Abstract
Murine gammaherpesvirus 68 (MHV68 [also known as γHV-68]) is distinguished by its ability to replicate to high titers in cultured cells, making it an excellent candidate for studying gammaherpesvirus virion composition. Extracellular MHV68 virions were isolated, and abundant virion-associated proteins were identified by mass spectrometry. Five nucleocapsid protein homologues, the tegument protein homologue encoded by open reading frame (ORF) 75c, and envelope glycoproteins B and H were detected. In addition, gene products from MHV68 ORF20, ORF24, ORF28, ORF45, ORF48, and ORF52 were identified in association with virions, suggesting that these gammaherpesvirus genes are involved in the early phase of infection or virion assembly and egress.
The herpesvirus virion is composed of an icosahedral nucleocapsid surrounded by a proteinacious layer of tegument, which in turn is enclosed by a glycoprotein-containing lipid envelope (50). The structure and protein composition of the nucleocapsid have been shown to be conserved among the three subfamilies (α−, β−, and γ−) of herpesviruses (11, 14, 62-64, 72, 74). The icosahedral nucleocapsid contains at least four integral structural proteins (the major capsid protein, triplex-1 protein, triplex-2 protein, and small capsid protein) surrounding a core of viral DNA (11, 14, 27, 42, 56, 62, 72, 76). The other components of the virion, the envelope and the tegument in particular, are less well understood (38). The envelope contains viral glycoproteins critical for virion binding, entry, and signaling upon infection of a host cell (4, 15, 26, 34, 55, 67). The tegument is the electron-dense component of the virion surrounding the capsid and interacting with the envelope (14, 38, 75). While the tegument component of alphaherpesviruses and betaherpesviruses is known to contain a number of gene products involved in assembly and egress of infectious virus (38) or modulation of the host cell environment upon initial infection (10, 13, 21, 25, 30, 40), little is known about the protein composition of the gammaherpesvirus tegument nor about the functions of gammaherpesvirus tegument proteins immediately after infection of the cell.
Study of the functions of tegument proteins in the two human gammaherpesviruses, Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV), is hampered by the lack of cell culture systems capable of supporting productive replication of these viruses. However, murine gammaherpesvirus 68 (MHV68, or γHV-68) is not constrained in this manner, replicating to high titers in conventional tissue culture systems. MHV68 is a model for studying de novo gammaherpesvirus infection and pathogenesis (16, 20, 36, 66, 73). The virus is found in wild murid rodents and is capable of infecting laboratory strains of mice (8, 39, 48). MHV68 establishes productive infection in lung epithelia and a latent infection in splenocytes, macrophages, dendritic cells, and lung epithelial cells (23, 48, 57, 61, 69).
The MHV68 virion exhibits morphological similarity to the virion organization of other gammaherpesviruses (35, 48, 59). The viral genome encodes canonical capsid, tegument, and glycoprotein homologues found in gammaherpesviruses (66). The transcriptome of predicted MHV68 genes has been studied (3, 20, 36); however, the proteins encoded by most of these genes have not yet been identified in infected cells or in association with virions. In addition, the functional roles of conserved gammaherpesvirus virion proteins can be addressed by mutagenesis of the corresponding viral genes (2). These features make MHV68 an excellent model for studying gammaherpesvirus virion structure, composition, and assembly. However, these studies cannot proceed without a systematic identification of the viral proteins associated with virion particles. Therefore, we set out to identify and characterize proteins associated with the MHV68 virion.
Purification of extracellular MHV68.
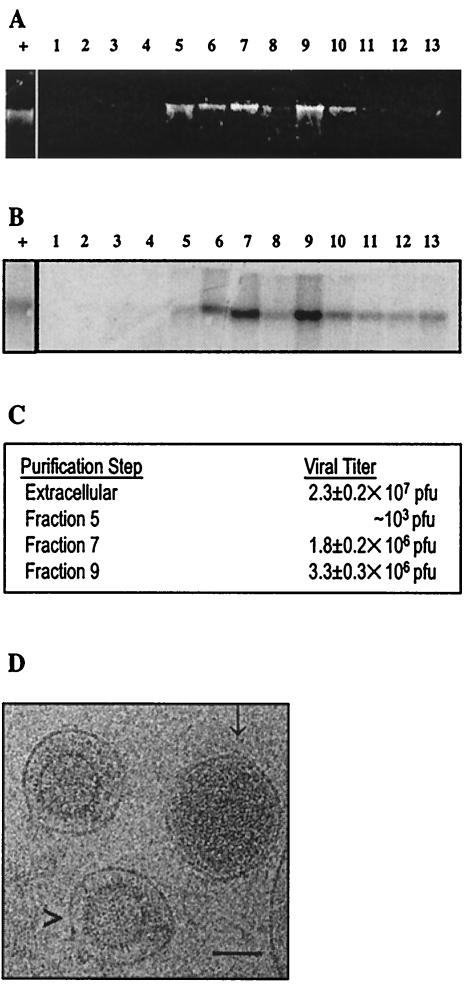
To obtain extracellular MHV-68 virions, 293T or NIH 3T3 cells were infected with wild-type MHV68 at a multiplicity of infection of 0.1. Supernatants were collected when cultures exhibited 90% cytopathic effect and were cleared of large cellular debris twice by centrifugation (1,000 × g, 15 min, 4°C). Extracellular virus was pelleted by ultracentrifugation through a 5% sucrose cushion (65,000 × g, 1 h, 4°C). The pellet was resuspended in 50 mM Tris (pH 7.5)-5 mM MnCl2 and digested with 0.03 U of DNase I (Invitrogen)/μl at 37°C for 30 min. Virus was then purified by 5 to 55% discontinuous sucrose density gradient ultracentrifugation (25,000 × g, 4.5 h, 4°C) in an SW41Ti rotor (Beckman). Thirteen fractions were isolated from top to bottom of the sucrose gradient. Three fractions (fractions 5, 7, and 9) contained visible bands of material. Nucleic acids extracted from the gradient fractions (Fig. 1A) were tested for viral DNA by Southern blotting with a PCR-generated probe to ORF67 in the MHV-68 genome (Fig. 1B). Fractions 5 through 13 contained elevated signals. Fractions 7 and 9 contained the highest concentrations of viral DNA and were examined for the presence of infectious virus by plaque assay (Fig. 1C). Extracellular virus input of 2.3 × 107 ± 0.2 × 107 PFU showed an approximately twofold loss of infectivity during purification. Fraction 7 contained on average 1.8 × 106 ± 0.2 × 106 PFU, fraction 9 contained 3.3 × 106 ± 0.3 × 106 PFU, while fraction 5 contained approximately 103 PFU of infectious virus. An aggregated pellet at the bottom of the ultracentrifuge tube contained 8.0 × 106 ± 2.4 × 106 PFU. These results are similar for virus isolated from both NIH 3T3-infected and 293T-infected cell media (data not shown). Infectivity was directly proportional to viral DNA content in fractions 5, 7, and 9. This indicates that extracellular virus is concentrated in fractions 7 and 9. Fractions 5, 7, and 9 were pelleted for further study of virus particle and protein content.
