Abstract
Escherichia coli mRNA translation is facilitated by sequences upstream and downstream of the initiation codon, called Shine–Dalgarno (SD) and downstream box (DB) sequences, respectively. In E.coli enhancing the complementarity between the DB sequences and the 16S rRNA penultimate stem resulted in increased protein accumulation without a significant affect on mRNA stability. The objective of this study was to test whether enhancing the complementarity of plastid mRNAs downstream of the AUG (downstream sequence or DS) with the 16S rRNA penultimate stem (anti-DS or ADS region) enhances protein accumulation. The test system was the tobacco plastid rRNA operon promoter fused with the E.coli phage T7 gene 10 (T7g10) 5′-untranslated region (5′-UTR) and DB region. Translation efficiency was tested by measuring neomycin phosphotransferase (NPTII) accumulation in tobacco chloroplasts. We report here that the phage T7g10 5′-UTR and DB region promotes accumulation of NPTII up to ∼16% of total soluble leaf protein (TSP). Enhanced mRNA stability and an improved NPTII yield (∼23% of TSP) was obtained from a construct in which the T7g10 5′-UTR was linked with the NPTII coding region via a NheI site. However, replacing the T7g10 DB region with the plastid DS sequence reduced NPTII and mRNA levels to 0.16 and 28%, respectively. Reduced NPTII accumulation is in part due to accelerated mRNA turnover.
INTRODUCTION
In Escherichia coli translation is facilitated by mRNA–rRNA interactions between the Shine–Dalgarno (SD) sequence upstream of the AUG translation initiation codon and the anti-Shine–Dalgarno sequence (ASD) at the 3′-end of the small (16S) rRNA. A second mRNA element is the downstream box (DB). DB is defined as a 15 nt sequence downstream of the AUG translational initiation codon complementary to the penultimate stem of the 16S rRNA or the anti-downstream box (ADB) (nt 1469–1483) (1,2). In E.coli increasing complementarity of the DB sequence to the 16S rRNA ADB region resulted in a significant increase in protein accumulation (3,4).
In higher plant plastids mRNA sequences in the 5′-untranslated region (5′-UTR) were shown to be important for translation. These sequences are complementary to the 16S rRNA 3′-end and may be SD-like (GGA) (5) or distinct from SD, such as RBS1 (AAG) and RBS2 (UGAUGAU) (6). Signals for light-dependent mRNA translation (7,8) and mRNA stability (9) are also localized in the 5′-UTR. The sequence context around the translation initiation codon is conserved (10) and is probably important for efficient translation. Silent mutations downstream of the rbcL AUG dramatically reduced (35-fold) translation efficiency without an effect on mRNA stability (11). Since both the 5′-UTR and sequences downstream of AUG affect protein accumulation they will collectively be referred to as the translation control region (TCR). Sequences enhancing translation downstream of the AUG in plastids have been termed downstream sequences (DS) to distinguish them from the E.coli DB sequence (11). The role of the 5′-UTR in regulating translation and mRNA stability is best characterized in the chloroplasts of the unicellular alga Chlamydomonas (12–14). General rules deduced from the Chlamydomonas studies may accelerate the understanding of translational regulation in higher plants.
To explore the feasibility of using heterologous translation control signals in combination with plastid promoters we have fused the strong σ70-type plastid rRNA operon promoter (Prrn) with the T7 phage gene 10 (T7g10) TCR. The T7g10 TCR was shown to promote high level protein accumulation in bacteria (15) and has a well-characterized DB sequence (2). Specifically, we tested if increasing complementarity between the plastid 16S rRNA penultimate stem (ADS) and sequences downstream of the AUG (DS sequence) facilitates enhanced neomycin phosphotransferase (NPTII) accumulation.
We report here that the phage T7g10 TCR promotes accumulation of NPTII up to ∼16% of total soluble leaf protein (TSP). Thus, the heterologous phage TCR, when fused to a strong plastid promoter, is suitable for high level expression of recombinant proteins in chloroplasts. Enhanced mRNA stability and an improved NPTII yield (∼23% of TSP) was obtained from a construct in which the T7g10 5′-UTR was linked with the NPTII coding region via a NheI site. However, replacing the T7g10 DB region with the cognate plastid DS sequence dramatically reduced NPTII accumulation (to ∼1% of the T7g10+DB/Ec control) and accelerated mRNA turnover.
