Abstract
The human thrombopoietin (TPO) gene, which codes for the principal cytokine involved in platelet maturation, shows a peculiar alternative splicing of its last exon, where an intra-exonic 116 nt alternative intron is spliced out in a fraction of its mRNA. To characterize the molecular mechanism underlying this alternative splicing, minigenes of TPO genomic constructs with variable exon–intron configurations or carrying exclusively the TPO cDNA were generated and transiently transfected in the Hep3B cell line. We have found that the final rate of the alternative intron splicing is determined by three elements: the presence of upstream constitutive introns, the suboptimal splice sites of the alternative intron and the length of the alternative intron itself. Our results indicate that the recognition of suboptimal intra-exonic splice junctions in the TPO gene is influenced by the assembly of the spliceosome complex on constitutive introns and by a qualitative scanning of the sequence by the transcriptional/splicing machinery complex primed by upstream splicing signals.
INTRODUCTION
Alternative splicing of pre-mRNAs represents a regulatory mechanism for qualitative and quantitative control of gene expression and of protein function (1). Although there is a growing body of evidence of the mechanisms of splice site selection and interaction among cis-acting elements and trans-acting factors, in most cases these mechanisms remain to be determined.
Patterns of alternative splicing can be very complex as a result of alternative 5′ splice site or alternative 3′ splice site usage, exon inclusion/skipping, mutually exon exclusion and intron retention. Most alternative splicing outcomes involve the competition among potential splice sites, so splicing patterns can be controlled by any mechanism that alters the relative rates of splice site recognition (2).
Among the different mechanisms of constitutive and alternative splicing, the possible exclusion of sequences where both the 5′ and the 3′ intronic junctions are located within an exon represents a further pattern of alternative splicing. Human fibronectin is an example of a gene exhibiting this kind of splicing within the IIICS exon (3). The human IIICS exon is alternatively spliced in a complex pattern, generating up to five variants of fibronectin mRNA, by the use of three alternative 3′ splice sites and an internal alternative 5′ splice site (4,5). In particular, the presence of suboptimal 5′ and 3′ intra-exonic splice sites leads to the generation of the IIICS splice variant C through the splicing out of an intra-exonic 93 nt sequence (6,7). The expression levels of all the IIICS mRNA variants involve the interaction of cis-acting elements and cellular factors leading to a developmental and cell type-specific regulation (6–8).
Another example is represented by the human thrombopoietin gene (TPO), whose sixth and last exon shows a peculiar alternative splicing resembling that of the fibronectin-IIICS exon. The last exon of TPO undergoes alternative splicing generating up to six variants of mRNA, by the use of four alternative 3′ splice sites and an internal alternative 5′ splice site (9,10). Of the six splice variants, the most peculiar is the one arising from the use of 5′ and 3′ intra-exonic splice sites. These suboptimal splice sites define a 116 nt sequence of the sixth exon, which is spliced out in a fraction of the TPO mRNA.
In transient transfection experiments we have observed that the 116 nt sequence is not recognized as an intron if TPO cDNA is used. Conversely, the 116 nt sequence is efficiently spliced out following the expression of a construct carrying the entire TPO gene. This observation indicates that the assembly of the splicing machinery on a constitutive intron is somehow essential for the processing of the 116 nt intra-exonic sequence. Starting from this initial observation, our interest was aimed at determining which and how different splicing signals act in a coordinated fashion to achieve recognition of suboptimal splice sites when an intra-exonic region is subjected to alternative splicing. Therefore, we have addressed the study of suboptimal splice site recognition, characterizing the alternative splicing of the intra-exonic 116 nt sequence of the human TPO pre-mRNA, through the comparison of its behavior both in the context of the TPO gene and of the TPO cDNA.
MATERIALS AND METHODS
Constructs
The human TPO genomic region spanning exons 2–6 (3800 bp) was amplified with DyNAZyme Ext Polymerase (Finnzymes), according to the manufacturer’s instructions, using genomic DNA extracted from human peripheral blood leukocytes as template and the following oligos: TPO–SNKX, 5′-ttttgtcgacgctagcggtaccttactcgaggccaccccggccagaatggagctgact-3′ and TPO–NHX, 5′-tttgcggccgcaagcttctagacttacccttcctgagacagattctgg-3′. The human TPO cDNA (1109 bp) was amplified by RT–PCR using as template total RNA extracted from human peripheral blood leukocytes. Poly(dT) cDNA was synthesized using MMLV reverse transcriptase (Gibco BRL) and was amplified with the above mentioned oligos for 35 cycles (30 s at 94°C, 30 s at 62°C, 1 min at 72°C), using 2 U of Taq DNA polymerase (Roche Diagnostic). Both the TPO gene and the TPO cDNA were digested with KpnI and NotI restriction enzymes and ligated into pcDNA 3 eukaryotic expression vector (Invitrogen) KpnI/NotI cut, generating the parental constructs TPO gene and TPO cDNA. The two minigenes were sequenced using the CEQ 2000 sequencer machine (Bekman-Coulter) to exclude the presence of mutations.
To study the strength of splice sites on the splicing efficiency of the 116 nt sequence, we used an overlap extension system (11) to mutagenize the 5′-splice site towards consensus with primer 5′C-for, tccgaggacaggtaagtatcctgat and 5′C-rev, atcaggatacttacctgtcctcgga and the 3′ splice site, which was improved partially with primers 3′I-for, acgagctcccttgtttaaacaggacttct and 3′I-rev, tccgaggacaggtaagtatcctgat or fully with primers 3′C-for, agtcctcacactgaacgttttttttttcaggacttct and 3′C-rev, agaagtcctgaaaaaaaaaacgttcagtgtgaggact, respectively.
To mutagenize the TPO gene minigene away from the consensus in order to delete the 5′ splice site, the following primers were used: Delta 5′-for, tgctccgaggaaacctaagcttcctgatgctt and Delta 5′-rev, aagcatcaggaagcttaggtttcctcggagca. To delete the 3′ splice site, the primers used were: Delta 3′-for, tgaacgagctcccacgaattccttctggatt and Delta 3′-rev, aatccagaaggaattcgtgggagctcgttca. The primers used as external oligos were 331sense, caactgggacccacttgcct and TPO–NHX. The resulting mutagenized PCR fragments were digested with BamHI and NotI restriction enzymes and were used to replace the wild-type BamHI–NotI sequence into the parental constructs. TPO cDNA Δ5′/3′C was generated using primers Delta 5′-for and Delta 5′-rev and TPO cDNA+3′C as the template. The external primers were TPO–SNKX and TPO–NHX and the mutagenized PCR product was KpnI/NotI digested and cloned into pcDNA 3 vector.
