Abstract
Mammalian MBNL (muscleblind-like) proteins are regulators of alternative splicing and have been implicated in myotonic dystrophy, the most common form of adult onset muscular dystrophy. MBNL3 functions as an inhibitor of muscle differentiation and is expressed in proliferating muscle precursor cells but not in differentiated skeletal muscle. Here we demonstrate that MBNL3 regulates the splicing pattern of the muscle transcription factor myocyte enhancer factor 2 (Mef2) by promoting exclusion of the alternatively spliced β-exon. Expression of the transcriptionally more active (+)β isoform of Mef2D was sufficient to overcome the inhibitory effects of MBNL3 on muscle differentiation. These data suggest that MBNL3 antagonizes muscle differentiation by disrupting Mef2 β-exon splicing. MBNL3 regulates Mef2D splicing by directly binding to intron 7 downstream of the alternatively spliced exon in the pre-mRNA. The RNA binding activity of MBNL3 requires the CX7CX4–6CX3H zinc finger domains. Using a cell culture model of myotonic dystrophy and myotonic dystrophy patient tissue, we have evidence that expression of CUG expanded RNAs can lead to an increase in MBNL3 expression and a decrease in Mef2D β-exon splicing. These studies suggest that elevating MBNL3 activity in myogenic cells could lead to muscle degeneration disorders such as myotonic dystrophy.
Keywords: Muscular Dystrophy, RNA-binding Protein, RNA Splicing, Skeletal Muscle, Transcription Factors, Muscle Differentiation, Muscleblind, Myocyte Enhancer Factor, Myotonic Dystrophy
Introduction
Myotonic dystrophy (DM)2 is an autosomal dominant neuromuscular degenerative disease that is characterized by myotonia (muscle hyperexcitability), skeletal muscle weakening and wasting, cardiac conduction defects, as well as non-muscle-related symptoms including insulin resistance, cataract formation, testicular atrophy, and mental impairment (1). There are two genetic forms of myotonic dystrophy. Type 1 (DM1) accounts for nearly 98% of cases and is caused by CTG repeat expansions in the 3′-UTR of the Dmpk (dystrophic myotonic protein kinase) gene (2–4). DM2 results from a CCTG expansion in the first intron of the Znf9 (zinc finger 9) gene (5). Because the expanded repeats occur in the noncoding region of two completely unrelated genes, the current thinking of DM pathogenesis revolves around the expression and accumulation of the mutant RNA transcripts in the nuclei of diseased cells.
Mbl (muscleblind) is a gene required for terminal muscle differentiation in Drosophila melanogaster (6). Mbl mutant flies are unable to properly organize the Z-bands in the sarcomeric apparatus, resulting in paralysis and embryonic lethality (6). In vertebrates, the MBNL (muscleblind-like) proteins are encoded by three genes: Mbnl1 (formally Mbnl/Exp42), Mbnl2 (Mbll), and Mbnl3 (Chcr/Mbxl) (7). MBNL proteins are regulators of alternative splicing and contain four CX7CX4–6CX3H zinc finger domains that confer RNA binding activity (8, 9). MBNL1 is thought to promote muscle differentiation (10). We have reported that MBNL3 functions in an opposing manner and inhibits muscle formation (11, 12). More recently, MBNL2 was discovered to participate in integrin α-3 subcellular localization (13).
The intersection of MBNL proteins with myotonic dystrophy became evident when endogenous MBNL1 and MBNL2 were shown to colocalize with CUG expanded transcripts that are genetically linked to the disease (14–16). Subsequent studies in mouse models demonstrated that inactivation of MBNL1 resulted in splicing defects of different pre-mRNAs that could account for the myotonia, cardiac conduction defects, and insulin resistance characteristic of DM1 (17–19). However, mice lacking functional MBNL1 or MBNL2 protein do not experience the muscle weakening and wasting characteristic of DM patients (20, 21). Therefore, it remains to be determined what is responsible for the DM-associated skeletal muscle degeneration, one of the major causes of patient mortality.
We have found that expression of MBNL3 in proliferating C2C12 myoblasts inhibits transcription of muscle-specific genes, including a number of myocyte enhancer factor 2 (Mef2) target genes (12). Mef2 is an important family of transcription factors that work in concert with MyoD to regulate muscle gene transcription (22). There are four Mef2 proteins in vertebrates, known as Mef2A, Mef2B, Mef2C, and Mef2D, each encoded by a unique gene (23–26). All of the Mef2 proteins contain a common DNA-binding and dimerization domain, but the transcripts are subject to alternative splicing within their transactivation domain. Between exons 6 and 7, each Mef2 gene has a short and highly conserved β-exon that is alternatively spliced into the mature message (27). Inclusion of the β-exon produces a Mef2 isoform that is more robust in activating Mef2-responsive promoters (27). It has been reported that Mef2 mRNAs containing the β-exon are expressed predominantly in striated muscle and in the brain and that inclusion of the β-exon into the Mef2 transcript is promoted during muscle differentiation (27). Therefore, proper control of the activation potential and expression level of the Mef2 myogenic transcription factors plays a critical role in muscle differentiation.
We report here that MBNL3 selectively binds to Mef2D intron 7 sequences and functions as a silencer of β-exon splicing during muscle differentiation. In a cell culture model of myotonic dystrophy and DM skeletal muscle tissue, a decrease in Mef2D β-exon splicing was accompanied by an increase in MBNL3 expression. Interestingly, no change in MBNL1 protein expression was observed. These data suggest that an increase in MBNL3 activity may be a hallmark of DM muscle and may play a role in the skeletal muscle degeneration experienced by myotonic dystrophy patients.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 cells were maintained in DMEM with 10% FBS, penicillin/streptomycin, 2 mm l-glutamine in a humidified incubator at 37 °C in 5% CO2. C2C12 control myoblasts and C2C12-MBNL3 myoblasts expressing Myc-tagged MBNL3 are stable cell lines that have been described previously (11). The stable cell lines were cultured under the same conditions as C2C12 cells but in the presence of 800 μg/ml G418. CTG5 and CTG200 cells were generously provided by the Mahadevan lab and have been characterized previously (28). Maintenance of the CTG5 and CTG200 cells required the addition of 800 μg/ml of G418 to the culture medium. Differentiation of myoblasts into myotubes was achieved by culturing cells, which had reached 70–80% confluency, into DMEM containing 2% horse serum, penicillin/streptomycin, l-glutamine, and ITS liquid supplement (10 ng/ml insulin, 5.5 μg/ml transferrin, and 10 ng/ml selenium). The cells were maintained in differentiation medium for up to 5 days, with medium changes every 2 days.
Retroviral Infection
Mouse Mef2D8 (without β-exon) or Mef2D5 (with β-exon) in pclBabe vector (gift from Dr. Stephen Tapscott) were packaged into retrovirus by transfection into Phoenix Ampho cells (derived from T293 cells). Viral infection of C2C12-MBNL3 stable cells was carried out at a multiplicity of infection of 5 with 8 μg/ml polybrene. Colonies of virally infected C2C12-MBNL3 cells were selected by culturing in medium containing 4 μg/ml of puromycin. Drug-resistant cells were maintained in 2 μg/ml puromycin and 600 μg/ml G418.
