Abstract
Biomarker discovery approaches in urine have been hindered by concerns for reproducibility and inadequate standardization of proteomics protocols. In this study, we describe an optimized quantitative proteomics strategy for urine biomarker discovery, which is applicable to fresh or long frozen samples. We used urine from healthy controls to standardize iTRAQ (isobaric tags for relative and absolute quantitation) for variation induced by protease inhibitors, starting protein and iTRAQ label quantities, protein extraction methods, and depletion of albumin and immunoglobulin G (IgG). We observed the following: (a) Absence of protease inhibitors did not affect the number or identity of the high confidence proteins. (b) Use of less than 20 μg of protein per sample led to a significant drop in the number of identified proteins. (c) Use of as little as a quarter unit of an iTRAQ label did not affect the number or identity of the identified proteins. (d) Protein extraction by methanol precipitation led to the highest protein yields and the most reproducible spectra. (e) Depletion of albumin and IgG did not increase the number of identified proteins or deepen the proteome coverage. Applying this optimized protocol to four pairs of long frozen urine samples from diabetic Pima Indians with or without nephropathy, we observed patterns suggesting segregation of cases and controls by iTRAQ spectra. We also identified several previously reported candidate biomarkers that showed trends toward differential expression, albeit not reaching statistical significance in this small sample set.
With ongoing advances in mass spectrometry (MS) and proteomics technology, proteomics analysis is progressively occupying a central position in biomarker discovery platforms. Biofluids such as urine and blood are the preferred media for proteomics analysis because of their ease of collection and extensive history of use in clinical laboratory practice. Urine, in particular, is an information-rich fluid that can be collected non-invasively and in large quantities. Many urine proteins are produced or shed in the kidney and urogenital tract (1), making urine a promising proximal source of biomarkers for diseases affecting these structures.
However, proteomics-based biomarker discovery in urine faces multiple challenges. Urine proteomics is complicated by low urine protein concentration, variations in pH, and high concentrations of salts and urea or other urine components that interfere with sample processing. The urine proteome can also change with individual variables such as hydration, diurnal change, diet, and physical activity as well as variation in sample collection, processing, and storage. In addition, urine proteomics shares the usual challenges of biomarker discovery in other biofluids such as throughput, cost, and the need for a reproducible and quantitative work flow.
Isotopic or isobaric labeling methods to reduce variation, increase throughput, and enable quantitative analysis have been developed to address some of these challenges. One such method, isobaric tags for relative and absolute quantitation (iTRAQ)1 (2), combines relative and absolute peptide quantification with multiplexing ability to enable an increased throughput as well as simultaneous comparison of up to eight samples within one experimental run. Variations induced by urine sample processing have been systematically evaluated for proteomics analyses using two-dimensional gel electrophoresis (3–6), differential gel electrophoresis (7), and liquid chromatography-coupled mass spectrometry (LC-MS) (5, 8, 9). However, no systematic analyses of urine sample collection and processing have been reported for iTRAQ.
Before utilizing iTRAQ-based quantitative proteomics for urine biomarker discovery, we evaluated the impact of variation in several processing steps (addition of protease inhibitors, the starting protein quantities, quantity of the iTRAQ label, protein extraction methods, and depletion of abundant proteins) on iTRAQ protein identification and quantitation. Applying this optimized biomarker discovery protocol to small quantities of long frozen urine samples from the Pima longitudinal study of diabetic nephropathy, we observed patterns suggestive of segregation of cases and controls by iTRAQ spectra. We also observed trends toward differential expression in several proteins that had been identified as putative biomarkers in previous studies. However, given the small sample size, none of these proteins retained statistical significance after multiple testing correction.
EXPERIMENTAL PROCEDURES
Normal and Diabetic Urine Collection and Storage
The experiments were preformed using urine samples from healthy volunteers or Pima diabetic subjects with or without diabetic nephropathy. The urine specimens from two Caucasian male healthy volunteers were second voids of the day, collected midstream in sterile 15- or 50-ml conical centrifuge tubes. These healthy volunteers were 25 and 27 years old, had no medical history, and did not take any prescription medicines. When indicated, a complete protease inhibitor mixture was added to the samples immediately upon collection. This protease inhibitor mixture contained 104 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 80 μm aprotinin, 4 mm bestatin, 1.4 mm E-64, 2 mm leupeptin, and 1.5 mm pepstatin A (product P8340, Sigma-Aldrich). The collected urine samples were immediately centrifuged at 5000 rpm for 5 min at room temperature to remove particulate matter and cell debris. The pellets were discarded, and supernatants were divided into 1-ml aliquots and stored in 1.5-ml Eppendorf tubes at −80 °C until further processing. The storage time at −80 °C ranged from 12 h to 2 weeks. The urine samples used in any given experiment were from the same batch, i.e. collected at the same time and stored at −80 °C for the same duration of time.
To process the samples, aliquots were thawed and underwent protein extraction and iTRAQ processing over the subsequent 48 h. For samples that were depleted of albumin/IgG, the processing time was 72 h. One individual (M. A.) processed all the samples up to iTRAQ labeling. Subsequent fractionation and mass spectrometry steps were performed by two people (S. T. D. and M. C. G.) jointly. This study was approved by the Institutional Review Board overseeing human subject research at Massachusetts General Hospital. Written informed consents were obtained from participants.