To insert single introns within the TPO cDNA in their native position, the primers used were as follows. Intron 2: 5′s, cggccagaatggagctgactggtgagaacacacctgaggg; 5′as, ccctcaggtgtgttctcaccagtcagctccattctggccg; 3′s, ctcacctctcctcatctaagaattgctcctcgtggtcatg; 3′as, catgaccacgaggagcaattcttagatgaggagaggtgag. Intron 3: 5′s, atgtccttcacagcagactggtgagaactcccaacattat; 5′as, ataatgttgggagttctcaccagtctgctgtgaaggacat; 3′s, ccaccaatctttttcaacagagccagtgcccagaggttca; 3′as, tgaacctctgggcactggctctgttgaaaaagattggtgg. Intron 4: 5′s, gagaatggaaaacccagatggtaagaaagccatccctaac; 5′as, gttagggatggctttcttaccatctgggttttccattctc; 3′s, ctcttccatctctttctcaggaggagaccaaggcacagga; 3′as, tcctgtgccttggtctcctcctgagaaagagatggaagag. Intron 5: 5′s, agagcctccttggaacccaggtaagtccccagtcaaggga; 5′as, tcccttgactggggacttacctgggttccaaggaggctct; 3′s, ggaccatcctctgccctcagcttcctccacagggcaggac; 3′as, gtcctgccctgtggaggaagctgagggcagaggatggtcc.
To delete intron 4 and/or 5 from the TPO gene construct, a multi-step overlapping PCR was carried out with the following primers and templates. Deletion of intron 4: -4-5′s, ccaccaatctttttcaacagagccagtgcccagaggttca and -4-3′as, tcccttgactggggacttacctgggttccaaggaggctct with cDNA as template; -4-5′as, tgaacctctgggcactggctctgttgaaaaagattggtgg and TPO–SNKX with TPO gene as template; -4-3′s, agagcctccttggaacccaggtaagtccccagtcaaggga and TPO–NHX with TPO gene as template. Deletion of intron 5: -5-5′as, tcctgtgccttggtctcctcctgagaaagagatggaagag and TPO–SNKX with TPO gene as template; -5-5′s, ctcttccatctctttctcaggaggagaccaaggcacagga and TPO–NHX with TPO cDNA as template. Deletion of introns 4 and 5: -4,5-5′as, tgaacctctgggcactggctctgttgaaaaagattggtgg and TPO–SNKX with TPO gene as template; -4,5-5′s, ccaccaatctttttcaacagagccagtgcccagaggttca and TPO–NHX with TPO cDNA as template.
The mutagenized constructs were generated through the subsequent PCR overlap of the amplified fragments. Both for single intron insertion and for intron 4/5 deletion the primers used as external oligos were TPO–SNKX and TPO–NHX and the mutagenized PCR amplicons were KpnI/NotI digested and cloned into pcDNA 3 vector. To delete the splice sites of intron 2 in the TPO cDNA+IVS2 Δss construct, the multi-step overlapping PCR was carried out with the following primers. IVS2 Δ5′, agacccaagcttggtaccttactcgaggccacaccccggccagaatggagctgactggggagaa; IVS2 Δ3′s, tcctcatctattaattgctcctcgt; IVS2 Δ3′as, acgaggagcaattaatagatgagga. To delete the splice sites of intron 3 in the TPO cDNA+IVS3 Δss, the following primers were used. IVS3 Δ5′s, ttcacagcagactggggagaactccca; IVS3 Δ5′as, tgggagttctccccagtctgctgtgaa; IVS3 Δ3′s, tctttttcaacttaaccattgcccacttgt; IVS3 Δ3′as, acaagtgggcaatggttaagttgaaaaaga. For each mutagenesis, the primers used as external oligos were TPO–SNKX and TPO–NHX and the mutagenized PCR amplicons were KpnI/NotI digested and cloned into pcDNA 3 vector.
To insert the intronic spacer into the middle of the 116 nt sequence of the TPO cDNA construct, a three-step overlapping PCR was carried out with the following primers and templates. TPO cDNA+Spacer S (sense): TPO–SNKX and TPO–IVS4 spacer 5′as, cccacatgtagaagtttggtgggtggggcccgcct, with TPO cDNA as template. TPO–IVS4 spacer 5′s, aggcgggccccacccaccaaacttctacatgtggg and TPO–IVS4 spacer 3′as, ttctgctggggacagctgttttcttgctttaagaatagca with TPO gene as template. TPO–IVS4 spacer 3′s, tgctattcttaaagcaagaaaacagctgtccccagcagaa and TPO–NHX with TPO cDNA as template. TPO cDNA+spacer AS (anti-sense): TPO–SNKX and TPO–IVS4 spacer AS 5′as, attcttaaagcaagaaaggtgggtggggcccgcct with TPO cDNA as template. TPO–IVS4 spacer AS 5′s, aggcgggccccacccacctttcttgctttaagaat and TPO–IVS4 spacer AS 3′as, tctgctggggacagctgtaaacttctacatgtggg, with TPO gene as template. TPO–IVS4 spacer AS 3′s, cccacatgtagaagtttacagctgtccccagcaga and TPO–NHX, with TPO cDNA as template. For each mutagenesis, the primers used as external oligos were TPO–SNKX and TPO–NHX and the mutagenized PCR amplicons were KpnI/NotI digested and cloned into pcDNA 3 vector. All the constructs were sequenced to exclude the presence of mutations.
Transfections
Human hepatocarcinoma Hep3B cell line was grown in Dulbecco’s modified Eagle’s medium supplemented with 4.5g/l glucose, 10% fetal calf serum, 50 µg/ml gentamicin and 4 mM glutamine.
The DNA used for transfections was purified with JetStar columns (Genomed). Liposome-mediated transfections of 4 × 105 Hep3B cells with 2.5 µg of each construct were performed using DOTAP (Roche Diagnostic), according to the manufacturer’s instructions. After 12 h the medium was replaced with fresh medium and 24 or 48 h later the cultures were terminated. The RNA was extracted with the RNAzol B (Biotecx Laboratories) method. Each transfection experiment was repeated at least four times.
All the RNA samples were treated with DNase RNase free (Roche Diagnostic) to remove DNA contamination from the RNA preparations. Total RNA (10 µg) was incubated for 1 h at 37°C with 1 U of DNase in 50 mM Tris buffer pH 8 and 5 mM MgCl2 (reaction volume, 40 µl). At the end of the reaction the RNA was extracted once with phenol-acid solution and chloroform and precipitated by the addition of 0.1 vol 3 M sodium acetate pH 4 and 2 vol 100% ethanol. The mRNA was synthesized with MMLV reverse transcriptase using a primer annealing specifically within the pcDNA constructs. TPO–SNKX and TPO–NHX were used for PCR amplifications (35 cycles: 30 s at 94°C, 30 s at 62°C, 30 s at 72°C), using 2 U of Taq DNA polymerase. Normalization of transfection efficiency was performed amplifying the neomycin gene of the pcDNA vector using the primers Neo 706 AS acgaggaagcggtcagccca and Neo 254 S, agctgtgctcgacgttgtca (30 cycles: 30 s at 94°C, 30 s at 62°C, 30 s at 72°C). PCR products were fractionated on 0.8% agarose gels. PCRs were optimized to be in the exponential phase of amplification. Cold PCRs were performed for 30 cycles and then 0.1 µl of [α-32P]dCTP (1 µCi) were added to each sample along with 1 U of Taq and were amplified for five more PCR cycles. Radioactive PCR products were run on vertical 0.8% agarose gel for 1 h at 80 V (thickness, 1.5 mm). Agarose gels were dried for 2 h at room temperature. A phospoimager (Instant Imager, Packard Instrument Co.) was used to quantitate PCR amplifications normalized for the C content in the PCR-product sequence. Statistical analysis of the results was carried out using Mac version 2.0 of PRISM program.