MATERIALS AND METHODS
Plasmid construction
Prrn is the σ70-type promoter of the plastid rRNA operon (16,17). The chimeric Prrn promoters are contained in SacI–NheI fragments in pBSIIKS+ derivatives. The PrrnLT7g10+DB/Ec cassette (Prrn promoter with the T7g10 leader and E.coli DB) is carried by plasmid pHK18. The PrrnLT7g10+DS/pt cassette (Prrn promoter with the T7g10 leader and plastid DS) is carried by plasmid pHK19. The PrrnLT7g10 cassette (Prrn promoter with the T7g10 leader and NheI site) is carried by plasmid pHK20. The promoter fragments were constructed by PCR. Construction details are available upon request.
In the pHK plasmids listed above the neo coding region from plasmid pSC1 (18) is expressed in a cassette consisting of the Prrn promoter fragment and TrbcL (the plastid rbcL gene 3′-UTR) (19). The Prrn promoter derivative and the neo coding region are linked via the engineered NheI site. Plastid vectors pHK38, pHK39 and pHK40 were obtained by cloning the neo gene from plasmids pHK18, pHK19 and pHK20 as SacI–HindIII fragments into plastid vector pPRV111A (20).
Plastid transformation and regeneration of transgenic plants
Transforming DNA (Plasmid Maxi Kit; Qiagen) was introduced into leaf chloroplasts on the surface of tungsten particles (1 µm) using the Du Pont PDS1000He Biolistic gun. Transplastomic plants were selected on RMOP medium containing 500 mg/l spectinomycin dihydrochloride. A uniform population of transformed plastid genome copies was confirmed by DNA gel blot analysis (17). The transgenic plants were grown on MS (Murashige–Skoog) medium (21) containing 3% sucrose and 0.6% agar under sterile culture conditions.
RNA gel blot analysis
Total cellular RNA was prepared from the leaves of plants grown in sterile culture (22). The RNA (4 µg/lane) was electrophoresed on 1.2% agarose/formaldehyde gels and then transferred to Hybond N membranes (Amersham) using the PosiBlot Transfer apparatus (Stratagene). Hybridization to the probe was carried out in Rapid Hybridization Buffer (Amersham) overnight at 65°C. Double-stranded DNA probe was prepared by random primed 32P-labeling. The template for probing neo was a gel purified NheI–XbaI fragment excised from plasmid pHK30. The template for probing the tobacco cytoplasmic 25S rRNA was a PCR fragment amplified from total tobacco cellular DNA with primers 5′-TCACCTGCCGAATCAACTAGC-3′ and 5′-GACTTCCCTTGCCTACATTG-3′. RNA hybridization signals were quantified using a Molecular Dynamics PhosphorImager and normalized to the 25S rRNA signal.
SDS–PAGE and immunoblot analysis
Leaves for protein extraction were taken from plants grown in sterile culture. To obtain TSP, leaves (200 mg) from plants grown in sterile culture were homogenized in 1 ml buffer containing 50 mM HEPES–KOH pH 7.5, 10 mM potassium acetate, 5 mM magnesium acetate, 1 mM EDTA, 1 mM dithiothreitol and 2 mM phenylmethylsulfonyl flouride. The homogenate was centrifuged to remove the insoluble material. Protein concentrations were determined with a Bio-Rad Protein Assay reagent kit (Bradford). The proteins were separated by SDS–PAGE (15% acrylamide, 6 M urea) (23), followed by either staining with Coomassie Brilliant Blue R250 or immunoblotting. For immunoblot analysis protein was transferred onto nitrocellulose membranes using a semi-dry transfer apparatus (Bio-Rad). After blocking, the membrane was incubated with 4000-fold diluted polyclonal rabbit antiserum raised against NPTII (5Prime→3Prime Inc.). 20 000-fold diluted HRP-conjugated secondary antibody and ECL chemiluminescence reagent (Amersham) were used for immunoblot detection on X-ray film (Kodak). NPTII was quantified on the immunoblots by densitometric analysis with the DensoSpot program of Alpha Imager 2000 (Alpha Innotech) by comparison of the experimental samples with a dilution series of commercial NPTII (5Prime→3Prime Inc.).
