Abstract
Three different nitrile-containing amino acids, para-cyanophenylalanine, meta-cyanophenylalanine and S-cyano homocysteine, have been introduced near the active site of the semi-synthetic enzyme Ribonuclease S (RNase S) to serve as probes of electrostatic fields. Vibrational Stark spectra, measured directly on the probe-modified proteins, confirm the predominance of the linear Stark tuning rate in describing the sensitivity of the nitrile stretch to external electric fields, a necessary property for interpreting observed frequency shifts as a quantitative measure of local electric fields that can be compared with simulations. The X-ray structures of these nitrile-modified RNase variants and enzymatic assays demonstrate minimal perturbation to the structure and function, respectively, by the probes and provide a context for understanding the influence of the environment on the nitrile stretching frequency. We examine the ability of simulation techniques to recapitulate the spectroscopic properties of these nitriles as a means to directly test a computational electrostatic model for proteins, specifically that in the ubiquitous Amber ′99 force field. Although qualitative agreement between theory and experiment is observed for the largest shifts, substantial discrepancies are observed in some cases, highlighting the ongoing need for experimental metrics to inform the development of theoretical models of electrostatic fields in proteins.
Keywords: Vibrational Probes, Stark, Electrostatic Fields, Infrared spectroscopy, Nitriles, Molecular Dynamics Simulation, Proteins
INTRODUCTION
Vibrational transitions, measured by infrared (IR) or Raman spectroscopy, provide a useful tool with which to study the structural and dynamic properties of biological molecules. Most studies to date make use of vibrational probes that are naturally present, such as the amide stretch in peptides, the hydroxyl stretch of water or unique features of bound prosthetic groups; however, introducing a non-natural chemical moiety has many potential advantages over the natural chromophores. The ideal probe would employ a transition that absorbs strongly and in an uncluttered region of the spectrum. The probe should have a strong coupling between the spectroscopic observable and the property of interest (e.g. between observed frequency and electric field), and, to the extent possible, that coupling should be understood on a theoretical basis and independently verified by experiment. Our laboratory and many others have sought to incorporate non-natural moieties into biological molecules that circumvent the limitations of the intrinsic vibrational chromophores by synthesis1, semi-synthesis2,3, biosynthesis (with non-natural4,5 or isotopically labeled6,7 amino-acids), post-translational modification2 or via the binding of an inhibitor that contains the probe8-10. Ideally, introduction of an extrinsic chromophore should minimally perturb the structure and properties under study, a condition that must be independently verified by techniques such as NMR, crystallography and/or functional assays.
Our focus has been on probes that can report upon the magnitude, direction and fluctuations in the electric field present at different sites in the interior of biological molecules and to compare these measured fields with predictions from simulations. These fields are the consequence of the charged, polar, and polarizable groups that comprise the molecule and are anticipated on theoretical grounds11 to be quite large and vary from site-to-site (± 10's of MV/cm). Such large variations are therefore expected to dramatically influence the functions of biomolecules, including catalysis involving charge-separation in the transition state12-14, electron transfer in redox processes15, and the folding and association of biological polymers16,17. In order to find IR transitions that are best suited to report on these fields, the sensitivity of a number of vibrational chromophores to external electric fields was ascertained by measuring the vibrational Stark effect18 (VSE). The nitrile stretching transition, which exists in a relatively uncluttered region of the spectrum, has a relatively large extinction coefficient and is quite sensitive to an electric field19-21, emerged as a promising target, and since then, examples of nitriles as probes have proliferated1,2,9,22-30. The VSE experiment provides a quantitative calibration of the sensitivity of the nitrile to electric fields in the form of the linear Stark tuning rate, |Δμ⃗CN| (in units of cm−1/(MV/cm)), a measure of the sensitivity of the transition frequency to an electric field, which can in turn be used to translate observed frequency shifts, (in cm−1), into variations in the projection of the protein electrostatic field, ΔF⃗protein (in MV/cm) on Δμ⃗CN using:
| (1) |
where h is Planck's constant and c is the speed of light.
In order to use observed frequency shifts in response to changes in the field due to changes in pH, exposure to solvent and/or mutations to extract values of ΔF⃗protein there are four criteria that must be demonstrated. First, the value of |Δμ⃗CN|, determined previously only for model compounds in organic glasses, must be known and must be shown to dominate the spectral response to electrostatic fields. Second, since the -CN ground state dipole moment is substantial, this could affect the environment it is meant to probe, thus it is necessary to show that the structure and function of the protein are minimally impacted by the presence of the probe. Third, Δμ⃗CN is typically parallel to the C-N bond20, thus in order to extract the orientation of ΔF⃗protein with respect to the protein's molecular axes, it is necessary and sufficient to determine the position and orientation of the C-N bond axis relative to the protein framework through structure determination. And finally, the possibility of hydrogen bonds to the nitrile represents an example of a specific chemical interaction that may complicate the simple behavior implied by Eqn. 1, where ΔF⃗protein is due to the collective electrostatic properties of the environment around the probe; the possibility of such specific interactions must be ascertained by independent means31,32.
In this paper we focus on these issues using the classic semi-synthetic enzyme, ribonuclease S (RNase S)33. RNase S is the limited proteolysis product of subtilisin acting on RNase A, and consists of a small fragment including amino acids 1-20, the S-peptide, and a larger fragment including residues 21-124, the S-protein. When separated, the S-protein is non-functional because the active site is split; the S-peptide is disordered34, but when combined non-covalently with the S-protein, nearly native enzymatic activity is restored33. Because the S-peptide can be readily prepared by solid-phase peptide synthesis, it is straightforward to introduce non-natural amino acids. Here we have used this strategy to introduce para- or meta-cyanophenylalanine (pCN- and mCN-RNase, respectively) by replacing phenylalanine at position 8 or S-cyano-homocysteine (SCN-RNase) by replacing methionine at position 13. We examine the propensity of the nitrile to perturb the RNase structure by X-ray crystallography and measure its effect on catalytic function by standard assays. The vibrational Stark spectra of the probe-modified proteins are acquired, providing a quantitative calibration of the sensitivity of these probes to electrostatic fields and demonstrating the transferability of these same parameters determined for model compounds. X-ray crystal structures of these probe-modified proteins allowed us to rule out the existence of hydrogen bonds to the nitriles, which could complicate the otherwise simple behavior of the probes. Then we compare the experimentally measured frequency shifts at different sites using Eqn. 1 and interpreted as differences in the electrostatic fields using Eqn. 1, to computationally simulated fields. We employ a simple modification to the GROMACS molecular dynamics program to output electrostatic fields at the probe location, at every time-step, over many nanoseconds of a molecular dynamics (MD) trajectory. This provides a means to directly and quantitatively test the electrostatic models employed in molecular dynamics.
RESULTS AND DISCUSSION
Enzyme Reconstitution and Kinetics
Small, nitrile-containing molecules such as acetonitrile and benzonitrile have large ground state dipole moments (3.9 and 4.2 Debye, respectively35) and so we first address whether a peptide containing such polar nitrile probes can be can be accommodated within the native structure and how it perturbs enzymatic activity. The sites of probe incorporation in pCN-, mCN- and SCNRNase, are such that the nitrile is within 4-8 Å of the catalytic histidines (His 12 or His 119, Table 1). As with the native S-peptide, all three probe-modified S-peptide species restore catalytic activity to the isolated S-protein, as measured by initial rates of cyclic cytidine monophosphate hydrolysis (see Table 1). The combined steric and electrostatic effects of the nitrile on the catalytic rate constants is small in all cases, comparable to that seen for mutations in non-active site residues in other enzymes36 and much smaller than the effect on RNase A catalysis37 of a change of one pH unit above or below pH 7; thus we conclude that the nitrile does not significantly perturb the catalytic property it is meant to probe. Similar observations have been made for thiocyanate-modifed ketosteroid isomerase incorporating a similar probe deep in the active site pocket30,32. The existence of a stable complex capable of catalysis demonstrates that the steric and electrostatic perturbations due to the nitrile are not great enough to exceed the driving force for peptide-protein association in any of the three cases studied.