The detection of TPO mRNA variants after transient transfection of TPO cDNA+IVSs and TPO gene –IVS4/5 constructs was carried out by RT–PCR using the primers specific for the –12 mRNA isoform A4, 5′-gctgtggtcctgccctgggt-3′ and S2, 5′-ttcaccctttgcctacacct-3′ as described by Sasaki et al. (10). –197 mRNA isoform was specifically amplified using oligo –197 AS 5′-aacaatccagaagtcctgggt-3′ and S2 primer. +60 mRNA isoform was specifically amplified using oligo +60 AS 5′-tcaggcctcccttgtctgggt-3′ and S2 primer. The PCR conditions for detection of splice variants were as follows: 35 cycles, 94°C for 30 s, 68°C for 30 s, 72°C for 30 s.
In order to sequence splicing products, the PCR amplicons were excised from agarose gel and cloned in pCR II-TOPO vector (TOPO TA cloning, Invitrogen).
RESULTS
TPO gene and TPO cDNA constructs differ in their ability to splice out a 116 nt intra-exonic sequence
In order to investigate the molecular mechanisms underlying the alternative splicing of TPO 116 nt intra-exonic sequence, we generated two TPO minigenes that were cloned into the eukaryotic pcDNA3 expression vector (Fig. 1A). The first carries the genomic region of the human TPO gene spanning exons 2–6 (TPO gene). The second carries the TPO cDNA, including the 116 nt intra-exonic sequence within the exon 6 (TPO cDNA). These two constructs were transiently transfected in Hep3B cells and the splicing pattern was compared with that of the endogenous gene.
Figure 1.
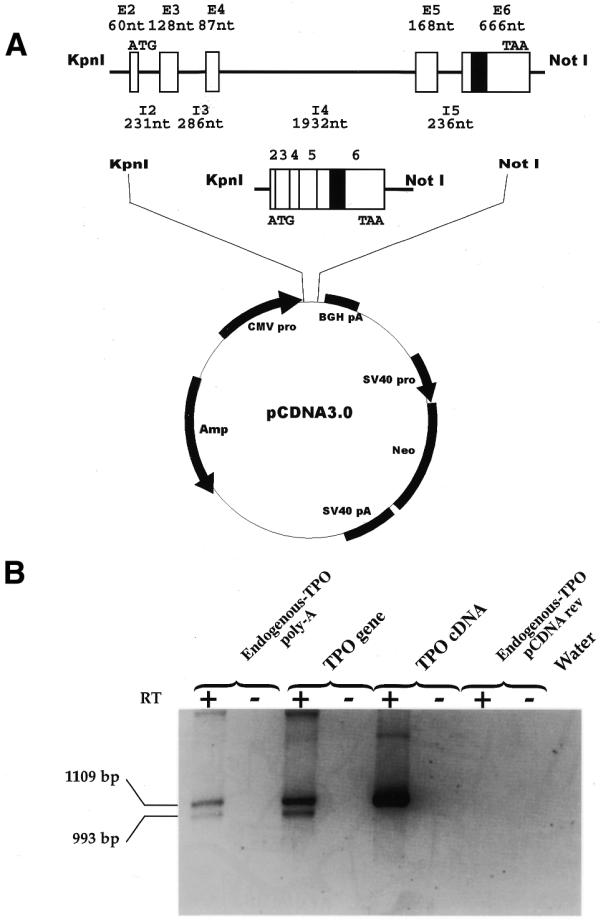
Transient transfection of the TPO gene and TPO cDNA constructs into the Hep3B cell line. (A) Schematic representation of the human TPO gene (GenBank accession no. D32046) and TPO cDNA minigenes cloned in pcDNA 3 vector. E (white boxes), exons; I (lines), introns; black box, the 116 nt alternative intron. Sizes of exons and introns are indicated. KpnI and NotI are the cloning sites. ATG and TAA, start and stop codon, respectively. (B) Analysis of pre-mRNA splicing of human constructs in Hep3B cells. The upper band corresponds to the full-length TPO cDNA (1109 bp) and the lower band to the TPO-3 cDNA lacking the116 nt sequence (993 bp). Amplicons were separated on 0.8% (w/v) agarose gel. Endogenous TPO poly(A), RT–PCR of RNA derived from non-transfected Hep3B cells retrotranscribed with poly(dT) primer. TPO gene and TPO cDNA, RT–PCR of RNA from transfected Hep3B cells retrotranscribed with pcDNA reverse primer, specific for TPO constructs. Endogenous TPO pcDNA rev, RT–PCR of RNA from non-transfected Hep3B cells retrotranscribed with pcDNA reverse primer, specific for TPO constructs. The use of pcDNA reverse primer for cDNA synthesis allows the specific retrotranscription of transfected constructs and not of the endogenous TPO mRNA. Water, control PCR for reagents. RT +/–, presence/absence of MMLV enzyme in the RT mixture.
The transfected cells were harvested, the RNA extracted and treated with DNase to free RNA preparations from residual plasmid. After reverse transcription using a primer specific for the constructs, the splicing pattern was determined by radioactive PCR amplification and phospoimager analysis. An identical strategy was employed in all the following experiments using mutated versions of these minigene constructs.
Figure 1B shows that the transient transfection of the TPO gene construct resulted in the recognition of the 116 nt sequence as an alternative intron, producing two mRNA species including and excluding that sequence in a manner identical to the endogenous gene. In contrast, the transient transfection of TPO cDNA resulted in the expression of only the mRNA species that includes the 116 nt sequence. None of the other possible splice variants was detected indicating that they are expressed at a lower level.
Role of constitutive introns for the splicing of the 116 nt alternative intron
These results suggested that the gene context in the TPO gene construct, rather than exonic sequences, was required to obtain the 116 nt alternative intron.