RESULTS
Experimental design
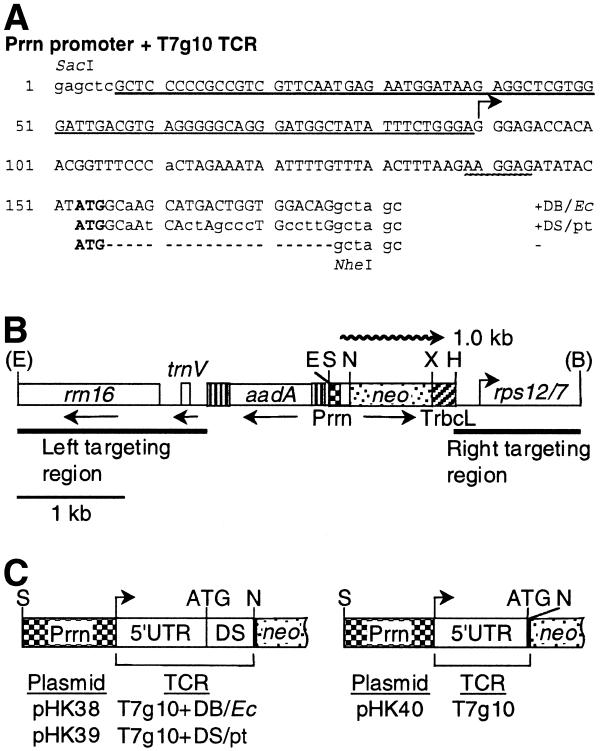
In E.coli the DB was defined as a 15 nt sequence downstream of the AUG translational initiation codon complementary to the penultimate stem (ADB region) of the 16S rRNA. The penultimate stem–loop structure of the 16S rRNA is conserved between E.coli and plastids, although the actual mRNA sequence is different (Fig. 1; 2,24). The chimeric genes have the same promoter (Prrn), 5′-UTR (T7g10), coding region (neo) and 3′-UTR (TrbcL) and differ only with respect to the neo coding region N-terminus. The design of the three chimeric promoter/TCR constructs is as follows. In plasmid pHK38 the Prrn derivative has the phage T7g10 5′-UTR and DB (Figs 1B and 2A). In plasmid pHK39 the Prrn promoter derivative has the phage T7g10 5′-UTR and the plastid DS sequence. The plastid DS sequence is defined as the sequence complementary to the tobacco plastid 16S rRNA penultimate stem corresponding to the E.coli ADB region (ADS sequence; Figs 1B and 2A). In plasmid pHK40 the Prrn promoter is fused with the phage T7g10 5′-UTR and the leader is linked to the neo coding region via a NheI site (Figs 1B and 2A).
Figure 1.
The E.coli mRNA DB and plastid mRNA DS regions. (A) The 15 nt ADB regions in the E.coli (Ec) 16S rRNA (nt 1469–1483) (2,45) and the cognate DS region in the tobacco plastid (pt) 16S rRNA (nt 1416 and 1430) (46). (B) Complementarity of the mRNA DB and DS regions with the plastid 16S rRNA penultimate stem ADS region. Watson–Crick (lines) and G-U (filled circles) pairs are marked. Nucleotides corresponding to the NheI site and neo coding region are in lower case. The numbers of potential pairs formed with the 15 nt DS region are in parentheses.
Figure 2.