Table 1.
Catalytic parameters for RNase variants (measured at 0.4 μM) determined by the method of initial rates of cyclic cytidine monophosphate hydrolysis, monitored by UV-Vis spectroscopy49. Also shown are the distances from the nitrile nitrogen to the nearest heavy atom of the imidazole ring of the catalytic His 12. Wild-type refers to intact RNase A.
| Wild-Type | pCN-RNase a | mCN-RNase a | SCN-RNase a | |
|---|---|---|---|---|
| kcat (s−1) | 3.0 | 1.7 | 1.5 | 2.5 |
| Km (mM) | 1.8 | 1.1 | 4.0 | 1.7 |
| distance (Å) | -- | 5 | 8 or 4b | 8 |
pCN and mCN are at position 8; -SCN is at position 13
mCN has two alternative conformations (see text and Fig 2); both distances are given.
X-ray Crystallography
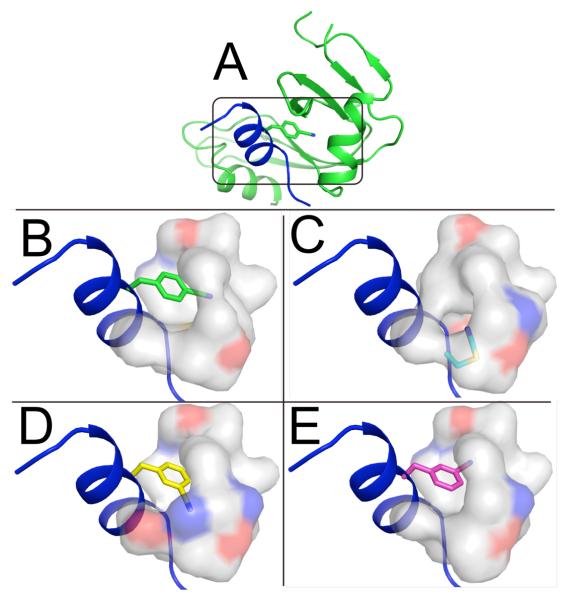
The structures of all three probe-modified proteins were determined by X-ray crystallography (Fig. 1). All three crystals belong to the C2 space group, with two molecules per asymmetric unit. The structures of pCN-RNase, SCN-RNase and mCN-RNase were refined using data collected to 1.5, 2.3 and 2.5 Å resolution, respectively. Non-crystallographic symmetry constraints were relaxed in the later refinement steps of the structure-solving process, and thus the two molecules in the asymmetric unit serve as mutual controls for random variation. Conservative cut-offs were chosen to select the highest resolution shell (see Table 2), and in all cases the region around the nitrile is well resolved. In all three cases, the nitrile is buried in the hydrophobic interior, well inside the solvent-accessible surface-area, and importantly, none of the structures exhibit hydrogen bond donating groups within 3.5 Å or less from the nitrile nitrogen. Structures were deposited in the RCSB Protein Data Bank and given PDB accession codes 3OQY, 3OQZ and 3OR0 for pCN-, mCN- and SCN-RNase, respectively.
Figure 1.
Structural models of X-ray data for RNase variants. (A) Ribbon diagram for pCN-RNase with the S-peptide in blue and S-protein in green. (B-E) Expanded view of the region around each nitrile from four aligned structures, with S-peptide in a blue cartoon representation, a semi-transparent surface representation of neighboring residues colored grey for carbon, blue for nitrogen and red for oxygen, and stick representations of the nitrile-modified residues: pCN-RNase (B; carbons in green), SCN-RNase (C; cyan), and the two independently modeled structures from the asymmetric unit of mCN-RNase: (α) carbons in yellow (D) and (β) carbons in magenta (E).
Table 2.
X-ray data. Refinement statistics for the three nitrile-modified RNase S variants with data for the highest resolution shell in brackets, all three solved in the C2 space group.
| Molecule | pCN-RNase | mCN-RNase | SCN-RNase |
|---|---|---|---|
| Resolution [high res. shell] | 1.5 Å [1.49-1.54 Å] | 2.5 Å [2.50-2.56 Å] | 2.3 Å [2.30-2.36 Å] |
| # of reflections [high res. shell] | 36722 [2652] | 7887 [560] | 9888 [686] |
| % possible [high res. shell] | 95.8 [92.4] | 99.9 [100] | 95.8 [95] |
| Redundancy [high res. shell] | 4.5 [3.2] | 3.2 [3.2] | 1.8 [1.9] |
| Rworka | 0.205 | 0.209 | 0.218 |
| Rfreea | 0.225 | 0.271 | 0.271 |
| Cα RMSDb (A chain / B chain) | 0.37 / 0.32 Å | 0.42 / 0.37 Å | 0.37 / 0.46 Å |
| Local RMSDc (A chain / B chain) | 0.23 / 0.21 Å | 0.31 / 0.10 Å | 0.52 / 0.26 Å |
Rfree and Rwork are the R-factors ( where F are the observed or calculated X-ray structure factors) for two randomly selected subsets of the reflection data. 95% of the total Fobs data points are used to guide refinements to the model (minimizing Rwork), while 5% are set aside to evaluate the improvement (Rfree).
The average root-mean-square deviation for all backbone carbon atoms for one molecule of the two molecule asymmetric unit, calculated against a set of published RNase structures of different space groups (7RSA and 1DY5 = P21, 1RNU = P312 ). The average RMSD for the members of the comparison set relative to each other is 0.36 Å.
RMSD of all the atoms in the 5 Å neighborhood of the nitrile (excluding the substituted amino-acid) relative to the same atoms in the comparison set. The average RMSDs for the comparison set relative to each other are 0.22, 0.24 / 0.13 and 0.17 Å for pCN, mCN (A /B) and SCN neighborhoods respectively.
To assess the degree to which the nitrile is disruptive to the native structure, root mean square deviations (RMSDs) were calculated between the solved structures and a control set consisting of four representative published structures of native RNase (7RSA, 1RNU and the A and B monomers of the two-molecule asymmetric unit of 1DY5) from the Protein Data Bank (PDB). Comparing only the alpha carbons of the peptide backbone (Cα), the probe-modified proteins show comparable RMSD's when compared with the control set as the control set does compared to itself, ruling out gross structural changes as a result of nitrile incorporation (Table 2). Choosing a more focused comparison of all the atoms in the 5 Å neighborhood of the nitrile (excluding the substituted amino-acid) a statistically significant large RMSD (i.e. much larger than the average RMSD for members of the control set compared to each other) is observed only for SCN-RNase and, of this structure, only for one of the two monomers of the asymmetric unit (Table 2); these differences between monomers could either be an artifact of the model due to low resolution of the data or a consequence of asymmetric packing interactions in the crystal. Close visual inspection of the structural models does reveal some subtle rearrangements for both mCN- and SCN-RNase not large enough to exceed the statistical uncertainty of the RMSD analysis, but large enough to have an effect on the highly local properties probed by the nitrile bond. Nonetheless, in each case the perturbation of the structure by the nitrile is small relative to the accuracy of the structural models routinely obtained from X-ray data.