Urine samples from four type 2 diabetic patients with and four without nephropathy were obtained from the National Institute of Diabetes and Digestive and Kidney Diseases longitudinal study of diabetes in Pima Indians described in detail elsewhere (8, 9). Briefly, Pima and the closely related Tohono O'odham (Papago) Indians of the Gila River Indian Community (Phoenix, AZ) participated in a comprehensive longitudinal diabetes study from 1965 through 2007. The study consisted of a biennial assessment of diabetes with an oral glucose tolerance test and an evaluation of diabetic complications. An untimed urine collection was obtained at each examination, and measurement of urinary albumin excretion was performed at all examinations conducted on or after July 1, 1982. No protease inhibitors were added at collection, and these samples have been stored at −80 °C since collection. This study was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases, and written informed consent was obtained from all participants. For our studies, the cases were type 2 diabetic Pima Indians who developed diabetic nephropathy, which was defined by macroalbuminuria (urine albumin ≥20 mg/dl) and/or abnormal serum creatinine concentration (≥1.2 mg/dl). Control subjects were type 2 diabetic Pima Indians who were normoalbuminuric (urine albumin <2 mg/dl) and had a normal serum creatinine concentration (<1.2 mg/dl). Cases and controls were matched by sex. Although adequate matching by age was not possible in this small sample size (see Table II), given the pilot nature of this study, we accepted matching only by sex. Samples were shipped on dry ice and, upon arrival, were immediately stored at −80 °C until processing. The total −80 °C storage times for these samples are shown in Table II. As with samples from the healthy volunteers, processing of these urine samples included protein extraction and iTRAQ labeling over 48 h. Sample processing up to iTRAQ labeling was done by one person (M. A.); the subsequent fractionation and mass spectrometry was performed by two people (S. T. D. and M. C. G.) jointly. Use of Pima samples for this study was approved by the Institutional Review Board overseeing human subject research at Massachusetts General Hospital.
Table II. Demographic and clinical characteristics of Pima cases and controls.
Status, case/control (Ctrl) status; freezing time, sample storage time in −80 °C since collection; SBP/DBP, systolic/diastolic blood pressure at sample collection; Cr, serum creatinine at sample collection; HBA1C, hemoglobin A1C at sample collection; ACR, albumin-to-creatinine ratio in urine at sample collection; F, female; M, male.
| Status | Diabetes duration | Freezing Time | Collection date | At time of urine collection |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age | Sex | SBP | DBP | Cr | HBA1C | ACR | ||||
| yr | yr | yr | mg/dl | mg/dl | ||||||
| Case 1 | 17.3 | 17 | 11/14/1991 | 66.8 | F | 186 | 72 | 2.0 | 10.9 | 5658.7 |
| Case 2 | 24.8 | 14 | 11/17/1994 | 61.8 | F | 164 | 90 | 0.8 | 11.4 | 3173.4 |
| Case 3 | 14.8 | 11 | 12/4/1997 | 59.5 | F | 118 | 60 | 0.9 | 6.1 | 9350.0 |
| Case 4 | 12.8 | 13 | 7/25/1995 | 43.2 | M | 130 | 90 | 0.9 | 11.6 | 527.7 |
| Ctrl 1 | 10.3 | 14 | 1/25/1994 | 58.5 | F | 106 | 68 | 0.8 | 8.1 | 15.9 |
| Ctrl 2 | 15.0 | 13 | 8/2/1995 | 50.5 | F | 110 | 70 | 0.7 | 10.4 | 21.9 |
| Ctrl 3 | 15.1 | 9 | 11/18/1999 | 57.8 | F | 118 | 64 | 0.7 | 8.5 | 26.8 |
| Ctrl 4 | 13.5 | 13 | 7/18/1995 | 44.0 | M | 122 | 78 | 0.8 | 8.7 | 9.2 |
Urine Protein Extraction
The following protein extraction methods were tested on 0.3–0.4 ml of urine from healthy volunteers described above. Each precipitation method was repeated 10–12 times on urine samples from each individual, and the final experiment was conducted in duplicate or triplicate to assess reproducibility.
Organic Solvent Precipitation
Urine samples were precipitated with 100% stock solutions of an organic solvent (methanol, ethanol, acetonitrile, or trichloroacetic acid) at a ratio of 1:9 (v/v), incubated at −20 °C for 12–16 h, and centrifuged at 14,000 × g for 30 min at 4 °C. The pellet was washed once with the solvent used for initial precipitation, air-dried, and resuspended in ultrapure water or iTRAQ dissolution buffer. Protein concentration was determined using the Bradford assay with a standard curve derived from bovine serum albumin (BSA) (10). Of note, the assessment of protein concentration by the Bradford assay was not affected by background interferences in crude urine as was shown by a comparison of BSA standards diluted in urine or ultrapure water (supplemental Fig. 1).
Ultrafiltration
Amicon centrifugal devices with a 3-kDa molecular mass cutoff (Millipore, Billerica, MA) were used for ultrafiltration following the manufacturer's instructions. Each urine sample was added to the reservoir of the device, centrifuged at 12,000 × g for 60 min at 4 °C, washed twice with ultrapure water, and recentrifuged (12,000 × g for 30–60 min at 4 °C). The ultrafiltrate was recovered by a reverse spin (1000 × g for 3 min at 4 °C) and concentrated by vacuum centrifugation. The resulting pellet was resuspended in iTRAQ dissolution buffer, and protein concentrations were determined using the Bradford assay as described above.
The ultrafiltration method had a yield of only 25–30% (i.e. the percentage of the total urine protein that was recovered) (Table I). We tested a manufacturer-recommended pretreatment step to determine whether it could increase this low protein recovery. Following the manufacturer's recommendations, centrifugal devices were pretreated with Tween 20 or Triton X-100 to determine whether pretreatment would increase the yield of protein recovery by blocking nonspecific protein adsorption to the membrane and plastic components. The yield for protein recovery was not changed by pretreatment at least as measured by the Bradford assay. In the absence of appreciable change in total protein recovery in pretreated samples, we chose to eliminate pretreatment to keep sample processing as simple as possible. Therefore, the samples used for spectrometry in this study were processed without this pretreatment step.
Table I. Yield of protein extraction methods.
The percent yield was calculated as the percentage of total urine protein that was recovered by the extraction method. Protein concentration was measured in the urine sample and the resulting protein extract by the Bradford assay. Each protein extraction method was tested 10–12 times with similar results. The ranges shown are representative of the observed values.
| Protein extraction method | Yield |
|---|---|
| % | |
| Organic solvent precipitation | 60–70 |
| Methanol | 60–70 |
| Ethanol | 30–40 |
| Acetonitrile | <30 |
| TCA | |
| Ultrafiltration | 25–30 |
| Dialysis, lyophilization | 70–80 |
Dialysis and Lyophilization
Using the Slide-A-Lyzer dialysis cassettes with a 3.5-kDa molecular mass cutoff (Thermo Scientific, Waltham, MA), 0.3 ml of urine was dialyzed against >500× volume of dialysis solution (20 mm NH4HCO3, pH 7.8) following the manufacturer's instructions. The dialysate was snap frozen using liquid nitrogen and then lyophilized. The resulting powder was resuspended in iTRAQ dissolution buffer, and protein concentrations were determined using the Bradford assay as described above.