FIG. 1.
Purification of extracellular MHV68. (A) Nucleic acids extracted from MHV68 sucrose gradient fractions. Extracellular virus was purified by 5 to 55% sucrose density gradient ultracentrifugation. Numbers shown on top of the panel are fractions collected from top (1) to bottom (13) of the gradient. Nucleic acids were extracted and electrophoretically separated in a 0.75% agarose-Tris-acetate-EDTA gel. (B) Southern blot analysis of extracellular virus. DNA shown in panel A was transferred to a positively charged nylon membrane and probed with random-primed [α32P]dCTP-labeled virus-specific probe (a 760-bp PCR product of ORF67). + is intact viral genomic DNA. (C) Infectivity of sucrose gradient-purified virus. BHK cell monolayers infected with fractions 5, 7, or 9 were incubated in methylcellulose overlay medium at 37°C in 5% CO2 for 5 days, and numbers of PFU were calculated. Plaque assays were performed twice. (D) Electron cryomicrograph of MHV68 virions and enveloped capsid particles. MHV68 particles from fraction 9 were embedded in vitreous ice and recorded at 100 kV on a JEOL JEM1200 electron cryomicroscope at magnification ×30,000 at a dosage of 6 electrons/angstrom2. A representative image is shown, with putative virions (with DNA) indicated by an arrow (↓) and noninfectious enveloped particles (no DNA) indicated by an arrowhead (>). Bar, 100 nm.
MHV68 virion morphology.
Extracellular MHV68 particle morphology was studied by electron cryomicroscopy, which reveals the intact forms of the viral particles by transmission projection without staining or dehydration (14, 63, 75). Two predominant morphologies of particles are present in fractions 7 and 9 (Fig. 1D). Enveloped icosahedral capsids devoid of visible viral DNA and containing only a low-density tegument region (i.e., noninfectious enveloped particles) were present with denser enveloped, tegumented nucleocapsids with characteristic herpesvirus virion morphology (14, 50, 75), including the “fingerprint” pattern of close-packed double-stranded DNA (9, 74). While these particles were present in approximately equal ratios in fraction 7, fraction 9 contained predominantly virions. A small number of heterogeneous particles were also present, including naked capsids, which most likely resulted from the loss of the viral envelope from virions or noninfectious enveloped particles during purification. The existence of two or more kinds of enveloped extracellular particles has been documented for other herpesviruses, including human cytomegalovirus, herpes simplex virus type 1 (HSV-1), and pseudorabies virus (5, 29, 37).
Virion-associated fractions contain MHV68 virion proteins.
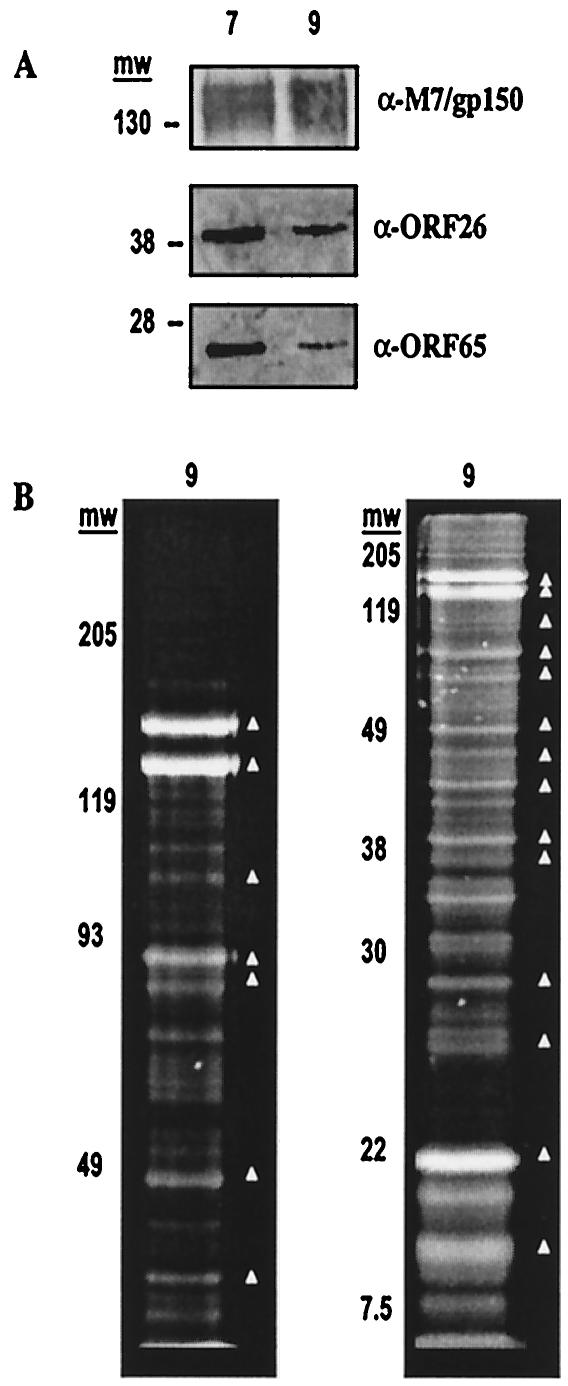
Proteins in fractions 7 and 9 were examined for the presence of MHV-68 structural protein homologues and envelope glycoproteins. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting, using polyclonal antisera raised in rabbits against bacterially expressed ORF26 (triplex-2/capsid protein homologue) (66), ORF65/M9 (small capsid protein homologue) (42, 66), or virion-associated glycoprotein-150, a product of the M7 gene (59). ORF26, ORF65, and gp150 antigens were found in both fractions (Fig. 2A) at molecular masses of 39, 26, and 130 to 150 kDa, respectively. The presence of MHV68 capsid antigens and virion-associated glycoprotein is in accordance with the electron cryomicroscopy observation revealing enveloped viral particles in both fractions 7 and 9 (Fig. 1D). In contrast, fraction 5 showed only minimal levels of the three antigens (data not shown). No signal was detected upon reprobing the blot with monoclonal antibody against actin (Sigma), excluding copurification of the abundant 42-kDa cellular form of this protein with the virion-associated fractions (not shown).
