Abstract
Identification of missing genes or proteins participating in the metabolic pathways as enzymes are of great interest. One such class of pathway is involved in the eugenol to vanillin bioconversion. Our goal is to develop an integral approach for identifying the topology of a reference or known pathway in other organism. We successfully identify the missing enzymes and then reconstruct the vanillin biosynthetic pathway in Aspergillus niger. The procedure combines enzyme sequence similarity searched through BLAST homology search and orthologs detection through COG & KEGG databases. Conservation of protein domains and motifs was searched through CDD, PFAM & PROSITE databases. Predictions regarding how proteins act in pathway were validated experimentally and also compared with reported data. The bioconversion of vanillin was screened on UV-TLC plates and later confirmed through GC and GC-MS techniques. We applied a procedure for identifying missing enzymes on the basis of conserved functional motifs and later reconstruct the metabolic pathway in target organism. Using the vanillin biosynthetic pathway of Pseudomonas fluorescens as a case study, we indicate how this approach can be used to reconstruct the reference pathway in A. niger and later results were experimentally validated through chromatography and spectroscopy techniques.
Keywords: Metabolic Pathway, reconstruction, eugenol, vanillin, Pseudomonas, Aspergillus, GC-MS
Background
The identification of metabolic pathway's genes and proteins is an emerging area of great interest. The accumulation of data from different origins and the development of methods to mine that data create an opportunity to bridge the gap between the fragmentary view of genes and proteins and the more integrated approach of Bioinformatics [1]. Moreover, increasing amounts of experimental data that can be mined for information about how proteins in cells assemble as metabolic and signal transduction pathways circuits are generated each day [2–6]. Datasets available for such tasks include the primary literature, whole genome two hybrid screenings, large scale microarray experiments, full genome sequences and the patterns of conserved/non-conserved homologues and orthologues in them. Theoretical and experimental methods are being developed and used to analyze these different types of data and infer networks of proteins or genes that are involved in the similar type biological processes [7]. In general, the networks derived by the computational analysis of these data are static, in the sense that they provide little information [8]. This can be an important problem while assembling the network structure of either novel pathways or complex pathways with an unclear reaction network. Thus, it is a challenge to identify the correct bioconversion pathway that allows for the creation of sequence synteny based pathway network whose mechanism can be analyzed and tested against experimental observations. To achieve such a goal, strategies that combine the different theoretical and wet lab confirmations to identify proteins and generate a set of plausible pathway network for the process of interest are needed. Such a process integrates homologous data and provides testable predictions and information about unexplored pathway mechanism in other organisms. In the present work, we studied eugenol to vanillin bioconversion pathway in Pseudomonas fluorescens as a reference pathway and successfully reconstructed in Aspergillus niger on the basis of participating enzyme's protein sequences synteny map. However, vanillin (4- hydroxy-3- methoxybenzaldehyde) is one of the high value aided aromatic product of the flavor, food, cosmetic and pharmaceutical industry. Moreover, flavors and fragrances find wide applicability in food, cosmetic and pharmaceutical industries and represent a worldwide market of about 18 billion US dollars per year. Abundantly the flavors in the world market are chemically synthesized which is environmentally unfriendly and the production process too lacks the substrate specificity resulting in the formation of undesirable reaction mixtures, hazardous to health and increase the downstream cost. Less than 55 of the flavors are extracted from the plants and are natural products [9]. However, the biotechnological means using naturally originated product as substrate for the production of flavors in high quantities are also considered as natural and is an emerging approach for the flavor production [9]. On the other hand, production of natural vanillin from the orchid Vanilla planifolia is very low and unable to fulfill the demand of the world market of 12,000 tons per year. The microbial conversion of various phenyl propanoids such as ferulic acid, isoeugenol, eugenol, coniferyl aldehyde etc. to produce vanillin at a cost effective rate and free from health hazardous chemicals, has been an area of great interest [10–13]. The bioconversion of eugenol to vanillin has been studied in various bacterial systems including different species of Pseudomonas, Rhodococcus, Corynebacterium and recombinant strains of Amycolatopsis so as to establish the pathways in the prokaryotic system. The drawback lies within the eukaryotic fungal system where no single fungus has been reported since now to be able to biotransform eugenol to vanillin [14–17]. A combination of two fungal systems A. niger and Pycnoporus cinnabarinus were reported to convert ferulic acid to vanillin by catalysis in two different steps [18,19]. The establishment of the bioconversion pathway in fungal systems would lead to hypothesize the metabolic fate of eugenol in eukaryotic systems.