Depletion of Abundant Proteins
Albumin and IgG were depleted from urine using Sigma ProteoPrep® immunoaffinity columns (Sigma-Aldrich) following the manufacturer's instructions with the exception that the Tris-based equilibration buffer was replaced by phosphate-buffered saline (PBS) because Tris would interfere with iTRAQ labeling.
iTRAQ Labeling and Subsequent Fractionation
iTRAQ Labeling
The samples compared in each figure were run in one iTRAQ 8-plex, and normalization was done by keeping the protein quantity constant in all the samples run in one 8-plex. For example, for the experiment in Fig. 3, two methanol-extracted, three ethanol-extracted, and three dialyzed samples were labeled in one 8-plex at 30 μg per sample (please see supplemental Fig. 2 for details of replicates and sample pooling for Figs. 1–5). Depending on the experiment, 10–50 μg of protein was labeled following the iTRAQ manufacturer's instructions (Applied Biosystems, Foster City, CA). Briefly, the extracted protein was resuspended in dissolution buffer (0.5 m triethylammonium bicarbonate at pH 8.5) at a concentration of 1–2.5 μg/μl, depending on the protein amount, and denatured with 0.1% (v/v) SDS. The protein samples were then reduced by addition of 4 mm tris(2-carboxyethyl)phosphine for 1 h at 56 °C. Disulfide bonds were blocked by incubation with a final concentration of 8 mm methyl methanethiosulfonate at room temperature for 10 min. The sample was then digested by addition of trypsin (Promega; 1 μg/μl; exclusive cleavage C-terminal to arginine and lysine) at a ratio of 1 μg of trypsin/10 μg of protein followed by a 12–16-h incubation at 37 °C. Trypsin digests from up to eight samples were labeled with the 8-plex iTRAQ isobaric tags (113, 114, 115, 116, 117, 118, 119, and 121) according to the manufacturer's protocol. Before pooling, success of labeling was confirmed by evaluating five of the highest intensity peaks on a mass spectrometer. The eight samples labeled with iTRAQ tags 113, 114, 115, 116, 117, 118, 119, and 121 were then pooled and composed one experimental run (8-plex). The details of technical replicates and the pooling scheme for Figs. 1–5 are detailed in supplemental Fig. 2.
Fig. 3.
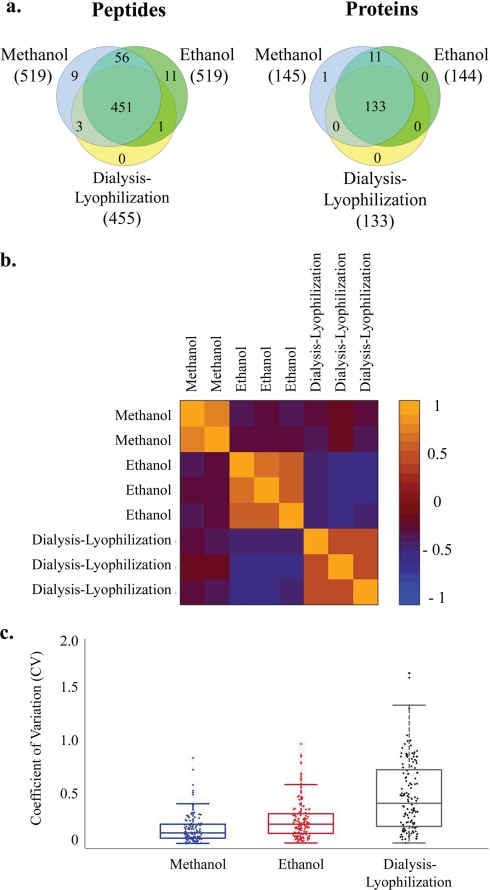
Protein extraction methods. a, number and overlap in identity of peptides and proteins identified in urine extracted with methanol versus ethanol versus dialysis-lyophilization. The numbers in parentheses represent the number of identified proteins (or peptides) for each sample type. The peptide Venn diagram displays the number of unique peptides with ≥20% confidence that are used in identifying and quantitating the proteins. b, correlation in iTRAQ intensities between samples extracted with methanol, ethanol, or dialysis-lyophilization. The values represent the Pearson correlation for normalized iTRAQ values ((iTRAQ value − mean)/S.D.) of all proteins between two samples. c, Box-and-whisker plots depicting the observed range for the coefficients of variation (CV) for iTRAQ ratios between replicate samples extracted with methanol, ethanol, or dialysis-lyophilization. Boxes reflect 25%–75% range of CV's observed in three replicate experiments; whiskers reflect the 2%–98% range of CV's observed in three replicate experiments.
Fig. 1.
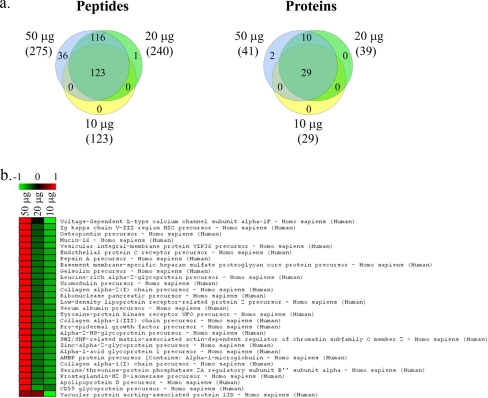
Titration of starting protein quantity. a, Venn diagrams represent numbers and overlap in identities of peptides or proteins identified in samples with starting protein quantities of 50, 20, and 10 μg. The numbers in parentheses represent the number of identified proteins (and peptides) for each sample type. The peptide Venn diagram displays the number of unique peptides with ≥20% confidence that are used in identifying and quantitating the proteins. b, colorgram depicting iTRAQ ratios for proteins identified in samples with starting protein quantities of 50, 20, and 10 μg. The columns represent individual samples; rows represent identified proteins. iTRAQ ratios are shown on a pseudocolor scale with red denoting high ratios and green denoting low ratios.
Fig. 2.