FIG. 2.
Fractions 7 and 9 contain virion antigens. (A) Western blot using polyclonal antiserum raised in rabbits against bacterially expressed and purified viral capsid proteins (ORF26, middle panel, and ORF65, lower panel) and envelope protein (gp150, upper panel). (B) Proteins from fraction 9 are separated on SDS-8% (left) or 15% (right) PAGE and stained with SYPRO-Ruby (Molecular Probes). Bands were excised, digested in sequence-grade modified trypsin, and analyzed by liquid chromatography with tandem mass spectrometry. Proteins matching the MHV68 proteome (▵) correspond to Table 2 from high to low apparent molecular weight (mw); cellular protein matches and unidentified bands are not marked.
Identification of abundant virion-associated proteins by mass spectrometry.
Abundant MHV68 proteins were further analyzed in the virion-associated fractions by mass spectrometry. Fraction 9, which contains high levels of viral DNA, virion antigens, and infectious virus, was selected for proteomic analysis. Bands containing proteins were excised individually from a denaturing SDS-polyacrylamide gel (Fig. 2B) and digested in-gel with sequence-grade modified trypsin (Promega), and peptides were extracted for analysis by micro-liquid chromatography with tandem mass spectrometry (LCMSMS) using an ion-trap mass spectrometer (LCQ-DECA; ThermoFinnegan, San Jose, Calif.) (54, 70). Fragment ionization was performed on abundant peptides in each sample. LCMSMS-generated peptide mass and sequence tag data were collected using Excalibur software and then matched to the predicted MHV68 proteome using the program Sequest (Table 1). Of 23 prominent protein bands excised from the gel (Fig. 2B), 14 contained peptides positively identifying proteins in the viral genome (Table 2). Proteins predicted to be structural components of the gammaherpesvirus capsid, tegument, and envelope as well as several putative novel virion-associated proteins were identified.
TABLE 1.
Peptide identification by mass spectrometrya
| Accession no. | Gene product | Predicted function | Size (aa) | Distinct peptides identifiedb | Accession no. | Gene product | Predicted function | Size (aa) | Distinct peptides identifiedb |
|---|---|---|---|---|---|---|---|---|---|
| NP_044848 | ORF8 | gB | 849 | 69-80/VCGVAATGETFR | NP_044885 | ORF48 | Unknown | 333 | 5-29/MDLETREGGGCQQVSILMVTSDREK |
| 188-198/PVDGLTGNIQR | 159-174/RPHTDTCVSLKTLQR | ||||||||
| 267-276/RADMRVREVK | 177-210/TAMLGNPETLSIWTLDDLVEDPVVFK | ||||||||
| 488-497/DTLMWYELSK | GYESAIRR | ||||||||
| 638-661/TVELYSSTERKLASSVFDIESMFR | 287-306/SHDQEFYTEAFKEDLHIGLK | ||||||||
| 307-332/RLVAMVLGQNHSWLDGFLTDTIVTGK | |||||||||
| NP_044858 | ORF20 | Fusion protein | 254 | 1-16/MNELGAKQLLNKLPKR | NP_044889 | ORF52 | Unknown | 135 | 5-19/KPDKTYEEMVKEVER |
| VTAHLIFK | 9-19/TYEEMVKEVER | ||||||||
| 111-144/VEGSNQLRDSAKALAVLAPVGTDPCR | 33-48/SSGAVSSDDSILTAAK | ||||||||
| NP_044860 | ORF22 | gH | 730 | 95-118/DSIFVTNDTIHIDSDSLFVCPVGR | 49-58/RESIIVSSSR | ||||
| 374-382/FLSGVQIER | 49-66/RESIIVSSRALGAVAMR | ||||||||
| 508-514/FDFSANK | 50-58/ESIIVSSSR | ||||||||
| 59-66/ALGAVAMR | |||||||||
| NP_044862 | ORF24 | Unknown | 717 | 104-133/IDTCTYVPVIYSFEQTDAHYDGMGPGKLR | 79-95/AVTEQELTSLLQSLTLR | ||||
| 253-262/LMSVCEIQLR | 96-120/VDVSMEETTVGASGGIGPSSQTETK | ||||||||
| 347-365/PSNGEMLMNLYRRIDYLPK | 96-121/VDVSMEETTVGASGGIGPSSQTETKK | ||||||||
| 680-688/NSNYLSFNK | |||||||||
| NP_044900 | ORF62 | Capsid | 380 | 49-66/LTRVVIAMDRYGGLLGGFLR | |||||
| NP_044863 | ORF25 | Major capsid | 1,373 | 185-203/QAPPTFILQSINDPSAGR | 154-162/LLTPISLTR | ||||
| 239-259/DYVLAVLSDAVTAVNSESVFK | 202-215/NLHEGLVFVGPLIK | ||||||||
| 325-332/NFDSFLSR | 231-246/GDTVLNESLSHGLVLK | ||||||||
| 374-382/VFAIESLQR | 247-266/LPVEQFMDFETTNTFHYTGR | ||||||||
| 738-752/VSFYIGDELYDNQER | 298-307/TPIEGPEFTR | ||||||||