Analysis of Mef2 β-Exon Splicing
Total RNA was purified from cells cultured under growth and differentiation conditions using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Total RNA was extracted from human tissues using TRIzol according to the manufacturer's protocol. The RNA yield was determined by measuring A260. RNA samples were subjected to RT-PCR using the StrataScript one-tube RT-PCR system (Stratagene). The splicing of the β-exon into endogenous mouse Mef2 transcripts was determined using the following primers: forward, 5′-gatctgcgggtcatcacttc-3′; and reverse, 5′-cgagtgggtagactgggaga-3′. Transcripts derived from C2C12 cells transfected with pcDNA3.0/Mef2D(6-b-7) minigene construct (kindly provided by Dr. A. Berglund) were detected using the T7 primer as the forward primer and the reverse primer for mouse Mef2 described above. The primers for detecting human Mef2D were: forward, 5′-tacccacagcacccagctt-3′; and reverse, 5′-tagactgggagacccaagg-3′. The Mef2DΔT5 mutant minigene was constructed as follows. A unique ClaI restriction site was introduced into the WT minigene using primers 5′-cacaacattggaatcgatgacacaagcctaatc-3′ and 5′-gattaggcttgtgtcatcgattccaatgttgtg-3′. The mutant plasmid was linearized with ClaI, blunt-ended with DNA polymerase Klenow fragment, and digested with PmlI to drop out the T5 segment of intron 7. The 7-kb fragment lacking intron 7 sequence was gel-purified and recircularized. The boundaries of the T5 deletion was confirmed by DNA sequencing.
Immunocytochemistry
The cells were fixed with 2% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 1% normal goat serum solution in PBS. Primary and secondary antibodies were diluted in 1% goat serum, PBS solution. The primary antibody against myosin heavy chain, mAb MF20 (gift from Dr. S. Hauschka), was used at a dilution of 1:100. The primary antibody reactions were incubated overnight at 4 °C. Fluorescein-conjugated secondary antibodies were diluted 1:1000 and applied for 2 h at room temperature. DAPI staining was used to visualize nuclei. The samples were mounted with Vectashield and visualized using a Nikon Eclipse E600 microscope. Two different filters (UV-2A and FITC-HYQ) were employed to capture the stained images.
Ribonucleoprotein Immunoprecipitation
pIRES-puro Glue vector for expression of TAP-tag proteins was obtained from R. Moon (University of Washington). Full-length cDNA for mouse MBNL3 was PCR-amplified and cloned downstream of the tandem affinity tags. Three 10-cm plates of C2C12 cells were transfected with TAP-tag MBNL3 expression plasmid. Forty-eight hours post-transfection, formaldehyde was added to the medium at a final concentration of 1% and incubated at 37 °C for 10 min. The cells were washed with ice-cold PBS three times and lysed in 1 ml of lysis buffer (50 mm Tris, pH 7.5, 1% Nonidet P-40, 0.05% sodium deoxycholate, 0.05% SDS, 1 mm EDTA, 150 mm NaCl) containing protease inhibitors and RNasin (20 units; Promega). The cells were sonicated three times 20 s (amplitude setting was 5). The resulting lysates were cleared by centrifugation and incubated with streptavidin-agarose (Novagen) overnight at 4 °C. The beads were washed 10 times with washing buffer (50 mm Tris, pH 7.5, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 1 m NaCl, 2 m urea) and then resuspended in 100 μl of 50 mm Tris, pH 7.5, 5 mm EDTA, 10 mm DTT, and 1% SDS and incubated at 70 °C for 45 min to reverse the cross-linking. The released RNA was purified using TRIzol according to the manufacturer's protocol (Invitrogen). Isopropanol precipitation was then performed in the presence of Glycoblue (Ambion). The precipitated RNA was resuspended in RNase-free water and digested with DNase I for 45 min at 37 °C. DNase I was inactivated by the addition of 2.5 mm EDTA and a 10-min incubation at 65 °C. RNA samples were subjected to RT-PCR using StrataScript kit (Stratagene). T2, T4, T5, and T6 primer sets are provided in Table 1.
TABLE 1.
Sequence of forward and reverse primers for Mef2D pre-mRNA
| mMef2D primers | Sequence |
|---|---|
| T2 (forward) | 5′-CGG AAG CTT GGG AGG TGG GAA TAA-3′ |
| T2 (reverse) | 5′-AAT GCG GCC GCG CAG AGT GCT CTT T-3′ |
| T3 (forward) | 5′-CGG AAG CTT CAA AGA GCA CTC TGC-3′ |
| T3 (reverse) | 5′-AAT GCG GCC GCA CTC AGC TCT ATG-3′ |
| T4 (forward) | 5′-CGG AAG CTT GTG CAT AGA GCT GAG T-3′ |
| T4 (reverse) | 5′-AAT GCG GCC GCT AGG CTT GTG TC-3′ |
| T5 (forward) | 5′-CGG AAG CTT CCA AGA CAC AAG CCT A-3′ |
| T5 (reverse) | 5′-CCC GCG GCC GCT CAC TTG TTT ATT C-3′ |
| T6 (forward) | 5′-CGG AAG CTT GGA ATA AAC AAG TGA CTC-3′ |
| T6 (reverse) | 5′-AAT GCG GCC GCA CAT GCA CAC ACA-3′ |
In Vitro Transcription of RNA Probes for UV Cross-linking
Sense and antisense oligonucleotides (Table 1) were synthesized to PCR-amplify fragments that collectively spanned the Mef2D β-exon and intron 7. PCR fragments were subcloned into pcDNA3.0 as HindIII-NotI fragments. The resulting plasmids were linearized by NotI and gel-purified. Linearized plasmids were transcribed in the presence of [α-32P]UTP (specific activity, 800 Ci/mmol) using T7 RNA polymerase to generate uniformly radiolabeled RNA. In vitro synthesized RNAs were electrophoresed through 6% denaturing polyacrylamide gels (0.5× TBE) at 200 volts for 30 min. Radiolabeled RNAs of the expected size were gel-purified and used in UV cross-linking experiments.
Expression and Purification of His-tagged MBNL3 Proteins
The cDNA for WT-MBNL3 and zinc finger mutant were cloned into pET28 bacterial expression vector. Protein expression was carried out in BL21 cells as follows. Overnight cultures (5 ml) were used to inoculate 200 ml of LB + 30 μg/ml kanamycin, and the cultures were grown at 37 °C to A590 = 1.0. Protein expression was induced with 0.1 mm isopropyl β-d-thiogalactopyranoside, and the cells shifted to 16 °C for 16–18 h. For protein purification, the cells were pelleted, frozen with liquid N2, resuspended, and thawed in 10 ml of 0.4 m HEMG buffer (25 mm HEPES, pH 7.9, 0.5 mm EDTA, 12.5 mm MgCl2, 10% glycerol, 0.4 m KCl) containing 5 mm β-mercaptoethanol and protease inhibitors. The cells were lysed by sonication and centrifuged at 10,000 rpm for 15 min at 4 °C. The clarified supernatant was incubated with nickel-agarose beads. The bound beads were washed with 10 column volumes of 0.4 m HEMG + 10 mm imidazole followed by 10 column volumes of 25 mm imidazole in 0.4 m HEMG. Bound proteins were eluted with 0.4 m HEMG + 200 mm imidazole, and peak fractions were dialyzed against 0.2 m HEMG. Purified proteins were aliquoted and stored at −80 °C.
UV Cross-linking Experiments
RNA binding reaction (10 μl) containing 200 ng of purified His-MBNL3 and 32P-labeled RNA in 20 mm Tris-HCl, pH 7.5, 175 mm NaCl, 5 mm MgCl2, 1.25 mm β-mercaptoethanol, 12.5% glycerol, 0.2 mg/ml BSA, and 2 μg of tRNA was incubated for 30 min at room temperature. For UV cross-linking, the binding reactions were transferred to 72-well microtiter dish, precooled on ice, and exposed to 254-nm UV light for 15 min. The cross-linked samples were transferred to 1.5-ml tubes containing 1.5 μl of 10 mg/ml RNase A and incubated at 37 °C for 10 min. The samples were subjected to 12% SDS-PAGE after heat denaturation. The resulting SDS-polyacrylamide gel was dried, and cross-linked RNA-protein complexes were detected by autoradiography.