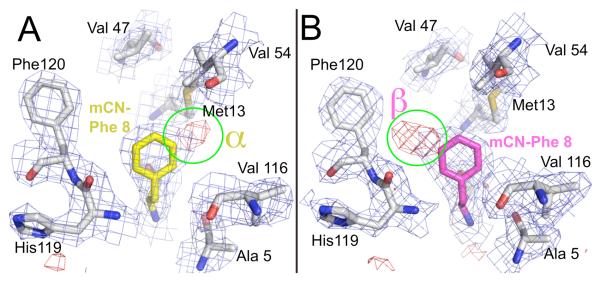
For mCN-RNase, two possible orientations of the nitrile with respect to the surrounding structure are possible for the same position of the phenyl ring. In order to avoid model bias during the refinement, the residue was modeled only as phenylalanine, revealing very clear regions of electron density for the nitrile missing in the Fo-Fc difference electron density map (see Fig 2). This resulted in a surprising observation: the map is consistent with a nitrile occupying only one of the two possible orientations in one of the molecules of the asymmetric unit and occupying strictly the other conformation in the second molecule. The conformer with the nitrile close to the sulfur of Met 13 will be referred to throughout as the α conformer (the one observed in the first molecule of the two molecule asymmetric unit, denoted ‘chain A’) and the one with the nitrile near the phenyl ring of Phe 120 as β (‘chain B’38).
Figure 2.
Electron density maps for mCN-RNase. (A) and (B) correspond to the chain names for the two monomers in the asymmetric unit in the PDB file. The experimental electron density map represented with wire mesh (2Fobs-Fcalc contoured at 1 sigma in blue mesh, 1Fobs-Fcalc contoured at 3 sigma in red mesh) for a model with the mCN-Phe residue modeled as Phe, showing missing density for the nitrile (see text). The names α and β are given to the conformer with the nitrile nearest Met13 and Phe120, respectively. (Two foreground residues removed from view for clarity).
FTIR spectroscopy of RNase Variants
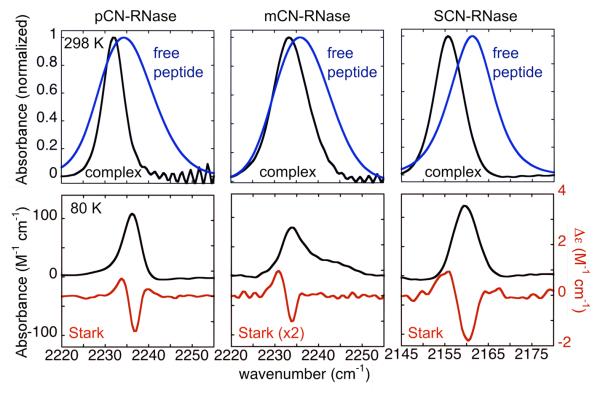
We measured the IR absorption spectra of each S-peptide free in solution and the RNase S complex by FTIR. In all three cases, substantially different peak positions and linewidths are observed for the free peptides in buffer vs. the folded RNase S complex at room temperature (Fig. 3, top panels). All three nitrile probes display a red-shift in their absorption frequency and a narrowing of the absorption linewidth upon sequestration from water into the protein interior. It has been well established that nitriles, when exposed to bulk water, accept a hydrogen bond3,39,40; based on the peak frequencies, linewidths and thermochromic behavior of these nitrile-modified peptides this same conclusion applies to the free peptides (see Supplemental Info. Part I for details). Hydrogen bonds to nitriles have the effect of blue-shifting the nitrile stretching frequency39,39, in opposition to the red-shift expected were this a strictly dipole-dipole interaction in the direction observed for aprotic dipolar solvents; this conclusion is based upon broad surveys of solvent induced frequency shifts25,40-43 and ab initio quantum chemical calculations40,44,45. As noted in the crystallography section above, in the RNase S complex, none of the nitriles are within hydrogen bonding distance to any potential donors, either from the solvent or protein, providing an explanation for the red-shift upon complex formation (Fig. 3): in each case a solvent-nitrile hydrogen bond is occluded upon sequestering the nitrile in the protein interior and is not replaced by a protein-nitrile hydrogen bond.
Figure 3.
FT-IR spectra of nitrile-modified RNase S and S-peptide. Top: normalized spectra of nitrile-modified S-peptide, free (blue) and bound to S-protein to form the RNase S complex (black) in 50% (vol/vol) glycerol in water, pH 7, 298 K. Bottom: Absorption spectra of the same set of RNase S complexes as above, taken at 80 K in black (left axis), and Stark spectra scaled to an applied field of 1 MV/cm in red (right axis), see Table 3.
Vibrational Stark spectroscopy
We calibrated the response of the nitrile in the three protein complexes to external electric fields by vibrational Stark spectroscopy, yielding the Stark tuning rate, or f•|Δμ⃗CN| where f is the local field correction factor (see Bublitz and Boxer46 for more detail). To date, the Stark tuning rates, |Δμ⃗CN|, for nitriles have (with only one exception2) been determined only for small model compounds in glass-forming solvents18-20, leaving open the extent to which the protein environment influences the tuning rate itself (this not to be confused with the influence of the electrostatic field of the protein on the frequency, via Eqn. 1). While it is desirable to recalibrate in situ, this will not be possible in most cases because the analysis requires high signal-to-noise data for the absorption both in the presence and absence of an external electric field, and protein samples cannot typically be prepared at the high concentrations of small-molecule experiments. The vibrational Stark measurement requires that the sample be frozen. Nearly all X-ray structures, including those reported here, are now obtained at cryogenic temperatures, comparable to those used for Stark spectroscopy, even though most functional data are obtained at or near room temperature. With RNase, where concentrations as high as 20 mM are accessible, we were able to measure the Stark tuning rates, demonstrating that the values in the protein and for small nitriles in a frozen glass of 2-methyl-tetrahydrofuran are indeed similar (Table 3). In both environments, the Stark effect is dominated by |Δμ⃗CN|, i.e. this is a linear Stark effect in which the Stark spectrum for an isotropic, immobilized sample is dominated by the second derivative of the absorption19,21 (see Stark spectra in Figure 3). This provides evidence that the parameters determined for model compounds are generally transferable, i.e. it is sufficient to calibrate small molecule probes and then use the Stark tuning rates thus determined to interpret properties of protein-bound probes.
Table 3.
Comparison of Stark tuning rates measured at 80 K for nitriles in RNase complexes in 50% glycerol/water (self buffered at pH 7) and for relevant model compounds in 2-methyl-tetrahydrofuran. Error reported as standard deviation over 3 measurements.
| Molecule | pCN-RNase | FMOC- pCN-Phea |
mCN-RNase | FMOC- mCN-Phea |
SCN-RNase | EtSCN |
|---|---|---|---|---|---|---|
|
f•|ΔμCN| [cm−1/(MV/cm)] |
0.59 ± 0.05 | 0.67 ± 0.01 | 0.60 ± 0.03b | 0.61 ± 0.01 | 0.62 ± 0.04c | 0.72 ± 0.03 |
FMOC stands for the fluorenylmethyloxycarbonyl group, attached to the amine
determined for the main (low energy) band only
reproduced from previous study54
Temperature Dependence of the IR Spectra
To better understand the origins of the widths and asymmetries of the room temperature lineshape and what relation if any they bear to the underlying electrostatic field distributions, we employed temperature modulation. Depending on the mechanism of line broadening, acquiring spectra at lower temperature often has the effect of narrowing the transition envelop; if a peak is composed of spectroscopically distinct, overlapping subpopulations, it is sometimes possible to resolve their separate contributions to the spectrum at low temperature.