| 1041-1054/TDELLTENILYSNR | 359-374/GGGLSLIEIPDFTVSR | ||||||||
| 1073-1101/ADAVGFDVSNEIASLDTAMGYSSTLIPAR | |||||||||
| NP_044903 | ORF65 | Small capsid | 186 | 8-18/APAFHPEPHNK | |||||
| NP_044864 | ORF26 | Capsid | 298 | 127-151/LDSNDVNLVFPSVVPAGLAQMGIQK | 30-51/DSIGKDPEEAPVPLLLHTCAVR | ||||
| 152-166/ILMYNLYSNLLAAER | 52-61/FYEEYKEKTR | ||||||||
| 62-71/DNTLPTLVNK | |||||||||
| NP_597855 | ORF28 | Unknown | 75 | 50-58/CFFWVMYKR | |||||
| 50-72/CFFWVMYKRAQIMGLPAQALLSR | NP_044915 | ORF75c | Tegument | 1,310 | 4-30/HFAFIYFGDSQYNETEKELIEDTEAGR | ||||
| 59-72/AQIMGLPAQALLSR | 21-30/ELIEDTEAGR | ||||||||
| 79-88/LPVTATPIDK | |||||||||
| NP_044866 | ORF29 | DNA package | 348 | 29-51/DANEKMVNVVSYVCSEHMEDFNK | 831-843/LTLAGTIFQQISK | ||||
| 300-339/NTKCIYHKNKTTTFQSKTHTMSDDVLIACV | 1011-1021/TDLGLMGPGMR | ||||||||
| MTCYVMTTNK | 1240-1248/NLEAAHYPR | ||||||||
| NP_044882 | ORF45 | IRF7-b. hom. | 206 | 1-9/MDPFKKPVR | |||||
| 1-14/MDPFKKPVRMLPIK | |||||||||
| 90-100/VSESSTSEDSD | |||||||||
| 160-177/ETQSDSSSDSSGNSHKKR | |||||||||
| 177-199/RRVQEEESSRILKTPAPISGNGK | |||||||||
| 190-199/TPAPISGNGK | |||||||||
| 190-206/TPAPISGNGKYNWPWLD |
Supplementary table of LCMSMS peptide matches: NCBI accession number, predicted MHV68 gene product, function, and size (amino acids [aa]). Distinct matching peptides were identified in mass spectrometry experiments using Sequest. More than 1,200 peptides were detected in experiments, the top 2.5% of which matched to the predicted MHV68 proteome with X-correlation factors, 1.4 < X < 6.3, under conditions of static modification of cysteine (+57 Da) and differential modification of methionine (+16 Da). All gene products reported include at least one match with an X value of > 1.4. Match data was curated using Excel.
Position/sequence.
TABLE 2.
Proteomic identification of MHV-68 virion-associated proteinsa
| Mass excised (kDa) | Accession no. | Gene product | Predicted function | Predicted mass (kDa) | No. of peptides | Most confident peptideb |
|---|---|---|---|---|---|---|
| 160 | NP_044863 | ORF25 | Major capsid protein | 153.2 | 7 | 738-752/VSFYIGDELYDNQER |
| 130 | NP_044915 | ORF75c | Tegument/FGARAT | 145.7 | 6 | 831-843/LTLAGTIFQQISK |
| 105 | NP_044860 | ORF22 | Glycoprotein H | 82.9 | 3 | 374-382/FLSGVQIER |
| 88 | NP_044848 | ORF8 | Glycoprotein B | 96.6 | 4 | 638-661/TVELYSSTERKLASSVFDIESMFR |
| 80 | NP_044866 | ORF29 | DNA packaging | 73.9 | 3 | 300-339/NTKCIYHKNKTITFQSKTHTMSDDVLIACVMTCYVMTTNK |
| 48 | NP_044882 | ORF45 | IRF7-binding homologue | 22.4 | 4 | 1-9/MDPFKKPVR |
| 45 | NP_044858 | ORF20 | Fusion protein | 28.3 | 2 | 111-144/VEGSNQLRDSAKALAVLAPVGTDPCRVTAHLIFK |
| 42 | NP_044900 | ORF62 | Triplex-1 (capsid) | 42.7 | 8 | 247-266/LPVEQFMDFETTNTFHYTGR |
| 39 | NP_044864 | ORF26 | Triplex-2 (capsid) | 33.4 | 2 | 152-166/ILMYNLYSNLLAAER |
| 39 | NP_044885 | ORF48 | Unknown | 37.9 | 6 | 307-332/RLVAMVLGQNHSWLDGFLTDTIVTGK |
| 38 | NP_044862 | ORF24 | Unknown | 82.9 | 4 | 104-133/IDTCTYVPVIYSFEQTDAHYDGMGPGKLR |
| 28 | NP_044882 | ORF45 | IRF7-binding homologue | 22.4 | 3 | 190-199/TPAPISGNGK |
| 26 | NP_044889 | ORF52 | Unknown | 14.8 | 2 | 79-95/AVTEQELTSLLQSLTLR |
| 26 | NP_044903 | ORF65 | Small capsid protein | 19.9 | 4 | 30-51/DSIGKDPEEAPVPLLLHTCAVR |
| 22 | NP_044889 | ORF52 | Unknown | 14.8 | 16 | 79-95/AVTEQELTSLLQSLTLR |
| 22 | NP_044903 | ORF65 | Small capsid protein | 19.9 | 4 | 30-51/DSIGKDPEEAPVPLLLHTCAVR |
| 15 | NP_597855 | ORF28 | Unknown | 8.5 | 3 | 59-72/AQIMGLPAQALLSR |
For each band excised (at approximately the molecular mass listed) and digested, peptide masses and sequence data are matched to the predicted MHV68 proteome using Sequest. Summary table of matches: NCBI accession number, predicted MHV68 gene product, function, and molecular mass. The number of matching peptides and sequence of best matching peptide to predicted gene product are shown.