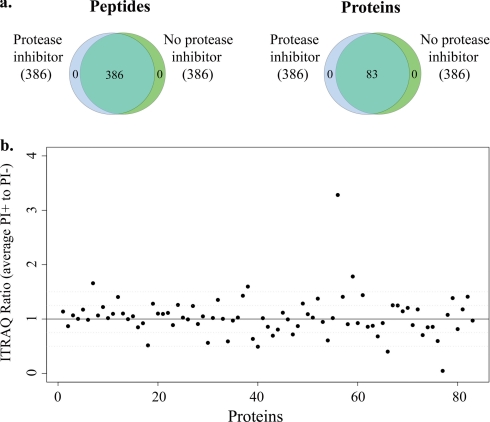
Protease inhibitors. a, number and identity of peptides and proteins identified in urine samples collected with or without addition of protease inhibitors. The numbers in parentheses represent the number of identified proteins (and peptides) for each sample type. The peptide Venn diagram displays the number of unique peptides with ≥20% confidence that are used in identifying and quantitating the proteins. b, ratio of iTRAQ intensity for each of the 83 identified proteins in PI+ samples (with protease inhibitors) to PI− samples (without protease inhibitors).
Fig. 4.
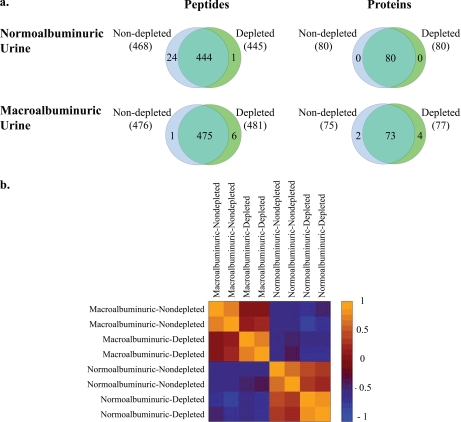
Depletion of albumin and IgG. a, number and identity of peptides and proteins identified in normoalbuminuric or macroalbuminuric urine samples either depleted or non-depleted of albumin and IgG. The numbers in parentheses represent the number of identified peptides or proteins for each sample type. The peptide Venn diagrams display the number of unique peptides with ≥20% confidence that are used in identifying and quantitating the proteins. b, correlation in iTRAQ intensities between normal and proteinuric samples either depleted or left undepleted of albumin/IgG. The values represent the Pearson correlation for normalized iTRAQ values ((iTRAQ value − mean)/S.D.) of all proteins between two samples.
Fig. 5.
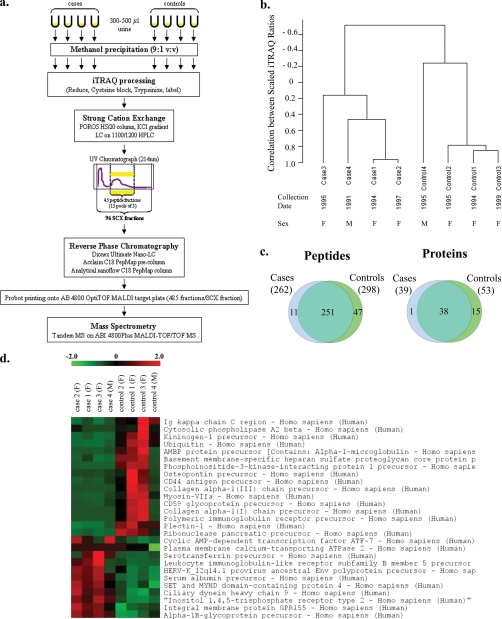
Pima cases and controls. a, proteomics work flow for urine sample processing. b, unsupervised hierarchical clustering analysis of the scaled iTRAQ values of the 54 identified proteins segregates cases and controls into two separate groups. The urine collection dates and subject genders are listed under sample names. c, number and overlap in identity of peptides and consensus proteins identified in Pima cases and controls. The consensus proteins are those that occur in 75% (three of four) of cases (or controls). The numbers in parentheses represent the number of identified peptides or proteins for each sample type. The peptide Venn diagram displays the number of unique peptides with ≥20% confidence that are used in identifying and quantitating the proteins. d, colorgram depicting the iTRAQ intensities of the 27 proteins whose levels were more than 1.5-fold different (lower or higher) in cases compared with controls and had statistically significant raw p values for differential expression. The columns represent individual samples with case/control status and gender (F, female; M, male) denoted above the column. The rows represent the identified proteins. iTRAQ ratios are shown on a pseudocolor scale (−2 to 2) with red denoting high ratios and green denoting low ratios. SCX, strong cation exchange.
Two-dimensional Liquid Chromatography
To maximize the number of identified peptides, we used two-dimensional peptide fractionation before mass spectrometry. Pooled samples were concentrated by vacuum centrifugation to dry off organic solvents. 80 μg of protein was added to 1 ml of buffer A (10 mm KH2PO4, 25% acetonitrile, pH 2.8) for strong cation exchange chromatography. Peptides were initially separated over a 4.6 × 100-mm POROS HS/20 column (Applied Biosystems) using a KCl gradient. Liquid chromatography was performed on an 1100/1200 HPLC system (Agilent Technologies, Santa Clara, CA) using a two-step gradient at a flow rate of 0.5 ml/min over 50 min with collection of 96 fractions. Based on the chromatogram from the UV absorbance at 214 nm, 45 fractions spanning the entire eluted peptide peak were selected and pooled into 15 groups of three consecutive fractions each (supplemental Fig. 3). Each pooled fraction was dried by vacuum centrifugation and resuspended in 100 μl of reverse phase buffer A (2% acetonitrile, 0.1% trifluoroacetic acid). Pooled fractions underwent reverse phase chromatography by injection onto a Dionex UltiMate NanoLC system equipped with an Acclaim C18 PepMap 100 μ-Precolumn (300 μm × 5 mm, 5-μm beads, 100-Å pores) followed by an analytical nanoflow C18 PepMap 100 column (75 μm × 15 cm, 3-μm beads, 100-Å pores). Peptides were eluted with a 5–50% gradient of acetonitrile over 60 min. All fractions containing peptides, based on UV absorbance at 214 nm, were directly spotted onto ABI 4800 OptiTOF MALDI (matrix-assisted laser desorption ionization) target plates using a Probot printing robot (Dionex, Sunnyvale, CA). α-Cyano-4-hydroxycinnamic acid ionization matrix (Sigma-Aldrich) was mixed with the sample at a 1:2 ratio using an in-line mixing tee in the Probot. A total of 485 fractions per strong cation exchange fraction were collected.