Preparation of Whole Cell Lysates
The cells were washed twice with PBS and resuspended in cell lysis buffer (50 mm Tris-HCl, pH 7.5, 400 mm NaCl, 1% Nonidet P-40, 1 mm DTT, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin). Cell lysates were incubated on ice for 20 min, with vortexing every 5 min for 15 s. Whole cell extracts were collected after centrifugation at 14,000 rpm in a microcentrifuge for 10 min at 4 °C to remove insoluble material.
Western Blotting
Protein concentration of whole cell extracts was determined using the Bio-Rad protein assay kit according to the manufacturer's instructions. Fifty to one hundred micrograms of total protein were loaded per lane for SDS-polyacrylamide gel electrophoresis. The proteins were transferred onto nitrocellulose membranes and probed with the following primary antibodies at the indicated dilutions: 1:1000 myogenin mAb F5D (Developmental Studies Hybridoma Bank), 1:200 myosin heavy chain mAb MF20 (gift from Dr. S. Hauschka), 1:5 mouse MBNL3 mAb P1E7, 1:1000 MBNL1 mAb MB1a (kindly provided by Dr. G. Morris), 1:10,000 anti-Mef2D (Pharmigen), 1:10,000 GAPDH mAb 6C5 (Adv. ImmunoChem Inc.), and 1:5000 α-tubulin (Abcam). The appropriate horseradish peroxidase-conjugated secondary antibody was used at 1:5000 to 1:10,000 dilution. Proteins of interest were detected by chemiluminescence (GE Healthcare).
Analysis of MBNL3 Transcript Levels
Total RNA was extracted from human tissues using TRIzol according to the manufacturer's protocol. The RNA yield was determined by measuring A260. RNA samples were subjected to quantitative RT-PCR using Brilliant SYBR Green QPCR Master Mix (Stratagene). Quantitative RT-PCRs were carried out as described previously with the annealing temperature increased to 62 °C (12). MBNL3 transcript was detected using the following human specific primers: hMBNL3(F), 5′-aaacgaacgctcaaatgtcatcacttgg-3′; and hMBNL3(R), 5′-cctggcattgcaagaggtg-3′. The primers used to amplify GAPDH transcripts were: GAPDH(F), 5′-cagagactggctcttaaaaagtgc-3′; and GAPDH(R), 5′-gtccaccaccctgttgctgta-3′.
RESULTS
MBNL3 Inhibits Mef2 β-Exon Splicing during Muscle Differentiation
We previously compared the expression pattern of C2C12 cells that stably express MBNL3 (C2C12-MBNL3) to that of control C2C12 cells using DNA microarrays (12). We observed that seven of 31 genes down-regulated by MBNL3 contained potential Mef2 binding sites in their promoter region. Mef2 transcripts are subject to alternative splicing of exons coding for their transactivation domain (Fig. 1A) (26). Inclusion of the alternatively spliced β-exon occurs predominantly in skeletal muscle and produces Mef2 isoforms that are more robust in activating muscle gene transcription (27). MBNL proteins have been shown to be regulators of alternative splicing (30). Therefore, we asked whether MBNL3 could affect the pattern of Mef2a and Mef2d β-exon splicing during muscle differentiation.
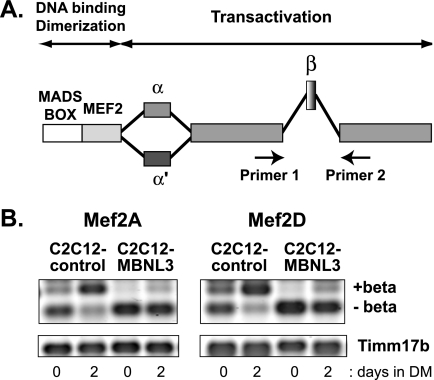
FIGURE 1.
MBNL3 promotes exclusion of β-exon from Mef2A and Mef2D transcript during muscle differentiation. A, a schematic diagram of Mef2 alternative splicing and the relative positions of primers used for RT-PCR are provided. The MADS box and the Mef2 domain comprise the highly conserved DNA-binding and dimerization domains. The remainder of the protein represents the transactivation domain. The alternatively spliced β-exon and two forms of the α-exon are shown. B, total RNA was isolated from control (C2C12-control) and MBNL3 (C2C12-MBNL3) expressing C2C12 cells maintained for 0 or 2 days in differentiation media (DM). Splicing pattern of Mef2A and Mef2D was examined by RT-PCR using primers that can distinguish between the (+)β and (−)β isoforms, as diagrammed in A. The levels of Timm17b were measured to control for variations in input RNA. The data shown are representative of four independent experiments.
In C2C12 control cells, we found that (+)β transcripts were detected at low levels under proliferation conditions but became the predominant transcript after 2 days in differentiation medium (Fig. 1B, n = 4). This trend was observed for both Mef2A and Mef2D. The increase in (+)β transcripts was accompanied by a reciprocal decrease in (−)β transcript levels. The splicing pattern observed in C2C12-MBNL3 cells was radically different, whereby nearly all the Mef2a and Mef2d RNAs consisted of the (−)β-exon isoform (Fig. 1B, n = 4). The profound bias against the β-exon was observed both in growth and differentiation media. These data suggest that MBNL3 promotes exclusion of the β-exon from Mef2 transcripts during muscle differentiation.
Mef2D(+)β Isoform Restores Muscle Differentiation to C2C12-MBNL3 Cells
It has been previously shown that Mef2 proteins encoded by the (+)β transcript are more potent activators of transcription (27). Therefore we wished to determine whether the predominance of the less active Mef2(−)β isoform was responsible for the down-regulation of muscle gene expression in C2C12-MBNL3 cells. If so, we would expect that selective overexpression of the Mef2D(+)β protein would overcome the muscle differentiation defects of C2C12-MBNL3 cells. To test this hypothesis, we established C2C12-MBNL3 cells that stably expressed either Mef2D(+)β, Mef2D(−)β, or the empty vector (pclBabe) by retroviral infection and examined Mef2D protein levels by Western blotting. A 2.5-fold increase was detected in virally infected C2C12-MBNL3 cells when compared with C2C12 or control infected cells (Fig. 2A, n = 3). The (+)β isoform of Mef2D differs from the (−)β isoform by only seven additional amino acids, and we were unable to resolve this small difference in molecular weight by SDS-polyacrylamide gel electrophoresis.
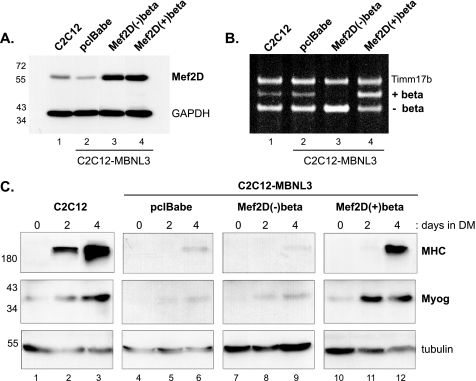
FIGURE 2.