Aspergillus genome indicates the presence of 13,071 predicted genes with average gene length of 1,384 bp spread across 8 chromosomes. Notably, a number of hypothetical/unknown proteins are annotated in A. niger [20]. This fact emphasizes the need to identify gene function by new approaches. In this work, we focus on eugenol to vanillin bioconversion pathway in A. niger. Currently, there are gaps in eugenol to vanillin bioconversion pathway of A. niger because of the insufficient annotation. We try to resolve such missing enzymes by using the protein local region sequence similarity in terms of conserved domain and motifs. Later these predicted domain and motifs were analyzed for their localization in orthologous sequences through KEGG and COG databases [21]. Results showed conserved protein domains and motifs in both the organisms on the basis of pathway specific enzyme's functional domain and motif sequences & structural similarity. These predicted proteins motifs are evolutionarily conserved and form a basis for the identification of missing or unexplored enzymes, which ultimately helped in reconstruction of eugenol to vanillin bioconversion pathway. However, most of the works have been done in prokaryotes only and the details of this pathway are not fully understood in any eukaryotic system. In P. fluorescens, the pathway has been more extensively studied and so far following pathway enzymes are reported:
- Eugenol hydroxylase cytochrome C subunit (EhyA),
- Eugenol hydroxylase flavoprotein subunit (EhyB),
Coniferyl alcohol dehydrogenase (CalA),
Coniferyl aldehyde dehydrogenase (CalB),
Feruloyl CoA synthase (Fcs) and
Enoyl CoA hydratase/aldolase (Ech) (Table 1 in supplementary material).
The reconstruction of biosynthetic pathway in A. niger indicates that functionally unexplored hypothetical proteins may have important role in the bioconversion of eugenol to vanillin. We also predicted the functional role of these hypothetical/unknown proteins as an enzyme for the development of high value aided aromatic product of the flavor, food, cosmetic and pharmaceutical industry.
Methodology
Biological sequence retrieval
Relevant information on the pathway specific genes and proteins of P. fluorescens involved in the bioconversion of eugenol to vanillin was studied through reported literature and later biological sequences were retrieved through nucleotide and protein sequence databases of both GenBank (NCBI) (www.ncbi.nlm.nih.gov/) & UniProtKB/Swiss-Prot (Expasy-EBI) (http://expasy.org/uniprot/) web servers.
Selection of datasets for pathway reconstruction
Two types of datasets were used for pathway construction:
Known or Reference data set - which includes protein sequences of five enzymes such as EhyA/EhyB, CalA, CalB, Fcs and Ech, participating in the vanillin biosynthetic pathway of model organism i.e., P. fluorescens with conserved protein domains & motifs,
Predictive or Target data set - which includes potential homologous protein sequences such as Cytochrome-c CYC_ASPNG or hypothetical protein (gi|2829474|sp|P56205.1 predicted as EhyA), hypothetical protein An09g01380 (gi|145241618|ref|XP_001393455.1 predicted as EhyB), hypothetical protein An15g01840 (gi|145250419|ref|XP_001396723.1 predicted as CalA), hypothetical protein An01g09260 (gi|145230075|ref|XP_001389346.1 predicted as CalB), hypothetical protein An14g05630 (gi|145249694|ref|XP_001401186.1 predicted as Fcs) and hypothetical protein An02g02820 (gi|145231906|ref|XP_001399422.1 predicted as Ech) predicted to be the pathway’s enzymes of target organism i.e., A. niger with conserved protein domains & motifs (Table 1 in supplementary material).
Sequence similarity and homology search
We first identified the number and details of enzymes participating in eugenol to vanillin bioconversion through literatures. Then, each of the all-against-all proteins in the list were checked for sequence homology. Here homologous clusters are computationally defined clusters of similar proteins having ≥ 50% similarity and E-value threshold ≤1.00E- 63 with similar conserved protein domain and motif local regions. Later this information was utilized to reconstruct the pathway reaction network in A. niger. Calculation of the statistical significance of matches was performed through Protein BLAST program (www.ncbi.nlm.nih.gov/BLAST/) at NCBI, USA webserver. Pairwise sequence alignment based studies indicates functional and evolutionary relationships between both the organisms as revealed by phylogenetic analysis and so helped in the identification of other family members which are not yet identified.
