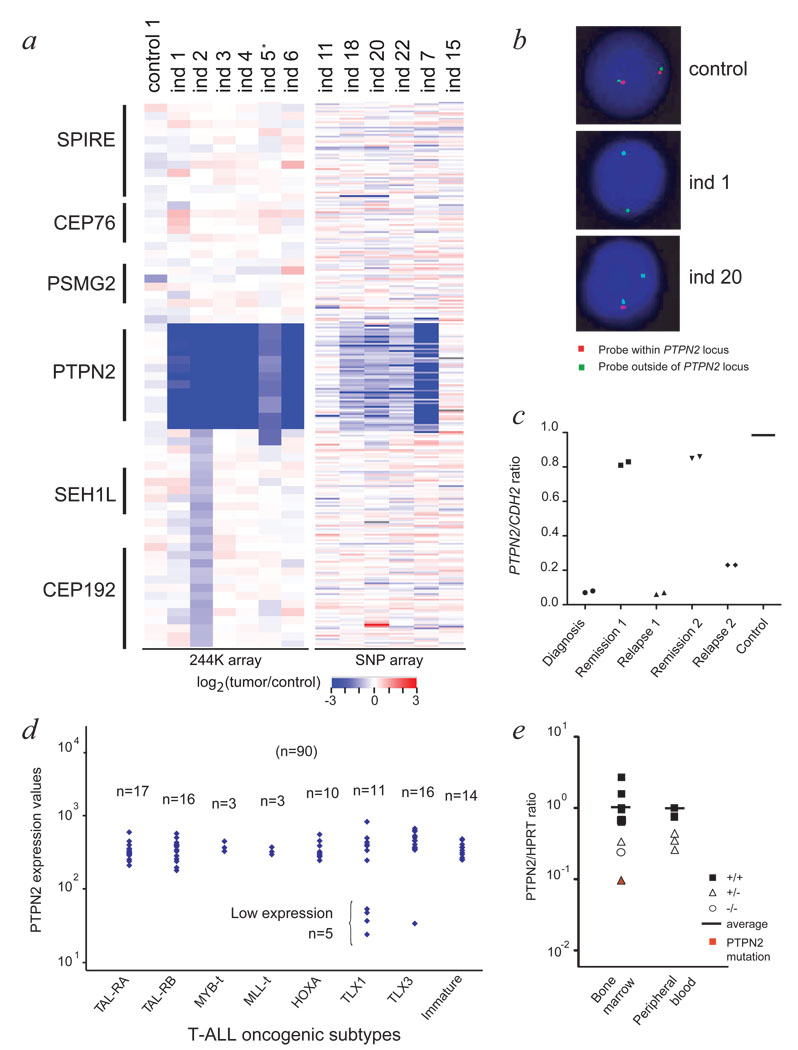
T-ALL is characterized by leukemic transformation of thymocytes caused by cooperative events altering proliferation, survival, cell cycle and differentiation of T-cells.8,9 Despite major improvements in our understanding of the molecular genetics of T-ALL, the mechanisms that lead to the abnormal proliferation and/or survival of T-lymphoblasts remain largely unknown.10 In order to identify novel oncogenes or tumor suppressor genes involved in the development of T-ALL, we performed microarray comparative genomic hybridization (array CGH) analysis using high density oligonucleotide arrays.11 In one individual we detected an acquired homozygous microdeletion at chromosome 18p11 (Fig. 1a, Supplementary Fig. 1 online). Fluorescence in situ hybridization (FISH) confirmed the presence of a homozygous deletion of PTPN2 in 90% of the bone marrow cells (Fig. 1b). Quantitative PCR on genomic DNA isolated from diagnosis, remission and relapse, confirmed that the deletion was acquired at diagnosis and again present at relapse (Fig. 1c).
Figure 1. Comprehensive analysis of T-ALL individuals featuring PTPN2 deletion.
- Agilent array CGH and Affymetrix SNP array data for 13 T-ALL cases indicating bi-allelic deletion of PTPN2 in individuals 1 to 7, and mono-allelic deletion of PTPN2 in individuals 18, 20 and 22. *blast cell content for individual 5 was only 50%.
- Interphase fluorescence in situ hybridization (FISH) confirmed the presence of PTPN2 deletion in >90% of the cells for individuals 1 (bi-allelic deletion) and 20 (mono-allelic deletion).
- Quantitative PCR analysis on genomic DNA isolated from diagnosis, remission and relapse samples of individual 1 confirmed that the deletion was acquired at diagnosis, and again present at relapse. A primer set for CDH2 (18q12) was used for normalization.
- PTPN2 expression level was analyzed in a T-ALL patient set (n=90) using Affymetrix gene expression arrays (probeset 213136_at) and categorized according to oncogenic T-ALL subtypes (x-axis). Y-axis displays PTPN2 expression levels as a logarithmic scale. N indicates number of patients analyzed.