Mef2D(+)β expression overcomes muscle differentiation defect of C2C12-MBNL3 cells. A, C2C12-MBNL3 cells that constitutively express MBNL3 were infected with no virus (C2C12), the empty control retrovirus (pclBabe), or retroviruses that express either the (−)β or (+)β isoforms of Mef2D. After puromycin selection for virally infected cells, drug-resistant cells were pooled, and whole cell lysates were prepared. Mef2D protein level was determined by Western blotting. GAPDH levels were used to control for variations in protein load. The positions of molecular mass markers (in kDa) are shown on the left. B, total RNA was isolated for RT-PCR analysis from C2C12 and different virally infected C2C12-MBNL3 cells. Positions of PCR products for (+)β and (−)β isoforms of Mef2D are indicated. Timm17b message levels were measured to control for input RNA. C, control C2C12 cells and virally infected drug-resistant cell pools were maintained in differentiation media (DM) for the indicated number of days. Whole cell lysates were prepared, and the ability of cells to execute the muscle differentiation program was determined by following two myogenic markers, myogenin (Myog) and MHC, by Western blotting. Tubulin protein levels were used to control for total protein load. The protein standards are indicated on the left. The data presented was observed in three independent experiments.
Unable to distinguish the two Mef2D isoforms at the protein level, we turned to monitoring mRNA levels by RT-PCR. As shown in Fig. 2B, C2C12 (lane 1) and control infected C2C12-MBNL3 (lane 2) cells primarily expressed the Mef2D(−)β transcript. Infection with the (−)β retrovirus increased Mef2D(−)β mRNA levels ∼2-fold (Fig. 2B, lane 3). Interestingly, the increase in (−)β levels was accompanied by a decrease in (+)β mRNA levels; however, the underlying mechanism remains to be determined. As expected, mRNA levels of the (+)β isoform increased more than 3-fold in cells infected with the (+)β retrovirus (Fig. 2B, lane 4).
Confident that the different cell populations were expressing the desired Mef2D isoforms, we examined the differentiation potential of each cell set. High levels of the muscle differentiation markers myogenin and MHC were detected in control C2C12 cells after 4 days of differentiation (Fig. 2C, lanes 1–3). Induction of myogenin and MHC was restored in C2C12-MBNL3 cells infected with the Mef2D(+)β expressing retrovirus (Fig. 2C, lanes 10–12). By contrast, expression of the Mef2D(−)β isoform had no effect on the differentiation potential of C2C12-MBNL3 cells (Fig. 2C, lanes 7–9) such that the level of myogenin and MHC was indistinguishable from that observed for the differentiation-deficient C2C12-MBNL3 cells infected with control virus (Fig. 2C, lanes 4–6).
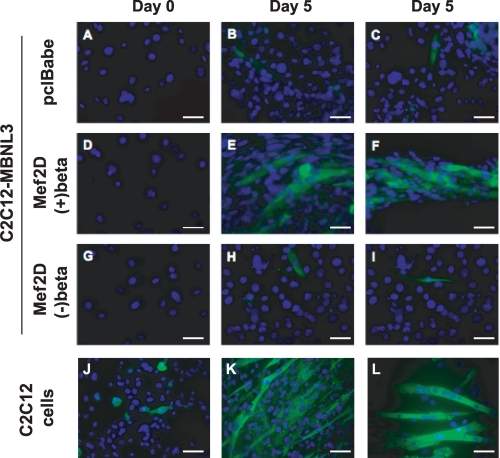
The ability of Mef2D(+)β to rescue muscle differentiation also was assessed by immunocytochemistry (Fig. 3). Consistent with the Western blot results, detection of multinucleated myotubes that stained positive for MHC was observed only in control C2C12 cells and in C2C12-MBNL3 cells infected with the (+)β isoform of Mef2D (Fig. 3, E, F, K, and L). These findings suggest that Mef2D(+)β is essential for muscle differentiation and that the inhibitory effect of MBNL3 on muscle differentiation is due in part to silencing of Mef2D β-exon splicing.
FIGURE 3.
Restoration of myotube formation by Mef2D(+)β expression in C2C12-MBNL3 cells. C2C12-MBNL3 cells (A–C), clonal pools of C2C12-MBNL3 cells expressing the indicated Mef2D isoform (D–I), and control C2C12 cells (J–L) were maintained in differentiation media for 0 or 5 days. The cells were fixed and analyzed by immunocytochemistry using an anti-myosin heavy chain mAb MF20 (green). The nuclei were visualized by DAPI staining (blue). Scale bars, 50 μm.
Association of MBNL3 with Intron Sequences Downstream of the Alternatively Spliced Mef2D β-Exon in Vivo
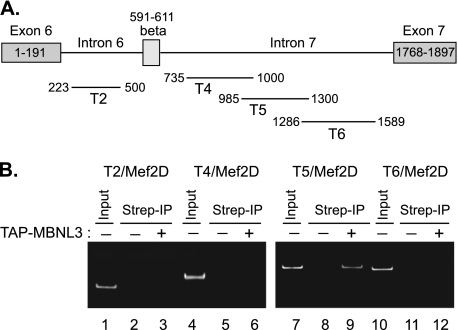
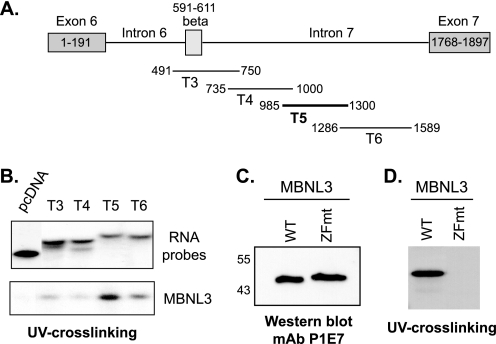
The rescue experiments described in Figs. 2 and 3 do not rule out the possibility that the (+)β isoform of Mef2D is overriding the muscle differentiation defect by a mechanism that is independent of MBNL3. Therefore, we set out to gather evidence that MBNL3 is directly involved in regulating Mef2D β-exon splicing. Regulators of alternative splicing often act by directly binding to the pre-mRNA. We carried out ribonucleoprotein immunoprecipitation, a variation of chromatin immunoprecipitation, followed by RT-PCR to determine whether MBNL3 binds to the endogenous Mef2D transcript in C2C12 cells (Fig. 4). Four different primer sets that spanned introns flanking the β-exon were used for RT-PCR analysis (Fig. 4A). A single PCR product of the expected size was detected only with the T5 primer set using the isolated RNAs as template (Fig. 4B, lane 9, n = 3). These results indicate that MBNL3 selectively associates with a 315-nucleotide region within intron 7, downstream of the β-exon, of the Mef2D pre-mRNA in vivo.
FIGURE 4.
MBNL3 associates with intron sequences downstream of Mef2D β-exon in vivo. A, schematic diagram of introns and exons surrounding the alternatively spliced Mef2D β-exon is provided. The relative positions of RT-PCR products for different intron primer sets are shown. B, C2C12 cells transiently transfected with TAP-tag MBNL3 expression plasmid (lane 3, 6, 9, and 12) were cross-linked with formaldehyde. Whole cell lysates were prepared, sonicated, and precipitated with streptavidin-agarose. The presence of Mef2D intron sequences in TAP-MBNL3-bound RNAs was determined by RT-PCR. The cells transfected with empty control vector (lanes 2, 5, 8, and 11) were subjected to the same protocol in parallel. Results representative of three independent experiments are shown. Lanes 1, 4, 7, and 12 (Input), RT-PCR signal for each primer set using C2C12 total RNA as template.