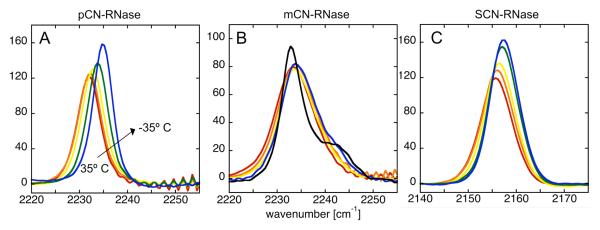
The temperature dependence of the nitrile stretching region of the absorption spectra of each probe-modified RNase variant in 50% glycerol/buffer47 was measured (Fig. 4) and contrasted with that of each free peptide in the same solution (Supplemental Info., Fig S3). For pCN-RNase and SCN-RNase the IR frequency increases with decreasing temperature over a range from 35 C to −35° C (negative thermochromism), and the line-width decreases; the spectral envelope is symmetric within experimental certainty at all temperatures studied. mCN-RNase, on the other hand, displays different behavior: a slightly asymmetric peak at room temperature evolves as the temperature is lowered into a peak with an increasingly distinct shoulder higher in energy than the main band. The main peak narrows substantially at low temperature, similar to the other two proteins, and both the peak and its shoulder shift with temperature. At cryogenic temperatures, the shoulder is very distinct. From fitting the main peak to a Gaussian of 5.2 cm−1 FWHM, the residuals show an asymmetric peak centered 8-9 cm−1 to the blue (see Supplemental Info., Fig S4). Thus, it appears that in this case one major contribution to the line-width comes from at least two spectroscopically distinct subpopulations. The X-ray crystal structure provides a plausible hypothesis for the nature of these distinct sub-states: they arise from the pair of possible orientations, α and β, with respect to the surrounding protein structure, of the nitrile substituent of the phenyl ring (see Fig. 2). A qualitative accounting for this spectroscopic splitting is suggested by inspecting the structures, where the β conformer places the nitrile near the partial positive charge of an aryl hydrogen of phenylalanine 120 (Fig. 2B) and the α conformer places it very near the partial negative charge of the sulfur of methionine 13 (Fig. 2A). As a control to insure that there is no appreciable fraction of unbound S-peptide, excess S-protein was added for each RNase variant, and no additional spectral changes were observed. For all three free peptides in glycerol/buffer, decreasing temperature leads to a monotonically increasing absorption frequency (negative thermochromism), but in contrast to the behavior in proteins, we observed a monotonically increasing linewidth (see Supplemental Info. for data and a model for these effects).
Figure 4.
Temperature dependence of the IR absorption spectra of nitrile-modified RNases in 50% glycerol/water, self-buffered at pH 7. Colors correspond to temperature: red, orange, yellow, green, blue, black (mCN-RNase only) equal, respectively, 23, 10, −5, −25, −35, −100° C.
Simulation of Stark shifts in the protein by electrostatics and molecular dynamics
The Stark model describes a quantitative relationship between electrostatic fields and environment-induced frequency shifts of a probe (Eqn. 1). Available methods for calculating the electric field in the environment of the probe (e.g. continuum electrostatics, molecular dynamics, hybrid QM/MD methods) involve approximations whose validity are subject to debate48; despite their widespread use, there are few experimental measures to directly test their theoretical underpinnings8. Having established the requisite structural and spectroscopic features of these nitrile-modified proteins, we now use the Stark effect as a uniquely discriminating metric to evaluate one very common electrostatic force model, that intrinsic to the Amber ′99 force field50.
The approach is as follows. Electric fields are calculated at every molecular dynamics step by summing the pair-wise electrostatic forces between a charged, mass-less, virtual particle located at the mid-point of the nitrile bond and all atoms of the protein and explicit water; the frequency shift at that instant due to this field is calculated according to Eqn. 1. To calculate absolute frequency, the shifts are added to the experimentally observed peak frequency of the appropriate model compound in n-hexane (see Methods for details). For each probe-modified protein, the probability distribution of the instantaneous frequency at every 2 fs step from multiple nanoseconds of MD sampling is calculated. To a first approximation, this distribution can be interpreted directly as a simulated absorption spectrum; a full treatment of the time-domain information in these trajectories and its use in more accurate simulations of line-shape is in progress.
The case of mCN-RNase requires additional attention due to the crystallographic observation of two conformations of the nitrile and the observed asymmetry at room temperature, which appears, based on low temperature spectra, to be due to a high-energy sub-population (see above). MD trajectories started in either the α or β conformation do not interchange in any trajectory. Thus for all mCN-RNase simulations separate trajectories are calculated for both conformers, and the results shown are for a combination of equal contributions from each distribution51.
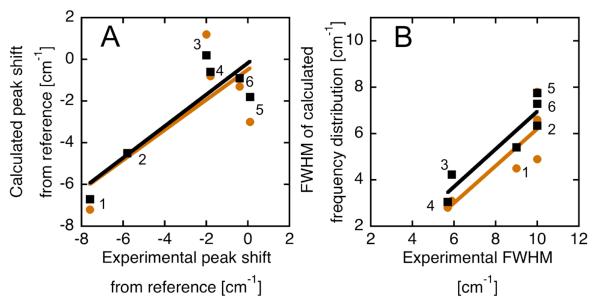
RNase A has four histidines, all of which are predominantly protonated at pH 4.5 and below and predominantly neutral at pH 8.0 and above according to direct titration by NMR52; His12 and His119 in the active site are in close proximity to the nitriles in all three RNase variants. Thus, collecting spectra at pH 4.5 and pH 8.0 and performing simulations with all histidines protonated or deprotonated respectively expands the set of observations and predictions to a total of six53 (data in Table 4). The agreement between measured and observed shifts, as represented by the root-mean-square (RMS) error for all six combinations of probe location and pH, is evaluated to test the electrostatic model of the Amber ′99 force field. A modest overall agreement is demonstrated by a RMS error of 2.0 cm−1 for conventional MD sampling that improves very slightly (to 1.5 cm−1) when employing Replica Exchange Molecular Dynamic sampling (REMD, Fig. 5 and Table 4; see Materials and Methods for details of MD). Examining the individual observations in more detail, the shift observed upon lowering the pH for SCN-RNase is the largest by far (− 1.8 cm−1) and the calculated shift (− 2.2 cm−1) comes close in value; however, for the much smaller shifts in pCN- and mCN-RNase (− 0.3 and + 0.5 cm−1, respectively), the predicted values are intermediate in magnitude and in the opposite direction from the observations (+ 0.7 and − 0.9 cm−1 respectively). Relative to the reference frequency in n-hexane, again the largest shift is for SCN-RNase, and the apparent correlation between measurement and simulation in Fig. 5 is dominated by the low and high pH observations of this construct; for pCN- and mCN-RNase the simulation gets the sign wrong on the low pH observations, but does recapitulate the observation that these two constructs, at either pH, are close in frequency to the reference states. Compared to the null-hypothesis, i.e. no shift due to electrostatic fields (Table 4, last column), the simulation does much better (RMS error of 4.1 vs 1.5 cm−1, respectively), though this is dominated by the observations for SCN-RNase. Note that the choice of reference is well justified by these results, since the average error is quite low (0.5 cm−1). Nonetheless, the RMS error of 1.5 cm−1 is comparable in magnitude to the observed shifts, and demonstrates that there is ample opportunity to improve the electrostatic model of this force field in order to achieve more quantitative agreement with experiment.