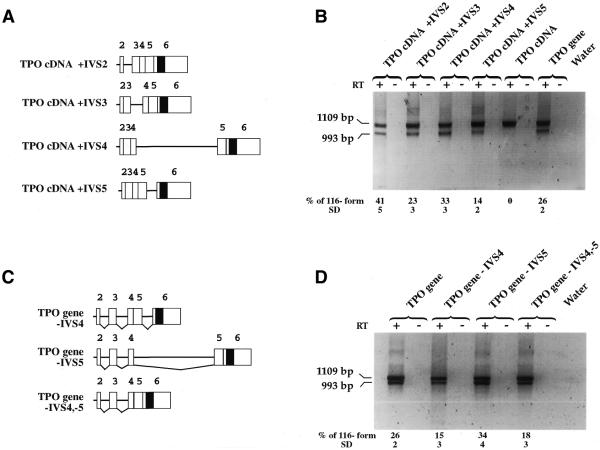
To investigate the role of each constitutive intron in the 116 nt sequence splicing, two additional sets of constructs were generated. The first set was based on the TPO cDNA construct with the insertion of each constitutive intron in its natural position (cDNA+IVS) (Fig. 2A). The transient transfection of all the cDNA+IVS constructs always resulted in the splicing of the 116 nt alternative intron. It was possible to conclude that all constitutive introns have a variable positive effect on recognition and splicing of 116 nt sequence and that the lower portion of 116 nt sequence splicing was associated with the presence of intron 5. The extension of transfection time to 48 h as well as variations in the amount of each construct used for transfection did not modify these observations (data not shown).
Figure 2.
Role of the constitutive introns for the splicing of the 116 nt alternative intron. (A) Schematic representation of the TPO cDNA constructs mutagenized by insertion of single introns in their native location. White boxes are TPO exons (exon number is shown); black box is the 116 nt sequence within exon 6; lines represent introns. (B) Analysis of pre-mRNA splicing of TPO cDNA+IVSs constructs in Hep3B cells. (C) Schematic representation of the TPO gene constructs mutagenized by deletion of intron 4 and/or 5. (D) Analysis of pre-mRNA splicing of TPO gene with intron 4/5 deletion. RT +/–, presence/absence of MMLV enzyme in the RT mixture. Water, PCR control. Relative amount of TPO –116 mRNA variant was quantified by phosphoimager analysis of radioactive PCRs as described in Materials and Methods. The proportion of the splicing of the 116 nt alternative intron was determined by densitometry using the total densitometric units of the full-length TPO cDNA and TPO cDNA 116 nt amplicons as 100%. The values were normalized for C contents of each PCR product. The data are the means of four independent experiments. SD, standard deviation.
These data were confirmed through a second set of constructs based on the TPO gene (Fig. 2C). Three mutants of the TPO gene were designed, the first two with the deletion of intron 5 (TPO gene–IVS5 and TPO gene–IVS4,-5) and the third, as a control, with the deletion of intron 4. Also in this experiment, the presence of intron 5 in the TPO gene–IVS4 construct was associated with a lower efficiency of 116 nt sequence splicing, compared with that resulting from the constructs lacking intron 5 (Fig. 2D).
To understand the reasons why the lower proportion of the 116 nt sequence splicing was associated with the presence of the intron 5, we considered the possible interference of the other alternative splicing variants arising from the last exon (Fig. 3A). Therefore, the presence of the splicing isoforms using three different 3′ splice sites was tested in all transfections using amplification primers specific for each splice variant (–12, –197, +60). These splicing isoforms were detected only in transfections with TPO cDNA+IVS5, TPO gene and TPO gene–IVS4 constructs, which are the constructs in which intron 5 was present (Fig. 3B). These results therefore suggest that the proximal donor splice site of intron 5 rescues at least four alternative acceptor splice site within intron 5 itself and within exon 6. The competition among all these acceptor splice site (even if at a low level) might account for the reduced proportion of the 116 nt sequence splicing.
Figure 3.
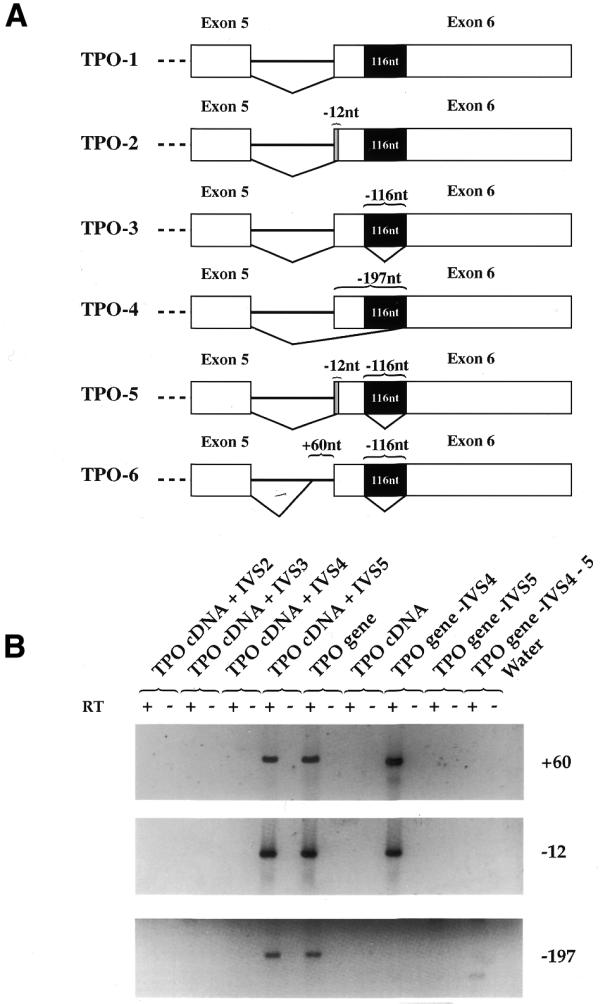
Detection of TPO mRNA isoforms arising from the alternative splicing of exon 6. (A) Diagram of the alternative splicing of the TPO sixth exon. White boxes are TPO exons 5 and 6; black box is the 116 nt sequence within exon 6; lines represent introns. TPO-2 and TPO-5 lack 12 nt (–12 nt); TPO-4 lacks 197 nt (–197 nt); TPO-6 lacks 56 nt (+60 nt). (B) The detection of TPO mRNA variants after transient transfection of TPO cDNA+IVSs and TPO gene –IVS4 and/or IVS-5 constructs was carried out by RT–PCR using primers specific for the –12, –197 and +60 mRNA isoforms, as described in Materials and Methods. The 253 bp PCR products were run on a 2% agarose gel. RT +/–, presence/absence of MMLV enzyme in the RT mixture. Water, control PCR for reagents.
In order to ascertain the possible influence of intronic sequences, we investigated the role of the splicing sites of the constitutive introns in the splicing of the 116 nt sequence. The cDNA+IVS2 construct was mutagenized to inactivate both the 5′ and the 3′ splice sites of intron 2 (Fig. 4A). To preserve the structure of each intron, only the splice junctions were mutated. Transient transfections of this construct showed that intron 2 was retained (Fig. 4B). An additional very faint band was observed below, which after cloning and sequencing was shown to present simultaneous retention of intron 2 and splicing of the 116 nt sequence. However, the proportion of the 116 nt sequence splicing was just 2% of the total amount of PCR products (Fig. 2B). We tried to confirm this observation by mutagenization of the splice sites of intron 3 in the cDNA+IVS3 construct. Essentially, the results were consistent with those of intron 2 splice-site mutagenesis, although, in this case, the splicing pattern was more complex. In fact, the major band was corresponding to the TPO cDNA-retaining IVS3 but additional bands were also present, arising from the activation of cryptic splice sites (Fig. 4B). These results suggest that the splicing of a constitutive intron is an event essential to promote the splicing of suboptimal splice sites.