Position/sequence.
Homologues of capsid proteins.
Five protein homologues to the capsid proteins of other gammaherpesviruses were identified (1, 42, 66), including ORF26 (triplex-2/capsid), ORF62 (triplex-1/DNA maturation/capsid), ORF25 (major capsid protein), ORF65/M9 (small capsid protein), and ORF29 (DNA packaging protein). Capsid proteins were detected in protein bands approximately corresponding to the predicted molecular masses of the polypeptides. For example, the major capsid protein encoded by ORF25 (predicted mass, 153.2 kDa) was found at approximately 160 kDa. Detection of peptides matching the major gammaherpesvirus capsid protein homologues by LCMSMS and detection of capsid proteins by Western blotting at similar molecular masses, confirmed for ORF26 (detected at 39 kDa) and ORF65 (detected at 26 kDa) in Fig. 2A, validated the efficacy of mass spectrometry for identifying other proteins associated with the MHV68 virion.
Tegument protein homologue.
One tegument protein homologue was identified in the virion-associated fraction. Peptides matching ORF75c, one of three KSHV ORF75/FGARAT homologues in the MHV68 genome (51, 66), were detected in a band close to the predicted mass of the full-length protein (145.7 kDa). ORF75c possibly represents the most abundantly packaged full-length ORF75 homologue in the MHV68 virion, since ORF75c is the most highly expressed of the three ORF75 homologues in MHV68 (20). A herpesvirus saimiri ORF75 homologue, the gene 75/EILF1 protein, is a virion protein (12).
Homologues of envelope proteins.
The MHV68 genome also encodes a number of genes highly homologous to conserved gammaherpesvirus glycoprotein genes thought to be associated with the virion envelope (1, 6, 41, 43, 58, 66). These include ORF8 (glycoprotein B) and ORF22 (glycoprotein H). A band excised from the SDS-polyacrylamide gel at approximately 88 kDa showed peptide matches to ORF8. KSHV gB is a virion envelope-associated protein (6) implicated in integrin-receptor-mediated signaling during virus entry (4). Previous study of MHV68 virions did not detect glycoprotein B as a virion-associated protein (58), although notably, the purification protocols used in this study and the previous study contain significant differences. Peptides matching ORF22 (glycoprotein H) were detected at a molecular mass of 105 kDa. Glycoprotein H is a virion-associated glycoprotein present in HSV-1 virions and is essential for infectivity (24, 26). The identification of putative MHV68 tegument and envelope homologues in the virion-associated fraction demonstrates that some proteins localized to these virion compartments exist in sufficient quantity to detect and identify by LCMSMS.
Novel virion-associated proteins.
Several predicted MHV68 gene products identified as virion-associated proteins have not been previously identified in the virions of gammaherpesviruses. These include ORF20, ORF24, ORF28, and ORF48 (Table 2). In addition, two unannotated proteins whose homologues have been suggested to be virion-associated proteins in other gammaherpesviruses, ORF45 (77) and ORF52 (53), were identified. ORF20, containing a predicted N-terminal domain homologous to HSV UL24 gene products (1), was detected at approximately 45 kDa by SDS-PAGE. Mutation of the HSV-1 UL24 gene impairs viral replication in tissue culture and in the mouse eye (31), and the HSV-2 UL24 protein is packaged into the HSV-2 virion (28). ORF24 encodes an uncharacterized gene product, detected in the virion-associated fraction at 38 kDa. ORF24 encodes a protein with a C-terminal domain with significant homology (36% identity) to the human cytomegalovirus (HCMV) UL87 protein family, whose function is unknown (1). ORF28 encodes a predicted 8.5-kDa gene product containing a transmembrane domain, which was detected at approximately 15 kDa in the virion-associated fraction. The ORF28 protein is uncharacterized. A transcript in the intergenic region between ORF27 and ORF29 of the MHV68 genome is reported to be expressed with late kinetics and is suggested to encode ORF28 (3). Peptides matching ORF45 were found at approximately 48 and 28 kDa in fraction 9 virions. The ORF45 gene is expressed as an early-late viral transcript, and the ORF45 protein is observed as a doublet at approximately 48 and 51 kDa in infected cells (32). Peptides at approximately 39 kDa matched to ORF48, encoding a 37.9-kDa polypeptide of unknown function. KSHV ORF48 is expressed as an immediate-early transcript (79), encoding a protein with 23% sequence identity to the predicted MHV68 ORF48 protein (66) and 19% identity to the hypothetical EBV BRRF2 protein (51). Peptides matching the primary gene product encoded by ORF52 were found at 26 and 22 kDa. ORF52 is a highly expressed late transcript suggested to encode a virion protein (3, 20). ORF52 encodes a protein with unknown function, homologous to KSHV ORF52 (28% identity) and EBV BLRF2 (40% identity) (66). BLRF2 putatively encodes the p21 protein component of the EBV viral capsid antigen complex and is detected in EBV virions at 21 and 23 kDa (53). The identification of predicted viral proteins associated with the MHV68 virion indicates that these proteins may be involved in virion morphogenesis, structure, or function during initial infection of the cell.