- Quantitative real-time PCR analysis of T-ALL patients detects lower expression levels for T-ALL cases with loss of PTPN2 compared to T-ALL patients with no deletion. Expression values were normalized for HPRT gene expression and blotted at the y-axis in logarithmic scale. Samples derived from bone marrow or peripheral blood were analyzed separately. Closed rectangles: no deletion (n=8); open triangles: mono-allelic deletion (n=5); open circle: bi-allelic deletion (ind 1). The individual with an additional mutation in PTPN2 is shown in red.
PTPN2 is highly expressed in thymocytes.12 In order to identify additional cases with PTPN2 deletion, we analyzed the gene expression profiles of 90 T-ALL cases,13 which identified 5 cases with low PTPN2 expression (Fig. 1d). Analysis of genomic DNA confirmed the presence of acquired homozygous deletions restricted to the PTPN2 locus specifically in the 5 cases with low expression levels (Fig. 1a, Supplementary Fig. 2 online). Strikingly, array CGH profiles suggested that in all cases with PTPN2 deletion, the breakpoints were highly similar. The deletion breakpoints were subsequently mapped within two Alu repeat regions flanking PTPN2 (Supplementary Fig. 3 online). SNP analysis excluded the presence of uniparental disomy at chromosome 18, providing evidence that deletion of the 2 different PTPN2 alleles occurred as 2 independent events.
We next screened an additional set of T-ALL cases (n=106) for copy number alterations (n=97), expression (n=9) or mutations (n=70) of PTPN2. Using this strategy, we identified 2 additional individuals with bi-allelic deletion, and 5 individuals with mono-allelic deletion of PTPN2 (Fig. 1a, Supplementary Fig. 4 online). Individuals with bi-allelic or mono-allelic deletion of PTPN2 showed significantly lower mRNA expression levels as compared to individuals with normal PTPN2 copy 3 number (Fig. 1e, Supplementary Fig. 5 online). One case with mono-allelic deletion of PTPN2 harbored a nonsense mutation in the residual PTPN2 allele (Table 1), but DNA methylation was not detected as a mechanism to silence PTPN2 gene expression in T-ALL (Supplementary Fig. 6 online). Deletion of PTPN2 was not detected in T-ALL cases with normal PTPN2 expression levels, nor in AML cases (n=60, data not shown), nor in published SNP array profiles of B-ALL.14 Strikingly, all individuals with homozygous deletion of PTPN2 belonged to the TLX1 (n=12) or TLX3 (n=1) positive T-ALL subgroup (Fig. 1d, Table 1, Supplementary Table 1 online). TLX1 positive cells accumulate at the CD4+CD8+CD3−/low differentiation stage,6 where PTPN2 expression is high (Supplementary Fig. 7 online). This observation suggests that CD4+CD8+CD3−/low thymocytes may be most sensitive to loss of PTPN2 function. Our data show that inactivation of PTPN2 occurs in approximately 6 % of T-ALL cases, associated with intermediate age (range 4–49 years, median 24 years), and the TLX1 positive subgroup (12 of 36 TLX1 positive cases, 33 %). Deletion of the entire PTPN2 gene is the most common inactivation mechanism.
Table 1.
Cytogenetic and molecular findings in individuals with PTPN2 deletion
| Ind | Sex | Age (years) |
Karyotypea | PTPN2 statusb |
TLX1/TLX3 expressionc |
NUP214- ABL1d |
|---|---|---|---|---|---|---|
| 1 | F | 30 | 46, XX, add(9)(p13), t(10;14) (q23;q11), del(12)(p12) / 46, XX, idic(9)(p13), t(10;14)(q24;q11), del(12)(p12) |
PTPN2 −/− | TLX1 | − |
| 2 | M | 24 | NA | PTPN2 −/− | TLX1 | + |
| 3 | F | 27 | 47,XX,del(6)(q21),t(7;10)(q35; q24),del(13)(q14), +mar |
PTPN2 −/− | TLX1 | − |
| 4 | M | 21 | 46,XY | PTPN2 −/− | TLX1 | − |
| 5 | M | 45 | 46,XY,t(10;14)(q24;q11) | PTPN2 −/− | TLX1 | + |
| 6 | M | 8 | 46,XY | PTPN2 −/− | TLX3 | − |
| 7 | M | 43 | 46,XY,del(6)(q15),t(7;10) (q34;q24),del(9)(p21) |
PTPN2 −/− | TLX1 | − |
| 8 | M | 20 | 46,Y,t(X;7)(p21;q36), del(6)(q23), t(10;14)(q24;q11), der(21)t(6;21)(q23;p11).ish del(9)(p21p21)x2 |
PTPN2 −/− | TLX1 | − |
| 9 | F | 4 | 46,XX | PTPN2 +/− | TLX1 | + |
| 10 | M | 11 | 46,XY,t(10,14)(q24,q11) | PTPN2 +/− | TLX1 | − |
| 18 | F | 35 | 49,XX,+7,t(7;10)(q34;q24),+13, +20 |
PTPN2 +/− | TLX1 | − |
| 20 | F | 9 | NA | PTPN2 +/− | TLX1 | + |
| 22 | F | 49 | 46,X,t(X;9)(q28;p22), del(5)(q23q33), t(7;10)(q34;q24),del(9)(p13) |
PTPN2 +/− PTPN2 mutation (862 C>T, R288Stop) |
TLX1 | − |
Summary of data on individuals with PTPN2 deletion; additional data is provided in supplementary table 1 online.