Figure 4.
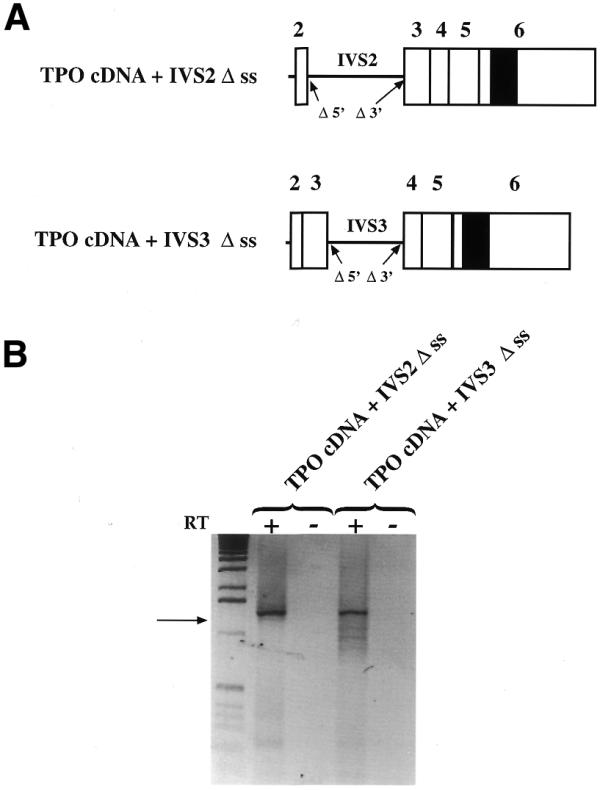
Role of the splice sites of the constitutive introns in the splicing of the 116 nt alternative intron. (A) Diagram of the TPO cDNA+IVS2 Δss and TPO cDNA+IVS3 Δss constructs in which the 5′ and the 3′ splice sites of introns 2 and 3 were deleted, respectively. (B) Analysis of pre-mRNA splicing of TPO cDNA+IVS2 Δss and TPO cDNA+IVS3 Δss constructs in Hep3B cells. TPO cDNA+IVS2 Δss transfections showed that the intron 2 was retained (upper band). The faint band observed below, after cloning and sequencing, was shown to present simultaneous retention of intron 2 and splicing of the 116 nt sequence (arrow). The proportion of the 116 nt sequence splicing was 2% of the total amount of PCR products (upper band + lower band). In TPO cDNA+IVS3, the splicing pattern was more complex with a main splicing product corresponding to the TPO cDNA retaining IVS3 (upper band). The additional bands present arise from the activation of cryptic splice sites. In the first lane 1 kb DNA molecular weight marker was loaded. RT +/–, presence/absence of MMLV enzyme in the RT mixture.
Role of 5′ and 3′ suboptimal splice sites in recognition of the 116 nt sequence
In order to understand the contribution of the cis-acting elements in the recognition of the 116 nt sequence, we have analyzed the composition of its 5′ and the 3′ splice sites and the role they play in this event. The comparison of the 5′ splice junction with the consensus sequence has shown that it differs only at positions +3 and +4, where the adenine bases of the consensus are replaced by a guanine and a cytosine, respectively (Fig. 5). The analysis of the 3′ splice site has shown a perfect match of the acceptor site with the consensus sequence; a short polypyrimidine tract made of four cytosines and a thymidine; and a good match with the consensus sequence, at the putative branch point (Fig. 5). The only deviation from consensus consists of the presence of an adenosine 3′-adjacent to the branch point, instead of a cytosine (Fig. 5).
Figure 5.
Comparison of splice sites of the 116 nt alternative intron of human TPO gene with consensus sequences for 5′ and 3′ splice sites. Organization of the 3′ splice site of the 116 nt sequence showing the potential branch point (underlined with consensus YNYTRAC), the polypyrimidine tract, (Y)n, and the 3′ splice junction, (consensus, NYAG/). The 3′ and 5′ splice sites of the 116 nt sequence of human TPO gene (upper line) are compared with consensus sequences (lower line). Points indicate identity between the two sequences. Respective base frequencies are also indicated for both consensus sequences and 116 nt sequence splice site junctions. N, any nucleotide; Y, pyrimidine; R, purine. Upper case, exons; lower case, introns.
Afterwards, we studied the effects of the mutagenization of the 116 nt sequence splice sites towards the consensus either in the genomic or cDNA context. The 5′ splice site of the 116 nt sequence was mutated towards consensus by correction of the nucleotides at positions –3, +3 and +4, both in the TPO gene and in the cDNA construct (Fig. 6A). In fact, although adenine at position –3 is highly represented in the consensus sequence, it is replaced with a cytosine in order to have complete complementarity of 5′ splice site with the U1 snRNA sequence. Interestingly, the improved 5′ splice site in the TPO cDNA induced partial recognition of 116 nt sequence, whereas in the TPO gene it was associated with a significant increase in the efficiency of 116 nt sequence splicing (Fig. 6B).
Figure 6.
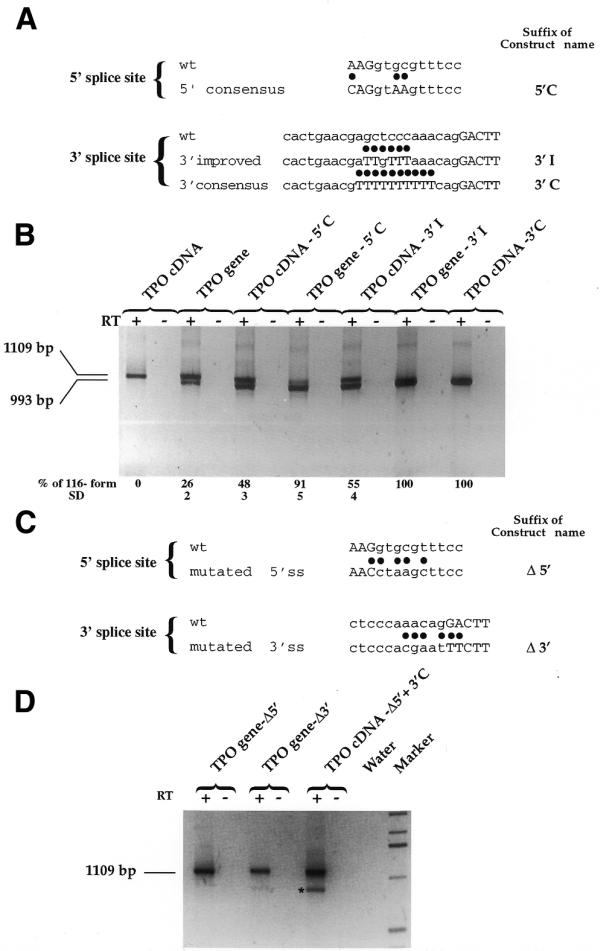
Effect of mutagenization of the splice sites of the 116 nt alternative intron. (A) Wild-type (upper lines) and mutagenized towards consensus (lower lines) 5′ and 3′ splice sites in the TPO gene and TPO cDNA constructs. Dots show the position of the mutagenized bases. Suffixes of constructs’ names are indicated. (B) Results of transfections with constructs mutated toward consensus. RT +/–, presence/absence of MMLV enzyme in the RT mixture. (C) Wild-type (upper lines) and mutagenized away from consensus (lower lines) 5′ and 3′ splice sites in the TPO gene construct. Dots show the position of the mutagenized bases. (D) Results of transfections with constructs mutated away from consensus. Transfection of TPO gene-Δ5′ and TPO gene-Δ3′ shows the expression of the full-length TPO cDNA. TPO cDNA-Δ5′+3′C construct presents a combination of deletion of 5′ splice site and full improvement of 3′ splice site. The asterisk indicates the PCR product arising from the activation of a cryptic 5′ splice site (cag/gtccgtctcc) 127 nt upstream of the wild-type in TPO cDNA-Δ5′+3′C lane. The last lane shows the 1 kb molecular weight marker (Gibco BRL). RT +/–, presence/absence of MMLV enzyme in the RT mixture. Water, PCR control. Relative amount of TPO –116 mRNA variant was quantified by phosphoimager analysis of radioactive PCRs as described in Materials and Methods. SD, standard deviation.