Plastid vectors to compare translation efficiency from neo mRNAs with the E.coli DB and plastid DS regions. (A) DNA sequence of Prrn promoters with T7g10 leader derivatives. The Prrn promoter region is underlined; the transcription initiation site is marked by a horizontal arrow; the translation initiation codon (ATG) is in bold; the SD sequence is underlined with a wavy line. The T7g10 DB (Ec) sequence is shown on top; the sequence with the plastid DS region (pt) is duplicated in the middle; the sequence with the NheI site is at the bottom. Plastid and T7g10 sequences are in capital letters; nucleotides added or modified during construction are in lowercase; gaps are marked with dashes. (B) The plastid targeting region of the transformation vectors. The spectinomycin resistance (aadA) and neo genes and plastid genes rrn16, trnV and rps12/7 (46) are shown. The positions of the neo promoter (Prrn) and 3′-UTR (TrbcL) are also marked. The wavy line represents 1.0 kb neo mRNA. Abbreviations for restriction sites: E, EcoRI; S, SacI; N, NheI; X, XbaI; H, HindIII; B, BglII. Restriction sites removed during plasmid construction are in parentheses. (C) Listing of plasmids and the schematic map of their promoter and N-terminal coding regions. For an explanation see (B).
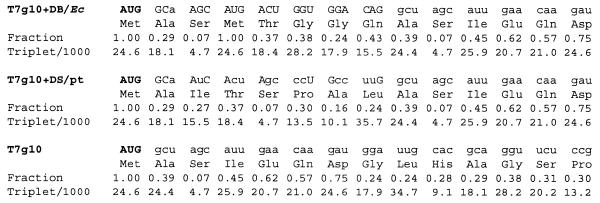
Replacement of frequently used codons with rare codons (1.4/2.1 per 1000 codons) in E.coli mRNAs results in reduced protein accumulation (25,26). To avoid the potential adverse effects of introducing rare codons into the neo coding region, codon usage frequencies were calculated for the chimeric genes. None of the changes made involved replacing a frequently used codon with a codon that would be considered rare by the E.coli criteria (≤4.6 per 1000 codons; Fig. 3). Codon usage frequencies shown in Figure 3 were calculated for the highly expressed photosynthetic genes rbcL, psaA, psaB, psaC, psbA, psbB, psbC, psbD, psbE and psbF (27).
Figure 3.
Codon frequency in the neo coding region N-terminus. Values are shown for the Prrn derivatives T7g10+DB/Ec (plasmid pHK38), T7g10+DS/pt (plasmid pHK39) and T7g10 (plasmid pHK40). Fraction is the relative usage frequency of a specific codon compared to the usage frequency of all codons encoding the same amino acid. Triplet/1000 is the frequency of occurrence of a certain triplet per 1000 triplets.
In the plastid transformation vectors the neo genes are physically linked with the selectable marker aadA and are flanked by plastid DNA to target insertion of aadA and neo into the trnV/rps12/7 intergenic region (Fig. 2B). A schematic map of the promoter regions is depicted in Figure 2C. The neo genes were introduced into the tobacco plastid genome by standard protocols and the leaves of transplastomic tobacco plants were tested for NPTII accumulation.
NPTII accumulation in plastids
NPTII was quantified by immunoblot analysis or staining of gels with Coomassie Brilliant Blue (Fig. 4A and B). The data are summarized in Table 1. Surprisingly, NPTII from the heterologous T7g10 5′-UTR (DB/Ec, Nt-pHK38) and the T7g10 leader combined with the NheI site (Nt-pHK40) accumulated to very high levels, ∼16 and ∼23% of TSP, respectively. These expression levels are comparable to the most highly expressed chloroplast proteins such as the Rubisco large and small subunits (Fig. 4B). This result indicates that highly efficient translation of NPTII from the neo mRNA occurred with the heterologous T7g10 5′-UTR combined with the T7g10 DB (DB/Ec) and the NheI site.
Figure 4.