MBNL3 Directly Binds to Intron 7 of Mef2D Pre-mRNA
To determine whether MBNL3 directly binds to the Mef2D pre-mRNA, we carried out UV cross-linking experiments. Fragments that collectively spanned the Mef2D β-exon and most of intron 7 were transcribed in vitro in the presence of α-32P-UTP, and the resulting uniformly radiolabeled RNA fragments were incubated with purified recombinant His-tagged mouse MBNL3 (Fig. 5A). The U content for the RNA fragments was calculated and did not differ dramatically, ranging from 18 to 26%. The amount of input RNA for the binding reactions was adjusted for the small differences in U content. Purified MBNL3 bound most efficiently to T5, a RNA fragment centrally located within intron 7, consistent with our in vivo ribonucleoprotein immunoprecipitation results (Fig. 5B, n = 4). The remaining Mef2D RNA probes showed much lower levels of interaction. MBNL3 did not bind to an unrelated RNA fragment synthesized from the pcDNA3.0 vector, suggesting that its RNA binding activity displays sequence specificity (Fig. 5B, pcDNA).
FIGURE 5.
MBNL3 directly binds to intron 7 of Mef2D pre-mRNA in vitro. A, the relative positions of uniformly radiolabeled RNA segments of Mef2D pre-mRNA transcribed in vitro for UV cross-linking experiments are shown. MBNL3 bound efficiently to the region indicated by the thick black line in vivo. B, bacterially expressed His-tagged MBNL3 was purified by nickel-agarose affinity chromatography. Purified MBNL3 was incubated with the indicated radiolabeled RNA fragments. Protein-RNA interactions were covalently cross-linked. The resulting stable complexes were separated on SDS-polyacrylamide, and protein bound to the radiolabeled probe was detected by autoradiography (lower panel). An aliquot of each RNA probe is shown (upper panel). The data presented are representative of four independent experiments. C, comparable amounts of purified WT MBNL3 and ZFmt were subjected to SDS-PAGE followed by Western blotting using the anti-MBNL3 mAb P1E7. The positions of molecular mass markers (in kDa) are shown on the left. D, RNA binding of WT and ZFmt MBNL3 proteins to the T5 fragment of Mef2D pre-mRNA was examined by UV cross-linking (n = 7) as described in B.
RNA Binding Activity of MBNL3 Requires the CX7CX4–6CX3H Zinc Finger Motifs
CX4–15CX4–6CX3H zinc finger domains are a feature characteristic of many cellular proteins involved in RNA processing. The high degree of amino acid identity within the zinc finger domains of the three mammalian MBNL proteins strongly supports the functional importance of these motifs. Mutation of the CX4–15CX4–6CX3H motifs in tristetraprolin and MBNL1, regulators of mRNA stability (31) and alternative splicing (30), respectively, disrupts their RNA binding ability. These findings suggest that the CX7CX4–6CX3H motifs may be required for MBNL3 binding to RNA. We constructed a bacterial expression vector for a MBNL3 zinc finger mutant (ZFmt) in which the third cysteine of each of the four CX7CX4–6CX3H motifs was changed to a serine. When comparable amounts of wild-type and mutant MBNL3, as determined by Western blotting (Fig. 5C), were examined in UV cross-linking assays, ZFmt displayed no interaction with the Mef2D-T5 intron 7 sequence (Fig. 5D, n = 4). These results have led us to conclude that one or more CX7CX4–6CX3H domains are required for MBNL3 RNA binding activity.
MBNL3 Directly Regulates Mef2D β-Exon Splicing
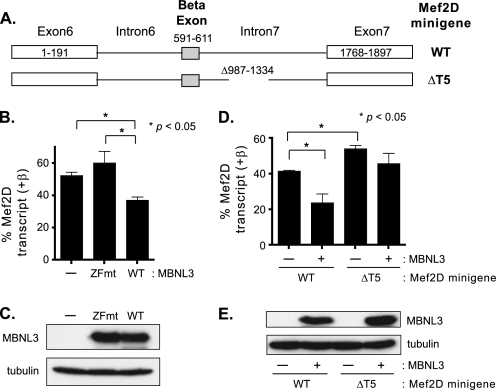
The observation that MBNL3 interacts with the Mef2D transcript in vivo and in vitro supports the idea that Mef2D is a direct target for the splicing function of MBNL3. To investigate this further, we used a minigene reporter construct that contains exons 6, β, and 7 and the intervening introns of the Mef2D gene (Fig. 6A). Minigene reporters have been instrumental in the identification of trans-acting factors that regulate pre-mRNA splicing (32). We took advantage of the T7 promoter sequence that was transcribed and remained attached to the 5′ end of the Mef2D minigene transcript. The presence of this short bacterial sequence enabled us to use the T7 primer as the forward primer and a Mef2D-specific reverse primer to uniquely amplify spliced products derived from the transfected plasmid. When the Mef2D minigene reporter was transiently transfected into C2C12 cells, we found that (+)β splice products represented ∼50% of the Mef2D transcripts detected 24 h post-transfection (Fig. 6B). No products were amplified when C2C12 cells were transfected with the control vector lacking any Mef2D sequence, demonstrating the specificity of the RT-PCR conditions for exogenous Mef2D minigene transcripts (data not shown).
FIGURE 6.
β-Exon splicing pattern of Mef2D minigene constructs in C2C12 cells. A, schematic diagrams of WT and mutant (ΔT5) Mef2D minigene constructs consisting of exons 6, β, and 7 and the intervening introns are shown. B, WT Mef2D minigene construct was cotransfected into C2C12 cells with either pcDNA3.0FLAG vector (−), an expression plasmid for the ZFmt, or the WT-MBNL3 expression plasmid (WT). Approximately 24 h post-transfection, total RNA was isolated, and the β-exon splicing pattern was determined by RT-PCR. Amplified products were loaded onto native polyacrylamide gels and detected by ethidium bromide staining. The intensity of (+)β and (−)β splice products was determined using National Institutes of Health Image J software and used to calculate the percentage of total Mef2D transcript for each isoform. Percentages from three independent experiments (n = 5) are provided. C, level of WT and ZFmt MBNL3 protein expressed from the transfected expression vector was determined by Western blotting. Tubulin protein levels were measured to control for variations in protein load. D, WT or ΔT5 Mef2D minigene was transfected into C2C12 cells in the absence (−) or presence (+) of WT-MBNL3 expression plasmid. The samples and resulting data were analyzed as described in B. E, expression levels of MBNL3 protein were determined by Western blotting. Tubulin protein levels were used to monitor for differences in protein loading. Statistically significant differences are indicated by the asterisks, and the p values are provided.
Next, we examined the consequences of expressing wild-type MBNL3 or the four zinc finger mutants on β-exon splicing of transcripts derived from the Mef2D minigene (Fig. 6B). Expression of wild-type MBNL3 led to a significant decrease in (+)β transcript levels in C2C12 cells compared with cells transfected with the empty control vector. By contrast, the zinc finger mutant (ZFmut), which lacks the ability to bind Mef2D intron sequences, had no detectable effect on the pattern of β-exon splicing. We confirmed by Western blotting that wild-type MBNL3 and ZFmut protein were expressed at comparable levels in the transfected cells (Fig. 6C). These data suggest that MBNL3 inhibits β-exon splicing by binding to Mef2D pre-mRNA.