In order to examine the possibility of cellular proteins associating with MHV68 virion fractions, LCMSMS peptide data not positively identified by matching against the predicted MHV68 proteome was used to search a database of mammalian proteins using the program Sonar MS/MS (22). Six peptides matched five cellular protein sequences with expectationvalues (e) of <0.035, and in two cases they were consistent with work on other herpesviruses. One peptide from approximately 40 kDa matched annexin I, and one matched annexin II. Annexin II has been reported to be associated with purified HCMV particles and to bind glycoprotein B (46, 47, 71). Two peptides matching a cytoplasmic β-actin homologue (CAA27369) were identified at 28 kDa in fraction 9 virions. It has been suggested that an immunologically distinct form of cytoplasmic actin is packaged into the HCMV virion (7). However, similar to the case with the HCMV-associated actin, commercially available monoclonal antibody to cytoplasmic actin (Sigma) does not recognize a protein at 28 kDa in MHV68 virions (not shown). Two more cellular proteins were identified by one match apiece: the hypothetical protein similar to BR-1 (NP_062810) at 44 kDa and the endomembrane protein MP70 (NP_542123) at 40 kDa. Mass spectrometry data for the remaining bands was of insufficient quality for positive identification. This does not preclude the existence of other viral or cellular components of the MHV68 virion. The presence of host cell proteins associated with MHV68 virions may provide insight into the pathway of virion egress, though the functional roles of these proteins, if any, are speculative.
ORF45 is a virion-associated protein.
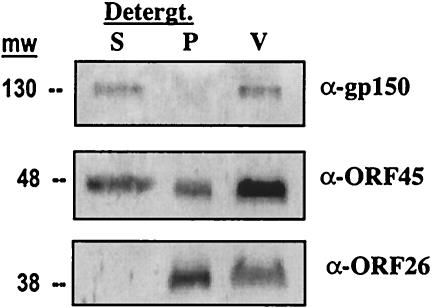
The gene product of MHV68 ORF45 was selected for further analysis as a putative virion-associated protein. We have previously identified MHV68 ORF45 as a gene important for viral replication. Inhibition of MHV68 ORF45 by RNA interference leads to a drastic reduction in the expression of lytic viral proteins and reduced production of virus progeny (32). We sought to study the association of ORF45 with MHV68 virions. Polyclonal antisera raised in rabbits against the full-length ORF45 gene product expressed in Escherichia coli detected a polypeptide in the virion-associated fractions at approximately 48 kDa on a Western blot (Fig. 3, lane V). This molecular weight range corresponds to one molecular weight (48,000) at which ORF45 is found by mass spectrometry analysis. Next, the sensitivity of virion-associated ORF45 to detergent was examined. Virions (2,000 PFU) from fraction 9 were treated with 2% Triton X-100, 0.01% SDS, and 22.5 mM EDTA at 37°C for 30 min, followed by 10 s of sonication and centrifugation (21,000 × g, 25 min, 23°C). Supernatant (detergent-sensitive) and pellet (detergent-resistant) phases were collected for analysis by Western blotting (Fig. 3, lanes S and P). The efficacy of this technique was demonstrated by solubilization of envelope protein (gp150) but not capsid protein (ORF26). ORF45 protein is partially solubilized, appearing in both detergent-sensitive and detergent-resistant phases. This observation can be reconciled by the hypothesis that an ORF45 gene product is packaged into the virion tegument. Partial sensitivity of the gene product to detergent implies that the ORF45 protein is bound to the capsid but less strongly than an integral capsid protein like ORF26. ORF45 is not as sensitive to detergent as envelope glycoprotein (gp150), which is almost completely removed by the detergent treatment. Thus, ORF45 may be associated with both the capsid and envelope as a component of the tegument. It has been recently suggested that KSHV ORF45 protein is a tegument protein (77). KSHV ORF45 protein has 33% sequence identity to MHV68 ORF45 protein and has been reported to interfere with interferon regulatory factor 7-mediated signaling (78), suggesting that the protein plays a role in modulation of innate immunity in infected cells. Experiments are under way to study the functional role of MHV68 ORF45 as a component of the virion.
FIG. 3.
ORF45 is a novel virion-associated protein partly resistant to detergent (Detergt.) treatment. Virions (lane V, approximately 2,000 PFU, fraction 9) were incubated with Triton X-100 (2%) and SDS (0.1%) for 30 min at 37°C and pelleted in a tabletop centrifuge at 23°C, 20,000 × g for 25 min. The supernatant (S) and pellet (P) were removed, denatured in Laemmli buffer, and separated on an SDS-15% polyacrylamide gel. Western blots were incubated with polyclonal antisera to recombinant viral proteins: glycoprotein 150 (upper panel), ORF45 (middle panel), and ORF26 (lower panel). mw, molecular weight.
The apparent molecular mass distribution and relative abundances of virion-associated polypeptides found in MHV68 (Fig. 2B) resembled that characterized in other gammaherpesviruses, including EBV (18, 19), herpesvirus saimiri (33), KSHV (42, 68), alcelaphine herpesviruses 1 and 2 (52), and murine herpesvirus 72 (49). The existence of glycoproteins with a high apparent mass and capsid proteins, particularly the major capsid protein, were observed in a number of these studies (6, 18, 19, 33, 42, 49, 68). However, the amino acid sequences of most of the virion proteins were not defined for these gammaherpesviruses. Using LCMSMS, we have identified a number of these proteins in MHV68, including homologues of capsid, tegument, and envelope proteins encoded in other gammaherpesvirus genomes (1, 6, 12, 42, 45, 51, 66). These include the structural components of the nucleocapsid identified in KSHV, the ORF29 packaging protein homologue, the putative tegument protein encoded by ORF75c, and glycoproteins B and H. We have also identified four proteins not previously predicted to be associated with gammaherpesvirus virions (ORF20, ORF24, ORF28, and ORF48), three of which have not been annotated. We also detected ORF52 protein, a homologue of a putative EBV virion protein of unknown function, and ORF45 protein in association with MHV68 virions. The identification of conserved MHV68 proteins previously not reported to be associated with the gammaherpesvirus virion, as well as cellular proteins, indicates possible functions of these proteins during the early stages of infection or in virion maturation and egress. In addition, the determinants of tissue tropism and the decision of whether to pursue a latent or lytic course of infection are not well understood. MHV68 is capable of infecting a number of tissues in the mouse, including lymphocytes, lung and intestinal epithelial cells, vascular endothelial cells, and several cell types in the brain (17, 44, 48, 60, 61, 65). Identification of virion components, particularly tegument and envelope proteins which may modulate the intracellular environment early in infection, is an essential step for understanding gammaherpesvirus pathogenesis in vivo. Determining the functional roles of these proteins in MHV68 infection awaits future studies.