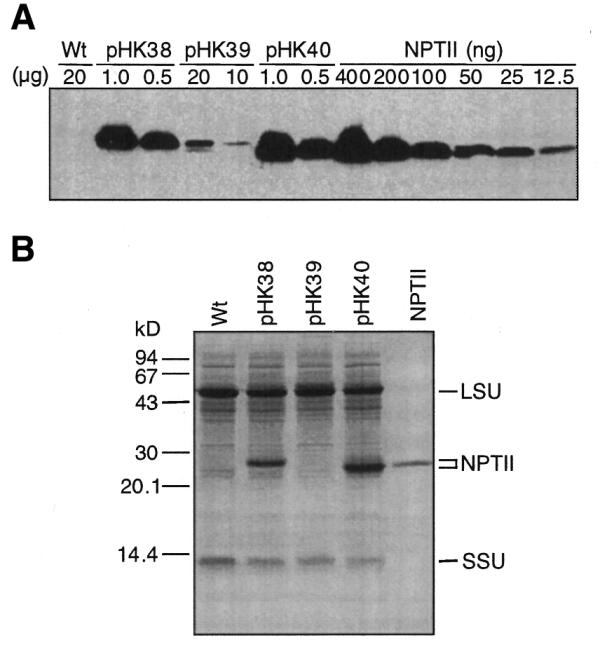
NPTII accumulation in tobacco leaves. (A) Immunoblot analysis to detect NPTII. The amount of TSP (µg) loaded on the SDS–PAGE gel is indicated above the lane. A NPTII dilution series was electrophoresed on the same gel. Lanes are marked with the plasmid name used for plant transformation. A protein sample from wild-type tobacco was also loaded (Wt). (B) Protein gel (20 µg/lane) stained with Coomassie Brilliant Blue R250. Control, NPTII (700 ng). The positions of NPTII and the Rubisco large (LSU) and small (SSU) subunits are marked.
Table 1. Levels of NPTII and neo mRNA.
| Plant linea |
DB/DSb |
neo mRNAc,d |
NPTII (%)d,e |
| Nt-pHK38-2E | Ec | 52.72 ± 21.11 | 16.39 ± 3.42 |
| Nt-pHK39-3B | pt | 14.53 ± 1.41 | 0.16 ± 0.13 |
| Nt-pHK40-12B | – | 100.00 | 23.00 ± 5.40 |
aPlasmid name (for example pHK38), number and letter combination identify plant lines.
bEc, E.coli; pt, tobacco plastid; –, absent.
cPercent of level in Nt-pHK40 plants.
dAverage of three to six measurements.
ePercent of total soluble cellular protein.
Based on the E.coli studies (2–4), we expected that incorporation of the plastid DS would further enhance NPTII levels. However, we found that plants transformed with the construct having the perfect plastid DS (pHK39) contained NPTII levels 100-fold lower than plants expressing NPTII from the T7g10 TCR (pHK38). This result indicates that, unlike in E.coli (2,4), 100% complementarity between the mRNA DS and 16S rRNA ADS region reduces protein accumulation in chloroplasts.
Perfect DS–ADS complementarity destabilizes plastid mRNAs
Steady-state mRNA levels also affect protein levels, as more stable mRNAs are available for translation for longer periods of time. Since the three neo genes are transcribed from the same promoter (Prrn), differences in steady-state mRNA levels directly reflect differences in mRNA turnover rates. Therefore, RNA gel blot analysis was carried out to determine steady-state neo mRNA levels in the transplastomic lines. Quantification of the RNA blots revealed significant differences in neo mRNA accumulation (Fig. 5 and Table 1). Highest levels were found in the Nt-pHK40 plants (T7g10 leader, NheI site), which were then used as the reference (100%). The neo mRNA levels were half as much (∼50%) in plants with the T7g10 DB (Nt-pHK38) and ∼7-fold less (∼14%) in plants with the perfect plastid DS (Nt-pHK39). Thus, incorporation of a NheI site slightly enhanced mRNA stability (Nt-pHK40 plants) while the perfect plastid DS downstream of the AUG significantly accelerated neo mRNA turnover (Nt-pHK39 plants).
Figure 5.