To investigate whether MBNL3 is directly functioning at the T5 segment of intron 7, we examined the effect of deleting this portion of intron 7 on β-exon splicing (Fig. 6A). In the absence of exogenous MBNL3 expression, removal of the 315-bp intron sequence from the minigene construct (Mef2DΔT5) resulted in an increase in Mef2D(+)β transcript levels when examined in C2C12 cells (Fig. 6D). These data suggest that we had removed a cis-element required for repressing β-exon splicing. Is MBNL3 a trans-factor that functions at the T5 splicing repressor element? To address this question, we examined the ability of MBNL3 to silence β-exon splicing of pre-mRNA derived from the WT and ΔT5 Mef2D minigene constructs (Fig. 6D). Expression of MBNL3 with the WT Mef2D minigene significantly decreased the level of (+)β transcripts generated. With the ΔT5 minigene, which lacks the MBNL3 binding site in intron 7, MBNL3 expression exhibited no significant effect on β-exon splicing in C2C12 cells. It was determined by Western blotting that comparable levels of MBNL3 were expressed in the presence of the WT and ΔT5 minigene constructs (Fig. 6E). Thus, we have mapped a silencer for β-exon splicing within intron 7 of Mef2D pre-mRNA and provide evidence that MBNL3 is a trans-acting factor that operates at this cis-element to promote exclusion of the β-exon from the mature Mef2D transcript.
Mef2D β-Exon Splicing Is Disrupted in a Cell Culture Model of Myotonic Dystrophy
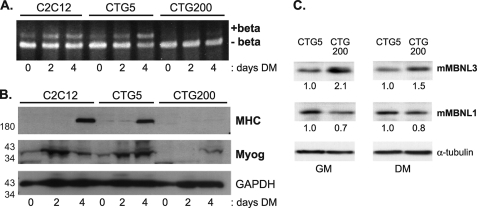
MBNL1-dependent changes in the splicing pattern of transcripts for the insulin receptor, cardiac troponin T, and ClC-1 chloride channel have been shown to contribute to the insulin resistance, cardiac conduction defects, and myotonia, characteristic of myotonic dystrophy, respectively (17–20). These findings led us to ask whether mis-splicing of Mef2D β-exon could be contributing to DM-associated skeletal muscle degeneration. We observed that in CTG200 cells, which represent a pool of clones that express the full-length DMPK 3′-UTR containing 200 CTG repeats downstream of the GFP coding region (28), the level of Mef2D(+)β mRNA was nearly undetectable under all conditions (Fig. 7A). This is in stark contrast to the increase in (+)β transcript levels that was detected in the parental C2C12 cells and in the differentiated CTG5 pool of cells, which express only five CTG repeats in the DMPK 3′-UTR (Fig. 7A). In addition to abnormal Mef2D splicing, defects in myogenesis were observed in CTG200 cells, with induction of the myogenic markers myogenin and MHC dramatically compromised (Fig. 7B). The phenotype of CTG200 cells is similar to that of C2C12-MBNL3 cells that constitutively express MBNL3. These data suggested that MBNL3 may be contributing to the skeletal muscle pathology of DM1 and could be overexpressed in the diseased cells.
FIGURE 7.
Increased MBNL3 protein levels accompany defects in Mef2D β-exon splicing and muscle gene expression in a cell culture model for myotonic dystrophy. A, C2C12 clonal cell pools that express GFP fused to the full-length DMPK 3′-UTR containing either 5 (CTG5) or 200 (CTG200) CTG repeats were kindly provided by Dr. Mani Mahadevan. These cells and control C2C12 cells were exposed to differentiation conditions, and the splicing pattern of Mef2D was examined by RT-PCR and polyacrylamide gel electrophoresis. B, expression of the muscle differentiation markers MHC and myogenin (Myog) were determined by Western blotting of whole cell lysates prepared from the indicated cell lines. GAPDH protein levels were monitored to control for total protein load. The positions of molecular mass markers (in kDa) are shown on the left. C, elevated levels of MBNL3 but not MBNL1 in C2C12 cells expressing CUG200 repeat transcripts. Whole cell lysates were prepared from C2C12 cells that expressed DMPK 3′-UTR containing either 5 (CTG5) or 200 (CTG200) CTG repeats. The cells were maintained in either growth (GM) or differentiation (DM) medium before the extracts were prepared. Approximately 100 μg of lysate was loaded onto SDS-polyacrylamide gels, transferred to nitrocellulose, and subjected to Western blotting with the MBNL3 mAb P1E7 and MBNL1 mAb MB1a. Tubulin levels were used to control for protein loading. The protein levels of MBNL3 and MBNL1 were determined using National Institutes of Health Image J software and used to calculate the relative levels, shown below each lane, under growth and differentiation medim conditions. The data presented were reproducibly observed in three independent experiments.
The expression level of the splicing regulator CUGBP is increased in DM patient tissue and in some mouse models of DM (33, 34). These findings prompted us to examine the expression levels of MBNL3 and MBNL1 in our cell culture model. We generated a monoclonal antibody, mAb P1E7, that specifically recognizes endogenous mouse MBNL3 in C2C12 whole cell lysates and does not cross-react with mouse MBNL1.3 mAb P1E7 was used to monitor MBNL3 expression levels in CTG5 and CTG200 cells under growth and differentiation conditions. By Western blotting, we found that MBNL3 was present at higher levels (1.5–2.0-fold) in C2C12 cells expressing the expanded DMPK 3′-UTR responsible for myotonic dystrophy (Fig. 7C). The elevated levels of MBNL3 were observed when the cells were cultured in either growth or differentiation medium. A similar analysis for MBNL1 indicated no change and even a slight decrease in MBNL1 upon expression of the expanded CUG repeat transcript (Fig. 7C). These findings suggest that MBNL3 differs from MBNL1 and may be actively involved in the pathogenesis of myotonic dystrophy.
MBNL3 Expression Levels and Mef2D β-Exon Splicing Are Altered in DM1 Skeletal Muscle
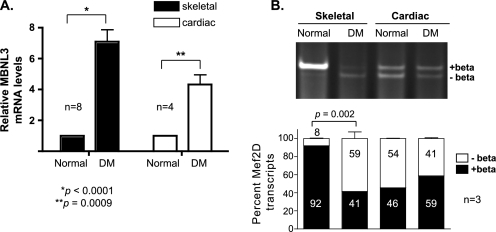
Whereas CTG200 cells display many of the properties of myotonic dystrophy cells, they are not equivalent to diseased cells. In addition, the inability to detect differentiation-dependent β-exon splicing in CTG200 cells in culture could be due to the fact that the cells fail to execute the muscle differentiation program and thus do not undergo the exon switch in Mef2 splicing. Therefore, we examined MBNL3 expression levels and Mef2D β-exon splicing in differentiated tissues obtained from myotonic dystrophy patients (Fig. 8). Total RNA was prepared from skeletal and cardiac muscle derived from DM and control matched patients. MBNL3 expression was monitored by quantitative RT-PCR and found to be ∼7-fold higher in DM skeletal muscle (Fig. 8A). A smaller increase (∼4-fold) in MBNL3 RNA levels was detected in DM heart tissue. The elevated expression of MBNL3 in the DM tissue prompted us to examine the pattern of Mef2D β-exon splicing. In normal skeletal muscle, nearly all the Mef2D transcript detected contained the β-exon (Fig. 8B). By contrast, very low levels of (+)β transcript were expressed in DM-derived skeletal muscle. The pattern of Mef2D β-exon splicing did not differ between normal and DM cardiac muscle, despite the observed increase in MBNL3 expression. These data further support our hypothesis that MBNL3 expression is up-regulated in myotonic dystrophy and disrupts Mef2D β-exon splicing, which contributes to the skeletal muscle weakening and wasting characteristic of the disease.
FIGURE 8.