Detection of orthologs based on conserved protein domain, motif & superfamily
Homologous proteins in A. niger were searched for conserved protein regions such as protein motifs, domains and superfamilies, having related functions to that of the P. fluorescens (Table 1 in supplementary material). For conserved protein functional domain identification Conserved Domain search tool (CDD-Search at NCBI) was used (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), while InterPro-EBI (www.ebi.ac.uk/interpro/) and Pfam-Sanger (http://pfam.sanger.ac.uk/) databases were used for conserved protein motifs identification. Cluster of Orthologous Group (COG) database (www.ncbi.nih.gov/COG/) was used for identification of orthologous cluster group. We used P. fluorescens gene entries from the KEGGGENES database (www.genome.jp/kegg/) [22, 23] and performed further annotation of motifs conservation to these genes in A. niger, by using the information about the gene ontology assignments through GenBank database at NCBI.
Experimental validation
Since protein sequence similarity or homology between reference and predicted pathway genes/proteins was lower therefore to confirm the theoretical predictions, localization of participating hidden and unexplored enzymes was further verified through chromatographic & mass spectroscopic techniques. The bioconversion of vanillin was screened on UV-TLC plates and later confirmed through GC and GCMS techniques.
Discussion
The proteins of P. fluorescens that are known to be involved in eugenol to vanillin biosynthetic pathway are shown in Table 1 (see supplementary material). Conserved common protein domain and motifs local regions in protein sequences of both the organisms are shown in Table 1 (see supplementary material). We first describe how the different large scale datasets are analyzed and combined, in order to inferred initial pathway network assembly in reference organism i.e., P. fluorescens. Then, we used local region sequence synteny mapping approach to analyze the pathway enzymes homolog in target organism i.e., A. niger. Finally, we hypothesized the most potential pathway enzymes based on the conserved protein motifs & experimental observations. Bibliographic analysis of published abstracts and papers were used to verify the different proteins/genes involved in bioconversion process (Figure 1 & Figure 2). We have used the genes known to be involved in eugenol to vanillin biosynthetic pathway inP. fluorescens to test this assumption in A. niger as an alternative organism for production of vanillin at industrial level.
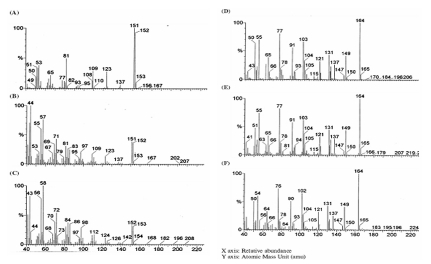
Figure 1.
Mass spectra (MS) of standard vanillin (A), Aspergillus mediated biotransformed extracellular vanillin (B), intracellular vanillin (C), standard eugenol (D), eugenol extracellular (E) and eugenol intracellular (F).
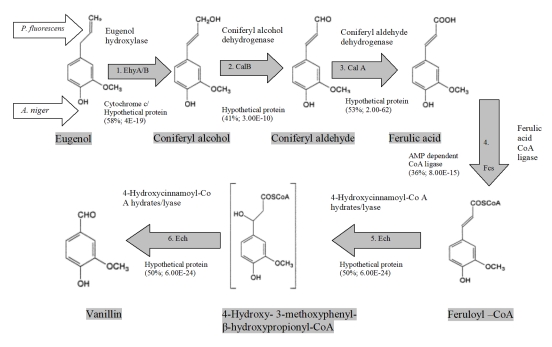
Figure 2.
Comparison of reference vanillin biosynthetic pathway of P. fluorescens with reconstructed pathway of A. niger. Values in parentheses showed protein sequence similarity and BLAST E-value.