The 3′ splice site of the 116 nt sequence was mutated toward consensus by improving the thymidine content of the polypyrimidine tract. Two sequential improvements were carried out: the first was obtained through the substitution of the wild-type polypyrimidine tract with a TTGTTT tract; and the second was obtained through the substitution of the wild-type polypyrimidine tract with 10 thymidines in a row (Fig. 6A). The partial improvement of the 3′ splice site induced the splicing of the 116 nt sequence in the cDNA context by 50%, whereas in the gene context the efficiency of the 116 nt sequence splicing reached 100%. Consistently, further improvement of the 3′ splice site induced 100% of splicing out of the the 116 nt sequence already in the cDNA context (Fig. 6B).
The relative strength of each splice site was also tested by the mutagenesis of splice sites away from the consensus in the gene context (Fig. 6C). This experiment showed that the deletion of the wild-type 5′ splice site completely abolished the 116 nt sequence splicing in the genomic construct (Fig. 6D).
On the other hand, the combination of deletion of the 5′ splice site and full improvement of the 3′ splice site in the cDNA context (TPO cDNA Δ5′+3′C construct) produced the full-length TPO cDNA retaining the 116 nt sequence and another splicing variant (deleted of 243 nt, due to the activation of a cryptic 5′ splice site 127 nt upstream of the donor site of the 116 nt alternative intron).
Effects of the distance between the splice sites of the 116 nt sequence on its rate of splicing
We also investigated the possibility that the distance between the splice junctions of the 116 nt sequence might be relevant for its splicing, because its short size might interfere with the splice site recognition.
In order to test this hypothesis, a 483 nt spacer deriving from the central part of the 1932 nt TPO intron 4 (from nucleotides +419 to +902) was inserted by mutagenesis in the middle of the 116 nt sequence within the cDNA both in the sense orientation (cDNA+spacer) and in the anti-sense orientation (cDNA+spacer AS) to increase the distance between the splice sites (Fig. 7A). The transfection of cDNA+spacer resulted in the expression of a cDNA where the 116 nt sequence was completely spliced out (Fig. 7B). The same result was obtained with the cDNA+spacer AS, excluding the possibility of some enhancer effect derived from the sequence used as a spacer. So, the occurrence of an accurate splicing of the 116 nt sequence when the space between the splice sites is increased may have two possible explanations. The first is that the proximity of the splice sites might cause a sterical hindrance that hampers the stable interaction of splicing factors with the splice sites. The second is that the increase of the distance between the splice sites might delay the progress of the RNA polymerase II–splicing factors complex and the transcriptional slowing down might, in turn, result in an improvement of the recognition of suboptimal 5′ and 3′ splice junctions.
Figure 7.
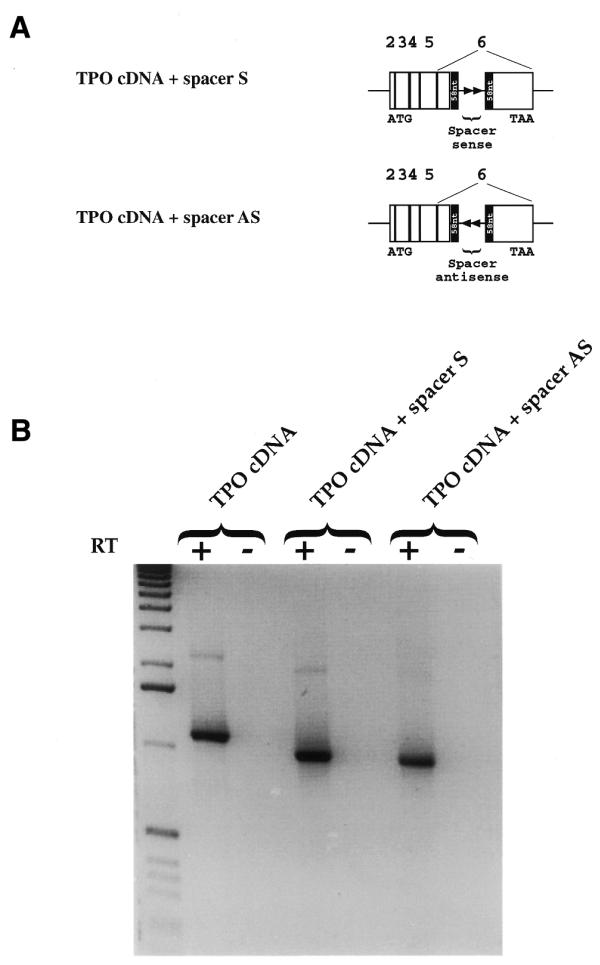
The insertion of a spacer in the middle of the 116 nt alternative intron results in its accurate splicing out. (A) Diagram of spacer cloned in sense (TPO cDNA+spacer S) and antisense (TPO cDNA+spacer AS) orientation within the 116 nt sequence. The black box shows the 116 nt sequence divided in half (58 nt) by the spacer. White boxes show exons. Arrows show the orientation of the spacer. (B) Analysis of pre-mRNA splicing of TPO cDNA+spacer constructs in Hep3B cells. Direct sequencing of the PCR products showed that the band corresponds to the TPO cDNA lacking 116 nt (993 bp). The first lane shows the 1 kb DNA molecular weight marker. RT +/–, presence/absence of MMLV enzyme in the RT mixture.