Table 4.
Comparison between measured and simulated frequency for six observations in RNase using different MD sampling strategies. Simulated shifts are calculated relative to the value for EtSCN, m-tolunitrile or p-tolunitrile in n-hexane as described in the text. The dielectric used in calculating the field at the nitrile was ε = 2. Results from either 3 ns of conventional MD sampling or 3 ns of Replica Exchange MD (REMD) sampling are shown.
| Experiment | MD | REMD | ||||||
|---|---|---|---|---|---|---|---|---|
| RNase- variant |
pH | peak [cm−1] |
FWHM [cm−1] |
peak [cm−1] |
FWHMb [cm−1] |
peak [cm−1] |
FWHMb [cm−1] |
Null shiftc [cm−1] |
| SCN | 4.5 | 2154.4 | 9.0 | 2154.8 | 4.5 | 2155.3 | 5.4 | 2162.0 |
| 8.0 | 2156.2 | 10 | 2157.5 | 4.9 | 2157.5 | 6.3 | ||
| pCN | 4.5 | 2231.2 | 5.9 | 2234.4 | 3.1 | 2233.4 | 4.2 | 2233.2 |
| 8.0 | 2231.5 | 5.7 | 2232.5 | 2.8 | 2232.7 | 3.1 | ||
| mCN | 4.5 | 2233.9 | 10 | 2230.8a | 7.8 a | 2232.0a | 7.8 a | 2233.8 |
| α only | 2234.4 | 8.2 | 2233.4 | 8.9 | ||||
| β only | 2230.3 | 5.6 | 2231.5 | 6.1 | ||||
| 8.0 | 2233.4 | 10 | 2232.5a | 6.6 a | 2232.9a | 7.3 a | ||
| α only | 2233.7 | 8.0 | 2233.2 | 8.5 | ||||
| β only | 2232.2 | 4.9 | 2232.9 | 5.6 | ||||
| avg. error |
0.3 | 0.5 | 2.9 | |||||
| RMS error |
2.0 | 1.5 | 4.1 | |||||
Calculated from a concatenated trajectory from both α and β conformers.
FWHM approximated as for a Gaussian distribution as where σ is the variance of the distribution of frequencies.
The null shift equals the reference frequency used and provides a comparison of how well the simulation does relative to no electrostatic perturbation.
Figure 5.
Comparison between measured and simulated frequency shifts and distributions for CN-modified RNase variants. (A) Measured shifts in the peak frequency for SCN-, mCN- and pCN-RNase at pH 4.5 and pH 8.0 vs. the peak of the distribution of frequencies from 3 ns of MD (orange circles) or 3 ns of REMD (black squares) calculated for these same six cases with a post-processing dielectric of 2 (see text); experimental and simulated shifts are calculated relative to the value for EtSCN, m-tolunitrile or p-tolunitrile in n-hexane respectively (see text). Numbering as follows (pH 4.5 and pH 8.0, respectively): SCN = 1, 2; pCN = 3, 4; mCN = 6, 5. Best-fit lines: slope = 0.76 or 0.72; R2 = 0.77 or 0.57 for REMD and MD respectively. (B) Widths of calculated frequency distributions versus experimental FWHM. Best-fit lines: slope = 0.81 or 0.80; R2 = 0.87 or 0.73 for REMD and MD respectively.
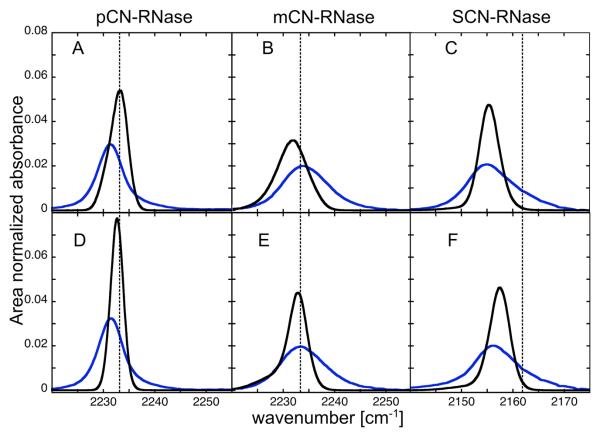
Besides the direction and magnitude of shifts, the accuracy of the model can be evaluated by examining the full width at half max (FWHM) of the calculated frequency distributions compared to the observed spectroscopic linewidths. The FWHM of the MD-derived frequency distributions are substantially narrower than the experimental FHWM of these IR transitions (Table 4, Figs. 5B and 6). An accurate treatment of the contributions of homogeneous broadening (which would reduce this discrepancy) and motional narrowing (which would increase it) cannot be made without knowledge of the vibrational excited state lifetime from time domain IR spectroscopy and these measurements are currently being pursued. However, if the time-domain effects are presumed to be similar, the relative experimental linewidths at the three probe sites can be compared to the calculated variances. Considering only the static properties, a too-narrow distribution could be the result of incomplete convergence of the simulated structural ensemble to the ensemble in the infinite time limit. For example, there is an average increase of 14% in the FWHM for 3 ns of REMD sampling relative to 3 ns of standard single replica MD, suggesting that REMD does a slightly better job at sampling the equilibrium ensemble, as expected. Experimentally, pCN-RNase has a markedly narrower FWHM than the other two constructs at both values of pH, and simulation recapitulates this relative difference; mCN- and SCN-RNase have similar spectroscopic FWHM to each other, and similar calculated FWHM as well (Table 4 and Fig. 5B). This provides a first hint that, at least at a qualitative level, the simulation captures the relative differences in the variances of electrostatic field distributions. Although this result bolsters the credibility of this electrostatic force model for MD simulation, the capability of the thoroughly characterized, nitrile-modified RNase variants to quantitatively evaluate the agreement with experiment will be more fully realized once the vibrational lifetime is known.
Figure 6.
Measured absorbance spectra (black) compared with calculated distributions of the instantaneous frequency (blue) both normalized to unit area with 0.2 cm−1 spacing between bins. (A-C) pH 4.5. (D-F) pH 8.0. The reference frequency for the simulations is based on, from left to right, p-tolunitrile, m-tolunitrile and EtSCN in n-hexane, and is shown by the dashed vertical line. A post-processing dielectric of 2 was employed (see text).
CONCLUSIONS
The X-ray crystal structures and Stark tuning rates for three nitrile-modified RNase S proteins were determined. To the best of our knowledge, these proteins constitute the first structures in the PDB to contain nitrile-modified amino acids, and join a very small number of other non-natural amino acid containing proteins in the database. No extrinsic chromophore can ever be a truly inert spectator, but the nitrile-modified enzymes studied here did not exhibit significantly altered catalytic or structural properties, consistent with this goal. Determination of the Stark tuning rate for these nitriles in the protein demonstrated the similarity of the Stark tuning rate determined using model compounds in organic glasses, justifying the latter, generally more practical approach of calibrating new probes. Through structure determination, we ruled out the possibility of hydrogen bond formation to the probe that would have otherwise complicated analysis of the electrostatic fields; furthermore, the orientation of the nitrile probes with respect to the protein makes possible the interpretation of the directional information contained in spectral shifts due to pH titration in the frame of the molecular axes of the protein.