Karyotype obtained from bone marrow culture at diagnosis.
Determined by FISH, Q-PCR or array-CGH.
Positivity for TLX1 or TLX3 expression or rearrangement.
Positivity for NUP214-ABL1 expression or rearrangement (+, positive for NUP214-ABL1; −, negative for NUP214-ABL1). −/−, bi-allelic deletion; +/−, mono-allelic deletion; NA, not available; F, female; M, male.
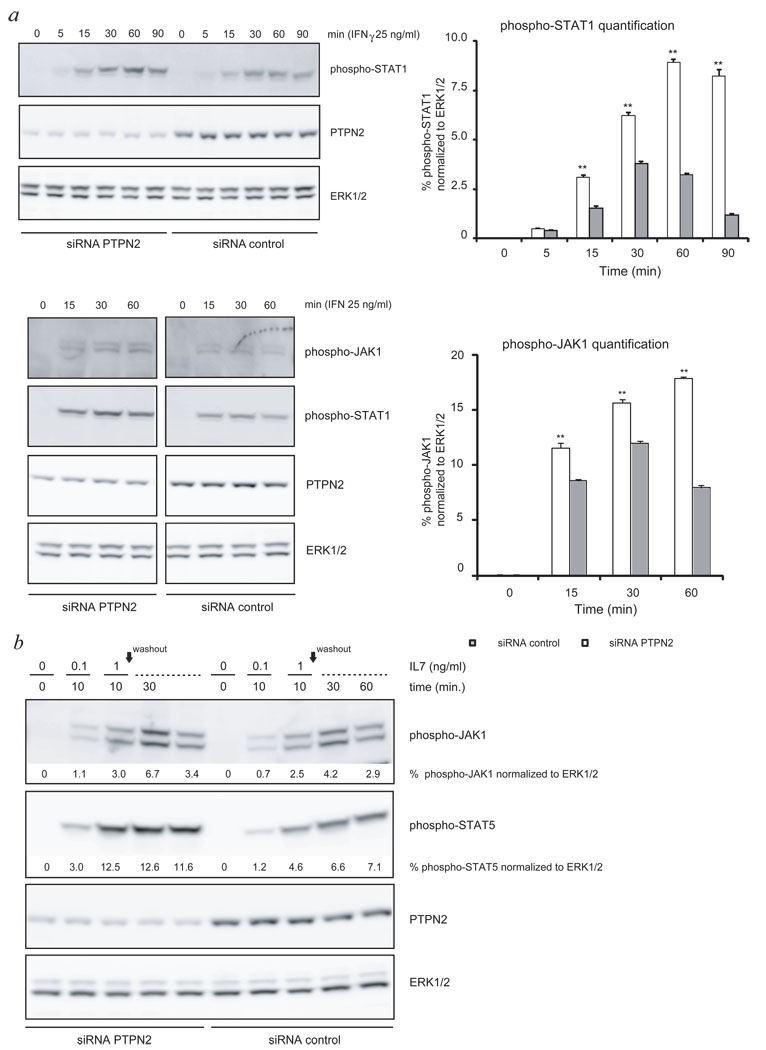
PTPN2 was described as a phosphatase for both JAKs and STATs,15–17 which are important signaling proteins downstream of cytokine receptors. To determine the effect of loss of PTPN2 on cytokine receptors implicated in T-cell development, we knocked down the expression of PTPN2 in human T-ALL cell lines and primary mouse T-ALL cells and measured the effect on IFNγ, IL2 and IL7 receptor signaling.18 JURKAT cells were electroporated with PTPN2 targeting siRNA or non-targeting siRNA and stimulated with IFNγ for various time periods. Knockdown of PTPN2 resulted in a significant increase of both the strength and the time of JAK1 and STAT1 phosphorylation in this cell line (Fig. 2a). HPB-ALL cells, another human T-ALL cell line, showed expression of the IL7 receptor and were responsive to exogenous stimulation with IL7. Knockdown of PTPN2 in these cells resulted in a significant increase of JAK1 and STAT5 phosphorylation in response to IL7 stimulation (Fig. 2b).
Figure 2. Knockdown of PTPN2 causes increased sensitivity of T-cell lines to cytokine stimulation.