Mass Spectrometry and Identification of Differentially Expressed Proteins
All fractions were analyzed on the ABI 4800Plus MALDI-TOF/TOF (time-of-flight/time-of-flight) MS system by tandem mass spectrometry. The 15 most abundant precursors of each spot were fragmented by MS-MS with collision-induced dissociation using medium gas pressure with ambient air. Mass tolerance for the precursor ions was set at 200 ppm; all fragment ions were considered regardless of mass. A minimum signal to noise ratio of 50 was applied as a filter for accepting individual spectra. This permissive threshold was selected to maximize sensitivity and consider a larger number of spectra for protein identification.
Statistical Analysis
Relative Abundance Quantitation and Peptide and Protein Identification
Relative abundance quantitation and peptide and protein identification were performed using ProteinPilot software 3.0 (Applied Biosystems, Software Revision 50861). The ProteinPilot v3.0 uses the Paragon algorithm (11) to perform database matching for protein identification, protein grouping to remove abundant hits, and comparative quantitation. A thorough identification search was conducted in ProteinPilot, i.e. a search that considers any selected identification, including those of much lower probability. All cleavage variants are searched, including those that do not conform to the selected digest agent. The Paragon algorithm searches a set of more than 80 biological modifications, including those that result from the specified sample preparation (e.g. iTRAQ labeling) and Cys alkylation. These modifications follow the Human Proteome Organisation's proteomics standards initiative modification nomenclature for mass spectrometry standard.
The Swiss-Prot Homo sapiens protein database (Swiss-Prot/UniProt version 55, released February 26, 2008) was used for all searches. The total number of protein entries searched in the database was 18,053. The data were normalized for loading error by bias correction and background correction using ProteinPilot. Bias correction is an algorithm in Protein Pilot that corrects for unequal mixing when combining the eight labeled samples of one experiment. It does so by comparing the total label amount (reflecting total protein amount) in each sample and assigning an autobias factor to it. Based on the assumption that the total protein is the same between samples, applying this factor can increase accuracy of quantification by normalizing unevenness in starting protein quantities.
The confidence value for each peptide was calculated based on agreement between the experimental and theoretical fragmentation patterns. Each protein was provided with a confidence score based on confidence scores of its constituent peptides with unique spectral patterns. The Protein Pilot software (11) calculates the iTRAQ ratio for each protein using the iTRAQ values of the unique peptides for that protein. The weighted average of the log ratio (i.e. the iTRAQ ratio) for each protein was calculated using Equation 1,
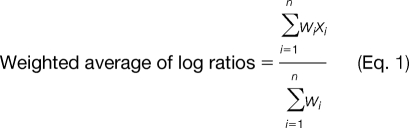 |
where xi is the peptide iTRAQ ratio, wi is 1/percent error, and n is the number of peptides. The final average ratio for each protein was calculated after bias correction according to the Pro Group algorithm (11). The high confidence proteins are defined as those with (a) >90% confidence as determined by ProteinPilot (ProtScore ≥ 1.0) and (b) two distinct peptides with different iTRAQ spectra with at least one peptide identified with ≥90% confidence. These high confidence proteins were identified using Perl and R scripts developed at the Beth Israel Deaconess Medical Center Genomics Center.
Hierarchical Clustering Analysis (HCA)
HCA was conducted with the uncentered correlation metric (for similarity calculation) and complete linkage using the Hclust package in R. The iTRAQ value for each protein was calculated relative to that of Control 4 (the reference); this iTRAQ value was then scaled to the mean iTRAQ value for that protein in the whole 8-plex (i.e. four cases and four controls). The scaling was done using the following formula: Scaled value = (x − μ)/σ where x is the iTRAQ value of a protein in a given sample and μ and σ are the mean and S.D. for the iTRAQ value of that protein in all cases and controls.
Identification of Differentially Expressed Proteins
Proteins that were identified in 75% of cases (or 75% of controls) and had a 1.5-fold or greater difference in iTRAQ ratio between cases and controls were evaluated for differential expression (see Fig. 5). The significance of differential expression in protein ITRAQ ratios was determined using the Wilcoxon rank sum test. The p values were adjusted for multiple testing using the Bonferroni method (see Table III).
Table III. -Fold change and significance of differential expression for case-overexpressed proteins.
The significance of differential expression in protein ITRAQ ratios was determined using the Wilcoxon rank sum test. After adjusting for multiple testing using the Bonferroni correction, none of the p values reach statistical significance (p values =1).
| Name | Gene | Swiss-Prot accession number | -Fold change | Raw p value |
|---|---|---|---|---|
| Ciliary dynein heavy chain 9 | DNAH9 | Q9NYC9 | 5.2 | 0.0209 |
| Inositol 1,4,5-trisphosphate receptor type 2 | ITPR2 | Q14571 | 5.0 | 0.0209 |
| α1B-Glycoprotein | A1BG | P04217 | 3.3 | 0.0209 |
| Integral membrane protein GPR155 | GPR155 | Q7Z3F1 | 2.9 | 0.0209 |
| SET and MYND domain-containing protein 4 | SMYD4 | Q8IYR2 | 2.6 | 0.0209 |
| Serum albumin | ALB | P02768 | 1.8 | 0.0209 |
| Cyclic AMP-dependent transcription factor | ATF-7 | P17544 | 16.5 | 0.0433 |
| Serotransferrin | TF | P02787 | 13.3 | 0.0433 |
| HERV-K_12q14.1 provirus ancestral envelope polyprotein | ENK1 | P61565 | 6.1 | 0.0433 |
| Leukocyte immunoglobulin-like receptor subfamily B member 5 | LRLRB5 | O75023 | 5.5 | 0.0433 |
| Plasma membrane calcium-transporting ATPase 2 | ATP2B2 | Q01814 | 25.5 | 0.0472 |
| Ankyrin and armadillo repeat-containing protein | ANKAR | Q7Z5J8 | 9.3 | 0.0833 |
| α1-Antitrypsin | A1AT | P01009 | 6.4 | 0.0833 |
| Calcium-activated potassium channel subunit α-1 | KCNMA1 | Q12791 | 4.1 | 0.0833 |
| Chloride channel protein ClC-Ka | ClC-Ka | P51800 | 2.1 | 0.3865 |
RESULTS
Urine from healthy controls was used to evaluate the impact of several sample processing steps on the iTRAQ spectra. Based on these results, an optimized protocol was developed and tested on a pilot set of long frozen samples from patients with and without diabetic nephropathy.