MBNL3 expression and Mef2D β-exon splicing in muscle of DM patients. A, MBNL3 transcript levels. Total RNA was isolated from psoas (skeletal) and heart (cardiac) muscle tissue obtained from two normal and two age/gender-matched DM patients. The level of MBNL3 expression was determined by quantitative RT-PCR. GAPDH transcript levels were measured to control for RNA input. Relative MBNL3 mRNA levels were calculated using the conventional 2−ΔΔCt method (29). The p values for significant differences are provided and indicated by the asterisks. B, Mef2D β-exon splicing. Mef2D(−)β and (+)β transcript levels in total RNA prepared from normal and DM patient tissues were determined by RT-PCR using human specific primers. Amplified products were separated on polyacrylamide and visualized by ethidium bromide staining. Relative intensity of (+)β and (−)β splice products was determined using National Institutes of Health Image J software and used to calculate the percentage of total Mef2D transcripts for each isoform (n = 3). Significant differences are indicated by the p value.
DISCUSSION
The MBNL proteins are regulators of alternative splicing and have been implicated in the splicing defects associated with the adult onset muscular dystrophy known as myotonic dystrophy. The majority of studies to date have focused on MBNL1, the founding member of the mammalian MBNL protein family. Inactivation of MBNL1 leads to misregulated splicing of the cardiac troponin T and muscle-specific chloride channel, which has been correlated with the cardiac conduction defects and myotonia observed in DM (19, 20, 35). Our studies with MBNL3 have demonstrated that MBNL3 can influence the splicing pattern of the Mef2 muscle transcription factor family, promoting exclusion of the β-exon from the mature message. The resulting transcripts encode a transcriptionally less active isoform of the Mef2 proteins. The activation of β-exon splicing occurs as proliferating myoblasts differentiate into multinucleated myotubes (27, 28). During muscle differentiation, the protein levels of MBNL3 decreased as formation of the Mef2(+)β transcripts increased. This inverse correlation supports our hypothesis that MBNL3 functions as an inhibitor of Mef2 β-exon splicing. To our knowledge, these findings are the first demonstration that an MBNL protein regulates the activity of a muscle transcription factor that acts during the early stages of myogenesis.
The ability of the splicing machinery to determine which splice sites to use is dictated by cis-acting elements within exons and introns that either enhance or silence the usage of adjacent splice sites. The two major classes of cis-regulatory elements are exonic splicing enhancers and exonic splicing silencers. These elements function by recruiting factors that interact favorably or interfere with the splicing machinery (36, 37). The discovery that MBNL3 association with intron sequences in the Mef2D transcript is required for inhibiting β-exon splicing suggests that MBNL3 may represent one such trans-acting factor. We have preliminary tandem affinity purification results suggesting that heterogeneous nuclear ribonucleoproteins are potential MBNL3 binding proteins.4 Heterogeneous nuclear ribonucleoproteins often interact with exonic splicing silencers to inhibit splicing (38, 39). It will be of interest to determine whether the interaction of MBNL3 with heterogeneous nuclear ribonucleoproteins is required for inhibition of Mef2D β-exon splicing during muscle differentiation.
MBNL1 is expressed predominantly in skeletal and cardiac muscle, whereas MBNL3 is most abundant in the placenta and is present at very low levels in muscle tissue (10, 16, 40). The discovery that MBNL3 is an inhibitor of muscle differentiation led us to propose that MBNL3 and MBNL1 have opposing functions during myogenesis. We found that MBNL3 directly interacts with and alters the splicing pattern of Mef2 pre-mRNA. Mammalian MBNL1, MBNL2, and MBNL3 have similar effects on the splicing pattern of cardiac troponin T and insulin receptor pre-mRNA (30). These results raise the question of whether the effect of MBNL3 on Mef2 splicing will be unique to this muscleblind family member and whether MBNL proteins have unique functions at select pre-mRNAs during muscle differentiation.
MBNL3 and MBNL1 each contain four CX7CX4–6CX3H zinc finger domains and differ from Drosophila Mbl, which contains only two CX7CX4–6CX3H motifs (10, 11, 16). The zinc fingers are highly conserved and are required for MBNL1 and MBNL3 RNA binding activity (41). In addition to the zinc fingers, MBNL3 also contains a proline-rich region located between the second and third CX7CX4–6CX3H motifs, which is conserved to varying degrees among the other MBNL proteins (7). Proline-rich regions have been extensively associated with protein-protein interactions (42). Differences in the positioning and spacing of the proline residues implicate different binding partners for the different MBNL proteins, which could account for the opposing functions of MBNL1 and MBNL3 in muscle differentiation.
Present in MBNL1 and MBNL2 but not in MBNL3 are alanine-rich regions. The functional relevance of these domains in MBNL1 and MBNL2 remains unknown. An alanine-rich region is required for dimerization of the RNA-binding protein AUFL and thus contributes to the binding affinity of AUFL for target RNA (43). Intriguingly, MBNL1 has been suggested to interact with RNA as a multimer (44).
The current model for the pathogenesis of myotonic dystrophy involves the sequestration of MBNL proteins by the repeat expanded mutant RNAs that accumulate in the nuclei of DM1 and DM2 cells (45). The inactivation of MBNL1 leads to splicing defects and the formation of fetal transcripts that encode for proteins that are functionally inadequate in the adult. Although MBNL3, when overexpressed as a GFP fusion protein, colocalized with CUG expanded transcripts in DM cells, the same behavior has not been demonstrated for the low levels of endogenous MBNL3 present in differentiated skeletal muscle. Western blot analysis of C2C12 cells expressing transcripts with 5 (CTG5) or 200 (CTG200) CUG repeats and skeletal muscle derived from DM patients revealed that MBNL3 protein levels were elevated under diseased conditions. We propose that the MBNL proteins are not equivalent in their contribution to DM pathogenesis. MBNL3 expression levels are increased in DM skeletal muscle, similar to what has been observed for the splicing factor CUGBP and thus may not be subject to inactivation by sequestration. We are currently performing immunocytochemistry experiments using MBNL3 mAb P1E7 to investigate the expression level and localization of MBNL3 in normal and DM cells.
There are many examples of members of the same protein family having functionally distinct and even opposing cellular roles. The use of positive and negative regulators provides greater flexibility in controlling a biological pathway. However, upsetting the delicate balance between these stop and go signals can lead to the development of human disease. Most likely both the inactivation of MBNL1 and the up-regulation of MBNL3 are contributing to the pathogenesis of myotonic dystrophy.
Acknowledgments
We thank Keri Lewis for excellent technical assistance. We also thank S. Hauschka for generously providing the myosin heavy chain mAb MF20, M. Mahadevan for the CTG5 and CTG200 cell lines, and A. Berglund for the Mef2D minigene plasmid. The normal and DM human tissue samples were obtained from the NICHD, National Institutes of Health Brain and Tissue Bank for Developmental Disorders. We are especially grateful to S. Kloet for critical reading of the manuscript and to other members of the Wang lab for valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AR049042. This work was also supported by University of Washington Royalty Research Fund Grant 4176.
K.-S. Lee, D. T. Campogan, K. Lewis, E. A. Wayner, and E. H. Wang, submitted for publication.
H. E. Witwicka and E. H. Wang, unpublished data.
- DM
- myotonic dystrophy
- Mef
- myocyte enhancer factor
- ZFmt
- zinc finger mutant.