- Western blot analysis of WCL showing increased sensitivity of JURKAT cells to cytokine stimulation (IFNγ) due to decreased PTPN2 expression. Stimulated cells expressing reduced levels of PTPN2 displayed an increased and prolonged phosphorylation of STAT1 as well as JAK1 (right panel). Results of two independent experiments are shown. ERK1/2 was used as loading control. Open bars: siRNA PTPN2; closed bars: siRNA control; bars show average ± s.e.m., n=3; error bars represent s.e.m. *p<0.05, **p<0.005
- HPB-ALL cells expressing reduced (siRNA PTPN2) or normal levels (siRNA control) of PTPN2 were stimulated with different concentrations of IL7 for indicated time points and WCL were analyzed for activation of STAT5 and JAK1 by western blot. Quantification of western blot experiments showed that knockdown of PTPN2 resulted in stronger activation of JAK1 and STAT5. Normalized quantification values calculated as fold- change compared to unstimulated cells are shown below respective immunoblots. ERK1/2 was used as loading control.
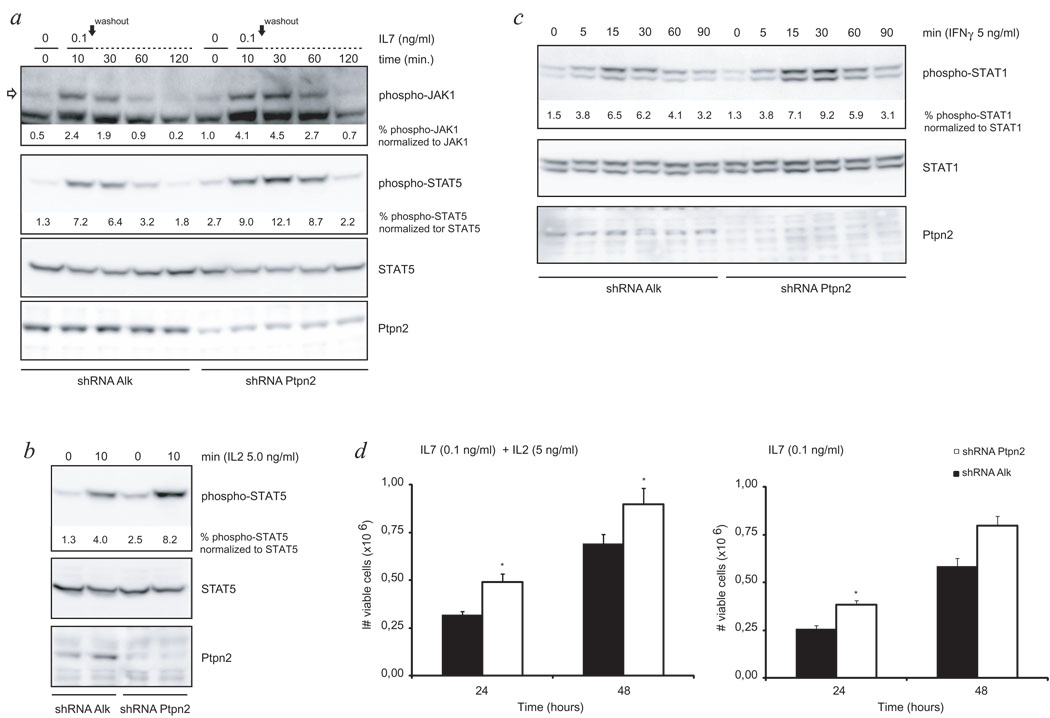
These data were further extended using ex vivo cultures of mouse primary T-cell leukemia cells, isolated from a spontaneous murine T-cell leukemia (CD4+CD8+), and expanded ex vivo in the presence of IL2 and IL7. Analysis of the response of these cells to IL2 and IL7 by monitoring phospho-JAK1 and phospho-STAT5 levels, revealed that knockdown of Ptpn2 caused an increased JAK/STAT signaling downstream of the IL2 and IL7 receptors (Fig. 3a, b). In addition, also for IFNγ a similar effect as observed in JURKAT cells, was observed in the mouse T-cell leukemia cells (Fig. 3c). To test if the augmentation in cytokine responsiveness also provided a proliferative advantage to these murine T-cells, we deprived cells of cytokines for 24h and followed their proliferation after re-stimulation with either low concentration of IL7 alone or a combination of IL2 and IL7. Knockdown of Ptpn2 caused a significant increase in the proliferation compared to control cells (Fig. 3d). Taken together, the data obtained in human and mouse T-cell leukemias indicate that loss of PTPN2 sensitizes T-ALL cells to cytokine stimulation resulting in enhanced activation of cytokine receptor pathways, which may support T-ALL development and proliferation under limiting cytokine concentrations in vivo.
Figure 3. Mouse T-ALL cells display altered cytokine receptor signaling in correlation with Ptpn2 expression levels.
- Knockdown of Ptpn2 enhanced responsiveness of the IL7 receptor pathway to ligand stimulation as displayed by increased strength and duration of the phosphorylation status of JAK1 and downstream protein STAT5 when compared to control cells. Open arrow points at the protein band corresponding to JAK1. Black arrow indicates time point of cytokine removal. STAT5 was used as loading control.
- Western blot analysis of WCL revealed increased sensitivity of primary mouse T-ALL cells to IL2 stimulation due to decreased Ptpn2 expression as analyzed by phosphorylation level of STAT5. STAT5 is shown as loading control.