Evaluating Impact of Starting Protein Quantity
First, the effect of variation in the starting protein quantity on the number and relative concentration of the identified proteins was assessed. Titration of the starting protein quantity showed that the number of identified proteins was comparable for starting protein quantities between 20 and 50 μg. However, use of less than 20 μg of protein led to a 29% drop in the number of proteins identified with high confidence (Fig. 1a). As expected, reduction of the starting protein quantity led to lower iTRAQ values (Fig. 1b). On a related note, with a starting protein quantity of 50 μg, titrating the iTRAQ label amount down to ¼ unit did not affect the number or identity of the proteins identified (supplemental Fig. 4).
Evaluating Role of Protease Inhibitors
To determine the impact of protease inhibitors on iTRAQ results, we compared duplicate urine samples from a healthy volunteer collected with or without immediate addition of protease inhibitors (PI) and stored at −80 °C for less than a week. The same 83 proteins were identified in samples with protease inhibitors (PI+) as in those without (PI−) (Fig. 2a). The iTRAQ ratios, representing protein concentrations in PI+ samples relative to PI− samples, were close to 1 (Fig. 2b).
Assessing Impact of Protein Extraction Method
Next, the protein extraction method was evaluated as a potential source of variation. We used urine samples from healthy controls to compare organic solvent precipitation, ultrafiltration, and dialysis-lyophilization for protein yield and quality of iTRAQ mass spectra. Among the tested protein extraction methods, methanol precipitation consistently led to the highest yield in terms of total extracted protein as measured by the Bradford assay (Table I). Acetonitrile precipitation generates the greatest number of independent protein spots visualized on two-dimensional gels (3, 4) and was therefore carried through to mass spectrometry despite its poor yield (i.e. total amount of protein extracted) (Table I). However, unlike two-dimensional gels (7, 11), with iTRAQ, the largest number of proteins were identified in the samples precipitated with methanol (Fig. 3a and supplemental Fig. 5a). The majority (>90%) of identified proteins were seen in all samples regardless of preparation method. However, methanol-precipitated samples contained all the proteins identified in other methods plus additional proteins identified only in these extracts (Fig. 3a and supplemental Fig. 5a). Relative protein concentrations were highly concordant between replicate samples with the same extraction method but quite discordant across extraction methods, highlighting the significance of keeping sample preparation protocols uniform between compared samples (Fig. 3b and supplemental Fig. 5b). Methanol precipitation had the lowest coefficients of variation for iTRAQ ratios between replicate samples (10–11%) (Fig. 3c). Representative spectra from this experiment are shown in supplemental Fig. 6.
Evaluating Role of Abundant Protein Depletion
To determine whether depletion of albumin and IgG would increase the depth of proteome coverage in normo- or macroalbuminuric urine, equal amounts of protein from duplicate depleted or non-depleted samples were compared using iTRAQ. This analysis was performed on urine from a healthy normoalbuminuric control as well as urine from a diabetic patient with macroalbuminuria. In the normoalbuminuric sample, the same 80 proteins were identified in both depleted and non-depleted samples (Fig. 4a, Venn diagram on the top right). In the macroalbuminuric sample, 75 and 77 proteins were identified in non-depleted and depleted samples, respectively. Of these, 73 were common between the two samples, four proteins were only identified in depleted samples, and two were only identified in non-depleted samples (Fig. 4a, Venn diagram on the bottom right). Relative protein concentrations were highly correlated between duplicates; depletion reduced the degree of correlation (Fig. 4b).
Development and Testing of an Optimized Protocol
Based on the above results, we developed a protocol for processing and analysis of urine samples (Fig. 5a). In a proof-of-principle experiment, this protocol was used to process urine samples from four diabetic Pima Indians with and four without overt nephropathy who were matched pairwise by gender. Matching by age was not possible in this small sample set (Table II). These samples were stored at −80 °C since collection 9–17 years ago (8, 9). A total of 54 unique proteins were identified in the four cases and four controls. To assess similarities and differences between the patients with and without overt nephropathy (cases and controls, respectively), an unsupervised HCA (12) was performed on the scaled iTRAQ values of the above 54 unique proteins. The unsupervised HCA of the scaled iTRAQ values was suggestive of segregation of cases and controls into two distinct clusters, showing that iTRAQ values may be able to separate cases from controls without prior definition of case/control status (Fig. 5b). Of the 54 identified proteins, 39 were present in 75% (three of four) of cases, and 53 proteins were present in 75% of controls. Of these consensus proteins, 38 were shared between cases and controls; one was unique to cases, and 15 were unique to controls (Fig. 5c). Of the 54 identified proteins, 15 were present in cases in concentrations 1.5-fold or higher than controls, and 22 were present in controls in concentrations 1.5-fold or higher than cases. Supplemental Fig. 7 illustrates the distribution of -fold change in expression among the 54 identified proteins. The 15 case-overexpressed proteins were evaluated for differential expression using the Wilcoxon rank sum test. Eleven proteins showed a trend for elevation in cases (Fig. 5d), although none reached statistical significance after Bonferroni correction for multiple testing (Table III). The accession numbers, number of unique peptides, percent sequence coverage, number of peptides used for quantitation, and mean and S.D. for the protein quantitation accuracy for each of the proteins identified in Figs. 1–5 and supplemental Figs. 4 and 5 are detailed in supplemental Tables 1–7, respectively.
DISCUSSION
The objective of this work was to optimize iTRAQ-based quantitative proteomics for comparative proteomics profiling of disease versus control urine samples. To do so, we evaluated the effect of several sample processing steps on the iTRAQ spectra. Based on these results, a urine processing protocol was developed and tested on a pilot set of urine samples from patients with and without diabetic nephropathy.