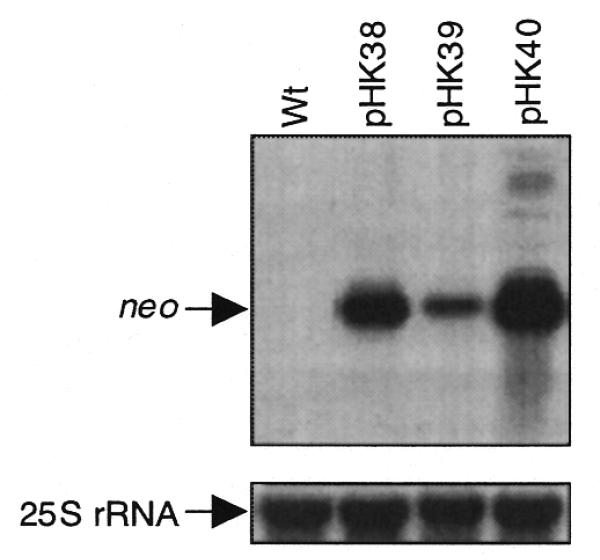
Steady-state levels of neo mRNA in the transplastomic leaves. An aliquot of 4 µg total cellular RNA was loaded per lane. The blots were probed for neo (top) and cytoplasmic 25S rRNA (bottom, loading control). The lanes are designated as in Figure 3.
DISCUSSION
We report here that increasing complementarity between sequences downstream of the AUG with the 16S rRNA 3′-region will reduce, rather than enhance, protein accumulation in chloroplasts. The chimeric neo genes are transcribed from the same promoter in the transplastomic lines. Thus, differences in the mRNA levels in the tobacco lines in part are due to differential mRNA turnover. While increased complementarity between the mRNA DS region and the 16S rRNA ADS region destabilized the neo mRNA in tobacco chloroplasts, it had no effect on mRNA stability in E.coli (2,4). There may be multiple alternative mechanisms that are the cause of accelerated mRNA turnover in chloroplasts. An obvious possibility would be formation of a stable mRNA–rRNA complex that interferes with translational elongation and ultimately triggers mRNA degradation. However, at least in E.coli, direct interaction between the mRNA DB box and 16S rRNA ADB sequence is unlikely (28). A more likely scenario is accelerated turnover via the general mRNA degradation pathway. RNA degradation in chloroplasts is initiated by an endonucleolytic cleavage followed by polyadenylation and degradation. The majority of the cleavage sites form in the coding region and near the start of the 3′-UTR (29,30). In an alternative mechanism, the endonucleolytic cleavage is followed by a 5′→3′ exonuclease activity (31,32). Furthermore, conformational changes of the mRNA may expose known cleavage sites in the 5′-UTR involved in regulating mRNA turnover in response to light and developmental signals (33,34).
Replacement of DB with DS sequences in the neo constructs resulted in an ∼4-fold reduction in mRNA levels and ∼100-fold reduction in tobacco NPTII levels. Therefore, it appears that accelerated mRNA turnover alone is insufficient to account for the reduction in NPTII accumulation. Reduced NPTII accumulation may in part be due to differential rates of neo mRNA translation, as RNA sequences downstream of the AUG were shown to affect translation efficiency in chloroplasts (11). Since the NheI site in Nt-pHK40 plants is directly downstream of the AUG, a region important for translation efficiency, it is possible that the choice of restriction site at this position is important for high-level protein expression. Furthermore, the chimeric neo gene products differ with respect to the amino acid sequence of their N-terminal regions (Fig. 3). Thus, it is also possible that differential NPTII levels are due in part to differential rates of NPTII turnover.
The promoter of the rRNA operon is one of the strongest plastid promoters (9,35). Products of the operon are non-translated RNA species. A general approach to obtain protein expression from the rrn operon promoter has been fusion with translation control signals of plastid genes (17,36,37). A more complex approach involves engineering the entire translation control region including the 5′-UTR and DS sequences (11). The 23% NPTII reported here is the highest protein level obtained from a chloroplast transgene transcribed by a plastid RNA polymerase. Expression of β-glucuronidase at about the same level (20–30%) was reported from plastid genes transcribed by a nuclear-encoded, plastid-targeted phage T7 RNA polymerase. However, expression of the phage polymerase was detrimental to the plants (38).