- Cytokine depleted cells were exposed to INF-γ for indicated time periods and subsequently analyzed for activation of STAT1 by western blot. Quantification of western blot experiments showed that reduction of Ptpn2 protein resulted in stronger activation of STAT1. STAT1 was assayed to ensure equal protein loading.
- Primary T-ALL cells were deprived from cytokines for 24 hours before re-stimulation with either IL7 alone or a combination of IL2 and IL7. Under both conditions, knockdown of Ptpn2 provided cells with a significant proliferative advantage in response to cytokine re-addition. Open bars: shRNA Ptpn2; closed bars: shRNA Alk (control); bars show average ± s.e.m. Error bars represent s.e.m. *p<0.05
Efficient knockdown of Ptpn2 was confirmed using anti-Ptpn2 (3E2) antibody. Differences in phosphorylation signal intensity were quantified for JAK1 and STAT proteins and corresponding values are shown below blots.
In addition to a general effect on cytokine receptor signaling, our genetic data indicated a possible link between PTPN2 deletion and expression of the NUP214-ABL1 tyrosine kinase. Out of nine NUP214-ABL1 positive T-ALLs analyzed in the described study, we have identified four NUP214-ABL1 positive individuals with a concomitant deletion of PTPN2. Two individuals featured bi-allelic (individuals 2 and 5) and two individuals (individuals 9 and 20) mono-allelic loss of PTPN2 (Table 1). The presence of NUP214-ABL1 was initially detected by array CGH for all four individuals and later confirmed by FISH analysis for individuals 5, 9 and 20 (data not shown). In case of individual 5, an extrachromosomal (episomal) amplification of ABL1 was detected in more than 80% of the bone marrow cells using the LSI BCR-ABL1 translocation probe. For individual 20, both FISH and array CGH data confirmed a local duplication of NUP214-ABL1. In conclusion, both deletion of PTPN2 and NUP214-ABL1 were present in the majority of cells and thus also occurred together within the same cells.
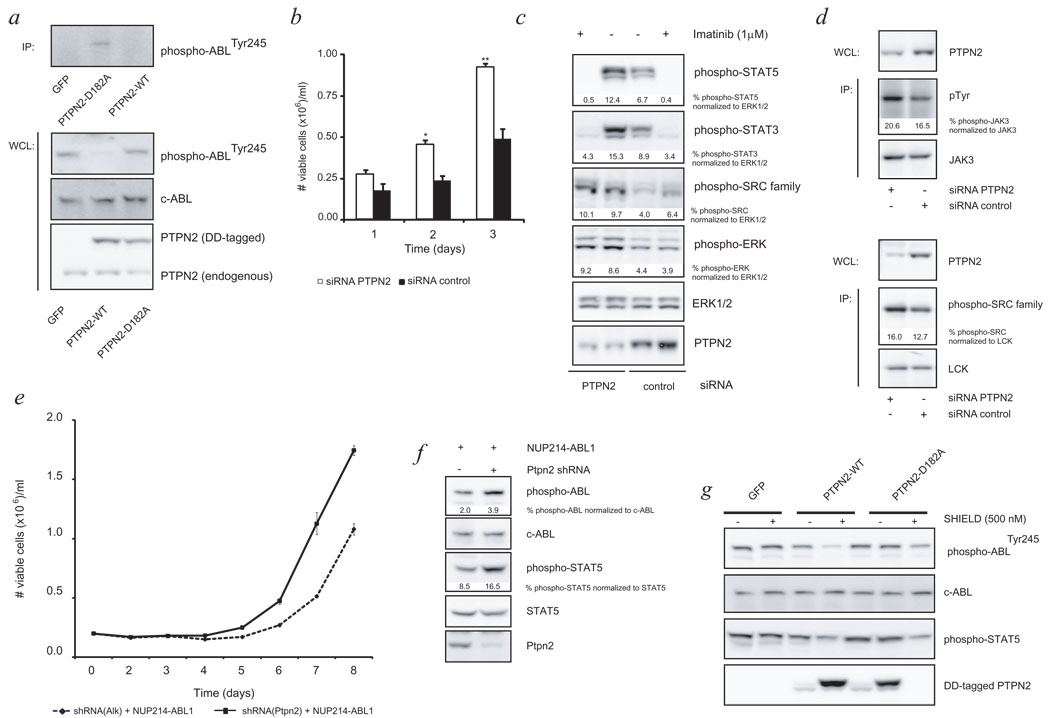
To determine if PTPN2 directly modulates NUP214-ABL1 signaling, we applied a retroviral system that allowed us to induce stable overexpression of either the wild type PTPN2 or a catalytic inactive mutant (PTPN2-D182A) in ALL-SIL cells.19 ALL-SIL is a cell line derived from a young adult T-ALL case expressing the NUP214-ABL1 fusion that is sensitive to the ABL1 kinase inhibitor imatinib.7 Overexpression of wild type PTPN2 in these cells resulted in a clear dephosphorylation of the NUP214-ABL1 protein, while this was not observed in cells expressing the catalytic inactive mutant (Fig. 4a). The PTPN2-D182A mutant is also known to form a complex with physiological substrates that is stable enough to allow purification upon interaction.20,21 Immune precipitation of the PTPN2-D182A from ALL-SIL cells resulted in co-precipitation of the NUP214-ABL1 protein, demonstrating a direct interaction between the tyrosine kinase and the phosphatase (Fig. 4a). Finally, downregulation of PTPN2 in ALL-SIL cells resulted in a significant increase in proliferation of the cells under normal culture conditions (Fig. 4b), which was in part attributed to increased signaling of NUP214-ABL1 through downstream proteins STAT5 and STAT3 (Fig. 4c). In addition, other signaling proteins such as LCK, JAK3 and ERK1/2 were also found to be constitutively active in ALL-SIL cells and more activated upon PTPN2 knockdown, therefore likely to contribute to the observed growth advantage (Fig. 4c, d).