Acknowledgments
We thank Ivo Atanasov and Rodrigo Aguilera for technical assistance in electron microscopy and mass spectrometry, respectively, and Tammy Rickabaugh-Zucker, Helen Brown, and Harumi Kasamatsu for helpful discussions.
This work was supported by NIH grants CA83525, CA91791, DE14153, and the STOP Cancer Foundation (R.S.); NIH grants AI12601 and AI29733 and DOE DE-FG03-01ER15253 (J.P.W.); NIH grants CA94809 and AI46420 (Z.H.Z); NIH grant CA90208, BBSRC (United Kingdom) grant 15/C12782, and a Royal Society (London) University Research Fellowship (J.P.S.). E.B. is supported by the Cellular and Molecular Biology Training Grant program at the University of California, Los Angeles (USPHS National Research Service Award GM07185).
REFERENCES
- 1.Albà, M. M., D. Lee, F. M. G. Pearl, A. J. Shepherd, N. Martin, C. A. Orengo, and P. Kellam. 2001. VIDA: a virus database system for the organisation of virus genome open reading frames. Nucleic Acids Res. 29:133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 5.Aleman, N., M. I. Quiroga, M. Lopez-Pena, S. Vazquez, F. H. Guerrero, and J. M. Nieto. 2003. L-particle production during primary replication of pseudorabies virus in the nasal mucosa of swine. J. Virol. 77:5657-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baghian, A., M. Luftig, J. B. Black, Y. X. Meng, C. P. Pau, T. Voss, P. E. Pellett, and K. G. Kousoulas. 2000. Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology 269:18-25. [DOI] [PubMed] [Google Scholar]
- 7.Baldick, C. L., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 9.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. USA 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher, S. J., J. Aitken, J. Mitchell, B. Gowen, and D. J. Dargan. 1998. Structure of the human cytomegalovirus B capsid by electron cryomicroscopy and image reconstruction. J. Struct. Biol. 124:70-76. [DOI] [PubMed] [Google Scholar]
- 12.Cameron, K. R., T. Stamminger, M. Craxton, W. Bodemer, R. W. Honess, and B. Fleckenstein. 1987. The 160,000-Mr virion protein encoded at the right end of the herpesvirus saimiri genome is homologous to the 140,000-Mr membrane antigen encoded at the left end of the Epstein-Barr virus genome. J. Virol. 61:2063-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castillo, J. P., and T. F. Kowalik. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19-34. [DOI] [PubMed] [Google Scholar]
- 14.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 15.Cheshenko, N., and B. C. Herold. 2002. Glycoprotein B plays a predominant role in mediating herpes simplex virus type 2 attachment and is required for entry and cell-to-cell spread. J. Gen. Virol. 83:2247-2255. [DOI] [PubMed] [Google Scholar]
- 16.Clambey, E. T., H. W. Virgin IV, and S. H. Speck. 2002. Characterization of a spontaneous 9.5-kilobase-deletion mutant of murine gammaherpesvirus 68 reveals tissue-specific genetic requirements for latency. J. Virol. 76:6532-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dal Canto, A. J., P. E. Swanson, A. K. O'Guin, S. H. Speck, and H. W. Virgin. 2001. IFN-gamma action in the media of the great elastic arteries, a novel immunoprivileged site. J. Clin. Investig. 107:R15-R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolyniuk, M., E. Wolff, and E. Kieff. 1976. Proteins of Epstein-Barr virus. II. Electrophoretic analysis of the polypeptides of the nucleocapsid and the glucosamine- and polysaccharide-containing components of enveloped virus. J. Virol. 18:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dolyniuk, M., R. Pritchett, and E. Kieff. 1976. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J. Virol. 17:935-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 21.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field, H. I., D. Fenyö, and R. C. Beavis. 2002. RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimizes protein identification, and archives data in a relational database. Proteomics 2:36-47. [PubMed] [Google Scholar]
- 23.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 24.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallina, A., L. Simoncini, S. Garbelli, E. Percivalle, G. Pedrali-Noy, K. S. Lee, R. L. Erikson, B. Plachter, G. Gerna, and G. Milanesi. 1999. Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J. Virol. 73:1468-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harman, A., H. Browne, and T. Minson. 2002. The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion. J. Virol. 76:10708-10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 28.Hong-Yan, Z., T. Murata, F. Goshima, H. Takakuwa, T. Koshizuka, Y. Yamauchi, and Y. Nishiyama. 2001. Identification and characterization of the UL24 gene product of herpes simplex virus type 2. Virus Genes 22:321-327. [DOI] [PubMed] [Google Scholar]
- 29.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 30.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 32.Jia, Q., and R. Sun. 2003. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 77:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keil, G., B. Fleckenstein, and W. Bodemer. 1983. Structural proteins of Herpesvirus saimiri. J. Virol. 47:463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomonte, P., P. Filee, J. R. Lyaku, M. Bublot, P. P. Pastoret, and E. Thiry. 1997. Glycoprotein B of bovine herpesvirus 4 is a major component of the virion, unlike that of two other gammaherpesviruses, Epstein-Barr virus and murine gammaherpesvirus 68. J. Virol. 71:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mackett, M., J. P. Stewart, S. Pepper, M. Chee, S. Efstathiou, A. A. Nash, and J. R. Arrand. 1997. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol. 78:1425-1433. [DOI] [PubMed] [Google Scholar]
- 37.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mistrikova, J., and D. Blaskovic. 1985. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 29:312-317. [PubMed] [Google Scholar]