There is significant interest in applying plant genetic engineering to the production of pharmaceutical compounds, vaccines and industrial enzymes (39–41). Thus far there are only a few examples of expression of economically important genes in chloroplasts (42–44). Transgenes utilizing the heterologous translation control signals described here are expected to perform better under field conditions than those utilizing native plastid signals as they are less likely to compete with native mRNAs for translation.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by NSF grants MCB 96-30763 and MCB 99-05043 to P.M.
References
- 1.Sprengart M.L., Fatscher,H.P. and Fuchs,E. (1990) The initiation of translation in E. coli: apparent base pairing between the 16s rRNA and downstream sequences of the mRNA. Nucleic Acids Res., 18, 1719–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprengart M.L., Fuchs,E. and Porter,A.G. (1996) The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J., 15, 665–674. [PMC free article] [PubMed] [Google Scholar]
- 3.Faxén M., Plumbridge,J. and Isaksson,L.A. (1991) Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471–1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res., 19, 5247–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etchegaray J.P. and Inouye,M. (1999) Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem., 274, 10079–10085. [DOI] [PubMed] [Google Scholar]
- 5.Hirose T., Kusumegi,T. and Sugiura,M. (1998) Translation of tobacco chloroplast rps14 mRNA depends on a Shine–Dalgarno-like sequence in the 5′-untranslated region but not on internal RNA editing in the coding region. FEBS Lett., 430, 257–260. [DOI] [PubMed] [Google Scholar]
- 6.Hirose T. and Sugiura,M. (1996) Cis-acting elements and trans-acting factors for accurate translation of chloroplast psbA mRNAs: development of an in vitro translation system from tobacco chloroplasts. EMBO J., 15, 1687–1695. [PMC free article] [PubMed] [Google Scholar]
- 7.Staub J.M. and Maliga,P. (1993) Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J., 12, 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staub J.M. and Maliga,P. (1994) Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J., 6, 547–553. [DOI] [PubMed] [Google Scholar]
- 9.Shiina T., Allison,L. and Maliga,P. (1998) rbcL transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell, 10, 1713–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonham-Smith P.C. and Bourque,D.P. (1989) Translation of chloroplast-encoded mRNA: potential initiation and termination signals. Nucleic Acids Res., 17, 2057–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuroda H. and Maliga,P. (2001) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol., 125, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern D.B., Higgs,D.C. and Yang,J.J. (1997) Transcription and translation in chloroplasts. Trends Plant Sci., 2, 308–315. [Google Scholar]
- 13.Bruick R.K. and Mayfield,S.P. (1999) Light-activated translation of chloroplast mRNAs. Trends Plant Sci., 4, 190–195. [DOI] [PubMed] [Google Scholar]
- 14.Nickelsen J., Fleischmann,M., Boudreau,E., Rahire,M. and Rochaix,J.D. (1999) Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell, 11, 957–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- 16.Vera A. and Sugiura,M. (1995) Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr. Genet., 27, 280–284. [DOI] [PubMed] [Google Scholar]
- 17.Svab Z. and Maliga,P. (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA, 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaudhuri S. and Maliga,P. (1996) Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. EMBO J., 15, 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- 19.Staub J.M. and Maliga,P. (1994) Extrachromosomal elements in tobacco plastids. Proc. Natl Acad. Sci. USA, 91, 7468–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zoubenko O.V., Allison,L.A., Svab,Z. and Maliga,P. (1994) Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res., 22, 3819–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murashige T. and Skoog,F. (1962) A revised medium for the growth and bioassay with tobacco tissue culture. Physiol. Plant., 15, 473–497. [Google Scholar]
- 22.Stiekema W.J., Heidekamp,F., Dirkse,W.G., van Beckum,J., de Haan,P., ten Bosch,C. and Louwerse,J.D. (1988) Molecular cloning and analysis of four potato tuber mRNAs. Plant Mol. Biol., 11, 255–269. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 24.Svab Z. and Maliga,P. (1991) Mutation proximal to the tRNA binding region of the Nicotiana plastid 16S rRNA confers resistance to spectinomycin. Mol. Gen. Genet., 228, 316–319. [DOI] [PubMed] [Google Scholar]