DISCUSSION
In this work we have investigated the molecular basis underlying the alternative splicing of a 116 nt sequence located within the last exon of the TPO gene. The 116 nt sequence is part of the TPO coding sequence that is normally included in the mRNA and translated into the functional TPO cytokine. However, it behaves as an alternative intron since it is spliced out in a fraction of the TPO mRNA in the liver as well as in a wide number of cell lines of different tissue origin (10). The resulting messenger is efficiently exported into the cytoplasm and translated into a truncated variant of TPO, whose function is presently unknown (9,12).
Since the 116 nt sequence is placed within the coding sequence the strategy to study its alternative splicing was based on the comparison of the behavior of this alternative intron within its gene context and within its cDNA context.
The initial observation of the different splicing efficiency of the 116 nt alternative intron within the TPO gene and the TPO cDNA constructs suggested that the gene context might be crucial because of the commitment of the splicing machinery. In order to identify the sequences that regulate the splicing efficiency of the 116 nt sequence we analyzed the gene elements that are possibly involved. The fact that the transfections with the cDNA construct without introns showed no alternative splicing suggested that there were no specific exonic sequences directly involved in initiating and enhancing the splicing of the 116 nt sequence, as indeed shown for other models (13–18). Therefore, the differences observed for the 116 nt sequence splicing pattern between the cDNA and the gene might be due to the spliceosome assembly on constitutive exon/intron structures, to the effect of intronic sequences or to a combination of both.
To address this issue, initially, we examined the effect of each preceding constitutive intron with the constructs carrying single or multiple introns. In this way, we have shown that the influence on the 116 nt sequence splicing is not restricted to the proximal intron but that single distant introns can also activate its splicing, reaching an efficiency even higher than that observed for the TPO gene. In this regard, intron 5 was the only intervening sequence associated with a slight decrease in the splicing efficiency of the 116 nt sequence, probably due to its rescuing of at least four alternative acceptor splice sites within intron 5 itself and within exon 6. Therefore, it is reasonable to argue that the competition among all the acceptor splice sites might account for the reduced proportion of the 116 nt sequence splicing. This hypothesis will be tested in future studies by selectively mutagenizing the additional 3′ splice sites in exon 6.
Furthermore, this finding suggests the existence of a priming effect of the splicing machinery by an upstream constitutive intron that permits the recognition of downstream suboptimal splice sites.
In other genes, it was shown that the presence of upstream introns might regulate the splicing of downstream introns conferring particular secondary structures to the transcripts (19–21). Moreover, in other models, sequences other than the splice sites were shown to increase the splicing rate of proximal introns (13,22–24). We investigated the presence of regulatory sequences other than the splice sites through the inactivation of the 5′ and 3′ splice sites of introns 2 or 3 in the cDNA constructs with single introns. Interestingly, the levels of 116 nt sequence splicing were virtually non-existent, demonstrating that the presence of strong splice sites, through the commitment of the splicing machinery, is necessary for an efficient splicing of suboptimal splice sites.
Further characterization of the TPO gene has shown that the low splicing rate of the 116 nt sequence is associated with the presence of both weak donor and acceptor splice sites. However, if the splice sites are mutated toward the consensus, the splicing of the alternative intron is not always complete. In the TPO gene, the strengthening of the 3′ splice site is more effective than of the 5′ splice site. In fact, whereas through the improvement of the 3′ splice site it is possible to observe the full intron splicing in the cDNA context, the strengthening of the 5′ splice site results in an increase of splicing efficiency of the 116 nt sequence up to 91% in the gene context, but up to 48% in the cDNA context. Our results suggest that a strong 3′ splice site can compensate for a weak 5′ splice site, whereas a strong 5′ splice site cannot counterbalance a weak 3′ splice site. Hence, our data are consistent with those observed for other splicing models, where it was shown that the balance of the strength of both splice sites and not the strength of a single splice site, influences the splicing rate (25,26).
Additionally, the 5′ splice site of the 116 nt sequence is in agreement with the consensus for vertebrate genes with high G+C content (≥0.5) (27). Interestingly, mutagenization of the 5′ splice site at positions +3 and +4 changed the profile towards a consensus that is generally associated with low G+C gene content (27). This resulted in an improvement of the splicing efficiency. This finding is indicative of a correlation between the G+C content, the presence of a G or an A at position +3 of 5′ splice sites and splicing efficiency.
On the other hand, the length of the 116 nt alternative intron might be invoked to explain its limited splicing efficiency associated with the presence of suboptimal splice sites. In fact, transfections with the TPO cDNA+spacer constructs, where accurate splicing of the 116 nt alternative intron was found, support the hypothesis that the proximity of weak splice sites might interfere with splicing factor interactions on those sites. This interaction might be weak when the splice sites are suboptimal and their distance is limited, whereas it would be stabilized when the splice sites are strong or when their distance is increased. This hypothesis is in keeping with studies showing that the simultaneous recognition of splice sites bordering the intron requires a minimal distance between the sites to prevent the sterical hindrance among the factors that interact with the individual sites (28–30).
Another intriguing explanation might come from the recent evidence that pre-mRNA splicing can be a co-transcriptional event (31–34), where RNA polymerase II and splicing factors interact dynamically to regulate splicing efficiency (35–37). The accurate splicing of the 116 nt sequence following the insertion of an intronic spacer within the alternative intron might support this hypothesis. We are tempted to speculate that these results might be the consequence of an increase in the time necessary for the RNA-polymerase II–splicing factor complex to fill the gap between the 5′ and 3′ splice sites. In this case, the transcription/splicing progress might be paused or delayed by the presence of the intronic spacer, so permitting a more efficient recognition and splicing of suboptimal splice sites that otherwise would be ignored.
Altogether, these results support the model relying on a dynamic interaction between proximal splice junctions. It has been suggested that the cooperation between introns might depend on direct interactions between the splicing complexes acting on individual exons or introns (38,39). These interactions and the concomitant interplay with the transcription machinery might modulate the progress of this process and, in turn, the splicing efficiency. Our data further extend this model indicating that the presence of constitutive introns can be capable of driving the recognition of suboptimal splice sites by the splicing machinery.
In conclusion, this study points to the assembly of the spliceosome complex on constitutive introns and to a qualitative scanning of the sequence by the transcriptional/splicing machinery primed by upstream splicing signals as the events essential for the recognition of downstream suboptimal intra-exonic splice junctions in the TPO gene.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Emanuele Buratti and Andres Muro for their helpful comments on the manuscript.
References
- 1.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 2.Krainer A.R. (1997) Eukaryotic mRNA processing. In Hames,B.D. and Glover,D.M. (eds), Frontiers in Molecular Biology. Oxford University Press, New York, NY.