Figure 4. Identification of NUP214-ABL1 oncogene as a novel substrate of PTPN2.
- Desphosphorylation of NUP214-ABL1 was observed upon induced overexpression of wild type PTPN2 (PTPN2-WT). Immune precipitation (IP) of PTPN2-D182A resulted in co-precipitation of NUP214-ABL1 protein, showing a direct interaction between kinase and phosphatase.
- Viable cell numbers were determined 3 days after electroporation with respective siRNA’s. Decrease in PTPN2 protein levels resulted in a significant proliferative advantage of ALL-SIL cells.
- Cells were exposed to imatinib at time point of maximal PTPN2 knockdown. Western blot analysis showed differential activation status of STAT family members downstream of NUP214-ABL1. ERK1/2 and a SRC-family kinase were detected to be constitutively active in ALL-SIL cells and knockdown of PTPN2 caused an augmented phosphorylation which stayed unchanged upon imatinib treatment. Loading control: ERK1/2
- IP experiments of ALL-SIL cells after electroporation with siRNA’s revealed an enhancing effect on JAK3 kinase activation (upper) and identified the differentially activated SRC-family kinase as LCK. Phosphorylation of JAK3 and LCK were detected with a phosphotyrosine (pTyr) or phospho-SRC family antibody, respectively. JAK3 or LCK were used to ensure equal protein loading.
- Ba/F3 cells carrying indicated constructs were grown in absence of IL-3 and mean growth ± s.e.m. was recorded in triplicate for 8 days. No s.e.m. is shown in case of values below 0.03.
- Western blot analysis of NUP214-ABL1 dependent Ba/F3 cells demonstrated stronger constitutive activation of NUP214-ABL1 and STAT5 in case of concomitant shRNA-mediated knockdown of Ptpn2. Loading control: c-ABL, STAT5
- Induced overexpression of PTPN2-WT in Ba/F3 cells carrying the NUP214-ABL fusion results in dephosphorylation of the oncogenic kinase and its downstream mediator STAT5. C-ABL is shown as loading control.
Values shown below immunoblots represent normalized (for loading control) quantitative values. Open bars: siRNA PTPN2; closed bars: siRNA control; bars show average ± s.e.m. Error bars represent s.e.m. *p<0.05, **p<0.005.
A similar relationship between NUP214-ABL1 and Ptpn2 was demonstrated in the IL3 dependent pro-B cell line Ba/F3. We have previously shown that NUP214-ABL1 can transform Ba/F3 to IL3 independent proliferation, but its oncogenic activity is less potent than the BCR-ABL1 kinase.22 shRNA-mediated downregulation of Ptpn2 expression made Ba/F3 cells more sensitive to NUP214-ABL1 transformation (Fig. 4e). NUP214-ABL1 showed stronger autophosphorylation when expressed in Ba/F3 cells with decreased Ptpn2 expression, and vice versa, induced overexpression of wild type PTPN2 decreased NUP214-ABL1 phosphorylation and downstream signaling (Fig. 4f, g).