- 25.Makrides S.C. (1996) Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev., 60, 512–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane J.F. (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol., 6, 494–500. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y., Gojobori,T. and Ikemura,T. (1999) Codon usage tabulated from the international DNA sequence databases; its status 1999. Nucleic Acids Res., 27, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connor M., Asai,T., Squires,C.L. and Dahlberg,A.E. (1999) Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of the 16S rRNA. Proc. Natl Acad. Sci. USA, 96, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kudla J., Hayes,R. and Gruissem,W. (1996) Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J., 15, 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 30.Lisitsky I., Klaff,P. and Schuster,G. (1996) Addition of destabilizing poly(A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc. Natl Acad. Sci. USA, 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drager R.G., Girard-Bascou,J., Choquet,Y., Kindle,K.L. and Stern,D.B. (1998) In vivo evidence for 5′→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J., 13, 85–96. [DOI] [PubMed] [Google Scholar]
- 32.Drager R.G., Higgs,D.C., Kindle,K.L. and Stern,D.B. (1999) 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J., 19, 521–531. [DOI] [PubMed] [Google Scholar]
- 33.Boudreau E., Nickelsen,J., Lemaire,S.D., Ossenbühl,F. and Rochaix,J.D. (2000) The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J., 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgs D.C., Shapiro,R.S., Kindle,K.L. and Stern,D.B. (1999) Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol., 19, 8479–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baumgartner B.J., Rapp,J.C. and Mullet,J.E. (1993) Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development: evidence for selective stabilization of psbA mRNA. Plant Physiol., 101, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrer H., Hockenberry,T.N., Svab,Z. and Maliga,P. (1993) Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet., 241, 49–56. [DOI] [PubMed] [Google Scholar]
- 37.Eibl C., Zou,Z., Beck,A., Kim,M., Mullet,J. and Koop,H.U. (1999) In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J., 19, 333–345. [DOI] [PubMed] [Google Scholar]
- 38.McBride K.E., Schaaf,D.J., Daley,M. and Stalker,D.M. (1994) Controlled expression of plastid transgenes in plants based on a nuclear DNA-encoded and plastid-targeted T7 RNA polymerase. Proc. Natl Acad. Sci. USA, 91, 7301–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J.K. and Vine,N.D. (1999) Plant expression systems for the production of vaccines. Curr. Top. Microbiol. Immunol., 236, 275–292. [DOI] [PubMed] [Google Scholar]
- 40.Hood E.E. and Jilka,J.M. (1999) Plant-based production of xenogenic proteins. Curr. Opin. Biotechnol., 10, 382–386. [DOI] [PubMed] [Google Scholar]
- 41.Arntzen C.J. (1998) Pharmaceutical foodstuffs—oral immunization with transgenic plants. Nature Med., 4 (suppl. 5), 502–503. [DOI] [PubMed] [Google Scholar]
- 42.McBride K.E., Svab,Z., Schaaf,D.J., Hogan,P.S., Stalker,D.M. and Maliga,P. (1995) Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology, 13, 362–365. [DOI] [PubMed] [Google Scholar]
- 43.Daniell H., Datta,R., Varma,S., Gray,S. and Lee,S.B. (1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nature Biotechnol., 16, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staub J.M., Garcia,B., Graves,J., Hajdukiewicz,P.T.J., Hunter,P., Nehra,N., Paradkar,V., Schlittler,M., Carroll,J.A., Ward,D., Ye,G. and Russel,D.A. (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nature Biotechnol., 18, 333–338. [DOI] [PubMed] [Google Scholar]
- 45.Dams E., Hendriks,L., Van de Peer,Y., Neefs,J.M., Smits,G., Vandenbempt,I. and De Wachter,R. (1988) Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res., 16 (suppl.), r87–r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shinozaki K., Ohme,M., Tanaka,M., Wakasugi,T., Hayashida,N., Matsabayashi,T., Zaita,N., Chungwongse,J., Obokata,J., Yamaguchi-Shinozaki,K., Deno,H., Kamogashira,T., Yamada,K., Kasuda,J., Takaiwa,F., Kato,A., Todoh,N., Shimada,H. and Sugiura,M. (1986) The complete sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J., 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]