- 3.Umezawa K., Kornblihtt,A.R. and Baralle,F.E. (1985) Isolation and characterization of cDNA clones for human liver fibronectin. FEBS Lett., 186, 31–34. [DOI] [PubMed] [Google Scholar]
- 4.Vibe-Pedersen K., Magnusson,S. and Baralle,F.E. (1986) Donor and acceptor splice signals within an exon of the human fibronectin gene: a new type of differential splicing. FEBS Lett., 207, 287–291. [DOI] [PubMed] [Google Scholar]
- 5.Kornblihtt A.R., Umezawa,K., Vibe-Pedersen,K. and Baralle,F.E. (1985) Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J., 4, 1755–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hershberger R.P. and Culp,L.A. (1990) Cell-type-specific expression of alternatively spliced human fibronectin IIICS mRNAs. Mol. Cell. Biol., 10, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardon H.J. and Sebastio,G. (1992) Regulation of alternative splicing in the IIICS region of human fibronectin pre-mRNA encoding cell binding sites CS1 and CS5. J. Cell Sci., 103, 423–433. [DOI] [PubMed] [Google Scholar]
- 8.Sarkissian M., Winne,A. and Lafyatis,R. (1996) The mammalian homolog of suppressor-of-white-apricot regulates alternative mRNA splicing of CD45 exon 4 and fibronectin IIICS. J. Biol. Chem., 271, 31106–31114. [DOI] [PubMed] [Google Scholar]
- 9.Chang M.S., McNinch,J., Basu,R., Shutter,J., Hsu,R.Y., Perkins,C., Mar,V., Suggs,S., Welcher,A., Li,L. et al. (1995) Cloning and characterization of the human megakaryocyte growth and development factor (MGDF) gene. J. Biol. Chem., 270, 511–514. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki Y., Takahashi,T., Miyazaki,H., Matsumoto,A., Kato,T., Nakamura,K., Iho,S., Okuno,Y. and Nakao,K. (1999) Production of thrombopoietin by human carcinomas and its novel isoforms. Blood, 94, 1952–1960. [PubMed] [Google Scholar]
- 11.Niksic M., Romano,M., Buratti,E., Pagani,F. and Baralle,F.E. (1999) Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet., 8, 2339–2349. [DOI] [PubMed] [Google Scholar]
- 12.Gurney A.L., Kuang,W.J., Xie,M.H., Malloy,B.E., Eaton,D.L. and de Sauvage,F.J. (1995) Genomic structure, chromosomal localization and conserved alternative splice forms of thrombopoietin. Blood, 85, 981–988. [PubMed] [Google Scholar]
- 13.Watakabe A., Tanaka,K. and Shimura,Y. (1993) The role of exon sequences in splice site selection. Genes Dev., 7, 407–418. [DOI] [PubMed] [Google Scholar]
- 14.Xu R., Teng,J. and Cooper,T.A. (1993) The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol. Cell. Biol., 13, 3660–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavigueur A., La Branche,H., Kornblihtt,A.R. and Chabot,B. (1993) A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev., 7, 2405–2417. [DOI] [PubMed] [Google Scholar]
- 16.Tian M. and Maniatis,T. (1994) A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev., 8, 1703–1712. [DOI] [PubMed] [Google Scholar]
- 17.Elrick L.L., Humphrey,M.B., Cooper,T.A. and Berget,S.M. (1998) A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol. Cell. Biol., 18, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muro A.F., Caputi,M., Pariyarath,R., Pagani,F., Buratti,E. and Baralle,F.E. (1999) Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol., 19, 2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong E.M. and Polisky,B. (1985) Alternative conformations of the ColE1 replication primer modulate its interaction with RNA I. Cell, 42, 959–966. [DOI] [PubMed] [Google Scholar]
- 20.Watakabe A., Inoue,K., Sakamoto,H. and Shimura,Y. (1989) A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res., 17, 8159–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clouet d’Orval B., d’Aubenton Carafa,Y., Sirand-Pugnet,P., Gallego,M., Brody,E. and Marie,J. (1991) RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science, 252, 1823–1828. [DOI] [PubMed] [Google Scholar]
- 22.Naeger L.K., Schoborg,R.V., Zhao,Q., Tullis,G.E. and Pintel,D.J. (1992) Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev., 6, 1107–1119. [DOI] [PubMed] [Google Scholar]
- 23.Seong J.Y., Park,S. and Kim,K. (1999) Enhanced splicing of the first intron from the gonadotropin-releasing hormone (GnRH) primary transcript is a prerequisite for mature GnRH messenger RNA: presence of GnRH neuron-specific splicing factors. Mol. Endocrinol., 13, 1882–1895. [DOI] [PubMed] [Google Scholar]
- 24.Yue B.G. and Akusjarvi,G. (1999) A downstream splicing enhancer is essential for in vitro pre-mRNA splicing. FEBS Lett., 451, 10–14. [DOI] [PubMed] [Google Scholar]
- 25.Grabowski P.J., Nasim,F.U., Kuo,H.C. and Burch,R. (1991) Combinatorial splicing of exon pairs by two-site binding of U1 small nuclear ribonucleoprotein particle. Mol. Cell. Biol., 11, 5919–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dirksen W.P., Sun,Q. and Rottman,F.M. (1995) Multiple splicing signals control alternative intron retention of bovine growth hormone pre-mRNA. J. Biol. Chem., 270, 5346–5352. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M.Q. (1998) Statistical features of human exons and their flanking regions. Hum. Mol. Genet., 7, 919–932. [DOI] [PubMed] [Google Scholar]
- 28.Wieringa B., Hofer,E. and Weissmann,C. (1984) A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell, 37, 915–925. [DOI] [PubMed] [Google Scholar]
- 29.Black D.L. (1991) Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev., 5, 389–402. [DOI] [PubMed] [Google Scholar]
- 30.Dominski Z. and Kole,R. (1991) Selection of splice sites in pre-mRNAs with short internal exons. Mol. Cell. Biol., 11, 6075–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beyer A.L. and Osheim,Y.N. (1988) Splice site selection, rate of splicing and alternative splicing on nascent transcripts. Genes Dev., 2, 754–765. [DOI] [PubMed] [Google Scholar]
- 32.Lewin A.S., Thomas,J.,Jr and Tirupati,H.K. (1995) Cotranscriptional splicing of a group I intron is facilitated by the Cbp2 protein. Mol. Cell. Biol., 15, 6971–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tennyson C.N., Klamut,H.J. and Worton,R.G. (1995) The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature Genet., 9, 184–190. [DOI] [PubMed] [Google Scholar]
- 34.Bauren G. and Wieslander,L. (1994) Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell, 76, 183–192. [DOI] [PubMed] [Google Scholar]
- 35.Cramer P., Pesce,C.G., Baralle,F.E. and Kornblihtt,A.R. (1997) Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA, 94, 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misteli T., Caceres,J.F. and Spector,D.L. (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature, 387, 523–527. [DOI] [PubMed] [Google Scholar]
- 37.Roberts G.C., Gooding,C., Mak,H.Y., Proudfoot,N.J. and Smith,C.W. (1998) Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res., 26, 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robberson B.L., Cote,G.J. and Berget,S.M. (1990) Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol., 10, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen I.T. and Chasin,L.A. (1994) Large exon size does not limit splicing in vivo. Mol. Cell. Biol., 14, 2140–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]