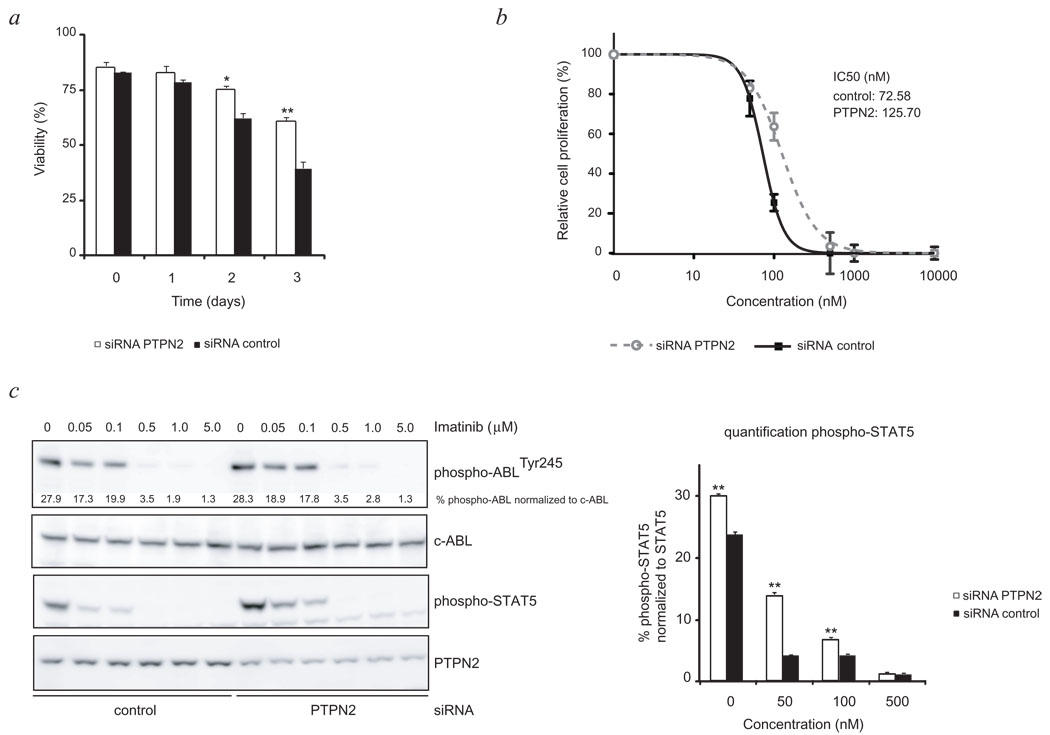
Next, we tested the effect of PTPN2 knockdown on the survival and proliferation of ALL-SIL cells upon treatment with the ABL1 kinase inhibitor imatinib. Downregulation of PTPN2 expression resulted in prolonged survival of ALL-SIL cells after imatinib treatment as compared to cells electroporated with a non-targeting siRNA (Fig. 5a). The proliferation of the cells was inhibited by imatinib, but with a shift of the cellular IC50 value from 73 to 126 nM (Fig. 5b). These data indicate that loss of PTPN2 affects response to treatment, which is likely to be caused by the direct effect on NUP214-ABL1 activation and its downstream signaling, as observed by an increased activation state of its important downstream mediator STAT5 (Fig. 5c). This is further supported by the enhanced phosphorylation of additional protein kinases such as LCK and ERK1/2 which persisted upon inhibitor treatment (Fig. 4c). A similar effect was observed in HSB-2, a cell line derived from a childhood T-ALL that is dependent on LCK for its proliferation and survival.23 Our results indicate a direct modulating effect of PTPN2 on LCK activation (Supplementary Fig. 8 online).
Figure 5. Reduction of PTPN2 expression levels affects treatment response of NUP214-ABL1 dependent cells to kinase inhibitor imatinib.
- Follow up of survival of ALL-SIL cells treated with imatinib (500 nM) demonstrated decreased sensitivity due to alteration of PTPN2 expression levels. Y-axis displays cell viability (%).Open bars: siRNA PTPN2; closed bars: control siRNA
- siRNA mediated knockdown of PTPN2 in ALL-SIL cells reduced activity of imatinib as depicted by an increased IC50 value (126 nM) compared to control cells (73 nM). Untreated cells were set to 100%. Graph displays relative cell growth (%) as a function of concentrations (nM, logarithmic scale). Open circles: siRNA PTPN2, closed rectangles: siRNA control.
- Western blot analysis of ALL-SIL cells exposed to increasing concentrations of imatinib revealed a stronger activation of NUP214-ABL downstream target STAT5 in knockdown cells (left panel). Bar graph displays calculated differences in STAT5 activation level due to alterations in PTPN2 expression levels (right panel). Loading control: c-ABL; open bars: siRNA PTPN2; closed bars: control siRNA
All values represent the average ± s.e.m. of three determinations. Significant differences compared to those of control siRNA are indicated. *p<0.05 and **p<0.005
Recently, mutational analysis of JAK1 resulted in the identification of activating point mutations in 10% of T-ALL cases,24 directly implicating deregulated JAK1 signaling in the pathogenesis of leukemia. In this study we identify inactivation of PTPN2 in T-ALL cells as another mechanism that results in increased JAK/STAT signaling downstream of cytokine receptors. PTPN2 is an important negative regulator of a variety of tyrosine kinases as well as other signaling proteins, which indicates that its loss in T-cells may affect a diverse set of signaling proteins thereby sensitizing T-cells to growth factors and increasing the effects of oncogenic kinases such as NUP214-ABL1. Our data identify the NUP214-ABL1 oncogene as a novel substrate of PTPN2 and reveal a correlation between activation status of the NUP214-ABL1 pathway, treatment response to kinase inhibitor imatinib and PTPN2 expression levels. This study adds further complexity to the genetics of T-ALL and shows that in addition to tyrosine kinase activation also inactivation of tyrosine phosphatases may provide a proliferation and survival advantage to these leukemias. Mutation and expression analysis of PTPN2 in various tumors is warranted.