- 40.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 41.Naranatt, P. P., S. M. Akula, and B. Chandran. 2002. Characterization of gamma2-human herpesvirus-8 glycoproteins gH and gL. Arch. Virol. 147:1349-1370. [DOI] [PubMed] [Google Scholar]
- 42.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oda, T., S. Imai, S. Chiba, and K. Takada. 2000. Epstein-Barr virus lacking glycoprotein gp85 cannot infect B cells and epithelial cells. Virology 276:52-58. [DOI] [PubMed] [Google Scholar]
- 44.Peacock, J. W., and K. L. Bost. 2000. Infection of intestinal epithelial cells and development of systemic disease following gastric instillation of murine gammaherpesvirus-68. J. Gen. Virol. 81:421-429. [DOI] [PubMed] [Google Scholar]
- 45.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietropaolo, R., and T. Compton. 1999. Interference with annexin II has no effect on entry of human cytomegalovirus into fibroblast cells. J. Gen. Virol. 80:1807-1816. [DOI] [PubMed] [Google Scholar]
- 47.Pietropaolo, R. L., and T. Compton. 1997. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J. Virol. 71:9803-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajcani, J., D. Blaskovic, J. Svobodova, F. Ciampor, D. Huckova, and D. Stanekova. 1985. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 29:51-60. [PubMed] [Google Scholar]
- 49.Reichel, M., J. Matis, M. Mistrikova, and J. Lesso. 1994. The analysis of polypeptides in the nuclei and cytoplasm of cells infected with murine herpesvirus 72. J. Gen. Virol. 75:1259-1265. [DOI] [PubMed] [Google Scholar]
- 50.Roizman, B., and P. E. Pellett. 2001. Herpesviridae: a brief introduction, p. 2381-2398. In B. N. Fields, D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Virology, 4th ed., vol. 2. Lippincott Williams and Wilkins Publishers, Philadelphia, Pa.
- 51.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seal, B. S., R. B. Klieforth, W. H. Welch, and W. P. Heuschele. 1989. Alcelaphine herpesvirus 1 and 2 SDS-PAGE analysis of virion polypeptides, restriction endonuclease analysis of genomic DNA and virus replication restriction in different cell types. Arch. Virol. 106:301-320. [DOI] [PubMed] [Google Scholar]
- 53.Serio, T. R., A. Angeloni, J. L. Kolman, L. Gradoville, R. Sun, D. A. Katz, W. Van Grunsven, J. Middeldorp, and G. Miller. 1996. Two 21-kilodalton components of the Epstein-Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21). J. Virol. 70:8047-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:50-58. [DOI] [PubMed] [Google Scholar]
- 55.Speck, P., K. M. Haan, and R. Longnecker. 2000. Epstein-Barr virus entry into cells. Virology 277:1-5. [DOI] [PubMed] [Google Scholar]
- 56.Spencer, J. V., B. L. Trus, F. P. Booy, A. C. Steven, W. W. Newcomb, and J. C. Brown. 1997. Structure of the herpes simplex virus capsid: peptide A862-H880 of the major capsid protein is displayed on the rim of the capsomer protrusions. Virology 228:229-235. [DOI] [PubMed] [Google Scholar]
- 57.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart, J. P., N. J. Janjua, N. P. Sunil-Chandra, A. A. Nash, and J. R. Arrand. 1994. Characterization of murine gammaherpesvirus 68 glycoprotein B (gB) homolog: similarity to Epstein-Barr virus gB (gp110). J. Virol. 68:6496-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart, J. P., N. J. Janjua, S. D. Pepper, G. Bennion, M. Mackett, T. Allen, A. A. Nash, and J. R. Arrand. 1996. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J. Virol. 70:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 61.Terry, L. A., J. P. Stewart, A. A. Nash, and J. K. Fazakerley. 2000. Murine gammaherpesvirus-68 infection of and persistence in the central nervous system. J. Gen. Virol. 81:2635-2643. [DOI] [PubMed] [Google Scholar]
- 62.Trus, B. L., J. B. Heymann, K. Nealon, N. Cheng, W. W. Newcomb, J. C. Brown, D. H. Kedes, and A. C. Steven. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trus, B. L., W. W. Newcomb, F. P. Booy, J. C. Brown, and A. C. Steven. 1992. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simples virus capsid. Proc. Natl. Acad. Sci. USA 89:11508-11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Berkel, V., B. Levine, S. B. Kapadia, J. E. Goldman, S. H. Speck, and H. W. Virgin IV. 2002. Critical role for a high-affinity chemokine-binding protein in gamma-herpesvirus-induced lethal meningitis. J. Clin. Investig. 109:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, F. Z., S. M. Akula, N. Sharma-Walia, L. Zeng, and B. Chandran. 2003. Human herpesvirus 8 envelope glycoprotein B mediates cell adhesion via its RGD sequence. J. Virol. 77:3131-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, Y.-C. J., Q. Zhang, and E. A. Montalvo. 1998. Purification of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) and analyses of the structural proteins. J. Virol. Methods 73:219-228. [DOI] [PubMed] [Google Scholar]
- 69.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitelegge, J. P., S. M. Gómez, R. Aguilera, R. W. Roberson, W. F. Vermaas, T. R. Crother, C. I. Champion, J. E. Nally, D. R. Blanco, M. A. Lovett, J. N. Miller, and K. F. Faull. 2002. Identification of proteins and intact mass measurements in proteomics. Appl. Genomics Proteomics 1:85-94. [Google Scholar]
- 71.Wright, J. F., A. Kurosky, E. L. G. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu, L., P. Lo, X. Yu, J. K. Stoops, B. Forghani, and Z. H. Zhou. 2000. Three-dimensional structure of the human herpesvirus 8 capsid. J. Virol. 74:9646-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou, Z. H., B. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in the herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]
- 75.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 77.Zhu, F. X., and Y. Yuan. 2003. The ORF45 protein of Kaposi's sarcoma-associated herpesvirus is associated with purified virions. J. Virol. 277:4221-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 99:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]