Quantitative modeling of Stark shifts using fields derived from molecular dynamics trajectories successfully recapitulated the largest shifts (>2 cm−1 ) but the random error was still large relative to the experimentally observed shifts in this set. The systematic error was found to be low for absolute frequency determination by this method suggesting an accurate choice was made for the reference frequency. The simulated variance in the frequency was also in qualitative agreement with experimental line-widths. The accuracy and the specificity of the information available by calibrated Stark probes, such as these nitriles, provides a stringent test of the electrostatic models in an MD-simulation. The hypothesis has been put forward that MD electrostatic models will improve significantly with introduction of atomic polarizibility to the force fields55; as these methods become more widely implemented, the system studied herein provides the opportunity for a rigorous test of this hypothesis.
MATERIALS AND METHODS
Synthesis
Type I-A Ribonuclease (RNase) A and Type VIII Subtilisin Carlsburg were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. RNase S was prepared according to standard procedures56. Briefly: 40 μL of 1% Subtilisin was added drop-wise with stirring to 5 mL of 2% RNase A on ice, both solutions in 100 mM Tris pH 8.0. After 12-16 hrs on ice the reaction was quenched with 8-10 drops of 2 M HCl to pH < 4 and immediately purified by HPLC on a C-18 reverse phase column using a gradient of 20-80% acetonitrile in water with 0.1% trifluoroacetic acid (TFA) over 60 minutes. The fractions containing the S-protein and the S-peptide were identified by LCMS.
Probe modified S-peptide variants were synthesized on rink amide resin using an ABI (Foster City, CA) peptide synthesizer employing standard routines for FMOC-protected amino acids, utilizing HBTU/HOBt activation (so-called FastMOC protocol). Residues 16-20 do not contribute to binding or catalysis57; we therefore utilized a construct consisting of residues 1-15. FMOC-protected meta- and para-cyano-phenylalanine were obtained from PepTech Corp (Burlington, MA). FMOC-S-trityl-homo-cysteine was obtained from Anaspec (San Jose, CA). Cleavage from the resin was achieved with 95% TFA / 2.5 % triethylsilane / 2.5 % water (plus 10 molar equivalents ethanedithiol for the homo-cysteine-containing peptide) for 2 hrs, followed by precipitation into cold diethyl ether. Purification was performed by HPLC, on a C-18 reverse phase column using a gradient of 5-35% acetonitrile in water with 0.1% TFA over 40 minutes.
Cyanylation of S-peptide with homo-cysteine at position 13 was carried out as previously reported2,58. The purified peptide with a free thiol was dissolved to 20 mM in 1M ammonium carbonate pH 9.5 and 1.2 equivalents of dithionitrobenzoic acid (DTNB; Sigma-Aldrich, St. Louis, MO) were added. The reaction was followed by absorbance at 412 nm until completion (less than one minute), and immediately lyophilized. The lyophilized intermediate peptide was dissolved in water to 2 mM, reacted with 10 equivalents of potassium cyanide (KCN; Mallinckrodt, Hazelwood, MO), and followed by absorbance at 412 nm to completion (less than one minute).
1.5 molar equivalents of S-protein was added to each S-peptide in 20 mM Bis-Tris pH 6.0 followed by FPLC on a quaternary ammonium anion-exchange column held at 4° C employing a gradient in the same buffer, ramping from 50-500 mM NaCl over 30 min. Fractions containing the S-peptide, the S-protein and the RNase S complex were identified by LCMS. The RNase S complex was then exchanged into pure water using a 10 kDa molecular weight cut-off centrifugal concentrator and lyophilized. The lyophilized product was then dissolved in the desired buffer for further use.
Enzymology
Catalysis was measured by monitoring rates of hydrolysis of cyclic cytidine monophosphate (cCMP) as described previously59. Initial rates of hydrolysis under substrate-saturating conditions were measured by adding 0.5 molar equivalents of the native or probe-modified S-peptide to S-protein in 20 mM bis-Tris pH 6.0, and adding this complex to solutions of varying cCMP concentration and monitoring the changes at 286 nm using a Perkin Ellmer UV-Vis spectrophotometer (Waltham, MA). kcat and Km were determined using the Michaelis-Menten equation60.
X-ray crystallography
FPLC purified complexes were used for crystal growth. The hanging drop method was applied in a 24-well plate format (Hampton Research, Aliso Viejo, CA), screening small variations around the previously published conditions61: 25% polyethylene glycol 4000, 0.1 M sodium citrate pH 5.5, 0.08 M ammonium sulfate, mixing equal volumes of crystallization buffer and purified RNase S (30 mg/mL) in water. Single crystals were mounted on a plastic loop and frozen in liquid nitrogen. X-ray diffraction was performed at the Advanced Light Source (LBNL) and at the Stanford Synchrotron Radiation Laboratory.
FT-IR
IR spectra were obtained on a Bruker Vertex 70 FT-IR spectrometer (Billerica, MA) equipped with an InSb detector. A band-pass (2000-2500 cm−1) interference filter from Spectrogon (Parsippany, NJ) was used. A gas-tight demountable liquid cell with sapphire windows and offset spacers (one 75 μm, and one 100 μm spacer on either side) was used. Absorbance spectra were measured relative to a background taken with bovine serum albumin in water at equivalent concentration in g/mL. Baselines were calculated using a polynomial fit (4th to 6th order, depending on curvature) with roots defined at least 15 cm−1 distant from the peak maximum. Peak positions were determined using a second-derivative-based method built into the OPUS FT-IR software (Bruker Photonics, Billerica, MA). Absorption spectra as a function of temperature were obtained using a variable temperature, vacuum-jacketed cell from Specac (London, UK). Ten minutes of equilibration at each temperature was allowed before beginning each spectrum of 256 scans at 1 cm−1 resolution. Solvatochromism studies were performed with methylthiocyanate (Acros, Geel, Belgium) or benzonitrile (Aldrich, St. Louis, MO) at 1% by volume in each solvent. 128 scans were taken at 1 cm−1 resolution. No effort was made to make solvents anhydrous. Vibrational Stark experiments were performed as described previously19 using a home-built liquid cell consisting of a pair of 1 mm thick, 13 mm diameter sapphire windows (Meller Optics, Providence, RI) with 40 Å of nickel vacuum deposited on the surfaces facing the sample. The nickel electrodes were connected to a high voltage DC power supply (Trek Instruments Inc., Medina, NY), whose output voltage was synchronized to the FTIR scan timing with a home-built control unit. The windows were separated from each other by a pair of 26 micron thick Teflon spacers and held in place with a metal clamp. The sample was rapidly frozen by immersing the cell in a custom liquid nitrogen cryostat62 to form a glass. 512 scans with an applied field of 1 MV/cm were alternated with 512 scans with no applied field. Field-off scans were averaged and subtracted from the averaged field-on scans. The Stark tuning rate reported is the average of three separate experiments.