REFERENCES
- 1.Harper P. (2001) Myotonic dystrophy, W. B. Saunders Co., London [Google Scholar]
- 2.Brook J. D., McCurrach M. E., Harley H. G., Buckler A. J., Church D., Aburatani H., Hunter K., Stanton V. P., Thirion J. P., Hudson T. (1992) Cell 68, 799–808 [DOI] [PubMed] [Google Scholar]
- 3.Fu Y. H., Pizzuti A., Fenwick R. G., Jr., King J., Rajnarayan S., Dunne P. W., Dubel J., Nasser G. A., Ashizawa T., de Jong P. (1992) Science 255, 1256–1258 [DOI] [PubMed] [Google Scholar]
- 4.Mahadevan M., Tsilfidis C., Sabourin L., Shutler G., Amemiya C., Jansen G., Neville C., Narang M., Barceló J., O'Hoy K. (1992) Science 255, 1253–1255 [DOI] [PubMed] [Google Scholar]
- 5.Liquori C. L., Ricker K., Moseley M. L., Jacobsen J. F., Kress W., Naylor S. L., Day J. W., Ranum L. P. (2001) Science 293, 864–867 [DOI] [PubMed] [Google Scholar]
- 6.Artero R., Prokop A., Paricio N., Begemann G., Pueyo I., Mlodzik M., Perez-Alonso M., Baylies M. K. (1998) Dev. Biol. 195, 131–143 [DOI] [PubMed] [Google Scholar]
- 7.Pascual M., Vicente M., Monferrer L., Artero R. (2006) Differentiation 74, 65–80 [DOI] [PubMed] [Google Scholar]
- 8.Teplova M., Patel D. J. (2008) Nat. Struct. Mol. Biol. 15, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He F., Dang W., Abe C., Tsuda K., Inoue M., Watanabe S., Kobayashi N., Kigawa T., Matsuda T., Yabuki T., Aoki M., Seki E., Harada T., Tomabechi Y., Terada T., Shirouzu M., Tanaka A., Güntert P., Muto Y., Yokoyama S. (2009) Protein Sci. 18, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller J. W., Urbinati C. R., Teng-Umnuay P., Stenberg M. G., Byrne B. J., Thornton C. A., Swanson M. S. (2000) EMBO J. 19, 4439–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Squillace R. M., Chenault D. M., Wang E. H. (2002) Dev. Biol. 250, 218–230 [DOI] [PubMed] [Google Scholar]
- 12.Lee K. S., Smith K., Amieux P. S., Wang E. H. (2008) Differentiation 76, 299–309 [DOI] [PubMed] [Google Scholar]
- 13.Adereth Y., Dammai V., Kose N., Li R., Hsu T. (2005) Nat. Cell Biol. 7, 1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fardaei M., Larkin K., Brook J. D., Hamshere M. G. (2001) Nucleic Acids Res. 29, 2766–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankodi A., Urbinati C. R., Yuan Q. P., Moxley R. T., Sansone V., Krym M., Henderson D., Schalling M., Swanson M. S., Thornton C. A. (2001) Hum. Mol. Genet. 10, 2165–2170 [DOI] [PubMed] [Google Scholar]
- 16.Fardaei M., Rogers M. T., Thorpe H. M., Larkin K., Hamshere M. G., Harper P. S., Brook J. D. (2002) Hum. Mol. Genet. 11, 805–814 [DOI] [PubMed] [Google Scholar]
- 17.Savkur R. S., Philips A. V., Cooper T. A. (2001) Nat. Genet. 29, 40–47 [DOI] [PubMed] [Google Scholar]
- 18.Mankodi A., Takahashi M. P., Jiang H., Beck C. L., Bowers W. J., Moxley R. T., Cannon S. C., Thornton C. A. (2002) Mol. Cell 10, 35–44 [DOI] [PubMed] [Google Scholar]
- 19.Mahadevan M. S., Yadava R. S., Yu Q., Balijepalli S., Frenzel-McCardell C. D., Bourne T. D., Phillips L. H. (2006) Nat. Genet. 38, 1066–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanadia R. N., Johnstone K. A., Mankodi A., Lungu C., Thornton C. A., Esson D., Timmers A. M., Hauswirth W. W., Swanson M. S. (2003) Science 302, 1978–1980 [DOI] [PubMed] [Google Scholar]
- 21.Hao M., Akrami K., Wei K., De Diego C., Che N., Ku J. H., Tidball J., Graves M. C., Shieh P. B., Chen F. (2008) Dev. Dyn. 237, 403–410 [DOI] [PubMed] [Google Scholar]
- 22.Potthoff M. J., Olson E. N. (2007) Development 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 23.Breitbart R. E., Liang C. S., Smoot L. B., Laheru D. A., Mahdavi V., Nadal-Ginard B. (1993) Development 118, 1095–1106 [DOI] [PubMed] [Google Scholar]
- 24.Martin J. F., Schwarz J. J., Olson E. N. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5282–5286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott J. C., Cardoso M. C., Yu Y. T., Andres V., Leifer D., Krainc D., Lipton S. A., Nadal-Ginard B. (1993) Mol. Cell. Biol. 13, 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin J. F., Miano J. M., Hustad C. M., Copeland N. G., Jenkins N. A., Olson E. N. (1994) Mol. Cell. Biol. 14, 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu B., Ramachandran B., Gulick T. (2005) J. Biol. Chem. 280, 28749–28760 [DOI] [PubMed] [Google Scholar]
- 28.Amack J. D., Paguio A. P., Mahadevan M. S. (1999) Hum. Mol. Genet. 8, 1975–1984 [DOI] [PubMed] [Google Scholar]
- 29.Bubner B., Gase K., Baldwin I. T. (2004) BMC Biotechnol. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho T. H., Charlet-B N., Poulos M. G., Singh G., Swanson M. S., Cooper T. A. (2004) EMBO J. 23, 3103–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999) Mol. Cell. Biol. 19, 4311–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper T. A. (2005) Methods 37, 331–340 [DOI] [PubMed] [Google Scholar]
- 33.Timchenko L. T., Miller J. W., Timchenko N. A., DeVore D. R., Datar K. V., Lin L., Roberts R., Caskey C. T., Swanson M. S. (1996) Nucleic Acids Res. 24, 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orengo J. P., Chambon P., Metzger D., Mosier D. R., Snipes G. J., Cooper T. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2646–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanadia R. N., Shin J., Yuan Y., Beattie S. G., Wheeler T. M., Thornton C. A., Swanson M. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11748–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J. Y., Maniatis T. (1993) Cell 75, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 37.Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Garcia-Blanco M. A., Manley J. L. (1994) Nature 368, 119–124 [DOI] [PubMed] [Google Scholar]
- 38.Mayeda A., Krainer A. R. (1992) Cell 68, 365–375 [DOI] [PubMed] [Google Scholar]
- 39.Amendt B. A., Hesslein D., Chang L. J., Stoltzfus C. M. (1994) Mol. Cell. Biol. 14, 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee K. S., Squillace R. M., Wang E. H. (2007) Biochem. Biophys. Res. Commun. 361, 151–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kino Y., Mori D., Oma Y., Takeshita Y., Sasagawa N., Ishiura S. (2004) Hum. Mol. Genet. 13, 495–507 [DOI] [PubMed] [Google Scholar]
- 42.Kay B. K., Williamson M. P., Sudol M. (2000) FASEB J. 14, 231–241 [PubMed] [Google Scholar]
- 43.DeMaria C., Sun Y., Wagner B., Long L., Brewer G. (1997) Nucleic Acids Symp. Ser. 12–14 [PubMed] [Google Scholar]
- 44.Yuan Y., Compton S. A., Sobczak K., Stenberg M. G., Thornton C. A., Griffith J. D., Swanson M. S. (2007) Nucleic Acids Res. 35, 5474–5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheeler T. M., Thornton C. A. (2007) Curr. Opin. Neurol. 20, 572–576 [DOI] [PubMed] [Google Scholar]