Starting Protein Quantity
The iTRAQ platform was developed for use with 50–100 μg of starting protein per label, which corresponds to ∼0.5–10 ml of normoalbuminuric urine (assuming ∼50% yield for protein extraction and a urine protein concentration of 1–10 mg/dl, which is within the normal range of 0–20 mg/dl). However, clinical samples are typically available in more limited quantities. It was therefore important to determine the lowest quantity of starting protein that could be used without sacrificing the sensitivity of this platform (as defined by the number of proteins identified with high confidence). Titration of the starting protein quantity demonstrated that reducing the starting protein below 20 μg led to a significant drop in the number of identified proteins, establishing that the minimum required quantity of starting protein is 20 μg per sample.
Role of Protease Inhibitors
To identify biomarkers for early diabetic nephropathy, we intend to use stored urine samples collected (usually without addition of protease inhibitors) many years ago as part of a longitudinal study. Whether protease inhibitors are necessary for proper storage of urine specimens is uncertain. Some investigators maintain that protease inhibitors are essential for preserving proteome integrity (6, 13). Others argue that in properly treated samples (i.e. samples that are frozen immediately upon collection and are not subject to repeated freeze-thaws), particularly of normoalbuminuric urine, addition of protease inhibitors is unnecessary (14) and may cause spectrometric artifacts by covalent binding to proteins (14, 15). Comparing the iTRAQ spectra from samples collected with and without protease inhibitors showed no difference in number or identity of high confidence proteins. Therefore, when handled properly (e.g. stored at −80 °C and without repeated freeze-thaws), samples that are collected without protease inhibitors remain informative for proteomics analysis.
Impact of Protein Extraction Method
iTRAQ labeling and subsequent MS steps are sensitive to pH and concentration of salts and contaminants (such as urea) and work best within a restricted range of starting protein amount and concentration. Furthermore, the variable and usually low protein concentrations in urine necessitate an additional concentration step. Therefore, proper protein extraction methods are critical to the success of iTRAQ-based proteomics. A number of methods are reported in the literature for desalting, concentrating, and removing contaminants from urine. Examples include protein precipitation by organic solvents, ultrafiltration by molecular weight, dialysis followed by lyophilization, and reverse phase chromatography (1, 4, 7, 16–18). These methods have been systemically evaluated for urine protein extraction prior to gel-based proteomics (4, 7) and some downstream mass spectrometry-based applications (17). However, there are currently no published reports on optimal urine protein extraction methods prior to iTRAQ. We compared several protein extraction methods, including ultrafiltration, organic solvent precipitation, and dialysis-lyophilization. Organic solvent precipitation with methanol consistently generated the highest total protein yield and largest number of proteins identified with high confidence. It was also the most reproducible (i.e. had the lowest coefficients of variation for iTRAQ ratios between replicate samples) of all methods tested.
Impact of Abundant Protein Depletion
Depletion of abundant urine proteins such as albumin increases the number of identified protein spots on a two-dimensional gel (1, 7, 16). However, the effect of such depletion on the urine proteome, as perceived by iTRAQ, has not been addressed. Furthermore, it is not clear whether depletion of abundant urine proteins changes the proteome differently in normoalbuminuric versus macroalbuminuric urine samples. We found that depletion of albumin and IgG from samples analyzed by iTRAQ did not increase the number of identified proteins in normoalbuminuric or macroalbuminuric urine samples. On the other hand, although relative protein concentrations were highly correlated between duplicates, depletion reduced the degree of this correlation. The drop in correlation after depletion may be due to the additional sample handling involved in the depletion step. Alternatively, removal of highly abundant proteins such as albumin and IgG may change the relative ratio of the remaining proteins by distorting the compositional stoichiometry of other urine proteins. Perhaps consistent with the latter hypothesis, the reduction in correlation was more notable in macroalbuminuric samples, which have a much larger component of albumin.
Developing and Testing an Optimized Protocol
Based on the above data, we developed an optimized urine processing protocol. To determine whether this protocol could be successfully applied to small quantities of long frozen urine samples, we used it to process a small test set of urine samples from Pima Indians with and without type 2 diabetic nephropathy (cases and controls, respectively). The iTRAQ spectra segregated cases and controls and highlighted several proteins that had been reported previously to be associated with diabetic nephropathy. Eleven proteins showed a trend for elevation in cases. However, as expected from the small sample size, none retained statistical significance after multiple testing correction (Table III).
As a reassuring internal positive control, albumin was among the proteins that were present in higher concentrations in cases. In addition, several previously reported candidate biomarkers showed trends toward differential expression, albeit not reaching statistical significance in this small sample set. Several of these proteins have been found previously to be up-regulated in diabetic nephropathy, e.g. transferrin, α1-antitrypsin, and α1B-glycoprotein. Serum transferrin, an iron transport protein, is up-regulated in the renal cortical proteome of diabetic db/db mice (19). In addition, urine transferrin levels are elevated in patients with diabetic nephropathy (20–22), and higher urine transferrin levels predict the onset of microalbuminuria in diabetic patients (20–22). In three independent proteomics scans, α1-antitrypsin, a serine protease inhibitor, was present at higher levels in urine and renal tissue of patients with diabetic nephropathy compared with controls (22–24). Higher levels of α1B-glycoprotein have also been reported previously in urine of patients with proteinuric diabetic nephropathy compared with normoalbuminuric diabetics (25).
In summary, we have optimized a quantitative biomarker discovery platform for application to small amounts of long frozen clinical urine samples. This method is simple, rapid, and reproducible and requires relatively small quantities of input protein (20 μg or less than 1 ml of urine), all essential criteria to success of a biomarker discovery strategy using limited quantities of clinical samples.
Supplementary Material
Acknowledgments
We thank Jun Ye (Thadhani laboratory, Massachusetts General Hospital) for assistance with graphics.
* This work was supported, in whole or in part, by the National Institutes of Health through the Intramural Research Program of the NIDDK. This work was also supported by a research grant from the Juvenile Diabetes Research Foundation (to R. T.).
 This article contains supplemental Figs. 1–7 and Tables 1–7.
This article contains supplemental Figs. 1–7 and Tables 1–7.