Supplementary Material
Acknowledgments
We thank Stein Aerts for assistance with data analysis. This work was supported by grants from the K.U.Leuven (concerted action grant to J.C., I.W., P.V.), the FWO-Vlaanderen (G.0287.07, J.C.), the Foundation against Cancer (SCIE2006-34, J.C.), an ERC-starting grant (J.C.), a José-Carreras fellowship grant from EHA (J.C.), the Interuniversity Attraction Poles (IAP) granted by the Federal Office for Scientific, Technical and Cultural Affairs, Belgium (J.C., P.V.), a grant from the French program Carte d'Identite des Tumeurs (CIT, Ligue Contre le Cancer) and from Canceropole d'Ile de France (F.S., J.S.); a Blood Disease Research Project research grant from The New York Community Trust (A.F.), and the National Institutes of Health (grants CA120196 and CA129382 to A.F.). A.F. is a Leukemia and Lymphoma Society Scholar. K.D.K. is a postdoctoral researcher and P.V. is a senior clinical investigator of FWO-Vlaanderen.
Footnotes
Author contributions
All authors contributed to the text; M.K. designed and performed experiments and analyzed data; I.L., T.E.C., K.D.K., N.M., C.G., K.V.R., I.W. performed experiments and analyzed data; A.A.F., A.L., J.M., F.S., T.H., P.V. collected samples, performed experiments and analyzed data; J.S., J.C. supervised the project and analyzed data.
References
- 1.Doody KM, Bourdeau A, Tremblay ML. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol Rev. 2009;228:325–341. doi: 10.1111/j.1600-065X.2008.00743.x. [DOI] [PubMed] [Google Scholar]
- 2.van Vliet C, et al. Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat Immunol. 2005;6:253–260. doi: 10.1038/ni1169. [DOI] [PubMed] [Google Scholar]
- 3.Shields BJ, Hauser C, Bukczynska PE, Court NW, Tiganis T. DNA replication stalling attenuates tyrosine kinase signaling to suppress S phase progression. Cancer Cell. 2008;14:166–179. doi: 10.1016/j.ccr.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Consortium W. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 7.Graux C, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–1089. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 8.De Keersmaecker K, Marynen P, Cools J. Genetic insights in the pathogenesis of T-cell acute lymphoblastic leukemia. Haematologica. 2005;90:1116–1127. [PubMed] [Google Scholar]
- 9.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–359. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 10.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 11.Lahortiga I, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39:593–595. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 12.Dik WA, et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J Exp Med. 2005;201:1715–1723. doi: 10.1084/jem.20042524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soulier J, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106:274–286. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 14.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 15.ten Hoeve J, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simoncic PD, Lee-Loy A, Barber DL, Tremblay ML, McGlade CJ. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 17.Lu X, et al. T-cell protein tyrosine phosphatase, distinctively expressed in activated-B-cell-like diffuse large B-cell lymphomas, is the nuclear phosphatase of STAT6. Mol Cell Biol. 2007;27:2166–2179. doi: 10.1128/MCB.01234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard WJ. Cytokines and immunodeficiency diseases. Nat Rev Immunol. 2001;1:200–208. doi: 10.1038/35105066. [DOI] [PubMed] [Google Scholar]
- 19.Blanchetot C, Chagnon M, Dube N, Halle M, Tremblay ML. Substrate-trapping techniques in the identification of cellular PTP targets. Methods. 2005;35:44–53. doi: 10.1016/j.ymeth.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiganis T, Flint AJ, Adam SA, Tonks NK. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- 22.De Keersmaecker K, et al. Kinase activation and transformation by NUP214-ABL1 is dependent on the context of the nuclear pore. Mol Cell. 2008;31:134–142. doi: 10.1016/j.molcel.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Wright DD, Sefton BM, Kamps MP. Oncogenic activation of the Lck protein accompanies translocation of the LCK gene in the human HSB2 T-cell leukemia. Mol Cell Biol. 1994;14:2429–2437. doi: 10.1128/mcb.14.4.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flex E, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clappier E, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110:1251–1261. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 26.Bene MC, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9:1783–1786. [PubMed] [Google Scholar]
- 27.Jaffe EHN, Stein H, Vardiman J, editors. Lyon: IARC; World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues, 84-86. 2001
- 28.De Keersmaecker K, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32) Blood. 2005;105:4849–4852. doi: 10.1182/blood-2004-12-4897. [DOI] [PubMed] [Google Scholar]
- 29.De Keersmaecker K, et al. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93:533–542. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- 30.Lorenzen JA, Dadabay CY, Fischer EH. COOH-terminal sequence motifs target the T cell protein tyrosine phosphatase to the ER and nucleus. J Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosinger B, Jr, Tillmann U, Westphal H, Tremblay ML. Cloning and characterization of a mouse cDNA encoding a cytoplasmic protein-tyrosine-phosphatase. Proc Natl Acad Sci U S A. 1992;89:499–503. doi: 10.1073/pnas.89.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.