Molecular Dynamics
Molecular dynamics simulations were performed in Gromacs63,64 3.3.1 using the AMBER ′99 force-field ported to Gromacs by Sorin and Pande65. Nitrile-containing amino acids were parameterized using Antechamber and Leap from the Amber 9 software suite66, employing the GAFF atom force field extension67 to AMBER. Calculated dipole moments of BenzCN and MeSCN parameterized with this same process are within 3% of their experimental gas-phase value. For all three structures, simulations were performed either with all 4 histidines neutral (pH 8.0) or all histidines protonated (pH 4.5). Simulations were equilibrated (20 ps energy minimization, followed by 20 ps of heavy-atom position restrained refinement) and run using the Nose-Hoover thermostat and the Parinello-Raman barostat ( i.e. the NPT ensemble). The Particle Mesh Ewald model for the calculation of long-range electrostatics was employed. Both standard single replica MD and Replica Exchange Molecular Dynamics68,69 (REMD), as implemented in Gromacs 3.3.1, were employed. The latter has been devised as a means to more efficiently sample configurational space and involves the parallel simulation of multiple replicas of the protein plus solvent conducted at different temperatures (in this case 24 replicas from 298 to 332.5 K, ordered in temperature with a spacing of 1.5 K per replica); exchanges are attempted every 2 ps between adjacent replicas according to the Metropolis Criteria (probability of exchange = minimum{ 1; exp(−βΔE)} where β is the inverse of the Boltzman constant times temperature and ΔE is the energy for the higher minus the lower temperature replica). Electric fields were sampled only from the final 3 ns of the 4 ns trajectory. Only the 298 K trajectory of the REMD simulation was sampled.
Electric fields at the midpoint of the nitrile bond were extracted from every time point of the simulation by evaluating the force on a mass-less particle with an infinitesimal charge and no Lennard-Jones parameters, defined between the C and N atoms of the nitrile (introduced using the virtual site facility in Gromacs). Additional lines of code must be added to md.c in the Gromacs source code to extract these forces (see Supporting Info). The convention is used where the vector pointing from the C to the N of the nitrile is taken to be positive, yielding a negative value for the difference dipole moment9,20; thus fields pointing in the negative direction along this axis lead to red-shifts according to Eqn 1. The simulations undertaken here were performed using a dielectric constant ε = 1, however, the dielectric used to evaluate pair-wise forces on the dummy-atom can be varied in the post-processing of the trajectory, allowing independent assessment of this aspect of the model for the electrostatic forces. The best agreement between experiment and simulation was observed for a dielectric ε = 2 (see Supporting Info for details) and is therefore used in all calculations of frequency shifts herein.
To report the results on an absolute frequency scale a reference must be chosen. The gas-phase frequencies are known for model compounds but there are multiple features of the gas-phase to condensed-phase frequency shift that the Stark model cannot capture, notably, the pressure dependent repulsive forces due to closed-shell atomic overlap70. The peak frequencies of model nitriles in n-hexane reflect all these forces, while having a minimal contribution from dipolar electric fields71, and thus the instantaneous Stark shift was added to the observed frequency for a 0.5 mM solution in anhydrous n-hexane of ethylthiocyanate (2161.8 cm−1), meta-tolunitrile (2233.6 cm−1) or para-tolunitrile (2233.0 cm−1) for SCN-, mCN- and pCN-RNase, respectively (n-hexane and nitriles all from Acros [Geel, Belgium]).
Supplementary Material
ACKNOWLEDGMENTS
We thank Hadar Feinberg and Bill Weis for their generous assistance in acquiring X-ray crystallographic data and their help in solving the structures. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Dept. of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Dept. of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Some diffraction data were also measured at the Advanced Light Source, Lawrence Berkeley National Laboratory. We thank Dan Ensign for help with coding the modifications to Gromacs and Vijay Pande for helpful suggestions on calculation strategies. This work was supported in part by a grant from the NIH (GM27738).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting studies on small molecule nitriles, peak fitting to the cryogenic absorption spectrum of mCN-RNase, details on the choice of dielectric used in electrostatic field calculations, modifications to the Gromacs source code and figures S1-S5. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Watson MD, Gai XS, Gillies AT, Brewer SH, Fenlon EE. J. Phys. Chem. B. 2008;112:13188–13192. doi: 10.1021/jp8067238. [DOI] [PubMed] [Google Scholar]
- 2.Fafarman AT, Webb LJ, Chuang JI, Boxer SG. J. Am. Chem. Soc. 2006;128:13356–13357. doi: 10.1021/ja0650403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 4.Kirshenbaum K, Carrico IS, Tirrell DA. ChemBioChem. 2002;3:235–237. doi: 10.1002/1439-7633(20020301)3:2/3<235::AID-CBIC235>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J. Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 6.Fang C, Wang J, Kim YS, Charnley AK, Barber-Armstrong W, Smith AB, Decatur SM, Hochstrasser RM. J. Phys. Chem. B. 2004;108:10415–10427. [Google Scholar]
- 7.Thielges M, Case D, Romesberg F. J. Am. Chem. Soc. 2008;130:6597–6603. doi: 10.1021/ja0779607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suydam I, Snow C, Pande V, Boxer S. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]
- 9.Webb LJ, Boxer SG. Biochemistry. 2008;47:1588–1598. doi: 10.1021/bi701708u. [DOI] [PubMed] [Google Scholar]
- 10.Das K, Remorino A, Arnold E, Hochstrasser RM. Proc. Natl. Acad. Sci. U.S.A. 2008 [Google Scholar]
- 11.Sharp K, Honig B. Annu. Rev. Biophys. Bio. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 12.Perutz MF. Proc. Royal Soc. London B Bio. 1967;167:448–449. [Google Scholar]
- 13.Vernon CA. Proc. Royal Soc. London B Bio. 1967;167:389–395. doi: 10.1098/rspb.1967.0036. [DOI] [PubMed] [Google Scholar]
- 14.Warshel A. Proc. Natl. Acad. Sci. U.S.A. 1978;75:5250–5254. doi: 10.1073/pnas.75.11.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parson W, Creighton S, Warshel A. Biophys. J. 1988;53:A205. [Google Scholar]
- 16.Ripoll DR, Piela L, Váasquez M, Scheraga HA. Proteins: Structure, Function, and Genetics. 1991;10:188–198. doi: 10.1002/prot.340100304. [DOI] [PubMed] [Google Scholar]
- 17.Stigter D, Alonso DO, Dill KA. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4176–4180. doi: 10.1073/pnas.88.10.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suydam IT, Boxer SG. Biochemistry. 2003;42:12050–12055. doi: 10.1021/bi0352926. [DOI] [PubMed] [Google Scholar]
- 19.Andrews SS, Boxer SG. J. Phys. Chem. A. 2000;104:11853–11863. [Google Scholar]
- 20.Andrews SS, Boxer SG. J. Phys. Chem. A. 2002;106:469–477. [Google Scholar]
- 21.Chattopadhyay A, Boxer SG. J. Am. Chem. Soc. 1995;117:1449–1450. [Google Scholar]
- 22.Kwang-Im Oh J, Lee J, Han J, Lee H, Cho M. J. Chem. Phys. 2008 [Google Scholar]
- 23.Lindquist BA, Corcelli SA. J. Phys. Chem. B. 2008;112:6301–6303. doi: 10.1021/jp802039e. [DOI] [PubMed] [Google Scholar]
- 24.Lindquist BA, Furse KE, Corcelli SA. Phys. Chem. Chem. Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]
- 25.Aschaffenburg DJ, Moog RS. J. Phys. Chem. B. 2009;113:12736–12743. doi: 10.1021/jp905802a. [DOI] [PubMed] [Google Scholar]
- 26.McMahon HA, Alfieri KN, Clark KAA, Londergan CH. J. Phys. Chem. Lett. 2010;1:850–855. doi: 10.1021/jz1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bas DC, Rogers DM, Jensen J. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee S, Chowdhury P, DeGrado WF, Gai F. Langmuir. 2007;23:11174–11179. doi: 10.1021/la701686g. [DOI] [PubMed] [Google Scholar]
- 29.Maienschein-Cline MG, Londergan CH. J. Phys. Chem. A. 2007;111:10020–10025. doi: 10.1021/jp0761158. [DOI] [PubMed] [Google Scholar]
- 30.Sigala PA, Fafarman AT, Bogard PE, Boxer SG, Herschlag D. J. Am. Chem. Soc. 2007;129:12104–12105. doi: 10.1021/ja075605a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decatur SM, Boxer SG. Biochem. Biophys. Res. Commun. 1995;212:159–164. doi: 10.1006/bbrc.1995.1950. [DOI] [PubMed] [Google Scholar]
- 32.Fafarman AT, Sigala PA, Herschlag D, Boxer SG. In Preparation [Google Scholar]
- 33.Richards FM, Vithayathil PJ. J. Biol. Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 34.Goldberg JM, Baldwin RL. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2019–2024. doi: 10.1073/pnas.96.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lide DR. CRC Handbook of Chemistry and Physics, 90th Edition. 90th ed. CRC Press; 2009. [Google Scholar]
- 36.Axe DD, Foster NW, Fersht AR. Biochemistry. 1998;37:7157–7166. doi: 10.1021/bi9804028. [DOI] [PubMed] [Google Scholar]
- 37.Eftink MR, Biltonen RL. Biochemistry. 1983;22:5123–5134. doi: 10.1021/bi00291a011. [DOI] [PubMed] [Google Scholar]
- 38.To assess the maximum possible partial occupancy of β in chain A and α in chain B, the refinement was repeated with both possible orientations of the nitrile present as partial occupancy sites in both molecules of the asymmetric unit and the refinement program was allowed to fit their relative contributions as an adjustable parameter, starting at 50/50. The resultant refinement showed α favored strongly in chain A and β favored strongly in chain B. The dominant conformer after occupancy refinement was favored approximately 70/30 in chain A and 80/20 in chain B, but the resolution is not good enough to give these occupancies more than qualitative significance. By eye, there is no evidence for the minor conformer in either chain.