Pathway reconstruction using Phylogenetic profiling
It is not possible to infer probable reaction network structure for the pathway just from the co-occurrence of genes. Additional data is necessary in order to translate the group of proteins that are identified to be involved in the biological process of interest into a structured network. Phylogenetic profiling of the genes assists in adding some structure to the network [24,25]. We searched for motif patterns of cooccurrence of sets of orthologs in other species of A. niger. The assumption behind the use of this approach is that, if the conserved pattern or motif present in two or more proteins of different species of Aspergillus is very close, this is an indication of co-evolution among the proteins. Such co-evolution can be taken as an indication that they are likely to be involved in the same cellular process [26,27]. Results from phylogenetic co-evolution analysis of the different genes involved in eugenol bioconversion are explained in Methodology section. This additional information assists in inferring some form of common motifs to the network structure of the studied pathway (Table 1 in supplementary material). For example, in P. fluorescens, conserved motifs viz. PS51007, IPR003088 & PF00034 of EhyA enzyme showed 100% homologues coincide with CYC_ASPNG gene of A. niger with 58% amino acid similarity and 38% identity. Similarly enzyme of P. fluorescens EhyB showed common evolutionary conserved motif viz., Pfam01565 and Pfam02913 in A. niger with 50% amino acid sequence similarity and 34% identity. This suggests that predicted hypothetical proteins are more likely to act directly on these reactions as participating unexplored enzymes, which are not yet functionally annotated.
Experimental validation through TLC and GC & GC-MS
Of five P. fluorescens genes, we annotated all five as putative enzymes of the eugenol to vanillin bioconversion pathway in A. niger with the information on common conserved protein motifs & domain. We predicted enzymatic reactions for all the participating enzymes on the basis of functionally conserved protein motif and domain homology and co-evolution. Later, we mapped them on target organism based on KEGG metabolic pathway database (Table 1 in supplementary material). We hypothesized cases where the reaction steps of missing enzymes can be catalyzed by other enzymes. Besides, to validate the above pathway, species of Aspergillus were screened experimentally in our lab for bioconversion capability from eugenol to vanillin. Two of the Aspergillus species namely, A. niger and A. flavus were found capable for bioconversion of eugenol to vanillin. The bioconversion of vanillin was screened on UV- TLC plates and later on confirmed through GC and GC-MS techniques (Figure 1). Experimental observations of UV- TLC and GC & GC-MS showed the likely roles of proteins that have been analyzed and also predicted through homology search and conserved domain and motif search tools. This eliminates some of the alternative network structures and assists in refining the remaining, by suggesting experiments that can further differentiate between them. As shown in this paper, bioinformatics tools like Interpro, ProDom, Pfam, COG, KEGG, BLAST etc. were successfully used along with experimental data through chromatographic and spectroscopic techniques so that to assist in the predictive analysis of the missing metabolic pathways. Another useful aspect of our approach is that it can be used in reassessing the interpretation of experimental chromatographic data. It is our goal to refine predictions based on updated sequence data and to apply this method to other organisms, thus assisting in the understanding of how the pathway of vanillin bioconversion has evolved. In principle, the combination of integrated approach presented here could be used in a flexible way to analyze similar problems in other biological system (Figure 2).
In our present work, we used a combination of experimental and theoretical methods to reconstruct the topology of the eugenol to vanillin bioconversion pathway in A. niger with the help of P. fluorescens reported data. The pathway enzymes were identified and a network of interactions between them was predicted using literature mining and computational analysis. Although bioinformatics tools do provide a synteny of the pathway network structure, human analysis remains necessary for curating all the relevant information. In fact, human curation of the pathway chain is a critical step for the derivation of possible alternative reaction schemes for the pathway. At this point, theoretical predictions are needed to validate experimentally in wet lab.
Conclusion
The eugenol to vanillin bioconversion pathway of P. fluorescens was reconstructed by using a flexible computational methodology that combines sequence analysis, literature search and bioinformatics applications. The roles of different P. fluorescens proteins in vanillin bioconversion were successfully predicted in A. niger. Some predictions are cross checked through published experimental data. Other predictions need further experimental work to validate it. The methodology used here is flexible and can be applicable in other systems. This methodology could be a step forward in integrating different types of data to obtain systemic knowledge about novel pathways and to clarify how current prediction of known pathways would work, thus generating rational hypothesis for testing. Besides, to validate the above pathway, different species of Aspergillus were screened experimentally in our lab for bioconversion capability from eugenol to vanillin. Two of the Aspergillus species namely, A. niger and A. flavus were found capable of vanillin bioconversion. Moreover, our hypothesis of vanillin biosynthetic pathway was later successfully confirmed through UV - TLC and GC & GC-MS data, which showed sign of vanillin synthesis in A. niger. Thus, vanillin production along with the formation of intermediates like ferulic acid strongly supports the significance of bioinformatics approaches in the reconstruction of unexplored metabolic pathway. We found mainly hypothetical proteins of A. niger which are in the same gene groups as real enzymes on the basis of conserved protein domain and motifs. There are so many missing enzymes in the biosynthetic pathways of eukaryotes, using such strategies we can find more hypothetical proteins that may replace missing enzymes information of important pathways. Validation of the results by using additional information, such as microarray and proteomic data analysis could be our future works.