1 The abbreviations used are:
- iTRAQ
- isobaric tags for relative and absolute quantitation
- HCA
- hierarchical clustering analysis
- PI
- protease inhibitor(s)
- SET
- Su(var)3-9, Enhancer-of-zeste, Trithorax
- MYND
- MYeloid Nervy & Deaf-1.
REFERENCES
- 1.Pieper R., Gatlin C. L., McGrath A. M., Makusky A. J., Mondal M., Seonarain M., Field E., Schatz C. R., Estock M. A., Ahmed N., Anderson N. G., Steiner S. (2004) Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics 4, 1159–1174 [DOI] [PubMed] [Google Scholar]
- 2.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 3.Khan A., Packer N. H. (2006) Simple urinary sample preparation for proteomic analysis. J. Proteome Res. 5, 2824–2838 [DOI] [PubMed] [Google Scholar]
- 4.Thongboonkerd V., Chutipongtanate S., Kanlaya R. (2006) Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J. Proteome Res. 5, 183–191 [DOI] [PubMed] [Google Scholar]
- 5.Zerefos P. G., Vlahou A. (2008) Urine sample preparation and protein profiling by two-dimensional electrophoresis and matrix-assisted laser desorption ionization time of flight mass spectroscopy. Methods Mol. Biol. 428, 141–157 [DOI] [PubMed] [Google Scholar]
- 6.Thongboonkerd V., McLeish K. R., Arthur J. M., Klein J. B. (2002) Proteomic analysis of normal human urinary proteins isolated by acetone precipitation or ultracentrifugation. Kidney Int. 62, 1461–1469 [DOI] [PubMed] [Google Scholar]
- 7.Sigdel T. K., Lau K., Schilling J., Sarwal M. (2008) Optimizing protein recovery for urinary proteomics, a tool to monitor renal transplantation. Clin. Transplant. 22, 617–623 [DOI] [PubMed] [Google Scholar]
- 8.Bennett P. H., Burch T. A., Miller M. (1971) Diabetes mellitus in American (Pima) Indians. Lancet 2, 125–128 [DOI] [PubMed] [Google Scholar]
- 9.Otu H. H., Can H., Spentzos D., Nelson R. G., Hanson R. L., Looker H. C., Knowler W. C., Monroy M., Libermann T. A., Karumanchi S. A., Thadhani R. (2007) Prediction of diabetic nephropathy using urine proteomic profiling 10 years prior to development of nephropathy. Diabetes Care 30, 638–643 [DOI] [PubMed] [Google Scholar]
- 10.Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 11.Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 12.Alexander R. W. (1976) Hierarchical grouping analysis and skeletal materials. Am. J. Phys. Anthropol. 45, 39–43 [DOI] [PubMed] [Google Scholar]
- 13.Zhou H., Yuen P. S., Pisitkun T., Gonzales P. A., Yasuda H., Dear J. W., Gross P., Knepper M. A., Star R. A. (2006) Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 69, 1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thongboonkerd V. (2007) Practical points in urinary proteomics. J. Proteome Res. 6, 3881–3890 [DOI] [PubMed] [Google Scholar]
- 15.Rai A. J., Gelfand C. A., Haywood B. C., Warunek D. J., Yi J., Schuchard M. D., Mehigh R. J., Cockrill S. L., Scott G. B., Tammen H., Schulz-Knappe P., Speicher D. W., Vitzthum F., Haab B. B., Siest G., Chan D. W. (2005) HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics 5, 3262–3277 [DOI] [PubMed] [Google Scholar]
- 16.Oh J., Pyo J. H., Jo E. H., Hwang S. I., Kang S. C., Jung J. H., Park E. K., Kim S. Y., Choi J. Y., Lim J. (2004) Establishment of a near-standard two-dimensional human urine proteomic map. Proteomics 4, 3485–3497 [DOI] [PubMed] [Google Scholar]
- 17.Zerefos P., Prados J., Kossida S., Kalousis A., Vlahou A. (2007) Sample preparation and bioinformatics in MALDI profiling of urinary proteins. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 853, 20–30 [DOI] [PubMed] [Google Scholar]
- 18.Lee R. S., Monigatti F., Briscoe A. C., Waldon Z., Freeman M. R., Steen H. (2008) Optimizing sample handling for urinary proteomics. J. Proteome Res. 7, 4022–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilton R. G., Haidacher S. J., Lejeune W. S., Zhang X., Zhao Y., Kurosky A., Brasier A. R., Denner L. (2007) Diabetes-induced changes in the renal cortical proteome assessed with two-dimensional gel electrophoresis and mass spectrometry. Proteomics 7, 1729–1742 [DOI] [PubMed] [Google Scholar]
- 20.Narita T., Hosoba M., Kakei M., Ito S. (2006) Increased urinary excretions of immunoglobulin g, ceruloplasmin, and transferrin predict development of microalbuminuria in patients with type 2 diabetes. Diabetes Care 29, 142–144 [DOI] [PubMed] [Google Scholar]
- 21.Kanauchi M., Akai Y., Hashimoto T. (2002) Transferrinuria in type 2 diabetic patients with early nephropathy and tubulointerstitial injury. Eur. J. Intern. Med. 13, 190–193 [DOI] [PubMed] [Google Scholar]
- 22.Varghese S. A., Powell T. B., Budisavljevic M. N., Oates J. C., Raymond J. R., Almeida J. S., Arthur J. M. (2007) Urine biomarkers predict the cause of glomerular disease. J. Am. Soc. Nephrol. 18, 913–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma K., Lee S., Han S., Lee S., Francos B., McCue P., Wassell R., Shaw M. A., RamachandraRao S. P. (2005) Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics 5, 2648–2655 [DOI] [PubMed] [Google Scholar]
- 24.Inoue W. (1989) Immunopathological analysis of acute phase reactant (APR) proteins in glomeruli from patients with diabetic nephropathy. Nippon Jinzo Gakkai Shi 31, 211–219 [PubMed] [Google Scholar]
- 25.Rao P. V., Lu X., Standley M., Pattee P., Neelima G., Girisesh G., Dakshinamurthy K. V., Roberts C. T., Jr., Nagalla S. R. (2007) Proteomic identification of urinary biomarkers of diabetic nephropathy. Diabetes Care 30, 629–637 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.