- 39.Eaton G, Pena-Nunez ES, Symons MCR. J. Chem. Soc. Faraday Trans. 1. 1988;84:2181–2193. [Google Scholar]
- 40.Riemers JR, Hall LE. J. Am. Chem. Soc. 1999;121:3730–3744. [Google Scholar]
- 41.Purcell KF, Drago RS. J. Am. Chem. Soc. 1966;88:919–924. [Google Scholar]
- 42.Fawcett WR, Liu G, Kessler TE. J. Phys. Chem. 1993;97:9293–9298. [Google Scholar]
- 43.Eaton G, Pena-Nunez AS, Symons MCR, Ferrario M, McDonald IR. Faraday Discuss. 1988;85:237–253. [Google Scholar]
- 44.Reimers JR, Zeng J, Hush NS. J. Phys. Chem. 1996;100:1498–1504. [Google Scholar]
- 45.Malaspina T, Fileti EE, Riveros JM, Canuto S. J. Phys. Chem. A. 2006;110:10303–10308. doi: 10.1021/jp062780p. [DOI] [PubMed] [Google Scholar]
- 46.Bublitz G, Boxer S. Annu. Rev. Phys. Chem. 1997;48:213–242. doi: 10.1146/annurev.physchem.48.1.213. [DOI] [PubMed] [Google Scholar]
- 47.The high glycerol content was necessary to explore low temperatures in the fluid state and to have an optical-quality glass at cryogenic temperatures. This concentration of glycerol causes a 0.5 cm− blue-shift in pCN-RNase and a 0.4 cm−1 red-shift in mCN-RNase relative to pure buffer, with no appreciable effect on the line-width in either case.
- 48.These include introducing dielectric boundaries, representing complex potential surfaces with simple atomic point charges, representing molecular polarizibility in common MD force fields with potentials of mean force, and treatment of the QM/MM boundary region. For a review see Warshel and Russel49.
- 49.Warshel A, Russel ST. Quart. Rev. Biophys. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 50.Wang JM, Cieplak P, Kollman PA. J Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 51.At early times in the trajectories, when the structures most resemble the X-ray structures, the α conformer begins several cm−1 higher in frequency than β, as expected based on proximity to the sulfur of Met13 (see text). At later times, the average structure drifts subtly from the starting conditions with the consequence that the two conformers have nearly the same frequency (within 1 cm−1) in REMD trajectories for pH 8 simulations. See Supplemental Info. for more discussion of this phenomenon.
- 52.Quirk DJ, Raines RT. Biophys. J. 1999;76:1571–1579. doi: 10.1016/S0006-3495(99)77316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.X-ray crystallography of these proteins was performed at pH 5.5, where both the neutral and ionized species are present to an appreciable extent at His 119 and His12; it is unlikely to matter structurally however, as X-ray structures of RNase as a function of pH have previously obtained54 been from pH 5 to 9 and the variations are quite small54 compared to the variations sampled by MD
- 54.Berisio R, Sica F, Lamzin V, Wilson K, Zagari A, Mazzarella L. Acta Crystallogr. D. 2002;58:441–450. doi: 10.1107/s0907444901021758. [DOI] [PubMed] [Google Scholar]
- 55.Schnieders MJ, Baker NA, Ren P, Ponder JW. J. Chem. Phys. 2007;126:124114. doi: 10.1063/1.2714528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doscher MS, Hirs CHW. Biochemistry. 1967;6:304–305. doi: 10.1021/bi00853a047. [DOI] [PubMed] [Google Scholar]
- 57.Goldberg J, Baldwin R. Biochemistry. 1998;37:2556–2563. doi: 10.1021/bi972403q. [DOI] [PubMed] [Google Scholar]
- 58.Doherty GM, Motherway R, Mayhew SG, Malthouse JPG. Biochemistry. 1992;31:7922–7930. doi: 10.1021/bi00149a025. [DOI] [PubMed] [Google Scholar]
- 59.Blackburn P. J. Biol. Chem. 1979;254:12484–12487. [PubMed] [Google Scholar]
- 60.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. 1st ed. W. H. Freeman; 1998. [Google Scholar]
- 61.Leonidas DD, Shapiro R, Irons LI, Russo N, Acharya KR. Biochemistry. 1997;36:5578–5588. doi: 10.1021/bi9700330. [DOI] [PubMed] [Google Scholar]
- 62.Andrews SS, Boxer SG. Rev. Sci. Instrum. 2000;71:3567–3569. [Google Scholar]
- 63.Berendsen HJC, van der Spoel D, van Drunen R. Comp. Phys. Comm. 1995;91:43–56. [Google Scholar]
- 64.Lindahl E, Hess B, van der Spoel D. J. Mol. Modeling. 2001;7:306–317. [Google Scholar]
- 65.Sorin EJ, Pande VS. Biophys. J. 2005;88:2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, DeBolt S, Ferguson D, Seibel G, Kollman P. Comp. Phys. Comm. 1995;91:1–41. [Google Scholar]
- 67.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. J Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 68.Sugita Y, Okamoto Y. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 69.Zhang W, Wu C, Duan Y. J. Chem. Phys. 2005;123:154105. doi: 10.1063/1.2056540. [DOI] [PubMed] [Google Scholar]
- 70.Ben-Amotz D, Lee M, Cho SY, List DJ. J. Chem. Phys. 1992;96:8781–8792. [Google Scholar]
- 71.We performed MD trajectories of MeSCN and BenzCN in a box of n atoms, evaluating the field at every time step and found that the average field was equal to −0.2 MV/cm, indeed negligibly small relative to the fields calculated below in the enzyme.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.