Supplementary material
Acknowledgments
We acknowledge Council of Scientific & Industrial Research, New Delhi Network Project'09 (CSIR-NWP09) for financial support at Central Institute of Medicinal & Aromatic Plants, Lucknow, India.
Footnotes
Citation:Srivastava et al, Bioinformation 4(7): 320-325 (2010)
References
- 1.Francke C, et al. Trends Microbiol. 2005;13:550–558. doi: 10.1016/j.tim.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Karp PD. Genome Biol. 2004;5:401. doi: 10.1186/gb-2004-5-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karp PD, et al. Bioinformatics. 2002;18:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 4.Karp PD, et al. Pac Symp Biocomput. 2004;9:190–201. doi: 10.1142/9789812704856_0019. [DOI] [PubMed] [Google Scholar]
- 5.Ideker T. Adv Exp Med Biol. 2004;547:21–30. doi: 10.1007/978-1-4419-8861-4_3. [DOI] [PubMed] [Google Scholar]
- 6.Ideker T, et al. Ann Biomed Eng. 2006;34:257–264. doi: 10.1007/s10439-005-9047-7. [DOI] [PubMed] [Google Scholar]
- 7.Alves R, et al. Nat Biotechnol. 2006;24:667–672. doi: 10.1038/nbt0606-667. [DOI] [PubMed] [Google Scholar]
- 8.Alves R, Sorribas A. BMC Systems Biol. 2007;1:10. doi: 10.1186/1752-0509-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vandamme EJ, Soetaert W. J Chem Tech Biotechnol. 2002;77:1323–1332. [Google Scholar]
- 10.Rabenhorst J. Appl Microbiol Biotechnol. 1996;46:470–474. [Google Scholar]
- 11.Furukawa H, et al. J Biosci Bioeng. 2003;96:401–403. doi: 10.1016/S1389-1723(03)90145-9. [DOI] [PubMed] [Google Scholar]
- 12.Yamada M, et al. Appl Microbiol Biotechnol. 2007;73:1025–1030. doi: 10.1007/s00253-006-0569-1. [DOI] [PubMed] [Google Scholar]
- 13.Yamada M, et al. Biotechnol Lett. 2008;30:665–670. doi: 10.1007/s10529-007-9602-4. [DOI] [PubMed] [Google Scholar]
- 14.Plaggenborg R, et al. Appl Microbiol Biotechnol. 2006;72:745–755. doi: 10.1007/s00253-005-0302-5. [DOI] [PubMed] [Google Scholar]
- 15.Hua D, et al. J Biotechnol. 2007;130:463, 470. doi: 10.1016/j.jbiotec.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Kasana RC, et al. Curr Microbiol. 2007;54:457–461. doi: 10.1007/s00284-006-0627-z. [DOI] [PubMed] [Google Scholar]
- 17.Tsujiyama S, Ueno M. Bioscience Biotechnol Biochem. 2008;72:212–215. doi: 10.1271/bbb.60606. [DOI] [PubMed] [Google Scholar]
- 18.Falcnnier B, et al. J Biotechnol. 1994;37:123–132. [Google Scholar]
- 19.Rao Sr, Ravishankar GA. J Sci Food Agric. 2000;80:289–304. [Google Scholar]
- 20.Priefert H, et al. Arch Microbiol. 1999;172:354–363. doi: 10.1007/s002030050772. [DOI] [PubMed] [Google Scholar]
- 21.Nikitin F, et al. Genome Inform. 2004;15:266–275. [PubMed] [Google Scholar]
- 22.Ogata H, et al. Biosystems. 1998;47:119–128. [Google Scholar]
- 23.Nakao M, et al. Genome Inform Ser Workshop Genome Inform. 1999;10:94–103. [PubMed] [Google Scholar]
- 24.Pellegrini M, et al. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, et al. Nat Biotechnol. 2005;23:253–260. doi: 10.1038/nbt1065. [DOI] [PubMed] [Google Scholar]
- 26.Singh IV, et al. Online J Bioinformatics. 2004;5:32–48. [Google Scholar]
- 27.Khan F, et al. J Integrative Bioinformatics. 2008;5(1):86. doi: 10.2390/biecoll-jib-2008-86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.